Abstract
Historically, king penguin populations on Macquarie Island have suffered greatly from human exploitation. Two large colonies on the island were drastically reduced to a single small colony as a result of harvesting for the blubber oil industry. However, recent conservation efforts have resulted in the king penguin population expanding in numbers and range to recolonize previous as well as new sites. Ancient DNA methods were used to estimate past genetic diversity and combined with studies of modern populations, we are now able to compare past levels of variation with extant populations on northern Macquarie Island. The ancient and modern populations are closely related and show a similar level of genetic diversity. These results suggest that the king penguin population has recovered past genetic diversity in just 80 years owing to conservation efforts, despite having seen the brink of extinction.
Keywords: King penguin, Macquarie Island, genetic diversity, ancient DNA, conservation
1. Introduction
King penguins (Aptenodytes patagonicus) are the second largest penguins, being surpassed in size only by emperor penguins (Aptenodytes forsteri). Individuals are approximately 90 cm in height and weight between 11 and 16 kg. King penguins have an estimated generation time of 14.7 years with growth rates for populations varying greatly, but the intrinsic growth rate is suggested to be approximately 9.7 per cent per annum [1,2]. The breeding cycle takes about 15 months with a maximum of two chicks per pair raised every 3 years [3]. The species has a circumpolar distribution, breeding on the Falkland Islands (which have been re-colonized after extermination in historical times) and a number of sub-Antarctic islands: South Georgia, Prince Edward, Crozet (over half of the world's population), Kerguelen, Heard (re-colonized after extermination) and Macquarie Island [4]. In 1911, the ornithologist Gregory Mathews proposed two subspecies on the basis of their size, which are currently recognized but whose status has been at times questioned. Aptenodytes patagonicus halli breeds on the Prince Edward, Crozet, Kerguelen, Heard and Macquarie Islands. Aptenodytes patagonicus patagonicus breeds on South Georgia and the Falkland Islands in the South Atlantic [5].
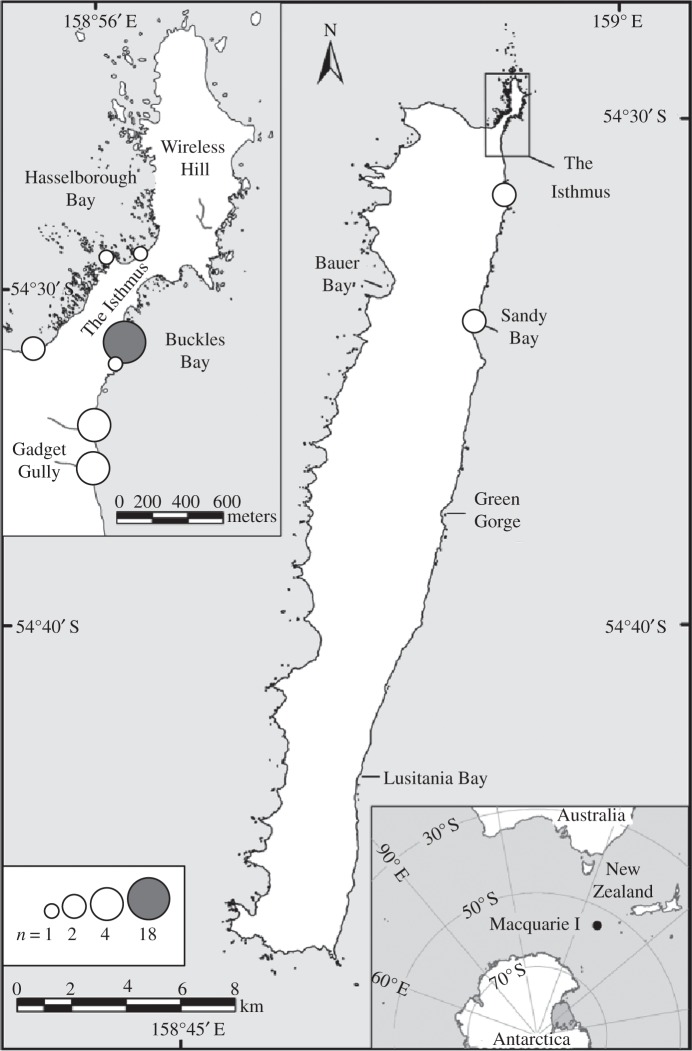
The subspecies on Macquarie Island has suffered, particularly from historical exploitation by humans. Two very large colonies were present when Macquarie Island was discovered in 1810: one at the Isthmus and one at Lusitania Bay [6] (figure 1). The first settlers sought to harvest blubber oil, among other products, initially from fur seals followed by elephant seals, king penguins and finally royal penguins. The effects of these activities devastated not only the Macquarie Island populations of the species but also populations on other islands, most of which are now on the rise again [7]. The king penguin colony at the Isthmus on Macquarie Island was extinct by 1894 and the colony at Lusitania Bay was reduced to ca 3400 birds by 1930 [8]. Macquarie Island was declared a wildlife sanctuary in 1933, a state reserve in 1972 and a World Heritage area in 1997 [9]. These protection efforts halted exploitation and excessive disturbance of the local wildlife, additionally fishing is now very limited and pests are being eradicated, enabling the king penguins to flourish once again. The current population now numbers 150 000–170 000 breeding pairs at Lusitania Bay (in 2000) and an estimated total of 400 000–500 000 king penguins on Macquarie Island as a whole (in 2006) [9]. From Lusitania Bay and possibly elsewhere, new breeding colonies were established by natural dispersal in Sandy Bay, Green Gorge and the Isthmus (around 1975, 1977 and 1995, respectively). The colony at Lusitania Bay may now be approaching its maximum carrying capacity and the colony at the Isthmus is increasing at an average 66 per cent per annum [8,10]. This indicates that the king penguin population on Macquarie Island is thriving once again. Sub-fossil remains show that colonies previously existed at Green Gorge, the Isthmus and Bauer Bay (previously dated ca 4000 BP), which is the only known colony occurrence on the west coast for this species [11,12].
