Abstract
With an increasing amount of data becoming available, comparative analyses have called attention to the associations between cooperative breeding, monogamy and relatedness. We focus here upon the association between allomaternal care and relatedness among females within a social unit. Previous studies found a positive association, but such results date back to before molecular tools were in common use, they considered only a few mammalian orders, neglected phylogenetic clustering and/or did not correct for group sizes. Here, we use molecular data on relatedness from 44 species of mammals to investigate the phylogenetic clustering of, and the association between, allomaternal care and relatedness among females within a social unit. We find (i) a strong phylogenetic signal for allomaternal care and a moderate one for relatedness and group size, and (ii) a positive association between relatedness and allomaternal care, even when correcting for the smaller than average group sizes in species with allomaternal care. We also find that, in species without allomaternal care, adult females often live with unrelated females even when groups are small. We discuss these results in the light of recent evidence for the role of kin selection and the monogamy hypothesis in cooperative breeding.
Keywords: cooperative breeding, allomaternal care, relatedness, kin selection, phylogenetic signal
1. Introduction
Cooperatively breeding vertebrate systems are characterized by individual ‘helpers’ that take care of young ones within the social group that are not their own offspring—a behaviour termed ‘alloparental care’. In mammals, care typically encompasses allolactation, pup-feeding, babysitting and carrying young [1].
Kin selection theory suggests that helping behaviour can evolve whenever the benefits multiplied by genetic relatedness outweigh the costs [2]. Even though the role of kin selection in explaining the evolution of alloparental care in vertebrates has now been confirmed empirically, its relative importance is still disputed [3]. One issue that remains unclear is whether the degree of relatedness is higher within the social groups of cooperative breeders than in other species that live in groups but do not breed cooperatively [3].
A previous study on mammals found that 88 per cent of 63 species that live in family groups have alloparental care [4]. This overview can now be improved upon in several ways. First, this study dates back to before molecular techniques were in common use. Molecular tools have greatly expanded our knowledge about the social lives of animals, for example, by revealing relatedness and the partitioning of reproduction among group members of species that are sometimes hard to observe in the wild. Second, ungulates and primates were excluded from the review. Recent molecular evidence suggests that quite a few of the non-cooperatively breeding species in these taxa live in family groups, even for the dispersing sex. For example, in the western gorilla, Gorilla beringei beringei, related females emigrate to the same groups [5]. Third, previous results were based upon data at the species level and neglected the shared ancestry of the taxa, which is considered an inappropriate approach [6,7]. In birds, it is known that the distribution of cooperative breeding is not random with respect to phylogeny [8]. This may also be the case for mammals, although a thorough investigation of this remains to be carried out.
A recent comparative analysis, based on molecular data found that there is a positive association between cooperative breeding and monogamy in mammals [9]. All else being equal, we therefore expect a positive association between allomaternal care and within-group relatedness. However, group size or dispersal may affect within-group relatedness irrespective of reproductive skew [10]. In addition, allomaternal care also occurs in species with lower reproductive skew, e.g. communal breeders, which are considered here to be cooperative breeders as well.
Based upon a dataset of 44 species of mammals, we test here whether (i) allomaternal care, relatedness and group size are randomly distributed with respect to phylogeny, and if (ii) the mean relatedness among adult females within groups is higher in species with allomaternal care than in other species that live in stable groups but do not show allomaternal care. We use estimates of relatedness measured on microsatellite markers and perform a comparative analysis on species level data.
2. Material and methods
We consider all studies of mammals living in the wild in a defined social unit (see electronic supplementary material, S1 and S2), an aggregation of at least two adult females with at least one of them having dependent young, for which there was an estimate of the mean relatedness for adult females based on microsatellite marker data. For the dataset and further information on data collection methods, see electronic supplementary material, S1 and S2, respectively.
All analyses are done in R, v. 2.14.1 [11]. We analyse the data, including within-species variation, using the generalized least-squares methods [12] with the function ‘gls’ in the package nlme, v. 3.1–102 [13]. We also analyse the data using Bayesian phylogenetic mixed models [14] with the function ‘MCMCglmm’ in the package MCMCglmm, v. 2.15 [15]. Both methods give consistent results (see electronic supplementary material, S2).
