Abstract
Mitogen-activated protein kinase kinase 4 (MKK4) is a critical mediator of stress-activated protein kinase signals that regulate apoptosis, inflammations, and tumorigenesis. Several polymorphisms have been identified in the MKK4 gene. We hypothesized that genetic variants in the MKK4 promoter may alter its functions and thus cancer risk. In the current, hospital-based case-control study of 471 cervical cancer cases and 600 sex and age frequency-matched cancer-free controls in an Eastern Chinese population, we genotyped two common polymorphisms in the MKK4 promoter region (-1304T>G, rs3826392 and -1044A>T, rs3809728)c and assessed their associations with the risk of cervical cancer. We found that compared with the most common -1304TT genotype, carriers of -1304G variant genotypes had a significantly decreased risk of cervical cancer [odds ratio (OR)=0.71; 95% confidence interval (CI)=0.53–0.92 for TG, and OR=0.52; 95%CI=0.30–0.91 for GG] in an allele dose-response manner (adjusted Ptrend=0.004). Moreover, the luciferase assay showed that the G allele in the promoter significantly increased the transcription activity of the MKK4 gene in vitro and that the MKK4 mRNA expression levels of the G variant carriers was significantly higher in tumor tissues than those of the -1304TT genotype. However, no significant association was observed between the -1044A>T polymorphism and risk of cervical cancer. Our data suggest that the functional -1304G variant in the MKK4 promoter contributes to a decreased risk of cervical cancer by increasing the promoter activity and that the G variant may be a marker for susceptibility to cervical cancer.
Introduction
As the second most common malignancy, cervical cancer is a serious health problem and a leading cause of cancer death in Chinese women. Nearly 500,000 new cases are diagnosed each year worldwide, 80% of which are in developing countries (Chen et al., 2011). It is well recognized that persistent infection of carcinogenic human papillomavirus (HPV) is the main cause of cervical cancer. The prevalence of HPV is popular. However, only a small portion of infected women develop cervical cancer, indicating that genetic factors may play an important role in the etiology of this malignancy. The role of genetic factors in the etiology of this malignancy has been documented (Magnusson et al., 1999; Sun et al., 2005; Jiang et al., 2010).
Epidemiological studies of cervical cancer have established many etiologic factors, including HPV infection (Galani and Christodoulou, 2009), smoking, air pollution, radiation, and long-term use of contraceptives (Mori and Sagae, 2001). These factors are all stressors to the cells that activate the mitogen-activated protein kinase (MAPK) pathways, which are known to regulate apoptosis, inflammations, and tumorigenesis (Maruyama et al., 2009). Mitogen-activated protein kinase kinase 4 (MKK4) belongs to a network of MAPK pathways, in which MKK4 in turn activates c-Jun NH2-terminal kinases and p38 (Cuenda, 2000), eliciting a variety of biological responses to extracellular signals that include growth factors, hormones, pro-inflammatory cytokines, and various stress stimuli (Cuenda, 2000). It has been found that MKK4 plays an important role in tumor formation and development (Chae et al., 2002; Cunningham et al., 2006; Spillman et al., 2007). Homozygous deletions of MKK4 exons were founded in pancreatic carcinoma cell lines, lung carcinoma cell lines, and other cancer cells (Teng et al., 1997; Su et al., 1998). Moreover, about 5% of tumors from various human tissues showed loss-of-function mutations in the MKK4 gene (Chae et al., 2002, Su et al., 2002). Further, MKK4 has been identified as a suppressor of metastasis of prostate and ovarian cancers (Spillman et al., 2007; Robinson et al., 2008), and lack of expression of the MKK4 gene in resected gastric adenocarcinoma was found to be highly associated with poor survival (Cunningham et al., 2006).
Human MKK4 gene encodes a 399-amino acid protein, spans over 120 kb on chromosome 17p11.2, and contains 11 exons. In its 1.6kb promoter region, four common polymorphisms (i.e., minor allele frequency [MAF]>5%) were identified according to our resequencing data of 30 normal Han Chinese subjects (Wei et al., 2009): -1304T>G [1304nt upstream to initiation transcription code ATG (A in bold indicate the boundary); rs3826392], -1044A>T [rs3809728], -641C>G [rs2190853], and -284T>C [rs9892151]. Because the pairs of three single-nucleotide polymorphisms (SNPs) (i.e., -1044A>T, -641C>G, and -284T>C) were in complete linkage disequilibrium (LD) (Biewenga et al., 2011) (r2=1.00, D'=1.00, for each pairs), two of our previous studies (Wei et al., 2009; Liu et al., 2010) analyzed the association between MKK4-1304T>G and -1044A>T and the risk of colorectal cancer and lung cancer. We found that the polymorphism -1304T>G have reduced risks for both colorectal cancer and lung cancer (Wei et al., 2009; Liu et al., 2010), by increasing the promoter activity (Liu et al., 2010). Because various environmental factors of cervical cancer can also activate MKK4 and MAPK pathways, we hypothesized that genetic variants in the MKK4 promoter are associated with the risk of cervical cancer.
