Abstract
The clock mechanism for circatidal rhythm has long been controversial, and its molecular basis is completely unknown. The mangrove cricket, Apteronemobius asahinai, shows two rhythms simultaneously in its locomotor activity: a circatidal rhythm producing active and inactive phases as well as a circadian rhythm modifying the activity intensity of circatidal active phases. The role of the clock gene period (per), one of the key components of the circadian clock in insects, was investigated in the circadian and circatidal rhythms of A. asahinai using RNAi. After injection of double-stranded RNA of per, most crickets did not show the circadian modulation of activity but the circatidal rhythm persisted without a significant difference in the period from controls. Thus, per is functionally involved in the circadian rhythm but plays no role, or a less important role, in the circatidal rhythm. We conclude that the circatidal rhythm in A. asahinai is controlled by a circatidal clock whose molecular mechanism is different from that of the circadian clock.
Keywords: Apteronemobius asahinai, circatidal clock, locomotor activity
1. Introduction
Circatidal rhythms are endogenous rhythms with a period of approximately 12.4 h corresponding to tidal flooding and ebbing, and have been documented in various organisms, mostly those living in the intertidal zone [1,2]. The clock mechanism for circatidal rhythms has long been controversial. There are two leading hypotheses: one is that circatidal rhythms are tidally adapted circadian rhythms [3,4], and the other is that a circatidal clock with a period of approximately 12.4 h produces the circatidal rhythm [5,6]. The former hypothesis is based on the premise that common molecular components control both the clock producing circatidal rhythms and the circadian clock. The oscillation of the circadian clock is regulated by cyclical expressions of circadian clock genes and proteins through feedback mechanisms [7]. However, the molecular mechanisms generating circatidal rhythms are completely unknown [2,8].
The mangrove cricket Apteronemobius asahinai, is endemic to mangrove forests, and forages on the forest floor during low tide and rests during high tide. Under constant darkness, adults of this species show a clear and persistent circatidal activity rhythm with a free-running period of approximately 12.6 h [9]. Moreover, a circadian rhythm modifying the circatidal rhythm by inhibiting activity during subjective day free-runs under constant darkness and can be entrained by light–dark cycles [9]. In insects, the molecular basis of the circadian clock has been extensively studied [10], and the predominant role of the gene period (per) is confirmed in some species, including a cricket, Gryllus bimaculatus [11]. Thus, the mangrove cricket is a suitable subject for examining the molecular basis of the circatidal rhythm.
In this study, we performed RNAi of per (per RNAi) to examine whether a clock possessing a common molecular basis with the circadian clock is involved in the circatidal rhythm in A. asahinai. The results showed that per RNAi disrupted the circadian modulation of the activity rhythm but did not affect the circatidal rhythm.
2. Material and methods
Adults of A. asahinai were collected from mangrove swamps in Kin (26°27′ N, 127°56′ E) and Ginoza (26°30′ N, 127°59′ E), Okinawa Prefecture, Japan in May–November, 2007–2009. A laboratory culture, originating from the collected crickets, was kept at 25.0 ± 1.0°C under 12 L : 12 D.
Total RNA was isolated from heads using Trizol (Invitrogen). cDNA synthesis and clone development for per and Elongation factor-1α (EF-1α) were performed as described by Ikeno et al. [12]. To obtain the complete sequence of per, 5′ and 3′ rapid amplification of cDNA ends were also performed using a Smart RACE cDNA Amplification Kit (Clontech). Nucleotide sequences of per and EF-1α were deposited in DDBJ/GenBank/EMBL (AB550300–AB00550302). Double-stranded RNAs (dsRNA) of approximately 500 bp for per (dsper) and for bacterial β-lactamase (dsbla), used as a control, were synthesized using T7 RiboMAX Express Large Scale RNA Production Systems (Promega). Primers are listed in the electronic supplementary material, table S1. The dsRNAs were suspended in water and their concentration was adjusted to 0.5 μg μl−1.
To verify gene silencing, quantitative real-time PCR with standard curve methodology was performed to measure per mRNA levels using GoTaq qPCR Master Mix (Promega) according to Ikeno et al. [12]. Male crickets within 10 days of adult emergence were collected from a laboratory colony, anaesthetized with CO2 and immediately placed on a slide glass, and injected with 1 μl of dsper or dsbla solution with a glass capillary into the abdomen. Five days after the treatment, survivors were individually collected at ZT 7, 13 and 19 (ZT stands for Zeitgeber time, and the onset of light is defined as ZT 0), and total RNAs were extracted from the head without antennae. EF-1α was used as a control gene for normalization.