Figure 1.
Map of Macquarie Island; lower right inset indicates location. Circle size represents the number of samples from different locations, shaded indicates ancient samples, white modern samples. The Isthmus is pictured in more detail in the top left inset. Map Courtesy Australian Antarctic Division Commonwealth of Australia 2006.
Using ancient DNA methodology, we aimed to compare past and present genetic diversity in order to assess the impacts of past anthropogenic pressures and present conservation efforts and, more specifically, to determine if the past levels of genetic diversity have been recovered in modern populations of king penguins on Macquarie Island.
2. Material and methods
Seventeen samples representing the modern population were taken from king penguin carcasses that were collected from northern Macquarie Island. A small 6 mm3 section was excised from the foot for DNA extraction. Sub-fossil bones representing the ancient king penguin population were collected stratigraphically from Landing Beach on the Isthmus, Macquarie Island. Approximately 300 mg of bone shavings from each of 20 sub-fossil bones were extracted in 200 µl of 0.5 M EDTA with 0.5 mg ml–1 Proteinase K and 0.1 per cent Triton X-100 incubating for 10 h at 56°C. The modern samples and the sub-fossil samples were then extracted and purified using the DNeasy Blood and Tissue Kit (Qiagen) according to manufacturer's instructions. All sub-fossil bones were handled in a dedicated ancient DNA laboratory where strict protocols apply.
The PCR reactions contained 5 µl template, 1 × PCR buffer, 1 mg ml–1 bovine serum albumin, 1.5 mM MgCl2, 0.4 µM of each primer, 0.2 mM of each dNTP and 1 unit of platinum Taq (Invitrogen); thermocycling was as follows: 1 min 94°C, 40 × (30 s 94°C, 30 s 60°C, 30 s 72°C), 5 min 72°C. Part of the mitochondrial genome's hyper variable region was amplified using one primer pair (PKHoF 5′-CACTTAATGTTAGCAAACATACTTACTTC-3′ + PKHoR 5′-CATTAAGACCGGGCTCTAAG-3′ 423 bp) for modern samples or two primer pairs (PKHoF + PKHiR 5′-GTTTAGTCCGAGGAATGAAGT-3′ 273 bp and PKHiF 5′-CGGACCATATCCKACTCTC-3′ + PKHoR 287 bp) for ancient samples.
PCR products were either isolated from an agarose gel (in case of unspecific by products) using the QIAquick Gel Extraction Kit (Qiagen) or cleaned with ExoSAP-IT (USB) according to manufacturer's instructions. The BigDye v. 3.1 (Applied Biosystems) kit was used according to manufacturer's instructions to sequence the DNA products. Three modern and two ancient products were reamplified and resequenced to confirm their authenticity [13]. Sequences are deposited in GenBank under accession numbers JQ256379–JQ256413.
Sequences were manually screened for errors before assembly and alignment. Haplotype networks were constructed using the program TempNet v. 1.4 [14]. Genetic diversity measures were calculated for both modern and ancient samples using the maximum composite likelihood model and pairwise deletion using the program MEGA 5 [15], variance was estimated using 500 bootstrap replicates.
Representative ancient samples were radiocarbon dated by the Rafter Radiocarbon Laboratory, National Isotope Centre, Institute of Geological and Nuclear Sciences Ltd (GNS Science), Lower Hutt, New Zealand. Radiocarbon ages were calibrated and corrected using the program Calib v. 6.1 [16] and the Marine09 calibration dataset with a ΔR value of 791 ± 121 years to account for the reservoir effect (electronic supplementary material, table S1) [17].