Based on this hypothesis, we carried out two independent, hospital-based case-control studies to investigate the relation between the polymorphisms in MKK4 promoter and risk of cervical cancer in Eastern Chinese population.
Materials and Methods
Study population
The present study included 471 cervical cancer patients and 600 healthy controls. All subjects were ethnically homogeneous Han Chinese. Patients with newly diagnosed cervical cancer were recruited consecutively from March 2001 to May 2010 at the First Affiliate Hospital of Soochow University (Suzhou, China). All eligible patients diagnosed at the hospital during the study period were invited to participate in the study, with 91% agreeing to do so. There were no restrictions in terms of age, stage of disease, or histology preventing people from participating in the study. The population controls consisted of cancer-free people living in the Suzhou region; they were selected from a nutritional survey conducted in the same period as the cases were collected. The control subjects were selected by random-digit dialing method from a database consisting of 3500 individuals based on a physical examination (convenience sample comprising laboratory staff, welders, students, farmers, nurses, and company staff). The selection criteria for control subjects included no history of cancer and the control population was matched in terms of age and sex with the cervical cancer patient group. At the time of recruitment, informed consent was obtained from each subject. This case-control set have been previously published (Zheng et al., 2011a; Zheng et al., 2011b; Jiang et al., 2011). This study was approved by the Medical Ethics Committee of The First Affiliate Hospital of Soochow University.
Genotyping analysis
Genomic DNA was extracted from a 5 mL peripheral blood sample, which was obtained from patients before they were given chemotherapy or radiotherapy to avoid any influence of these treatments on outcomes. Genotypes were analyzed using PCR-based methods as described below. Genotyping was performed without knowledge of the subject's case or control status. A 30% masked random sample of cases and controls was tested twice by different people, and the results were concordant for all masked duplicate sets. We developed a PCR-RFLP method to determine cervical cancer associated MKK4 polymorphisms. The primer pair designed to amplify the target DNA fragment containing the -1304T>G (rs3826392) polymorphism was 5′-CTT GTT CCA AAC CCA ATT TC-3′ (forward) and 5′-GGG CTA CTG ATT TCC AGA TG-3′ (reverse), which produced a 232-bp fragment. Similarly, the primer pair designed for -1044A>T (rs3809728) was 5′-CTA CGA TTT GTA AGC CAA CCA-3′ (forward) and 5′-CCA ACA TGC TGT GAA GAA CTC-3′ (reverse), which produced a 235-bp fragment. The PCR was performed in a 25-μL reaction system containing 5 mM MgCl2, 0.1 mM dNTPs, 3.0 units Taq polymerase, and the manufacturer's buffer (Fermentas). The PCR procedure consisted of an initial melting step at 94°C for 5 min, followed by 35 cycles of 94°C for 45 s, annealing at 58°C for -1304T>G and 60°C for -1044A/T for 45 s and 72°C for 45 s, with a final extension step at 72°C for 7 min. A native endonuclease AflII (Fermentas) site was present in the amplified fragment containing the -1304G>T (rs3826392) polymorphism. After digestion by AflII at 37°C for at least 3 h, the major T allele produced a single 232-bp band, whereas the minor G allele produced two bands (111 and 121 bp). The two bands could be easily separated by 3% agarose gel electrophoresis. The amplified fragment containing the -1044A>T (rs3809728) polymorphism could be cut using Tsp509I (Fermentas) at 65°C for at least 3 h. After digestion, the major A allele produced two bands (129 and 106 bp), whereas the minor T allele produced a single 235-bp band. Besides, 60 samples were selected to confirm the genotypes by DNA sequencing.
Construction of reporter plasmids
Because the MKK4-1304T>G polymorphism was found to be associated with significantly decreased risk of cervical cancer, we then determined whether this polymorphism had an effect on its gene expression in vitro. The T allelic reporter constructs were prepared by amplifying the 1612-bp MKK4 promoter region (from −1528 to +84 bp relative to the translation start site) from subjects homozygous for the T allele (-1304TT), including the artificial KpnI and Hind3 enzymes restriction sites with forward primer 5′-GCCGGTACCtaatctgtagtgctgcttcta-3′ and reverse primer 5′-TGGAAGCTTcgccggggaccctacggggc-3′. The amplified fragments were then cleaved with the KpnI and Hind3 enzymes (New England BioLabs). The pGL3-basic vector (Promega) was also cleaved with the KpnI and Hind3 enzymes, and the above-prepared fragment and pGL3-basic vector were then ligated by T4 DNA ligase (New England BioLabs). The p1304T-1044T, p1304G-1044A, and p1304G-1044T reporter constructs were obtained from the p1304T-1044A constructs by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were sequenced to confirm the allele, orientation, and integrity of each insert.