Forty male crickets collected in the field were used for activity recording from the day or next day following collection. They were individually housed in plastic chambers with unlimited food and water supply. An infrared beam (EE SPW-321, Omron) was set across the chamber, and the number of interruptions of the beam was recorded at 6 min intervals on a personal computer [13]. The recording chambers were set in an incubator kept at 25°C and equipped with a 15 W fluorescent lamp (FL 15W, Panasonic) with an irradiance of approximately 1.0–3.5 W m−2.
Circatidal rhythmicity was determined by using a chi-square periodogram [14]. If there was a single peak between 10 and 15 h in the periodogram above the 0.05 confidence level, then the activity was judged to be circatidal. Circadian rhythmicity was determined by comparison of activity levels between the adjacent circatidal cycles. In A. asahinai, a circadian rhythm modifies activity levels in circatidal active phases [9]. When a circatidal rhythm was detected, the number of infrared beam interruptions was counted for every circatidal period shown by the chi-square periodogram. The difference in the activity levels counted for each circatidal period of even and odd number cycles was examined by analysis of covariance (ANCOVA) on the assumption that the activity levels regress linearly. The activity was judged to be influenced by a circadian rhythm when the slope or the elevation was significantly different between even and odd number cycles (p ≤ 0.05; electronic supplementary material, figure S1).
3. Results
Quantitative real-time PCR verified that RNAi of per is effective in reducing the expression of per mRNA and abolishes its temporally variable expression in A. asahinai (electronic supplementary material, figure S2).
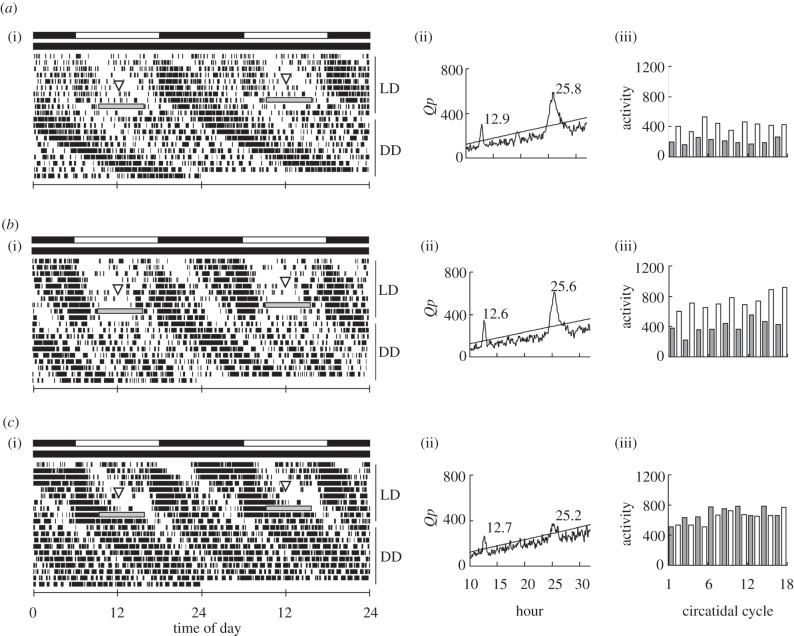
Locomotor activities were recorded under 12 L : 12 D for 10 days and under constant darkness for the subsequent 10 days. Crickets were anaesthetized, injected with dsbla or injected with dsper only at the middle of the photophase on the sixth day. All crickets except an intact one showed a circatidal rhythm irrespective of the treatment, and there was no significant difference in the period among the three groups (figures 1 and 2a). In the intact and dsbla-injected groups, a circadian rhythm was shown in approximately 90 per cent and 70 per cent of crickets, respectively (figures 1a,b and 2b). In the dsper-injected group, however, only one of 12 crickets showed a circadian rhythm and there were significant differences in the proportion of crickets with a circadian rhythm between the dsper-injected group and the other two groups (figures 1c and 2b).
Figure 1.
Examples of the locomotor activity rhythm in male adults of Apteronemobius asahinai (a) anaesthetized (control), (b) injected with double-stranded RNA for β-lactamase, and (c) injected with double-stranded RNA for per and transferred from light–dark cycles (LD) to constant darkness (DD). Injection was performed on the sixth day (triangles). (i) Double-plotted actograms, (ii) chi-square periodograms of activities in subsequent DD, and (iii) histograms of activity levels counted for each circatidal period in DD are shown. Black and white bars above the actograms indicate light and dark phases, respectively. Grey boxes indicate no data. The oblique line in the periodogram indicates the significance level of α = 0.05 and a peak value above the line is designated as significant. Grey and white histograms represent odd and even circatidal cycles, respectively.
Figure 2.
