Abstract
The biosynthetic gene cluster for laspartomycins, a family of 11 amino acid peptide antibiotics, has been cloned and sequenced from Streptomyces viridochromogenes ATCC 29814. Annotation of a segment of 88912 bp of S. viridochromogenes genomic sequence revealed the putative las cluster and its flanking regions which harbor 43 open reading frames. The lpm cluster, which spans approximately 60 kb, consists of 21 open reading frames. Those include four NRPS genes (lpmA/orf18, lpmB/orf25, lpmC/orf26 and lpmD/orf27), four genes (orfs 21, 22, 24 and 29) involved in the lipid tail biosynthesis and attachment, four regulatory genes (orfs 13, 19, 32 and 33) and three putative exporters or self-resistance genes (orfs 14, 20 and 30). In addition, the gene involved in the biosynthesis of the nonproteinogenic amino acid Pip was also identified in the lpm cluster while the genes necessary for the biosynthesis of the rare residue diaminopropionic acid (Dap) were found to reside elsewhere on the chromosome. Interestingly, the dabA, dabB and dabC genes predicted to code for the biosynthesis of the unusual amino acid diaminobutyric acid (Dab) are organized into the lpm cluster even though the Dab residue was not found in the laspartomycins. Disruption of the NRPS lpmC gene completely abolished laspartomycin production in the corresponding mutant strain. These findings will allow molecular engineering and combinatorial biosynthesis approaches to expand the structural diversity of the amphomycin-group peptide antibiotics including the laspartomycins and friulimicins.
Keywords: Lipopeptide antibiotics, laspartomycins, biosynthetic gene cluster, Streptomyces viridochromogenes, disrupted mutants
1. Introduction
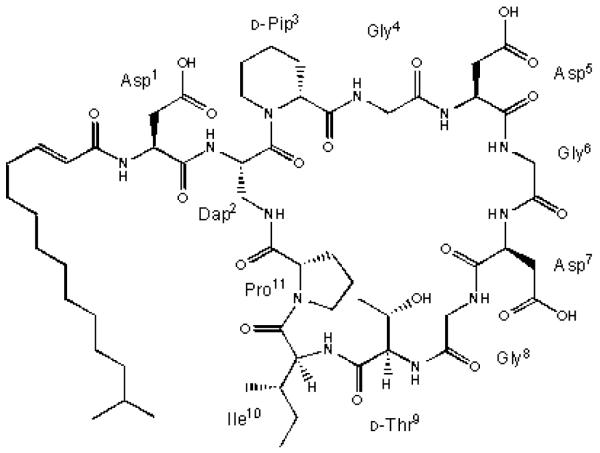
The laspartomycins were originally discovered by Naganawa et al. in 1968 from the soil bacterium Streptomyces viridochomogenes var. komabensis Hamada et Okami (ATCC29814) (Naganawa et al., 1968). They were produced as a mixture of at least three peptide compounds which differ in their attached fatty acid side chains (Borders et al., 2002). Laspartomycin C is the major component of this mixture and its structure was recently fully elucidated as a cyclic lipopeptide with a 2, 3-unsaturated C15-fatty acid side chain (Fig. 1) (Naganawa et al., 1970; Borders et al., 2007). As members of the amphomycin-group antibiotics, the laspartomycins differ from others in their peptide core and fatty acid side chains. The laspartomycins are comprised of 11 amino acid residues among which are ten residues forming a ring structure and an exocyclic Asp1 residue linked with an acyl group. Laspartomycin-related lipopeptide antibiotics include amphomycin (Heinemann et al., 1953; Cronk and Neumann, 1956), zaomycin (Hinuma, 1954), crystallomycin (Lomakina and Brazhnikova, 1959), aspartocin (Shay et al., 1959), glumamycin (Inoue, 1962; Shibata et al., 1962), tsushimycin (Shoji et al., 1968), glycinocin (Kong and Carter, 2003), A1437 and the friulimicins (Aretz et al., 2000; Vertesy et al., 2000). The friulimicins are produced by Actinoplanes friuliensis sp. nov. and glycinocin is produced by an unidentified Actinomyces species but all of the others are produced by Streptomyces spp. All of these antibiotics have similar structures with respect to their amino acid compositions and conserved residues, but they differ in the amino acid sequences, having one to three non-conservative residues, and varied lipid tails (chain length, position of the double bond and configuration). In comparison with the other lipopeptides mentioned above, the laspartomycins contain a unique cyclic peptide core and 2,3-unsaturated acyl side chains instead of 3, 4-unsaturated acyl groups, and have diaminopropionic acid (Dap2) instead of diaminobutyric acid (Dab2) as the amino acid for side chain attachment through proline (Pro1) (Fig. 1) (Borders et al., 2007).