Transient transfections and luciferase assays
HeLa and CaSki cells were grown in RPMI 1640 supplemented with 10% (v/v) heat-inactivated FCS, 2 mM L-glutamine, 100 units/mL penicillin, and 100 units/mL streptomycin at 37°C and 5% CO2 in a humidified incubator. For transient transfection experiments, 5×104 cells were plated in 10-mm 24-multiwell plates and grown to 60∼70% confluence. Transfection was carried out using LipofectAMINE Reagent (Life Technologies, Inc.) according to the manufacturer's protocol. Cells were co-transfected with 0.5 μg of reporter plasmid and 0.1 μg of pRL-SV40 (Luciferase Assay System; Promega); the latter was used to standardize transfection efficiency. Luciferase activity was determined according to the manufacturer's protocol using a luciferase assay system (Promega). Briefly, cells were scraped into lysis reagent, transferred to microfuge tubes, and centrifuged for 30s at 12,000×g. Luciferase activity was measured using a manual luminometer (Turner Designs; TD20/20) by mixing 100 μL of luciferase assay reagent with 20 μL of 1:10-diluted cell lysate, and the value for each sample was recorded three times at 10-s intervals. For each plasmid construct, three independent transfection experiments were performed, and each was done in triplicate. Results were expressed as a ratio of luciferase activity to pRL-SV40.
Real-time analysis of MKK4 mRNA
Thirty cervical cancer tissues were obtained from biopsy removed specimens of individual patients. The tissues were immediately placed in liquid nitrogen and then stored at −80°C before analysis. Total RNA was isolated from cervical cancer tissues using TRIzol reagent (Molecular Research Center Inc.). An aliquot of total RNA (2 μg) from each specimen was reverse transcribed into single-strand complementary DNA using oligo primer and SuperscriptII (Invitrogen). Relative gene expression quantitation for MKK4, with β-actin as an internal reference gene, was performed using the ABI Prism 7000 sequence detection system (Applied Biosystems) based on the SYBR-Green method. The primers used for MKK4 were 5′-aacaacactgggatttcact-3′ and 5′-tcactactccgcattactaca-3′ and for β-actin were 5′-ggcggcaccaccatgtaccct-3′ and 5′-aggggccggactcgtcatact-3′. The PCR reaction mixture consisted of 0.1 μM of each primer, 1× SYBR Premix EX-Taq (Perfect Real Time) premix reagent (TaKaRa), and 50 ng complementary DNA to a final volume of 20 μL. Cycling conditions were 95°C for 2 min, followed by 40 cycles at 95°C for 15s and 60°C for 1 min. The expression of individual MKK4 measurements was calculated relative to expression of β-actin using a modification of the method described by Lehmann and Kreipe (2001). All analyses were performed in a blinded fashion with the laboratory persons unaware of genotyping data.
Statistical analysis
Two-sided chi-square tests were used to assess differences in the distributions of age between cases and controls and the allele and genotypes. The Hardy-Weinberg equilibrium (HWE) was tested by a goodness-of-fit chi-square test to compare the expected genotype frequencies with observed genotype frequencies (p2+2pq+q2=1) in cancer-free controls. The association between case-control status and each SNP, measured by the odds ratio (OR) and its corresponding 95% confidence interval (CI), was estimated using an unconditional logistic regression model, with adjustment for age and body mass index (BMI). Logistic regression modeling was also used for the trend test (Lu et al., 2007; Zheng et al., 2011b).
Data were further stratified by age, BMI, family history of cancer, clinical stage, tumor grade, and histological type to evaluate the stratum variable-related ORs among the MKK4 genotypes. Homogeneity among stratum variable-related ORs was tested (Lu et al., 2007). The potential multiplicative and additive interactions among gene-gene and gene-environmental factors were also evaluated by means of logistic regression analysis (Liu et al., 2010). The 2LD program and the PROC ALLELE statistical procedure in SAS/Genetics (SAS Institute Inc.) software were used to detect the LD of the two SNPs. The statistical power was calculated by using the PS Software (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). The normalized expression values in cancer tissue of MKK4 between each genotype were analyzed by Kruskal-Wallis one way analysis of variance (ANOVA). We also used linear regression model to evaluate the main effect of MKK4 genotype on gene's expression with adjustment for age and sex. The tests were all two-sided and analyzed using the SAS software (version 9.1; SAS Institute). p<0.05 was considered statistically significant.
Results
Characteristics of the study population
The distributions of selected characteristics of cervical cancer patients and controls are summarized in Table 1. Overall, median age was 44 years (range 21–84) for case patients, and 45 years (range 16–84) for control subjects (p=0.647).
Table 1.