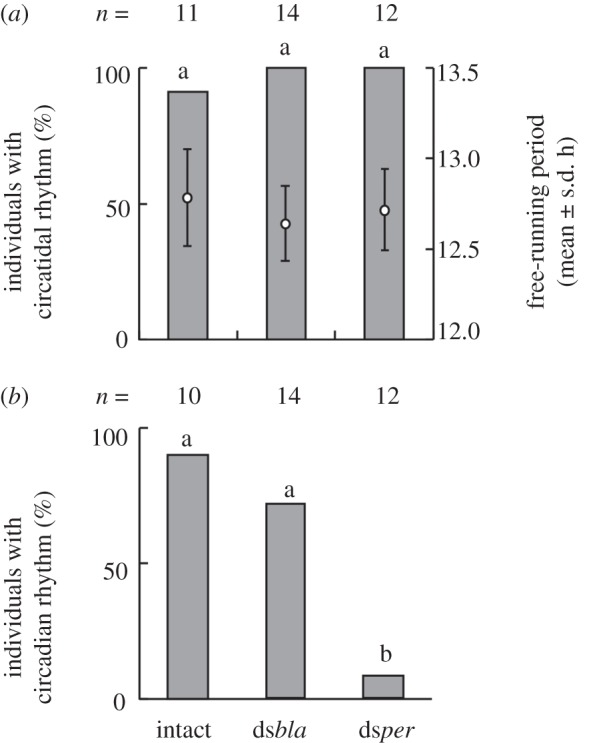
The effects of per RNAi on circatidal and circadian rhythms in male adults of Apteronemobius asahinai under constant darkness subsequent to light–dark cycles. Crickets were anaesthetized (intact control), injected with double-stranded RNA for β-lactamase (dsbla), or injected with double-stranded RNA for per (dsper). The proportions with the same letters in each panel are not significantly different (Tukey-type multiple comparisons for proportions, p > 0.05). (a) The proportion of crickets with a circatidal rhythm and its period determined by a chi-square periodogram. The circatidal periods were not significantly different among the three groups (p > 0.05, ANOVA). (b) The proportion of crickets with a circadian rhythm among those with a circatidal rhythm determined by ANCOVA for the activity levels of even and odd circatidal cycles (electronic supplementary material, figure S1).
4. Discussion
The hypothesis proposed by Palmer [1,3,4] that circatidal rhythms are tidally adapted circadian rhythms has been supported by the fact that circatidal rhythms are easily changed to circadian rhythms that can be entrained to light–dark cycles in a few species [15,16]. If this hypothesis is applicable to A. asahinai, then the circatidal rhythm should be disrupted by knockdown of per by RNAi because per is one of the key components of insect circadian clocks [10]. However, per RNAi disrupted the circadian modulation of activities but did not affect the circatidal rhythm. Thus, this hypothesis is not supported by present evidence.
Naylor [5,6] proposed the alternative hypothesis that a circatidal clock different from the circadian clock produces the circatidal rhythm. In A. asahinai, a clear and persistent circatidal activity rhythm is shown, and a circadian rhythm modifies the activity independently from the circatidal rhythm [9] as in some other intertidal animals [5,17,18]. In these species, therefore, existence of the circatidal clock independent of the circadian clock is probable. In A. asahinai, the phase-response curve for inundation pulses in the circatidal rhythm resembled that of circadian rhythms for light pulses, suggesting the existence of a circatidal clock with a period of approximately half that in circadian clocks [19]. Although the period is different between the circatidal and circadian clocks, the circatidal clock in A. asahinai could conceivably be driven by the same molecular mechanism as that of the circadian clock, because the circatidal clock has similar characteristics to a circadian clock in the phase response [19]. If this is the case, then knockdown of the circadian clock gene per should disrupt the circatidal rhythm in addition to the circadian modulation of activities. The present results, however, demonstrated that the circatidal rhythm of A. asahinai persisted even after disruption of the circadian modulation of activities by knockdown of per. It is, therefore, probable that the circatidal rhythm in A. asahinai is controlled by a circatidal clock in which per is not involved.
Acknowledgements
We thank Shuhei Hayaishi for collecting insects and Elizabeth Nakajima for linguistic corrections. This research was supported by a grant-in-aid (no. 20657017) from JSPS to H.N.
