Abstract
The genome size in turtles and crocodiles is thought to be much larger than the 1.2 Gb of the chicken (Gallus gallus domesticus, GGA), according to the animal genome size database. However, GGA macrochromosomes show extensive homology in the karyotypes of the red eared slider (Trachemys scripta elegans, TSC) and the Nile crocodile (Crocodylus niloticus, CNI), and bird and reptile genomes have been highly conserved during evolution. In this study, size and GC content of all chromosomes are measured from the flow karyotypes of GGA, TSC and CNI. Genome sizes estimated from the total chromosome size demonstrate that TSC and CNI are 1.21 Gb and 1.29 Gb, respectively. This refines previous overestimations and reveals similar genome sizes in chicken, turtle and crocodile. Analysis of chromosome GC content in each of these three species shows a higher GC content in smaller chromosomes than in larger chromosomes. This contrasts with mammals and squamates in which GC content does not correlate with chromosome size. These data suggest that a common ancestor of birds, turtles and crocodiles had a small genome size and a chromosomal size-dependent GC bias, distinct from the squamate lineage.
Keywords: genome size, chromosome size, GC content, reptile genome evolution
1. Introduction
The animal genome database provides genome size data for 4972 species, including the chicken (Gallus gallus domesticus, GGA) with a genome size of 1.25 pg equivalent to 1.2 Gb [1]. The same database gives the C-value of the red eared slider (Trachemys scripta elegans, TSC) as 1.93–2.65 pg (1.9–2.6 Gb) and that of the Nile crocodile (Crocodylus niloticus, CNI) as either 2.84 or 3.95 pg (2.8/3.9 Gb). Despite the large differences in genome size between these three species according to the database, GGA macrochromosomes, comprising about 70 per cent of the genome, show extensive homology in the karyotypes of TSC and CNI by chromosome painting [2]. This suggests that their genomes are well conserved after divergence from their common ancestor and questions the accuracy of genome size in the database.
Theoretically, genome sizes can be determined by DNA sequencing at the highest resolution at nucleotide level, however sequence data do not cover the whole genome owing to unresolvable gaps and the omission of centromeres and telomeres. Alternatively, genome size can be calculated from the sum of chromosome sizes determined by flow-karyotypic analysis using a flow cytometer [3–7]. For example, the sum of chromosome sizes in human (HSA) [4] and dog [5] by this method gives a value of 3.15 Gb and 2.75 Gb, respectively. It has been assessed by sequence data that the euchromatic regions in the human and dog are 2.85 Gb [8] and 2.45 Gb [9], respectively, and based on these data their total genome (TG) sizes are estimated to be 3.08 Gb [8] and 2.68 Gb [9], respectively, confirming that chromosome measurements are a reliable method for genome size analysis.
The GC content of chromosomes can also be determined from differences of base pair ratios in the flow karyotype [7]. Compared with the HSA flow karyotype which shows dispersed peaks [4], gecko chromosomes are aligned in the flow karyotype to form a linear array of peaks [10], reflecting similar base pair ratios in each gecko chromosome. This is consistent with the results of genome sequencing in the green anole lizard (Anolis carolinensis, ACA) [11]. Chromosomal size-dependent GC compartmentalization is unique to birds and other reptiles whose karyotypes consist of macro- and microchromosomes [12]. However, crocodile chromosomes are different from those of birds and turtles that have numerous indistinguishable microchromosomes, and the chromosome GC content is unknown in CNI.
In this study, we use flow karyotypes of GGA, TSC and CNI as representative species of birds, turtles and crocodiles to measure each chromosome size and GC content. Based on our more precise measurements, we find similar genome sizes in chicken, turtle and crocodile, and a relatively higher GC content in the smaller chromosomes of these three species.
2. Material and methods
TSC and CNI cells were grown from embryonic tissues obtained from La Ferme aux Crocodiles. Chromosome suspensions for sorting and painting were made according to conventional protocols [13].
