Abstract
ΦC31 integrase, a site-specific recombinase, can catalyze integration of circular DNA bearing attB site into pseudo attP sites in mammalian genomes. However, the integration efficiency mediated by integrase is relatively low. Our study centered on the investigation of the impact of the position, orientation, and number of attBs in the donor plasmid on the efficiency of ΦC31 integrase system. Donor plasmids bearing various types of attBs (including forward and reverse directions, tandem, and intersperse) and reporter enhanced green fluorescent protein (EGFP) were constructed. The plasmids plus helper plasmid encoding integrase were co-transfected into HeLa cells. After G418 selection, the resistant cell colonies were counted for calculating chromosomal integration frequency. EGFP expression was detected by fluorescence-activated cell sorter and enzyme-linked immunosorbent assay analysis. The results showed that efficiency of integration mediated by integrase accounted for 70%±7.1% of total integration events in the transfected HeLa cells. Compared with a forward orientation of attB in donor plasmid, a reverse direction of attB or interspersed attBs showed 1.5- or 2.8-fold increase in integration efficiency, respectively, while tandem attBs in donor plasmids caused a decreased efficiency of integration. We conclude that the adjustment of attB sites in donor plasmids may be of value for gene therapy and routine genetic engineering by using ΦC31 integrase system.
Introduction
ΦC31 integrase, a member of a serine-catalyzed superfamily, was first described as a recombinase of 613 amino acids (Kuhstoss and Rao, 1991). ΦC31 integrase mediates recombination between the 34-bp bacterial attachment site attB and the 39-bp phage attachment site attP (Groth et al., 2000), allowing integration of the phage genome into the bacterial host chromosome. Co-transfecting cells with donor plasmid bearing attB site and helper plasmid encoding ΦC31 integrase results in integration at preintegrated wild-type attP or native cellular pseudo attP sites in the mammalian genomes. Pseudo attP sites share partial sequence identity to the wild-type attP sequence. Previous studies found that integration of target genes at some pseudo attPs was predominant (Groth et al., 2000; Ou et al., 2008). In addition, many pseudo attP sites resided in open chromatins that might lead to a high expression of the integrated gene at these sites mediated with integrase (Chalberg et al., 2006). These features make ΦC31 integrase a useful tool for genetic engineering, and gene therapy.
However, ΦC31 integrase system showed relatively low efficiency compared with other nonviral system such as transposon system (Franz et al., 2011). Improved versions of ΦC31 integrase system were recently developed. Hyperactive integrase was made by synthesizing host codon-optimized ΦC31 integrase, or screening mutants for more efficient recombination (Raymond and Soriano, 2007; Keravala et al., 2009). Recently, Tasic et al. (2011) found that the efficiency of precise cassette exchange between attP and attB was significantly improved by inserting attP sites into a novel locus, H11 of the mouse genome.
The attB sequence is an important element in ΦC31 integrase system. We wonder whether the integration efficiency of ΦC31 integrase system can be improved by adjusting attB site in donor plasmid. The previous study showed that mutagenesis at the distal end of attB or close to the crossover site inhibits DNA-binding affinity and cleavage, indicating that sequences in attB affect the ability of ΦC31 integrase to synapse and to activate DNA cleavage (Gupta et al., 2007).
Accordingly, we investigated the impact of the direction, position, and number of attB upon the efficiency of ΦC31 integrase system. Our results demonstrated that manipulating the orientation and number of attB in the donor plasmid could provide an easy, rapid means for improving the efficiency of integration mediated by integrase.