Characteristics of Cervical Cancer Patients and Controls in Eastern Chinese Population Used for the Association Study
| |
Patients (n=471) |
Controls (n=600) |
|
||
|---|---|---|---|---|---|
| Characteristics | No. | (%) | No. | (%) | p |
| Age at diagnosis, year | |||||
| <38 | 112 | 23.78 | 177 | 29.50 | |
| 38–44 | 136 | 28.87 | 126 | 21.00 | |
| 45–51 | 112 | 23.78 | 148 | 24.67 | |
| ≥52 | 111 | 23.57 | 149 | 24.83 | 0.647 |
| Body Mass Index (BMI) | |||||
| ≤20 | 115 | 24.41 | 97 | 16.16 | |
| 20–28 | 336 | 71.34 | 436 | 72.67 | |
| ≥28 | 20 | 4.25 | 67 | 11.17 | 0.002 |
| Family History of Cancer | |||||
| Positive | 37 | 7.86 | 44 | 7.33 | |
| Negative | 434 | 92.14 | 556 | 92.67 | 0.743 |
| Stage | |||||
| 0 | 79 | 16.77 | |||
| I | 136 | 28.87 | |||
| II | 183 | 38.86 | |||
| III | 68 | 14.44 | |||
| IV | 5 | 1.06 | |||
| Tumor Grade | |||||
| Low | 43 | 9.13 | |||
| Intermediate | 216 | 45.86 | |||
| High | 132 | 28.02 | |||
| Unknown | 80 | 16.99 | |||
| Histological type | |||||
| Squamous cell carcinoma | 359 | 76.22 | |||
| Adenocarcinoma | 25 | 5.31 | |||
| Adenosquamous carcinoma | 7 | 1.48 | |||
| Carcinoma in situ | 80 | 16.99 | |||
| HPV infection status | |||||
| Positive | 395 | 83.86 | |||
| Negative | 76 | 16.14 | |||
HPV, human papillomavirus.
Genotypes and risk of cervical cancer
The genotyping results are given in Table 2. The allele frequencies for -1304G and -1044A were 0.228 and 0.818, respectively, in the control group and 0.172 and 0.820, respectively, in cervical cancer patients. The observed genotype frequencies of -1304T>G and -1044A>T polymorphisms in both controls and patients did not deviate from those expected based on HWE (p=0.175 for -1304T>G; and p=0.297 for -1044A>T).
Table 2.
Genotype Frequencies of the Two Single-Nucleotide Polymorphism s in Mitogen-Activated Protein Kinase Kinase 4 in Patients and Controls and Their Associations with Cervical Cancer
| |
Controls (n=600) |
Patients (n=471) |
|
||
|---|---|---|---|---|---|
| Genotypes | No. | (%) | No. | (%) | ORa(95% CI) |
| -1304T>G | |||||
| TT | 364 | 60.66 | 326 | 69.21 | 1.00 (Reference) |
| TG | 199 | 33.17 | 128 | 27.18 | 0.71 (0.53–0.92) |
| GG | 37 | 6.17 | 17 | 3.61 | 0.52 (0.30–0.91) |
| Ptrend | 0.004 | ||||
| TG+GG | 236 | 39.33 | 145 | 30.79 | 0.68 (0.52–0.87) |
| -1044A>T | |||||
| AA | 405 | 67.50 | 318 | 67.52 | 1.00 (Reference) |
| AT | 172 | 28.67 | 136 | 28.87 | 1.01 (0.75–1.31) |
| TT | 23 | 38.33 | 17 | 3.61 | 0.92 (0.45–1.62) |
| Ptrend | 0.943 | ||||
| AT+TT | 195 | 32.50 | 153 | 32.48 | 0.99 (0.75–1.31) |
Data were calculated by logistic regression analysis with adjustment for age and BMI.
OR, odds ratio; CI, confidence interval.
The frequencies for the -1304GG, TG, and TT genotypes in cervical cancer patients significantly differed from those in the control group (Ptrend=0.004). Relative to the -1304TT genotype, -1304TG and -1304GG were both associated with a significantly decreased risk of cervical cancer, with an OR of 0.71 (95%CI=0.53–0.92) and 0.52 (95%CI=0.30–0.91), respectively. However, no significant association was observed between the -1044A>T polymorphism and risk of cervical cancer in our study population.
LD analysis showed a weak linkage for -1304T>G and -1044A>T (r2=0.013, D'=0.064) in our study population. It suggested that these two polymorphism sites are relatively independent and unsuitable for haplotype analysis.
Stratification analysis of MKK4-1304 T>G genotypes and risk of cervical cancer
The risk of cervical cancer related to MKK4 genotype was further examined with stratification according to age, BMI, family history of cancer, clinical stage, tumor grade, histological type, and HPV infection status. The early stage (0 and I) cervical cancer patients have correlated with favorable prognosis, compared with patients with II, III, and IV stage cancer (Biewenga et al., 2011). Therefore, the clinic stage was categorized in two groups (0+I group and II+III+IV group). However, as indicated in Table 3, there was no significant association between the subgroups and MKK4-1304 T>G genotypes.
Table 3.