Normal male HSA lymphoblasts were used to provide a reference flow karyotype. For comparison, HSA (2n = 46), GGA (2n = 78), TSC (2n = 50) and CNI (2n = 32) chromosome samples were run on a flow cytometer (MoFlo, DAKO) separately but sequentially using the same settings. The DNA line was drawn from the origin through the peak position of HSA-4 [4] on GGA, TSC and CNI flow karyotypes. The chromosome sizes were calculated based on a value of 200 Mb for HSA-4 [4]. The result was calibrated by a separate estimation based on the peak positions of HSA-17 (89 Mb), which is comparatively GC-rich and, like chromosome 4, shows less polymorphism and relatively stable peak positions on the HSA flow karyotype. The calculation was confirmed by the size of HSA-19 (66 Mb). The GC content of each chromosome was calculated from the ratio obtained from the AT and GC fluorescence values based on the flow karyotype and the reference HSA GC content [14].
Each peak in the flow karyotype of GGA, TSC and CNI was sorted and the identity of chromosome-specific DNA was verified by hybridization to respective metaphases following the chromosomal idiogram for GGA [15] and karyotypes for TSC and CNI [2]. The identity of GGA chromosomes in the remaining peaks was determined by PCR analysis of chromosome-specific DNA using primer sets [16].
3. Results
(a). Flow karyotypes
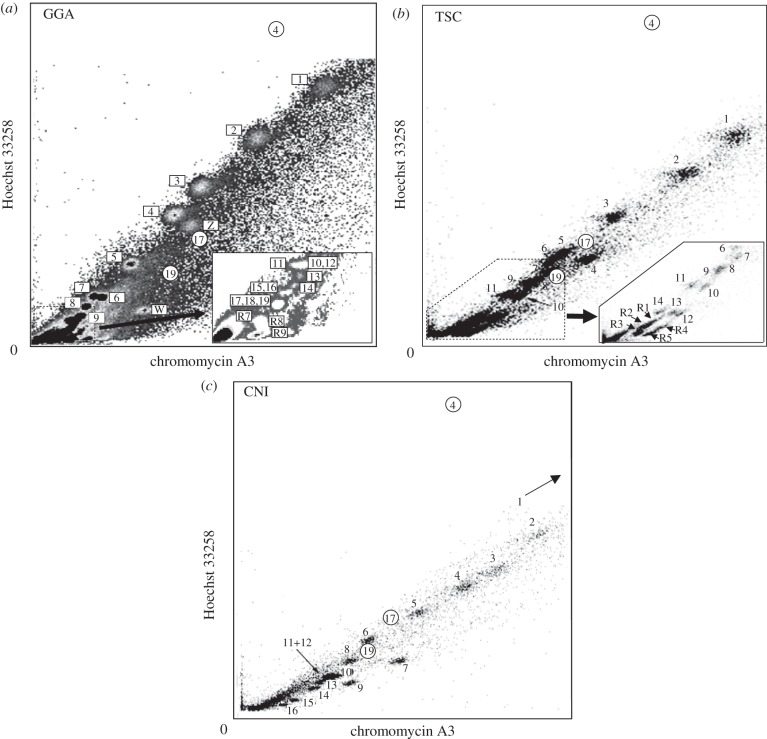
Bivariate flow karyotypes of GGA, TSC and CNI are shown in figure 1, including peak positions of HSA-4, -17 and -19 for reference. Small chromosomes have stronger chromomycin fluorescence intensity and thus a higher GC content. The GGA flow karyotype differs from previous reports [17,18] in which it shows the raw flow karyotype obtained after analysis of a large number of chromosomes without additional image processing.
Figure 1.
Bivariate flow karyotypes of (a) GGA, (b) TSC and (c) CNI. Smaller chromosomes in GGA and TSC were detected in higher resolution. CNI-1 is absent in this setting and appears in a higher position at lower resolution. Peak positions for HSA-4, -17 and -19 are marked by small circles.