Materials and Methods
Plasmid construction
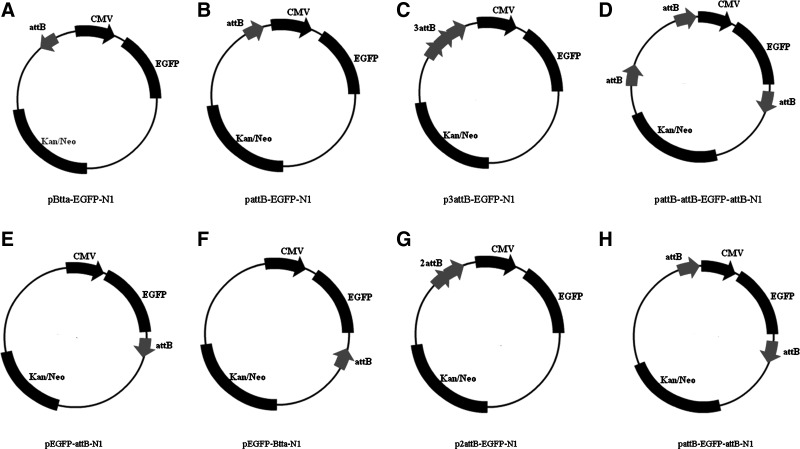
All restriction enzymes were purchased from New England Biolabs (Ipswich, MA) unless otherwise stated. The plasmids of pCMV-Int, pBCPB, and pInt were kindly provided by Dr. M.P. Calos, Stanford University. The construction of the donor plasmids is briefly described in the following procedures. The attB site was amplified using the primers attBPF and attBPR (Table 1), and the resulting polymerase chain reaction (PCR) fragments were inserted into the PciI sites of pEGFP-N1 (Clontech, Mountain View, CA). The plasmids of pBtta-EGFP-N1 (reverse direction for attB), pattB-EGFP-N1 (forward direction of attB), and p2attB-EGFP-N1 (tandem with two attBs) were then constructed (Fig. 1). The plasmid p3attB-EGFP-N1 (tandem with three attBs) was also generated, in which the other two tandem attBs from p2attB-EGFP-N1 were amplified using the primers attBAF and attBAR (Table 1) and the PCR fragment was inserted into the AseI site of pattB-EGFP-N1 (Fig. 1). For construction of the plasmid pEGFP-Btta-N1 (reverse direction of attB in downstream), attB was released from pEGFP-attB-N1 (Ma et al., 2006) by AflII digestion and then inserted into the same site of pEGFP-N1. For pattB-EGFP-attB-N1 (interspersed with two attBs), attB fragment was amplified using primers attBEF and attBER (Table 1), and the resulting fragment was inserted into the EcoO109I site of pEGFP-attB-N1. For pattB-attB-EGFP-attB-N1 (interspersed with three attBs), the amplification of attB was directed by primers attBAPF and attBAPR (Table 1), and the product was inserted into the AseI and PciI sites of pattB-EGFP-attB-N1. For pBCP2B and pBCP3B, a 318-bp fragment was amplified from pEGFP-attB-N1 using primers attBHF and attBHR (Table 1), and the amplified fragment was then ligated into the HindIII sites of pBCPB. For pBCP, pBCPB was cut by XhoI to remove attB and self-ligated afterward.
Table 1.
Primers Used in This Study
| Primers | Sequences 5′ to 3′ |
|---|---|
| attBPF | AGTAACATGTTCGAGGTCGACGATGTAGGT |
| attBPR | AGTAACATGTGAGGGGCCCAAGCTT |
| attBAF | AGTACTTAAGAACCGTATTACCGCCATGCAT |
| attBAR | AGTACTTAAGACCCC GTAATTGATTACTAT |
| attBEF | AAGGACCTTCGAGGTCGACGATGTAGGT |
| attBER | AAGGACCTGAGGGGCCCAAGCTTATC |
| attBAPF | AGTAATTAATTCGAGGTCGACGATGTAGGT |
| attBAPR | AGTAACATGTGAGGGGCCCAAGCTTATC |
| attBHF | ATGAAGCTTTCGAGGTCGACGATGTAGGT |
| attBHR | ATGAAGCTTGAGGGGCCCAAGCTTATC |
| attBRDF | CCCCTGAACCTGAAACAT |
| attBRDR | CAACACTCAACCCTATCTCG |
| attB8A1F | GTGGTTTGTCCAAACTCATC |
| attBA1R1 | ACATAAAATGAATGCAATTG |
| attBA1R2 | TTGCATGGCCTCATTTCCGTC |
FIG. 1.
Schematic diagram of eight plasmids used to investigate the relationship between attB sites and the efficiency of recombination in the ΦC31 integrase system. (A, B, E, and F) The four plasmids used for assessing attB position/orientation effects; (B, C, and G) plasmids with tandem attBs; (B, D, and H) plasmids with interspersed attBs.
Cell culture and integration assays
HeLa cells (ATCC, Cat. No. CCL-2) were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (Clontech). For integration assays, donor plasmids plus helper plasmids (encoding the integrase) were co-transfected into HeLa cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 24 h of transfection, cells were seeded on 100-mm dishes at a 1:20 dilution in 400 μg·mλ−1 G418 for 2 weeks. G418-resistant colonies were stained for 10 min with 1% methylene blue in 70% ethanol, washed by distilled water, and air-dried. Only colonies >0.3 mm in diameter were counted by the Tanon Colon Counter system (Tanon Technology, Shanghai, China).