3. Results and discussion
Seventeen specimens representing the modern population of king penguins on northern Macquarie Island, particularly the Isthmus population (figure 1), were characterized for 374 bp of the mitochondrial hyper variable region. Eighteen sub-fossil king penguin bones excavated from Landing Beach on the Isthmus yielded sufficient DNA to recover the homologous region. Radiocarbon dating revealed that seven of these bones aged on average 1002 years cal BP after calibration and correction for the marine reservoir effect (electronic supplementary material, table S1).
In addition, sub-fossil bones from Bauer Bay were analysed. Recently collected sub-fossil bones (D1 samples) and those from the McEvey museum collection (DE samples) failed to present amplifiable DNA, despite showing a remarkable degree of preservation. The reason for this failure is unlikely to be their older age, averaging 7927 years cal BP (electronic supplementary material, table S1), since other studies have amplified DNA from much older penguin remains [18]. It seems more likely that the absence of amplifiable DNA was due to the outcrop's deposition by riverine processes, which would have allowed for hydrolytic damage. This is attested by the wet-cardboard-like texture of the recently collected bones and the riverbank location of this site.
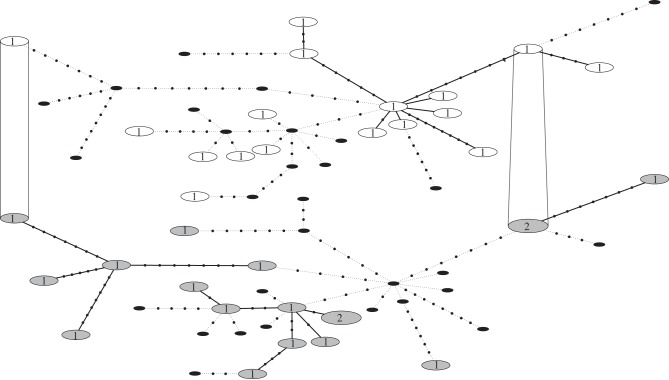
The haplotype network (figure 2) shows considerable genetic diversity in both ancient and modern populations. Most individuals possess unique haplotypes which do not cluster around a single ancestral haplotype, as is typically seen after strong population bottlenecks. Very few haplotypes are shared between the ancient and modern populations, although some are relatively closely related. Few corresponding ancient and modern haplotypes are expected since the original ancient Isthmus population went extinct, and the modern northern Macquarie Island population appears most likely to have been founded from the remnant Lusitania Bay population [10].
Figure 2.
Heterochronous haplotype network for ancient (shaded large ellipses) and modern (white large ellipses) samples; the two haplotypes that are shared between the haplotype networks are connected. Numbers represent the number of identical haplotypes. Small black ellipses are haplotypes that are represented on the opposing haplotype network. Black dots represent the hypothetical intermediate haplotypes.
Both the ancient and the modern populations on the Isthmus and surrounding areas show similar nucleotide diversity. Average nucleotide diversity estimates (nucleotide differences per site) of 0.0308 ± 0.0052 and 0.0268 ± 0.0046 were calculated for the ancient and modern dataset, respectively. These estimates show that the genetic diversity for both populations is very similar (Z-test = 0.5800, p = 0.5619). A relatively high level of genetic diversity must have survived in the founding Lusitania Bay population, as both the ancient and the modern populations are quite similar with the proportion of interpopulation nucleotide diversity estimated at 0.0050 ± 0.0054.
It is remarkable that a nearly extinct penguin population has been able to establish several new colonies and recover levels of past genetic diversity in only approximately 80 years. Conservation efforts have clearly aided the king penguin population to re-establish from the brink of extinction, also allowing for the expansion of considerable genetic diversity that had apparently survived in the last remaining individuals on Macquarie Island. Ancient DNA methodologies make it possible to monitor populations over time and, in combination with conservation genetics, allow for well-founded conservation and management strategies. Most studies combining ancient DNA with conservation genetics have shown a loss of genetic diversity in a wide range of species, be it because of anthropogenic or natural causes [19,20]. This study shows that the genetic diversity of a historically exploited population can recover to pre-human contact levels and exemplifies how ancient DNA studies can help evaluate conservation efforts and other human impacts on wildlife.
Acknowledgements
We thank Nancy Beavan, Leon Huynen and Sankar Subramanian for helpful discussions, Griffith University for providing T.H.H. with financial support and J.v.d.H. thanks Chris Oosthuizen and Ben Arthur for their efforts collecting the bone samples. Thanks also to the editor and two anonymous referees for helpful comments on an earlier draft.