Te 40 GGA chromosomes were resolved into 20 peaks (figure 1a). GGA peaks 6 and 7 in the flow karyotype [17] were assigned to chromosomes 7 and 6, respectively, to be consistent with the genome database. The identity of the 10 microchromosomes from GGA peaks R1–R6 [18] was determined in this paper. GGA-11 was detected in R1, GGA-10 and -12 in R2, GGA-13 in R3, GGA-14 in R4, GGA-15 and -16 in R5, GGA-17–19 in R6. Although, previously, GGA-R8 did not show specific signals [18], multiple signals were detected on microchromosomes in our experiments. However, for the remaining 19 microchromosomes, the GGA-R8 paint signals overlapped with R7 and R9, and the number in each peak could not be determined.
TSC has 14 pairs of macrochromosomes and 22 microchromosomes and the 25 chromosome pairs were resolved into 19 peaks (figure 1b). While TSC macrochromosomes showed a separate peak for each pair except for TSC-9 which included TSC-8, TSC-R1, -R2 and -R3 included three, four and two pairs of microchromosomes, respectively, and TSC-R4 and -R5 each contained one pair of microchromosomes. CNI chromosomes were resolved into 15 peaks (figure 1c), and each specific pair of chromosomes could be assigned to one peak, except for chromosomes 11 and 12 (figure 1c).
(b). Measurements on the flow karyotypes
Each chromosome size and GC content was estimated from the flow karyotype (table 1). GGA microchromosomes have a length of 8–21 Mb and a GC-content of 45–64%. CNI has a small chromosome CNI-16, 18 Mb in size, similar to microchromosomes in GGA and TSC. When they are grouped into lower (A) and higher (B) than average GC content, groups A of GGA, TSC, CNI and HSA consist of 10, 9, 6 and 13 chromosomes, respectively, which are relatively larger chromosomes in each species. Groups B of GGA, TSC, CNI and HSA consist of 29, 16, 10 and 10 chromosomes, respectively, which are smaller chromosomes with the exception of GGA-W, TSC-4 and HSA-1 (figure 2a). In the case of HSA chromosomes, the smaller chromosomes cannot be separated on the basis of GC content.
Table 1.
Chromosome size and GC content of each chromosome and TG.
| (a) GGA |
(b) TSC |
(c) CNI |
||||||
|---|---|---|---|---|---|---|---|---|
| size (Mb) | GC (%) | size (Mb) | GC (%) | size (Mb) | GC (%) | |||
| 1 | 186 | 41.6 | 1 | 179 | 45.0 | 1 | 244 | 45.9 |
| 2 | 147 | 41.2 | 2 | 147 | 45.2 | 2 | 169 | 46.5 |
| 3 | 111 | 41.0 | 3 | 108 | 44.8 | 3 | 141 | 47.8 |
| 4 | 92 | 41.2 | 4 | 83 | 48.6 | 4 | 125 | 47.7 |
| Z | 92 | 43.4 | 5 | 80 | 45.0 | 5 | 100 | 47.4 |
| 5 | 61 | 42.2 | 6 | 71 | 44.8 | 6 | 74 | 46.9 |
| W | 43 | 59.8 | 7 | 68 | 47.0 | 7 | 74 | 57.0 |
| 6 | 39 | 44.4 | 8 | 60 | 46.1 | 8 | 58 | 49.5 |
| 7 | 37 | 42.7 | 9 | 55 | 46.0 | 9 | 48 | 58.9 |
| 8 | 31 | 43.8 | 10 | 46 | 49.2 | 10 | 48 | 51.5 |
| 9 | 25 | 45.8 | 11 | 44 | 46.0 | 11 | 44 | 51.2 |
| 10 | 21 | 46.8 | 12 | 33 | 54.6 | 12 | 44 | 51.2 |
| 11 | 20 | 45.1 | 13 | 32 | 51.5 | 13 | 40 | 51.2 |
| 12 | 21 | 46.8 | 14 | 30 | 48.1 | 14 | 35 | 53.3 |
| 13 | 19 | 49.1 | R1 | 19 | 50.8 | 15 | 24 | 59.4 |
| 14 | 17 | 49.2 | R2 | 15 | 51.0 | 16 | 18 | 60.3 |
| 15 | 16 | 49.4 | R3 | 9 | 54.9 | TG | 1286 | 49.2 |
| 16 | 16 | 49.4 | R4 | 24 | 61.3 | |||
| 17 | 13 | 51.0 | R5 | 16 | 62.3 | |||
| 18 | 13 | 51.0 | TG | 1211 | 47.4 | |||
| 19 | 13 | 51.0 | ||||||
| R7 | 9 | 52.2 | ||||||
| R8 | 8 | 59.4 | ||||||
| R9 | 8 | 63.7 | ||||||
| TG | 1148 | 45.2 | ||||||
Figure 2.