Detection of genomic integration at pseudo attP sites
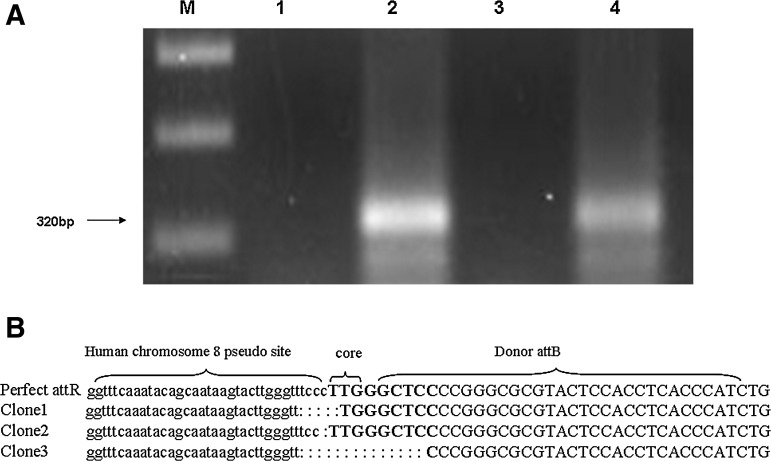
HeLa cells with 50%–80% confluency in 60-mm plates were co-transfected with 50 ng pEGFP-attB-N1 and 250 or 500 ng pCMV-Int or 200 ng pcDNA3.1/Zeo (+) using Lipofectamine 2000. Cells were harvested for DNA extraction 72 h after transfection. Integration at the pseudo attP sites in human genome was identified by PCR using primers attBRDF and attBRDR (Table 1) with the following conditions: 94°C for 5 min; 31 cycles of 94°C for 30 s, 62°C for 45 s, and 72°C for 45 s; and final extension at 72°C for 8 min. If integration was inserted at pseudo attP sites, no 630 bp of amplified fragment would be achieved due to the break at the “TT” core of attB site. Additionally, the most prevalent pseudo attP site (ψA) in the chromosome 8 (Thyagarajan et al., 2001) was also identified by nested PCR using primers attB8A1F and attBA1R1 from the vector sequence, and attBA1R2 (Table 1) from human genome. PCR was conducted as follows: 94°C for 5 min; 31 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 30 s; and final extension at 72°C for 8 min. A 320 bp of amplified fragment could be visualized as site-specific integration occurred at ψA. The integration at this site was further confirmed by sequencing analysis.
Enhanced green fluorescent protein expression assays in transfected cells
To measure enhanced green fluorescent protein (EGFP) expression, HeLa cells with 50%–80% confluency in 60-mm plates were co-transfected with 50 ng donor plasmid plus 500 ng pCMV-Int (ratio 1:10) or control plasmid pcDNA3.1/Zeo (+) (400 ng) using Lipofectamine 2000. Cells were harvested 24 h after transfection. Two-thirds of cells were analyzed by flow cytometry (BD FACS Calibur, Franklin Lakes, NJ). The remaining cells were seeded on 60-mm plates at an appropriate dilution. After 24 h, proteins were extracted from two-thirds of the left cells using a Cytosol Protein Extraction Kit (Beyotime, Haimen, China), and then stored at −80°C. The remaining cells were again plated on 60-mm plates. The process was repeated every 2–4 days, depending on the confluency of the cells. A sandwich enzyme-linked immunosorbent assay (ELISA) was used to measure EGFP expression in the protein extracts as described previously (Ou et al., 2008).
Statistical analysis
All experiments were performed at least twice, and all samples were tested in triplicate. Data from each group were expressed as mean±standard error or raw numbers with percentages. The Student's t-test was used for statistical analysis.
Results
Detection of integration at pseudo attP sites
PCR results from the DNA of the HeLa cells co-transfected by donor plasmid (pEGFP-attB-N1) and helper plasmid (pCMV-Int) showed that efficiency of integration mediated by integrase accounted for 70%±7.1% of total integration events. A prevalent integration site (ψA) in the human chromosome 8 was analyzed by nested PCR. About 320 bp of amplified fragment was present, indicating that the site-specific integration occurred at this site. The sequencing result of the region containing the genome–vector junction confirmed site-specific integration at the ψA site. However, the recombination events were slightly imprecise, accompanied by small deletion (Fig. 2). In addition, cells that were transfected with 50 ng pEGFP-attB-N1 and 500 ng pCMV-Int showed a greater amount of PCR product than cells transfected with 50 ng pEGFP-attB-N1 and 250 ng pCMV-Int (Fig. 2A), which may indicate a higher efficiency of site-specific integration with greater amounts of integrase-encoded plasmid transfected into cells.