One way to estimate if traits are constrained by the shared phylogenetic history of species, is to quantify the parameter lambda [16]. Lambda commonly ranges from 0 (independent from phylogeny) to 1 (covariance in trait(s) among species values fits a Brownian motion along the given tree). We estimate values of lambda using the functions ‘fitContinuous’ or ‘fitDiscrete’ (GEIGER v. 1.3-1 [17]). For model selection, we use the second-order Akaike Information Criterion (AICc [18]) and likelihood ratio (LR) tests. For more information, see the electronic supplementary material.
3. Results
In our dataset, 24 species out of 44 exhibited allomaternal care. The phylogenetic signal for the trait allomaternal care was strong and significantly different from 0 (λ = 1.00, AICc = 53.9, AICc for λ0 = 63.0, LR = 9.16, d.f. = 1, p = 0.002). Indeed, allomaternal care was common among the carnivores, but rare among the Chiroptera and primates (electronic supplementary material, S1). Mammalian social structures range from unrelated (on average) in several primates and bat species, up to related at the level of full-sibs (on average) in several carnivore species and the Damarland mole-rat (electronic supplementary material, S1). The phylogenetic signal for relatedness among adult females was moderate (λ = 0.50, AICc = −28.7, AICc for λ0 = −27.0, LR = 3.98, d.f. = 1, p = 0.046) and an analysis including intraspecific variation gives similar results (λ = 0.48, AICc = −25.2, AICc for λ0 = −23.5, LR = 3.96, d.f. = 1, p = 0.046). Average group size varied from two adult females up to almost 200 (electronic supplementary material, S1). It showed a moderate phylogenetic signal, but this failed to differ significantly from zero (λ = 0.72, AICc = −13.0 AICc for λ0 = −12.6, LR = 2.74, d.f. = 1, p = 0.098). The same is true when including intraspecific variation in the analysis (λ = 0.80, AICc = −7.5 AICc for λ0 = −6.8, LR = 2.96, d.f. = 1, p = 0.085).
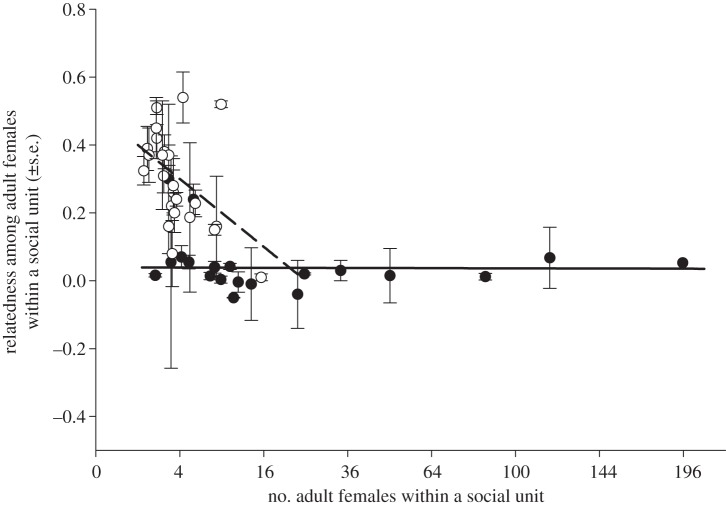
As the data displayed in figure 1 show, relatedness in species without allomaternal care did not differ significantly from zero (mean = 0.05, s.d. = 0.08, t = 1.9, p = 0.070), but was considerably lower than the relatedness in species with allomaternal care, in which females were related at the level of half-sibs on average (mean = 0.29, s.d. = 0.14). However, female group sizes were significantly smaller for species with allomaternal care than for species without (without: n = 20, mean = 31.1, s.d. = 48.9; with: n = 23, mean = 4.3, s.d. = 3.2). In a model including group size and allomaternal care as predictors (table 1), we found that species with allomaternal care had significantly lower relatedness than species without. We also found that the relatedness among adult females declines significantly with group size, but only for the species with allomaternal care. In species with allomaternal care, small groups are composed of relatives, while in species without allomaternal care groups consist mostly of unrelated individuals irrespective of group size (figure 1).
Figure 1.
Mean relatedness among adult females within a social unit (±s.e.) as a function of average number of females within a social unit. Species with allomaternal care (and significantly higher genetic relatedness values within social units) are shown as open symbols and broken regression lines. Species without allomaternal care (and lower relatedness within social unit) are shown as filled symbols and solid regression lines. Regression lines are the results of generalized least squares analysis. Values on the x-axis reflect that number of females within a social unit was square-root transformed prior to analysis.