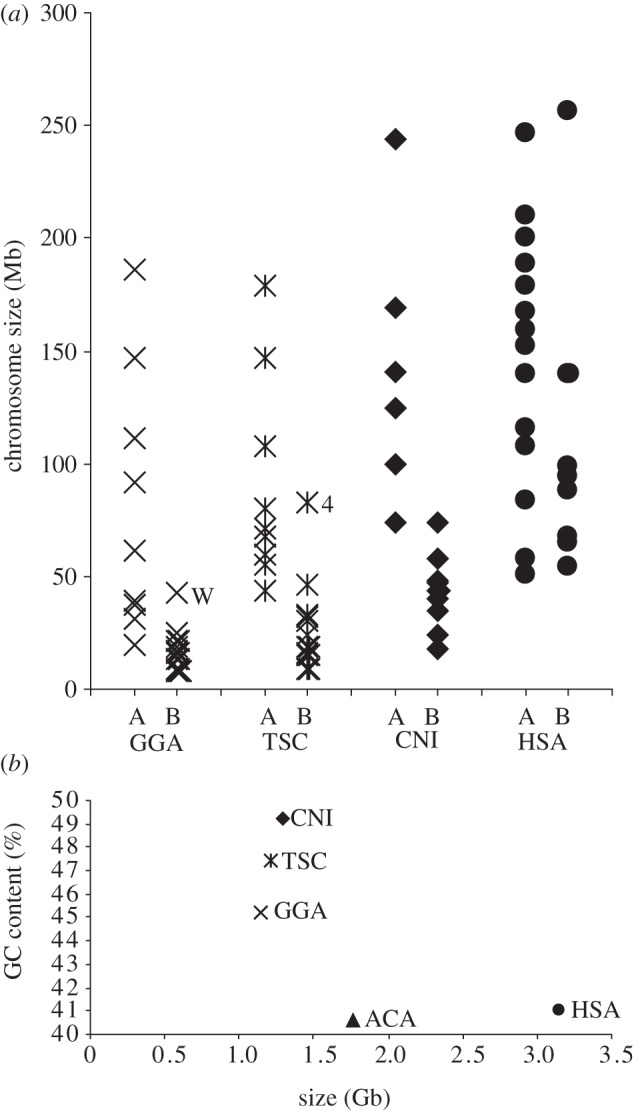
(a) GGA, CNI and TSC chromosomes classified into two groups based on GC content show strong relation to size. The CNI small chromosomes are larger than GGA and TSC microchromosomes, but have a higher GC content than the CNI large chromosomes. (b) The correlation between TG size and GC content is shown for the three species and compared with HSA.
TG size was calculated from the sum of individual chromosome sizes (table 1). The contribution to the TG size of the microchromosomes in GGA peaks R7–R9 was estimated from the average size of each peak. This gave a TG size of 1.148 Gb for GGA, approximately 37 per cent that of HSA, compared with 1.211 Gb for TSC and 1.286 Gb for CNI.
4. Discussion
TSC and CNI genome sizes based on the sum of chromosome sizes are close to GGA, but these sizes are smaller than those in the database [1]. The larger genome sizes were obtained from the measurement of nuclear DNA content at cellular level. Both methods differ from DNA sequencing in that they require a DNA reference point from another species. When the reference species used for nuclear measurement is distantly related from the target species, the result may be influenced by varying fluorescence intensity owing to species variation in cell or nuclear sizes. In contrast to cells, chromosome structure shows little variation between species, and human chromosomes can be used as a reference for many species, even for zebrafish [6]. Our measurement of 1.15 Gb for GGA is consistent with 1.05 Gb of the assembled sequence [19], showing that chromosome measurement has a high accuracy. Therefore, our method is to be preferred where whole-genome sequence is unavailable. Our data refine previous overestimations in turtles and crocodiles and may change earlier conclusions about genome evolution that are based on genome size using the database (see for example [20]).