FIG. 2.
Analysis of integration at the ψA site in the human chromosome 8 by nested polymerase chain reaction (PCR). (A) PCR analysis for site-specific integration at the ψA site. Lane M: 1 kb marker; lane 1: 50 ng pEGFP-N1 and 400 ng pcDNA3.1/Zeo (+); lane 2: 50 ng pEGFP-attB-N1 and 500 ng pCMV-Int; lane 3: 50 ng pEGFP-attB-N1 and 200 ng pcDNA3.1/Zeo (+); lane 4: 50 ng pEGFP-attB-N1 and 250 ng pCMV-Int. (B) Sequencing of PCR fragments reveals imprecise recombination between pseudo attP and attB, with a loss of 1–13 bp.
The orientation of attB, but not position, affects integration efficiency and EGFP expression
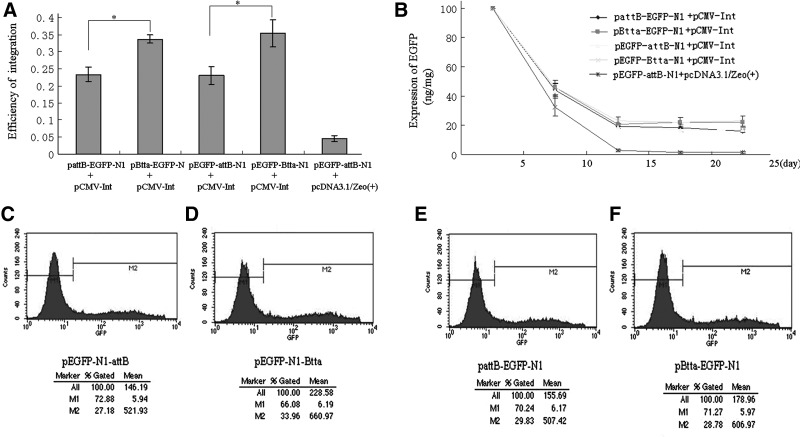
To evaluate whether position or orientation of the attB sites in donor plasmids influences the efficiency of integration as well as the expression of transgenes, four plasmids bearing different types of attB and EGFP reporter gene were constructed (Fig. 1A, B, E, F), followed by transfection into HeLa cells. After G418 selection, the resistant cell colonies were counted for calculating chromosomal integration rate and EGFP expression was detected by fluorescence-activated cell sorter (FACS) and ELISA analysis. The results showed that integration efficiency was not affected by the position of attB in relation to EGFP, while influenced by the orientation of attB (Fig. 3A). The number of colonies from cells transfected with reverse attB was 1.5-fold higher than that from cells transfected with forward attB (Fig. 3A; p<0.05). Additionally, four donor plasmids were, respectively, co-transfected into HeLa cells with pCMV-Int. FACS analysis demonstrated that cells receiving the reverse-oriented attB had stronger transient expression of EGFP than those with forward attB (Fig. 3C–F). ELISA results showed a stability of EGFP expression over 4-week time course, compared with its rapid extinction in the control group (Fig. 3B), suggesting that EGFP may integrate in the human genome. EGFP expression was comparable no matter whether the attB site was upstream or downstream of EGFP. However, the reverse-oriented attB site resulted in a 1.4-fold higher expression compared with the forward attB (Fig. 3B, p<0.05). Moreover, transient or stable expression of EGFP determined by FACS or ELISA showed that the direction rather than position of attB significantly influenced the expression efficiency of the target gene.
FIG. 3.
Effects of attB position and orientation on integration efficiency and enhanced green fluorescent protein (EGFP) expression. (A) Integration efficiency for the indicated plasmids. *Significant difference at p<0.05. (B) EGFP expression measured by enzyme-linked immunosorbent assay after transfecting the indicated plasmids into cells. (C–F) Time course of EGFP expression measured by flow cytometry 24 h after transfection with the indicated plasmids.