Stratification Analysis of the Mitogen-Activated Protein Kinase Kinase 4-1304T>G Genotypes by Selected Variables in Cervical Cancer Patients and Controls
| |
Patients (n=487) |
Controls (n=723) |
Adjusted OR (95% CI)a |
|
||
|---|---|---|---|---|---|---|
| TT N (%) | TG+GG N (%) | TT N (%) | TG+GG N (%) | TG+GG versus TT | p-valueb | |
| Age (years) | ||||||
| ≤48 | 220 (68.32) | 102 (31.68) | 244 (62.89) | 144 (37.11) | 0.79 (0.57–1.07) | |
| >48 | 106 (71.14) | 43 (28.86) | 120 (56.60) | 92 (43.40) | 0.53 (0.34–0.83) | 0.16 |
| Body mass index (BMI) | ||||||
| ≤20 | 76 (66.09) | 39 (33.91) | 63 (64.95) | 34 (35.05) | 0.95 (0.54–1.69) | |
| 20–28 | 234 (69.64) | 102 (30.36) | 262 (60.09) | 174 (39.91) | 0.66 (0.49–0.89) | |
| ≥28 | 16 (80.00) | 4 (20.00) | 39 (58.21) | 28 (41.79) | 0.35 (0.11–1.15) | 0.28 |
| Family history of cancer | ||||||
| Positive | 26 (70.27) | 11 (29.73) | 25 (56.82) | 19 (43.18) | 0.56 (0.22–1.40) | |
| Negative | 300 (69.12) | 134 (30.88) | 339 (60.97) | 217 (39.03) | 0.70 (0.54–0.91) | 0.64 |
| Clinical Stage | ||||||
| 0+I | 154 (71.63) | 61 (28.37) | 364 (60.67) | 236 (39.33) | 0.61 (0.44–0.86) | |
| II+III+IV | 172 (67.19) | 84 (32.81) | 364 (60.67) | 236 (39.33) | 0.75 (0.55–1.03) | 0.37 |
| Tumor grade | ||||||
| Low | 29 (67.44) | 14 (32.56) | 364 (60.67) | 236 (39.33) | 0.74 (0.39–1.44) | |
| Intermediate | 145 (67.13) | 71 (32.87) | 364 (60.67) | 236 (39.33) | 0.76 (0.54–1.05) | |
| High | 96 (72.73) | 36 (27.27) | 364 (60.67) | 236 (39.33) | 0.58 (0.38–0.88) | |
| Unknown | 56 (70.00) | 24 (30.00) | 364 (60.67) | 236 (39.33) | 0.66 (0.40–1.10) | 0.79 |
| Histological type | ||||||
| Squamous cell carcinoma | 248 (69.08) | 111 (30.92) | 364 (60.67) | 236 (39.33) | 0.69 (0.52–0.91) | |
| Adenocarcinoma | 17 (68.00) | 8 (32.00) | 364 (60.67) | 236 (39.33) | 0.73 (0.31–1.71) | |
| Adenosquamous carcinoma | 5 (71.43) | 2 (28.57) | 364 (60.67) | 236 (39.33) | 0.62 (0.12–3.21) | |
| Carcinoma in situ | 56 (70.00) | 24 (30.00) | 364 (60.67) | 236 (39.33) | 0.66 (0.40–1.10) | 0.97 |
| HPV infection status | ||||||
| Positive | 53 (69.74) | 23 (30.26) | 364 (60.67) | 236 (39.33) | 0.67 (0.40–1.12) | |
| Negative | 273 (69.11) | 122 (30.89) | 364 (60.67) | 236 (39.33) | 0.69 (0.53–0.90) | 0.92 |
ORs were adjusted for age in a logistic regression model.
P value of the test for homogeneity between stratum-related ORs for MKK4 gene (-1304 TG+GG vs. TT genotypes).
Effects of MKK4-1304 T>G polymorphism on transcriptional activity
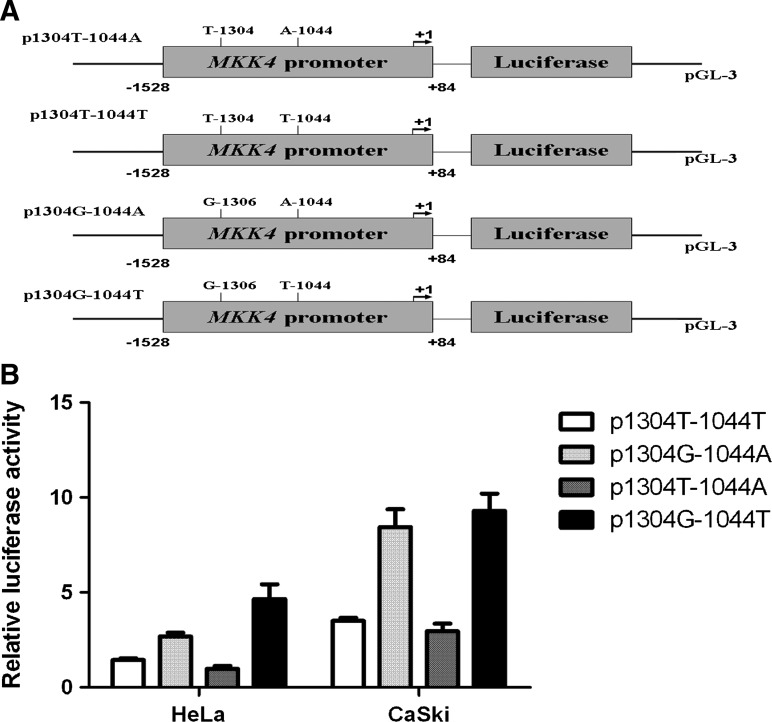
To directly determine the transcriptional activity of MKK4 native promoter, two luciferase reporter gene constructs were generated by PCR, spanning -1528 to +84 bp of the MKK4 promoter region, with a T or G at the -1304 polymorphic sites, and they were used to transiently transfect cervical cancer cell lines. As shown in Figure 1, we found that -1304G-containing MKK4 promoter drove a 1.8∼4.7-fold increased reporter expression compared with the -1304T-containing counterpart in both HeLa and CaSki cell line.