Table 1.
The association between mean relatedness among adult females (including intraspecific variation) and allomaternal care for mammals in the dataset (n = 44 species, number of parameters = 4). Results are shown (i) without phylogenetic correction (ordinary least squares), and (ii) with phylogenetic correction (generalized least squares). The coefficient of intercept shows mean relatedness for species without allomaternal care.
| coefficient (s.e.) | F | p | AICc | |
|---|---|---|---|---|
| (i) without phylogeny | −18.4 | |||
| intercept | 0.03 (0.04) | |||
| allomaternal care | 0.50 (0.08) | 37.7 | <0.0001 | |
| sqrt(number of females) | −0.002 (0.008) | 0.07 | 0.787 | |
| allocare×sqrt(number of females) | −0.10 (0.03) | 11.6 | 0.002 | |
| (ii) with phylogeny | −18.5 | |||
| intercept | 0.04 (0.04) | |||
| allomaternal care | 0.50 (0.08) | 35.3 | <0.0001 | |
| sqrt(number of females) | −0.0009 (0.008) | 0.01 | 0.917 | |
| allocare×sqrt(number of females) | −0.10 (0.03) | 11.7 | 0.002 |
4. Discussion
In this study, we are able to show that for mammals: (i) allomaternal care exhibits a strong phylogenetic signal, as do relatedness and group size to a more moderate extent; (ii) species with allomaternal care live in groups that are on average smaller than species without allomaternal care; (iii) there is a negative association between relatedness and group size, but only for species with allomaternal care; and (iv) species with allomaternal care that live in small groups do so in groups that consist of more related individuals relative to species without allomaternal care with similar group sizes.
The strong phylogenetic signal for allomaternal care found here means that closely related taxa are more similar in this behaviour than distantly related taxa, which is rather exceptional for a behavioural trait [7]. This indicates that the evolutionary transition from independent to cooperative breeding (and vice-versa) is not straightforward. This may be because the evolution of social monogamy and cooperative breeding are linked [9], or because cooperative breeding mammals share a common set of life-history or ecological constraints, as was shown for birds [8]. In birds, some families contain more cooperative breeding taxa than others and it would now be interesting to quantify the phylogenetic signal of cooperative breeding in birds using this impressive dataset [8].
Relatedness showed a moderate phylogenetic signal. This result indicates that some traits generating the genetic structure of groups (perhaps reproductive skew or dispersal) are at least moderately conserved across the mammal phylogeny. The next step is to explain which combination of life-history traits or ecological variables generate this signal. Group size also showed a moderate phylogenetic signal. This fits with previous results [6,7] that showed a similar or even stronger signal for some mammalian subgroups. Although our dataset is biased towards cooperative breeders, our results indicate that this holds for mammals in general.
The second result, that mammals with allomaternal care live in smaller social groups, is consistent with previous results [19]. This can be caused by, among other things, a higher reproductive skew [9] and perhaps a lower birth rate in these species. The third result, that relatedness declines with group size only for species with allomaternal care, clarifies the findings of a previous study [10]. This result is interesting since, owing to high skew and long tenure of dominants [9], one might expect cooperative breeders to produce almost exclusively full siblings. Overall, both results are consistent with the finding that, in smaller groups, subordinate reproduction can be more easily controlled and/or that, perhaps more generally, within-group competition is limited.
The fourth and most important result is the positive association between allomaternal care and relatedness. These results suggest that helpers gain indirect fitness benefits helping, although the importance of direct fitness benefits remains unclear. The higher within-group relatedness in species with allomaternal care co-varies with other life-history traits, such as dispersal, extra-pair paternity and reproductive skew [9].
Our dataset also reveals that allomaternal care can occur in species with low mean relatedness among adult females, as found for allosuckling wild boars (Sus scrofa) and for pup-guarding greater spear-nosed bats (Phyllostomus hastatus). If indirect benefits are important in these systems, they may have considerable levels of kin discrimination to cope with the presumably large variation in genetic relatedness among group members [20]. Overall, however, in cooperatively breeding mammals mean relatedness is high, group sizes are small and variance in relatedness is relatively low, and this may explain the low levels of kin discrimination found in the few species of cooperative breeding mammals that have been studied thus far [20].