As in mammals, the common amniote ancestor may have had a large genome [21] and a relatively low GC content [22]. ACA is 1.78 Gb in size with a 40.3 per cent total GC content according to the genome sequence data [11]. ACA has a few microchromosomes that are conserved between ACA and GGA, and that may have arisen in the reptile ancestor [11]. It is suggested that the formation of microchromosomes leads to a reduction of genome size [23]. We find that TSC and CNI genome sizes are closer to GGA than to ACA, and total GC content in GGA, TSC and CNI is higher than in ACA (figure 2b). This indicates that the reduction of genome size has occurred after the divergence from squamates and has involved the formation of additional microchromosomes. Consequently, a common ancestor of bird, turtle and crocodile might have a small genome size with a higher GC content when compared with the squamate lineage.
Most bird, turtle and squamate karyotypes consist of two types of chromosome based on size, macro- and microchromosomes, whereas CNI has only one considerably small chromosome. In contrast to HSA, whose smaller chromosomes sorted to both A and B groups, it is found that the larger chromosomes in GGA, TSC and CNI tend to have a lower than average GC content, whereas the smaller chromosomes have a higher than average GC content (figure 2a). ACA does not show this variation in GC content by chromosome size [11], suggesting that a common ancestor of bird, turtle and crocodile had two types of chromosomes correlated with chromosome size and GC content after divergence from the squamate ancestor.
References
- 1.Gregory T. R. 2012. Animal genome size database. See (http://www.genomesize.com) [Google Scholar]
- 2.Kasai F., O'Brien P. C. M., Martin S., Ferguson-Smith M. A. In press Extensive homology of chicken macrochromosomes in the karyotypes of Trachemys scripta elegans and Crocodylus niloticus revealed by chromosome painting despite the long divergence times. Cytogenet Genome Res. [DOI] [PubMed] [Google Scholar]
- 3.Harris P., Boyd E., Young B. D., Ferguson-Smith M. A. 1986. Determination of the DNA content of human chromosomes by flow cytometry. Cytogenet. Cell Genet. 41, 14–21 10.1159/000132190 (doi:10.1159/000132190) [DOI] [PubMed] [Google Scholar]
- 4.Trask B., van den Engh G., Mayall B., Gray J. W. 1989. Chromosome heteromorphism quantified by high-resolution bivariate flow karyotyping. Am. J. Hum. Genet. 45, 739–752 [PMC free article] [PubMed] [Google Scholar]
- 5.Langford C. F., Fischer P. E., Binns M. M., Holmes N. G., Carter N. P. 1996. Chromosome-specific paints from a high-resolution flow karyotype of the dog. Chromosome Res. 4, 115–123 10.1007/BF02259704 (doi:10.1007/BF02259704) [DOI] [PubMed] [Google Scholar]
- 6.Freeman J. L., et al. 2007. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genom. 8, 195. 10.1186/1471-2164-8-195 (doi:10.1186/1471-2164-8-195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renfree M. B., et al. 2011. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12, R81. 10.1186/gb-2011-12-8-r81 (doi:10.1186/gb-2011-12-8-r81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 10.1038/nature03001 (doi:10.1038/nature03001) [DOI] [PubMed] [Google Scholar]
- 9.Lindblad-Toh K., et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 10.1038/nature04338 (doi:10.1038/nature04338) [DOI] [PubMed] [Google Scholar]