Tandem attBs decrease site-specific integration efficiency in HeLa cells
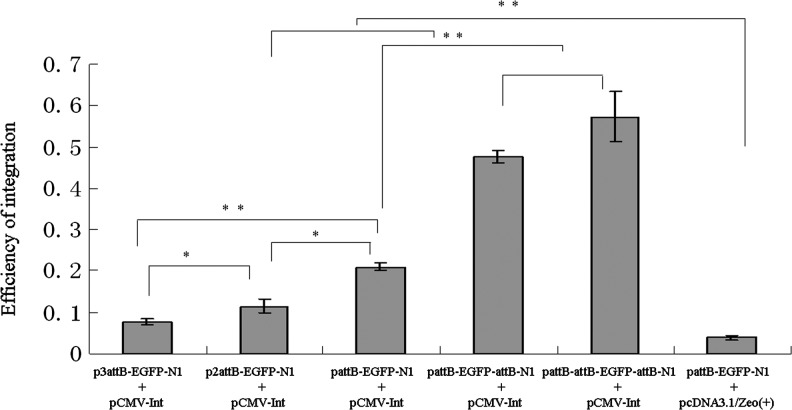
In the Cre system, multiple LoxP sites can increase the frequency of recombination events compared with a single allelic LoxP site (Liu et al., 2002). In the present study, plasmids containing two or three tandem attBs were investigated to determine their effect on integration efficiency in HeLa cells. Surprisingly, adjacent duplicate or triplicate attBs (Fig. 1C, G) decreased the efficiency of site-specific recombination (Fig. 4). Integration assay showed that plasmids containing one attB produced higher frequency of integration than those bearing two or three attBs in tandem (1.8- vs. 2.7-fold, respectively, p<0.05).
FIG. 4.
Effects of multiple tandem or interspersed attBs on integration efficiency. HeLa cells were transfected with the plasmid combinations indicated. After 2 weeks of selection, the number of G418-resistant colonies stained with methylene blue was used to calculate the integration efficiency. The data are indicated as mean±standard error from three independent experiments. Significant difference at *p<0.05 and **p<0.01.
Interspersed attBs increase site-specific integration efficiency
In addition to donor plasmids bearing attBs in tandem, vectors containing two or three interspersed attBs were also constructed for evaluating their influence on integration efficiency (Fig. 1D, H). The results showed that these plasmids had 2.2- and 2.8-fold increase in integration efficiency, respectively, as compared with the plasmid bearing one attB (p<0.01, Fig. 4). The integration efficiency of all experimental groups (tandem attBs and interspersed attBs) was significantly higher than that of the pcDNA3.1/Zeo (+) control group (p<0.01, Fig. 4).
Discussion
Although the efficacy and specificity shown by the wild-type ΦC31 integrase system appears successful in gene therapy (Calos, 2006), additional improvements to some parameters regarding this system are likely to make it even more powerful for gene delivery. The current study focused on the investigation of the effect of the orientation, position, and number of attB on the integration efficiency and expression of the integrated gene (reporter EGFP). The results demonstrated that the donor plasmids we constructed could specifically integrate into pseudo attP sites of human genome and accounted for 70%±7.1% of total integration events. Particularly, the reverse attB significantly enhanced the integration efficiency as well as the expression of reporter EGFP. The mechanism of difference in directions of attBs of the donor plasmids is unknown. A previous study assumed that a topological difference in a DNA sequence can affect the expression of a target gene (Li and Emery, 2008). In another experiment, we found that site-specific recombination mediated by integrase was undertaken efficiently between pseudo attP and reverse attB (data not shown). These observations made us speculate that, compared with forward attB, reverse attB might form more stable synapsis with attP mediated by integrase, which might account for the topological difference in integration efficiency. Watanabe et al. (2010) found that the reverse attB in donor plasmid could increase the expression of reporter luciferase in cells, suggesting that side effect may account for the topological difference. In spite of these probable explanations, the mechanism by which reverse attB improved the gene expression needs to be further investigated.
In this study, the number of attB in donor plasmids was also evaluated the effect on integration efficiency. As expected, interspersed attBs significantly improved the efficiency of integration (Fig. 4). It was possibly due to an increased opportunity for the binding of integrase with the independent attB sites within one plasmid. Effective use of this strategy would require further optimization of the distance between attB sites. However, tandem attBs significantly inhibited integrase-mediated integration in HeLa cells. The precise mechanism for this inhibition is unclear. ΦC31 integrase interacts with intracellular proteins, such as TTRAP (TRAF and TNF receptor-associated protein) and DAXX (death-associated protein 6), in eukaryotic cells (Chen et al., 2006; Wang et al., 2010). We suspected that the interaction between some intracellular proteins and integrase might block the formation of functional synapses between tandem attB sites and integrase, which may inhibit the efficiency of integration.