FIG. 1.
Transient reporter gene expression assays with constructs containing full length MKK4 promoter. (A) Schematic of reporter gene constructs having a full-length MKK4 promoter with the only difference between the four constructs being a T>G or A>T at the -1304 and -1044 polymorphic sites. (B) Luciferase expression of four constructs in HeLa and CaSKi cells co-transfected with pRL-SV40 to standardize transfection efficiency. Luciferase levels of pGL3-Basic and pRL-SV40 were determined in triplicate and standardized for transfection efficiency. Fold increase was measured by defining the activity of the empty pGL3-Basic vector as 1. Data shown are the mean fold increase 6 SD from three independent transfection experiments, each done in triplicate. MKK4, mitogen-activated protein kinase kinase 4.
Effects of MKK4-1304 T>G SNP on MKK4 mRNA levels
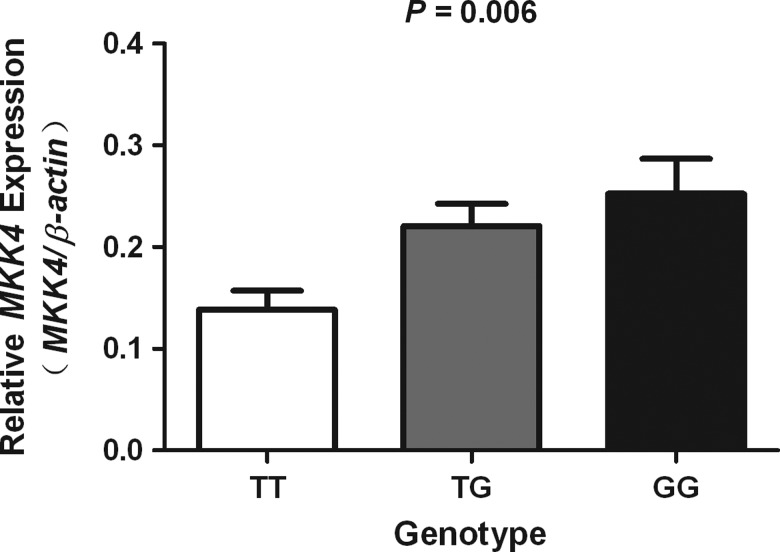
The effect of -1304T>G SNP on MKK4 expression was additionally examined by real-time PCR quantization of MKK4 mRNA in individual cervical cancer tissues. It was found that subjects with the -1304TG and -1304GG genotypes had significantly higher MKK4 mRNA levels (mean±SD) than those with the -1304TT genotype (TT, 0.138±0.019, n=15; TG, 0.221±0.022, n=11; GG, 0.253±0.034, n=4; one way ANOVA: p=0.006; linear regression test: p=0.0002), as shown in Figure 2.
FIG. 2.
MKK4 mRNA expression level in cervical cancer tissues as function of MKK4-1304T>G genotypes. Expression level among the −1304GG (n=15) genotype was significantly different from that among the TG (n=11) or TT (n=4) genotype (p=0.006).
Discussion
In the present study investigating 471 cervical cancer patients and 600 cancer-free controls, we found that the -1304G>T polymorphism in the MKK4 promoter was associated with cervical cancer risk, and the risk decreased as the number of -1304G allele increased in an allele dose-response manner. Moreover, in vitro assays, we found that the -1304G variant allele significantly increased the transcription activity of the MKK4 gene compared with the -1304T allele. However, for the -1044A>T polymorphism, no statistical evidence was found for any association between this polymorphism and cervical cancer risk.
Previous studies found that the carriers of MKK4-1304GG genotype had a 60% and 38% decreased risk of colorectal cancer and lung cancer, respectively, in Chinese Han cancer cases and cancer-free controls (Wei et al., 2009; Liu et al., 2010). Another study of our group also found that the carriers of -1304GG genotype had a 43% disease risk in 1237 Chinese Han nasopharyngeal carcinoma patients and 1328 cancer-free controls (Zheng et al., 2011b). Similarly, now we found that the carriers of -1304GG genotype also had a 47% decreased cervical cancer risk in Eastern Chinese population and we further found that the -1304G variant allele significantly increased the transcription activity of the MKK4 gene compared with the -1304T allele in vitro. Moreover, MKK4 belongs to a non-tissue specific network of MAPK pathways, which is thought to be a key modulator in cervical cancer pathogenesis just like lung cancer and colorectal cancer. Taken previous reports (Wei et al., 2009; Liu et al., 2010; Zheng et al., 2011b) and current findings together, our data suggest that the -1304G variant allele is a protective factor for different cancers. Nevertheless, further studies of this polymorphism and other cancers would be beneficial to confirm the findings.