- 10.Trifonov V. A., Giovannotti M., O'Brien P. C., Wallduck M., Lovell F., Rens W., Parise-Maltempi P. P., Caputo V., Ferguson-Smith M. A. 2011. Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res. 19, 843–855 10.1007/s10577-011-9241-4 (doi:10.1007/s10577-011-9241-4) [DOI] [PubMed] [Google Scholar]
- 11.Alföldi J., et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587–591 10.1038/nature10390 (doi:10.1038/nature10390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuraku S., Ishijima J., Nishida-Umehara C., Agata K., Kuratani S., Matsuda Y. 2006. cDNA-based gene mapping and GC3 profiling in the soft-shelled turtle suggest a chromosomal size-dependent GC bias shared by sauropsids. Chromosome Res. 14, 187–202 10.1007/s10577-006-1035-8 (doi:10.1007/s10577-006-1035-8) [DOI] [PubMed] [Google Scholar]
- 13.Rens W., Fu B., O'Brien P. C., Ferguson-Smith M. 2006. Cross-species chromosome painting. Nat. Protoc. 1, 783–790 10.1038/nprot.2006.91 (doi:10.1038/nprot.2006.91) [DOI] [PubMed] [Google Scholar]
- 14.Li W., Holste D. 2005. Universal 1/f noise, crossovers of scaling exponents, and chromosome-specific patterns of guanine–cytosine content in DNA sequences of the human genome. Phys. Rev. E. Stat. Nonlin. Soft. Matter. Phys. 71, 041 910 (doi:10.1103/PhysRevE.71.041910) [DOI] [PubMed] [Google Scholar]
- 15.Ladjali-Mohammedi K., Bitgood J. J., Tixier-Boichard M., Ponce De Leon F. A. 1999. International system for standardized avian karyotypes (ISSAK): standardized banded karyotypes of the domestic fowl (Gallus domesticus). Cytogenet. Cell Genet. 86, 271–276 10.1159/000015318 (doi:10.1159/000015318) [DOI] [PubMed] [Google Scholar]
- 16.Trukhina A. V., Smirnov A. F. 2010. Microsatellites from the linkage groups E26C13 and E50C23 are located on the Gallus gallus domesticus microchromosomes 20 and 21. Russ .J Genet. 46, 449–455 10.1134/S1022795410040101 (doi:10.1134/S1022795410040101) [DOI] [PubMed] [Google Scholar]
- 17.Griffin D. K., et al. 1999. Micro- and macrochromosome paints generated by flow cytometry and microdissection: tools for mapping the chicken genome. Cytogenet. Cell Genet. 87, 278–281 10.1159/000015449 (doi:10.1159/000015449) [DOI] [PubMed] [Google Scholar]
- 18.Nie W., O'Brien P. C., Ng B. L., Fu B., Volobouev V., Carter N. P., Ferguson-Smith M. A., Yang F. 2009. Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes): an atypical species with low diploid number . Chromosome Res. 17, 99–113 10.1007/s10577-009-9021-6 (doi:10.1007/s10577-009-9021-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Chicken Genome Sequencing Consortium 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 10.1038/nature03154 (doi:10.1038/nature03154) [DOI] [PubMed] [Google Scholar]
- 20.Organ C. L., Canoville A., Reisz R. R., Laurin M. 2011. Paleogenomic data suggest mammal-like genome size in the ancestral amniote and derived large genome size in amphibians. J. Evol. Biol. 24, 372–380 10.1111/j.1420-9101.2010.02176.x (doi:10.1111/j.1420-9101.2010.02176.x) [DOI] [PubMed] [Google Scholar]
- 21.Waltari E., Edwards S. V. 2002. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am. Nat. 160, 539–552 10.1086/342079 (doi:10.1086/342079) [DOI] [PubMed] [Google Scholar]
- 22.Shedlock A. M., Botka C. W., Zhao S., Shetty J., Zhang T., Liu J. S., Deschavanne P. J., Edwards S. V. 2007. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc. Natl Acad. Sci. USA 104, 2767–2772 10.1073/pnas.0606204104 (doi:10.1073/pnas.0606204104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burt D. W. 2002. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96, 97–112 10.1159/000063018 (doi:10.1159/000063018) [DOI] [PubMed] [Google Scholar]