Acknowledgements
Thanks to Ashleigh Griffin, Jan Komdeur, Simon Verhulst and Tom Wenseleers for useful discussions, and to two anonymous reviewers for valuable comments.
Reference
- 1.Lewis S. E., Pusey A. E. 1997. Factors influencing the occurrence of communal care in plural breeding mammals. In Cooperative breeding in mammals (eds Solomon N. G., French J. A.), pp. 335–363 New York, NY: Cambridge University Press [Google Scholar]
- 2.Hamilton W. D. 1964. Genetical evolution of social behaviour I. J. Theor. Biol. 7, 1–16 10.1016/0022-5193(64)90038-4 (doi:10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 3.Clutton-Brock T. 2002. Behavioral ecology—breeding together: kin selection and mutualism in cooperative vertebrates. Science 296, 69–72 10.1126/science.296.5565.69 (doi:10.1126/science.296.5565.69) [DOI] [PubMed] [Google Scholar]
- 4.Emlen S. T. 1995. An evolutionary theory of the family. Proc. Natl Acad. Sci. USA 92, 8092–8099 10.1073/pnas.92.18.8092 (doi:10.1073/pnas.92.18.8092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley B. J., Doran-Sheehy D. M., Vigilant L. 2007. Potential for female kin associations in wild western gorillas despite female dispersal. Proc. R. Soc. B 274, 2179–2185 10.1098/rspb.2007.0407 (doi:10.1098/rspb.2007.0407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freckleton R. P., Harvey P. H., Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726 10.1086/343873 (doi:10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 7.Blomberg S. P., Garland T., Ives A. R. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1111/j.0014-3820.2003.tb00285.x (doi:10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 8.Cockburn A. 2006. Prevalence of different modes of parental care in birds. Proc. R. Soc. B 273, 1375–1383 10.1098/rspb.2005.3458 (doi:10.1098/rspb.2005.3458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukas D., Clutton-Brock T. 2012. Cooperative breeding and monogamy in mammalian societies. Proc. R. Soc. B 279 10.1098/rspb.2011.2468 (doi:10.1098/rspb.2011.2468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukas D., Reynolds V., Boesch C., Vigilant L. 2005. To what extent does living in a group mean living with kin? Mol. Ecol. 14, 2181–2196 10.1111/j.1365-294X.2005.02560.x (doi:10.1111/j.1365-294X.2005.02560.x) [DOI] [PubMed] [Google Scholar]
- 11.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 12.Martins E. P., Hansen T. F. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667 10.1086/286013 (doi:10.1086/286013) [DOI] [Google Scholar]
- 13.Pinheiro J., Bates D., DebRoy S., Sarkar D. & R Core Team 2011. nlme: linear and nonlinear mixed effects models R package. See http://cran.r-project.org/web/packages/nlme/index.html.
- 14.Hadfield J. D., Nakagawa S. 2010. General quantitative genetic methods for comparative biology: phylogenies, taxonomies, and multi-trait models for continuous and categorical characters. J. Evol. Biol. 23, 494–508 10.1111/j.1420-9101.2009.01915.x (doi:10.1111/j.1420-9101.2009.01915.x) [DOI] [PubMed] [Google Scholar]
- 15.Hadfield J. D. 2010. MCMC methods for multi-response generalised linear mixed models: the MCMCglmm R package. J. Stat. Soft. 33, 1–22 [Google Scholar]
- 16.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884 10.1038/44766 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 17.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W. 2008. Geiger: investigating evolutionary radiations. Bioinformatics 24, 129–131 10.1093/bioinformatics/btm538 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 18.Burnham C. A., Anderson D. R. 2002. Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn New York, NY: Springer [Google Scholar]
- 19.Packer C., Lewis S., Pusey A. E. 1992. A comparative analysis of non-offspring nursing. Anim. Behav. 43, 265–281 10.1016/S0003-3472(05)80222-2 (doi:10.1016/S0003-3472(05)80222-2) [DOI] [Google Scholar]
- 20.Cornwallis C. K., West S. A., Griffin A. S. 2009. Routes to indirect fitness in cooperatively breeding vertebrates: kin discrimination and limited dispersal. J. Evol. Biol. 22, 2445–2457 10.1111/j.1420-9101.2009.01853.x (doi:10.1111/j.1420-9101.2009.01853.x) [DOI] [PubMed] [Google Scholar]