In recent years, it has been established that the JNK and p38 MAPK pathways both play a functional role in the pathogenesis and pathophysiology of cervical cancer (Liu et al., 2001; Kloth et al., 2005; Kang and Lee, 2008). MKK4 is a direct activator of both JNK and p38, and most of published studies supported the MKK4 gene as a candidate tumor suppressor (Teng et al., 1997; Whitmarsh and Davis, 2007). We hypothesized that dysfunction caused by MKK4 mutations may possibly affect susceptibility to the development of cervical cancer. Our hypothesis was supported by the results of our genotyping and transient transfection experiments. We found that the change of the -1304T to G allele in the promoter region of the MKK4 gene increased the transcriptional activity of the MKK4 gene as assessed by the luciferase assays in vitro. These results indicate that the -1304T>G polymorphism is a functional locus, which further supports our findings to be biologically plausible. The present study is the only study thus far to investigate the association between MKK4 mutations and susceptibility to cervical cancer.
Because many studies have investigated the MKK4-1304T>G polymorphism, it can provide a comparison for the tested frequencies of the genotypes. A previous study of colorectal cancer found that the frequency of the TT, TG, and GG alleles was 53.8%, 40.8%, and 5.4%, respectively, in 723 Chinese control subjects (Wei et al., 2009). Similarly, in another study of lung cancer in 1056 controls, the frequency of the TT, TG, and GG alleles was 57.3%, 35.9%, and 6.8%, respectively (Liu et al., 2010). These values are similar to the frequency of 60.6%, 33.2%, and 6.2% for the TT, TG, and GG alleles, respectively, determined in the 600 control subjects in the present study. The corresponding figures for these genotypes in the HapMap database are 63.1, 33.3, and 3.6% in 84 Chinese, 74.4, 23.3, and 2.3% in 86 Japanese, 54.0, 41.6, and 4.4% in 113 European descendents and 17.0, 42.9, and 40.2% in 112 Africans (http://hapmap.ncbi.nlm.nih.gov/:HapMap Genome Browser Phase 1, 2, and 3-merged genotypes and frequencies). These data suggest that the role of the MKK4-1304T>G polymorphism in cancer risk may vary with ethnicity, a possibility that warrants further investigation.
This study may have some limitations, because it was a hospital-based case-control study, restricted to Chinese Han subjects. The fact that the genotype frequencies among controls could fit the Hardy-Weinberg disequilibrium law suggested the randomness of subject selection; we have achieved a 84.5% study power (two-sided test, α=0.05) to detect an OR of 0.68 for the -1304G genetic genotypes compared with the -1304 TT genotype. Further, the finding of an association between the promoter -1304T>G polymorphism and risk of cervical cancer had been supported by additional functional assays. Therefore, these evidences suggest that our findings are sound and of note.
In conclusion, the present study indicates that, in eastern Chinese population, carriers of the -1304GG and GT genotypes have a decreased risk of cervical cancer compared with carriers of the MKK4-1304TT genotype. To the best of our knowledge, the present study is the first to demonstrate a significant association between the MKK4-1304G>T polymorphism and the risk of cervical cancer. Larger, and preferably population-based, case-control studies, and well-designed mechanistic studies are warranted to validate our findings.
Disclosure Statement
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Scientific Foundation of China grants 81001278 and 81171895, Jiangsu Provincial Natural Science Foundation (No. BK2011297), Jiangsu Province's Key Medical Department in 2011, and Beijing Nova Program.
References
- Biewenga P. van der Velden J. Mol B.W. Stalpers L.J. Schilthuis M.S. van der Steeg J.W., et al. Prognostic model for survival in patients with early stage cervical cancer. Cancer. 2011;117:768–776. doi: 10.1002/cncr.25658. [DOI] [PubMed] [Google Scholar]
- Chae K.S. Ryu B.K. Lee M.G. Byun D.S. Chi S.G. Expression and mutation analyses of MKK4, a candidate tumour suppressor gene encoded by chromosome 17p, in human gastric adenocarcinoma. Eur J Cancer. 2002;38:2048–2057. doi: 10.1016/s0959-8049(02)00147-8. [DOI] [PubMed] [Google Scholar]
- Chen X. Li H. Qiao Y. Yu D. Guo H. Tan W., et al. Association of CD28 gene polymorphism with cervical cancer risk in a Chinese population. Int J Immunogenet. 2011;38:51–54. doi: 10.1111/j.1744-313X.2010.00969.x. [DOI] [PubMed] [Google Scholar]
- Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) Int J Biochem Cell Biol. 2000;32:581–587. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- Cunningham S.C. Kamangar F. Kim M.P. Hammoud S. Haque R. Iacobuzio-Donahue C., et al. MKK4 status predicts survival after resection of gastric adenocarcinoma. Arch Surg. 2006;141:1095–1099. doi: 10.1001/archsurg.141.11.1095. discussion 100. [DOI] [PubMed] [Google Scholar]