References
- 1.Palmer J. D. 1995. The biological rhythms and clocks of intertidal animals. New York, NY: Oxford University Press [Google Scholar]
- 2.Naylor E. 2010. Chronobiology of marine organisms. Cambridge, UK: Cambridge University Press [Google Scholar]
- 3.Palmer J. D., Williams B. G. 1986. Comparative studies of tidal rhythms. II. The dual clock control of the locomotor rhythms of two decapod crustaceans. Mar. Behav. Physiol. 12, 269–278 10.1080/10236248609378653 (doi:10.1080/10236248609378653) [DOI] [Google Scholar]
- 4.Palmer J. D. 2000. The clocks controlling the tide-associated rhythms of intertidal animals. BioEssays 22, 32–37 (doi:10.1002/(SICI)1521-1878(200001)22:1<32::AID-BIES7>3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 5.Naylor E. 1958. Tidal and diurnal rhythms of locomotor activity in Carcinus maenas (L.). J. Exp. Biol. 35, 602–610 [Google Scholar]
- 6.Naylor E. 1996. Crab clockwork: the case for interactive circatidal and circadian oscillators controlling rhythmic locomotor activity of Caracinus maenas. Chronobiol. Int. 13, 153–161 10.3109/07420529609012649 (doi:10.3109/07420529609012649) [DOI] [PubMed] [Google Scholar]
- 7.Dunlap J. C. 1999. Molecular bases for circadian clocks. Cell 96, 271–290 10.1016/S0092-8674(00)80566-8 (doi:10.1016/S0092-8674(00)80566-8) [DOI] [PubMed] [Google Scholar]
- 8.Wilcockson D., Zhang L. 2008. Circatidal clocks. Curr. Biol. 18, R753–R755 10.1016/j.cub.2008.06.041 (doi:10.1016/j.cub.2008.06.041) [DOI] [PubMed] [Google Scholar]
- 9.Satoh A., Yoshioka E., Numata H. 2008. Circatidal activity rhythm in the mangrove cricket Apteronemobius asahinai. Biol. Lett. 4, 233–236 10.1098/rsbl.2008.0036 (doi:10.1098/rsbl.2008.0036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomioka K., Matsumoto A. 2009. A comparative view of insect circadian clock systems. Cell. Mol. Life Sci. 67, 1397–1406 10.1007/s00018-009-0232-y (doi:10.1007/s00018-009-0232-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriyama Y., Sakamoto T., Karpova S. G., Matsumoto A., Noji S., Tomioka K. 2008. RNA interference of the clock gene period disrupts circadian rhythms in the cricket Gryllus bimaculatus. J. Biol. Rhythms 23, 308–318 10.1177/0748730408320486 (doi:10.1177/0748730408320486) [DOI] [PubMed] [Google Scholar]
- 12.Ikeno T., Numata H., Goto S. G. 2008. Molecular characterization of the circadian clock genes in the bean bug, Riptortus pedestris and their expression patterns under long- and short-day conditions. Gene 419, 56–61 10.1016/j.gene.2008.05.002 (doi:10.1016/j.gene.2008.05.002) [DOI] [PubMed] [Google Scholar]
- 13.Shiga S., Numata H., Yoshioka E. 1999. Localization of photoreceptor and pacemaker for the circadian activity rhythm in the band-legged ground cricket, Dianemobius nigrofasciatus. Zool. Sci. 16, 193–201 10.2108/zsj.16.193 (doi:10.2108/zsj.16.193) [DOI] [Google Scholar]
- 14.Sokolove P. G., Bushell W. N. 1978. The chi-square periodogram: its utility for analysis of circadian rhythms. J. Theor. Biol. 72, 131–160 10.1016/0022-5193(78)90022-X (doi:10.1016/0022-5193(78)90022-X) [DOI] [PubMed] [Google Scholar]
- 15.Gibson R. N. 1973. Tidal and circadian activity rhythms in juvenile plaice, Pleuronectes platessa. Mar. Biol. 22, 379–386 10.1007/BF00391398 (doi:10.1007/BF00391398) [DOI] [Google Scholar]
- 16.Akiyama T. 1997. Tidal adaptation of a circadian clock controlling a crustacean swimming behavior. Zool. Sci. 14, 901–906 10.2108/zsj.14.901 (doi:10.2108/zsj.14.901) [DOI] [Google Scholar]
- 17.Hastings M. H., Naylor E. 1980. Ontogeny of an endogenous rhythm in Eurydice pulchra. J. Exp. Mar. Biol. Ecol. 46, 137–145 10.1016/0022-0981(80)90027-1 (doi:10.1016/0022-0981(80)90027-1) [DOI] [Google Scholar]
- 18.Gray D. R., Hodgson A. N. 1999. Endogenous rhythms of locomotor activity in the high-shore limpet, Helcion pectunculus (Patellogastropoda). Anim. Behav. 57, 387–391 10.1006/anbe.1998.0975 (doi:10.1006/anbe.1998.0975) [DOI] [PubMed] [Google Scholar]
- 19.Satoh A., Yoshioka E., Numata H. 2009. Entrainment of the circatidal activity rhythm of the mangrove cricket, Apteronemobius asahinai, to periodic inundations. Anim. Behav. 78, 189–194 10.1016/j.anbehav.2009.04.018 (doi:10.1016/j.anbehav.2009.04.018) [DOI] [Google Scholar]