In summary, the efficacy of integration and expression of a target gene mediated by integrase may be effectively improved by adjusting the orientation, position, and number of attB in the donor plasmids. The donor plasmids bearing one reverse-oriented attB or interspersed rather than tandem attBs promoted the efficiency of site-specific integration. We expect that the improvements of attB sites described in this study would provide valuable information as well as another potential tool for transgenesis application and therapeutic gene transfer by using ΦC31 integrase system.
Acknowledgments
This work was supported by grants from the National Science and Technology Major Project of China (No. 2009ZX08010-018B), and State and Shanghai Leading Academic Discipline (B204).
Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- Calos M.P. The ΦC31 integrase system for gene therapy. Curr Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- Chalberg T.W. Portlock J.L. Olivares E.C. Thyagarajan B. Kirby P.J. Hillman R.T. Hoelters J. Calos M.P. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Chen J.Z. Ji C.N. Xu G.L. Pang R.Y. Yao J.H. Zhu H.Z. Xue J.L. Jia W.P. DAXX interacts with phage PhiC31 integrase and inhibits recombination. Nucleic Acids Res. 2006;34:6298–6304. doi: 10.1093/nar/gkl890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A.W. Jasinskiene N. Sanchez-Vargas I. Isaacs A.T. Smith M.R. Khoo C.C. Heersink M.S. James A.A. Olson K.E. Comparison of transgene expression in Aedes aegypti generated by mariner Mos1 transposition and ΦC31 site-directed recombination. Insect Mol Biol. 2011;20:587–598. doi: 10.1111/j.1365-2583.2011.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C. Olivares E.C. Thyagarajan B. Calos M.P. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. Till R. Smith M.C. Sequences in attB that affect the ability of ΦC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007;35:3407–3419. doi: 10.1093/nar/gkm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keravala A. Lee S. Thyagarajan B. Olivares E.C. Gabrovsky V.E. Woodard L.E. Calos M.P. Mutational derivatives of ΦC31 integrase with increased efficiency and specificity. Mol Ther. 2009;17:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S. Rao R.N. Analysis of the integration functions of the streptomycete bacteriophage ΦC31. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- Li C.L. Emery D.W. The cHS4 chromatin insulator reduces gammaretroviral vector silencing by epigenetic modifications of integrated provirus. Gene Ther. 2008;15:49–53. doi: 10.1038/sj.gt.3303009. [DOI] [PubMed] [Google Scholar]
- Liu P. Jenkins N.A. Copeland N.G. Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells. Nat Genet. 2002;30:66–72. doi: 10.1038/ng788. [DOI] [PubMed] [Google Scholar]
- Ma Q.W. Sheng H.Q. Yan J.B. Cheng S. Huang Y. Chen-Tsai Y. Ren Z.R. Huang S.Z. Zeng Y.T. Identification of pseudo attP sites for phage phiC31 integrase in bovine genome. Biochem Biophys Res Comm. 2006;345:984–988. doi: 10.1016/j.bbrc.2006.04.145. [DOI] [PubMed] [Google Scholar]
- Ou H.L. Huang Y. Qu L.J. Xu M. Yan J.B. Ren Z.R. Huang S.Z. Zeng Y.T. A ΦC31 integrase-mediated integration hotspot in favor of transgene expression exists in the bovine genome. FEBS J. 2008;276:155–163. doi: 10.1111/j.1742-4658.2008.06762.x. [DOI] [PubMed] [Google Scholar]
- Raymond C.S. Soriano P. High-efficiency FLP and ΦC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B. Hippenmeyer S. Wang C. Gamboa M. Zong H. Chen-Tsai Y. Luo L. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci U S A. 2011;108:7902–7907. doi: 10.1073/pnas.1019507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B. Olivares E.C. Hollis R.P. Ginsburg D.S. Calos M.P. Site-specific genomic integration in mammalian cells mediated by phage ΦC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.Y. Xu G.L. Zhou C.H. Tian L. Xue J.L. Chen J.Z. Jia W. ΦC31 integrase interacts with TTRAP and inhibits NFkappaB activation. Mol Biol Rep. 2010;37:2809–2816. doi: 10.1007/s11033-009-9829-3. [DOI] [PubMed] [Google Scholar]
- Watanabe S. Nakamura S. Sakurai T. Akasaka K. Sato M. Improvement of a ΦC31 integrase-based gene delivery system that confers high and continuous transgene expression. N Biotechnol. 2010;28:312–329. doi: 10.1016/j.nbt.2010.11.001. [DOI] [PubMed] [Google Scholar]