- Galani E. Christodoulou C. Human papilloma viruses and cancer in the post-vaccine era. Clin Microbiol Infect. 2009;15:977–981. doi: 10.1111/j.1469-0691.2009.03032.x. [DOI] [PubMed] [Google Scholar]
- Jiang P. Liu J. Li W. Zeng X. Tang J. Role of p53 and p21 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim Biophys Sin (Shanghai) 2010;42:671–676. doi: 10.1093/abbs/gmq069. [DOI] [PubMed] [Google Scholar]
- Jiang L. Zhang C. Li Y. Yu X. Zheng J. Zou P., et al. A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population. Breast Cancer Res Treat. 2011;126:487–495. doi: 10.1007/s10549-010-1094-6. [DOI] [PubMed] [Google Scholar]
- Kloth J.N. Fleuren G.J. Oosting J. de Menezes R.X. Eilers P.H. Kenter G.G., et al. Substantial changes in gene expression of Wnt, MAPK and TNFalpha pathways induced by TGF-beta1 in cervical cancer cell lines. Carcinogenesis. 2005;26:1493–1502. doi: 10.1093/carcin/bgi110. [DOI] [PubMed] [Google Scholar]
- Kang Y.H. Lee S.J. The role of p38 MAPK and JNK in Arsenic trioxide-induced mitochondrial cell death in human cervical cancer cells. J Cell Physiol. 2008;217:23–33. doi: 10.1002/jcp.21470. [DOI] [PubMed] [Google Scholar]
- Lehmann U. Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- Liu B. Chen D. Yang L. Li Y. Ling X. Liu L., et al. A functional variant (-1304T>G) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis. 2010;31:1405–1411. doi: 10.1093/carcin/bgq126. [DOI] [PubMed] [Google Scholar]
- Liu B. Fang M. Lu Y. Mills G.B. Fan Z. Involvement of JNK-mediated pathway in EGF-mediated protection against paclitaxel-induced apoptosis in SiHa human cervical cancer cells. Br J Cancer. 2001;85:303–311. doi: 10.1054/bjoc.2001.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Wang L.E. Xiong P. Sturgis E.M. Spitz M.R. Wei Q. 172G>T variant in the 5' untranslated region of DNA repair gene RAD51 reduces risk of squamous cell carcinoma of the head and neck and interacts with a P53 codon 72 variant. Carcinogenesis. 2007;28:988–994. doi: 10.1093/carcin/bgl225. [DOI] [PubMed] [Google Scholar]
- Magnusson P.K. Sparen P. Gyllensten U.B. Genetic link to cervical tumours. Nature. 1999;400:29–30. doi: 10.1038/21801. [DOI] [PubMed] [Google Scholar]
- Maruyama J. Naguro I. Takeda K. Ichijo H. Stress-activated MAP kinase cascades in cellular senescence. Curr Med Chem. 2009;16:1229–1235. doi: 10.2174/092986709787846613. [DOI] [PubMed] [Google Scholar]
- Mori M. Sagae S. [Recent progress in epidemiologic research of uterine cancer] Gan To Kagaku Ryoho. 2001;28:174–178. [PubMed] [Google Scholar]
- Robinson V.L. Shalhav O. Otto K. Kawai T. Gorospe M. Rinker-Schaeffer C.W. Mitogen-activated protein kinase kinase 4/c-Jun NH2-terminal kinase kinase 1 protein expression is subject to translational regulation in prostate cancer cell lines. Mol Cancer Res. 2008;6:501–508. doi: 10.1158/1541-7786.MCR-07-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. Zhou Y. Li H. Han X. Shi Y. Wang L., et al. FASL -844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med. 2005;202:967–974. doi: 10.1084/jem.20050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G.H. Hilgers W. Shekher M.C. Tang D.J. Yeo C.J. Hruban R.H., et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- Su G.H. Song J.J. Repasky E.A. Schutte M. Kern S.E. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- Spillman M.A. Lacy J. Murphy S.K. Whitaker R.S. Grace L. Teaberry V., et al. Regulation of the metastasis suppressor gene MKK4 in ovarian cancer. Gynecol Oncol. 2007;105:312–320. doi: 10.1016/j.ygyno.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng D.H. Perry W.L., 3rd Hogan J.K. Baumgard M. Bell R. Berry S., et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- Wei Y. Wang L. Lan P. Zhao H. Pan Z. Huang J., et al. The association between -1304T>G polymorphism in the promoter of MKK4 gene and the risk of sporadic colorectal cancer in southern Chinese population. Int J Cancer. 2009;125:1876–1883. doi: 10.1002/ijc.24575. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. Davis R.J. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26:3172–3184. doi: 10.1038/sj.onc.1210410. [DOI] [PubMed] [Google Scholar]
- Zheng J. Zhang C. Jiang L. You Y. Liu Y. Lu J., et al. Functional NBS1 polymorphism is associated with occurrence and advanced disease status of nasopharyngeal carcinoma. Mol Carcinog. 2011a;50:689–696. doi: 10.1002/mc.20803. [DOI] [PubMed] [Google Scholar]
- Zheng J. Liu B. Zhang L. Jiang L. Huang B. You Y., et al. The protective role of polymorphism MKK4-1304 T>G in nasopharyngeal carcinoma is modulated by Epstein-Barr virus' infection status. Int J Cancer. 2011b doi: 10.1002/ijc.26253. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]