References
- 1.Delord K., Barbraud C., Weimerskirch H. 2004. Long-term trends in the population size of king penguins at Crozet archipelago: environmental variability and density dependence? Polar Biol. 27, 793–800 10.1007/s00300-004-0651-z (doi:10.1007/s00300-004-0651-z) [DOI] [Google Scholar]
- 2.Gales R., Pemberton D. 1988. Recovery of the king penguin, Aptenodytes patagonicus, population on Heard Island. Wildl. Res. 15, 579–585 10.1071/WR9880579 (doi:10.1071/WR9880579) [DOI] [Google Scholar]
- 3.Marchant S., Higgins P. J., Ambrose S., Davies S., Steele W. K. 1990. Handbook of Australian, New Zealand and Antarctic birds. Melbourne, Australia: Oxford University Press [Google Scholar]
- 4.Wilson G. J. 1983. Distribution and abundance of Antarctic and sub-Antarctic penguins: a synthesis of current knowledge. Cambridge, UK: SCAR and SCOR [Google Scholar]
- 5.Mathews G. M. 1911. The birds of Australia. London, UK: Witherby [Google Scholar]
- 6.Mawson D. 1943. Macquarie Island: its geography and geology . Australian Antarctic Expedition 1911–14 Sydney, Australia: Government Printing Office [Google Scholar]
- 7.Woehler E., Croxall J. 1997. The status and trends of Antarctic and sub-Antarctic seabirds. Mar. Ornithol. 25, 43–66 [Google Scholar]
- 8.Rounsevell D., Copson G. 1982. Growth rate and recovery of a king penguin, Aptenodytes patagonicus, population after exploitation. Wildl. Res. 9, 519–525 10.1071/WR9820519 (doi:10.1071/WR9820519) [DOI] [Google Scholar]
- 9.Parks and Wildlife Service 2006. Macquarie Island nature reserve and World Heritage area management plan. Hobart, Tasmania: Parks and Wildlife Service [Google Scholar]
- 10.van den Hoff J., McMahon C. R., Field I. 2009. Tipping back the balance: recolonization of the Macquarie Island isthmus by king penguins (Aptenodytes patagonicus) following extermination for human gain. Antarct. Sci. 21, 237–241 10.1017/S0954102009001898 (doi:10.1017/S0954102009001898) [DOI] [Google Scholar]
- 11.McEvey A., Vestjens W. 1973. Fossil penguin bones from Macquarie Island, southern ocean. Proc. R. Soc. Vic. 86, 151–174 [Google Scholar]
- 12.Colhoun E., Goede A. 1973. Fossil penguin bones, 14C dates and the raised marine terrace of Macquarie Island: some comments. Search 4, 499–501 [Google Scholar]
- 13.Winters M., Barta J. L., Monroe C., Kemp B. M. 2011. To clone or not to clone: method analysis for retrieving consensus sequences in ancient DNA samples. PLoS ONE 6, e21247. 10.1371/journal.pone.0021247 (doi:10.1371/journal.pone.0021247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prost S., Anderson C. N. K. 2011. TempNet: a method to display statistical parsimony networks for heterochronous DNA sequence data. Methods Ecol. Evol. 2, 663–667 10.1111/j.2041-210X.2011.00129.x (doi:10.1111/j.2041-210X.2011.00129.x) [DOI] [Google Scholar]
- 15.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 (doi:10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuiver M., Reimer P. J. 1993. Extended 14C database and revised CALIB radiocarbon calibration program. Radiocarbon 35, 215–230 [Google Scholar]
- 17.de Bruyn M., Hall B. L., Chauke L. F., Baroni C., Koch P. L., Hoelzel A. R. 2009. Rapid response of a marine mammal species to Holocene climate and habitat change. PLoS Genet. 5, e1000554. 10.1371/journal.pgen.1000554 (doi:10.1371/journal.pgen.1000554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian S., Denver D. R., Millar C. D., Heupink T., Aschrafi A., Emslie S. D., Baroni C., Lambert D. M. 2009. High mitogenomic evolutionary rates and time dependency. Trends Genet. 25, 482–486 10.1071/WR9820519 (doi:10.1071/WR9820519) [DOI] [PubMed] [Google Scholar]
- 19.Leonard J. A. 2008. Ancient DNA applications for wildlife conservation. Mol. Ecol. 17, 4186–4196 10.1111/j.1365-294X.2008.03891.x (doi:10.1111/j.1365-294X.2008.03891.x) [DOI] [PubMed] [Google Scholar]
- 20.Shepherd L., Lambert D. 2008. Ancient DNA and conservation: lessons from the endangered kiwi of New Zealand. Mol. Ecol. 17, 2174–2184 10.1111/j.1365-294X.2008.03749.x (doi:10.1111/j.1365-294X.2008.03749.x) [DOI] [PubMed] [Google Scholar]