Fig. 1.
Structure of laspartomycin C. The friulimicins’ amino acid composition differ from the laspartomycins at positions 2 (Dab), 4 (Me-Asp), 9 (Dab) and 10 (Val).
The laspartomycins were initially reported to be active against Staphylococcus aureus (Naganawa et al., 1968). Recent studies have shown that they are active against a broad spectrum of Gram-positive pathogens including MRSA, VRSA, vancomycin-intermediate S. aureus (VISA) and VRE (Borders et al., 2007; Curran et al., 2007). The MICs against methicillin-sensitive S. aureus are as low as 2 μg/mL. The laspartomycins were also reported to have antiherpes activity (Alarcon et al., 1984). The mechanism of antibiotic action of the laspartomycins is still unknown. Studies on the mode of action of the closely-related amphomycin and friulimicin have been reported. The biological target of amphomycin was identified as peptidoglycan biosynthesis, more precisely, inhibition of lipid intermediate I biosynthesis (Tanaka et al., 1977; Tanaka et al., 1979; Tanaka et al., 1982). The amphomycin is currently used as a topical antibacterial agent in the veterinary industry (Dini, 2005). Amphomycin was also shown to inhibit the formation of dolichol-phosphate-mannose (dol-P-Man) in eukaryotic N-linked glycoprotein biosynthesis (Elbein, 1981; Banerjee, 1989). The friulimicins were produced as a mixture of four lipopeptides (Vertesy et al., 2000). They were described as novel lipopeptide antibiotics with peptidoglycan synthesis-inhibiting activity (Aretz et al., 2000). The friulimicins differ from the laspartomycins in their fatty acid side chains and amino acid composition at positions 2, 4, 9 and 10 (Fig. 1). Friulimicin is an effective drug against Gram-positive pathogens including MRSA and methicillin-resistant S. epidermidis (Heinzelmann et al., 2005; Muller et al., 2007). Recent studies have demonstrated that friulimicin has a unique mode of action, which is different from that of daptomycin, a peptide antibiotic in current clinical use (Schneider et al., 2009; Wecke et al., 2009). Thus far, the only fully characterized amphomycin-type lipopeptide gene cluster is the one for friulimicin (Muller et al., 2007). We have cloned and sequenced the laspartomycin (lpm) gene cluster from S. viridochromogenes ATCC 29814. We report here the determination and annotation of the sequence for the entire lpm gene cluster and its flanking regions, disruption of the NRPS gene lpmC, and analysis of the metabolites produced by the disruptant.
2. Materials and methods
2.1. Bacterial strains, plasmids, fosmids and culture conditions
S. viridochromogenes ATCC 29814 and Escherichia coli S17-1 (ATCC 47055) were purchased from ATCC. EPI300 (Epicentre) and XL10-Gold (Stratagene) strains were routinely used as hosts for E. coli plasmids, cosmid and fosmid vectors. pMod3 and the pCC1Fos fosmid kit were purchased from Epicentre. The plasmid pIJ773 was provided by Professor K.F. Chater (Norwich, England). The SuperCos1 cosmid kit was purchased from Stratagene. Media and culture conditions for S. viridochromogenes ATCC 29814 were the same as previously described (Naganawa et al., 1968). All E. coli procedures were performed according to standard protocols (Sambrook and Russell, 2001).
2.2. DNA isolation and manipulation
Preparation of genomic DNA from the laspartomycin-producing S. viridochromogenes ATCC 29814 was conducted by using the cetyltrimethylammonium bromide (CTAB) procedure as described previously (Kieser et al., 2000). QIAprep Spin Miniprep kits (Qiagen) were used to prepare plasmids and cosmids from E. coli strains. Restriction endonucleases, DNA ligase, DNA polymerase and alkaline phosphatase were purchased from various sources and used according to the manufacturers’ recommendations. DNA fragments were purified using QIAquick Gel Extraction kits from Qiagen.
2.3. Generation of the peptide synthetase, acyl-CoA synthase and acyl-CoA dehydrogenase gene probes
PCR primers were designed to amplify the regions coding for the internal portion (motif A3 to T domain) of NRPS adenylation domains (Borchert et al., 1992; Marahiel et al., 1997). A pair of degenerate oligonucleotide primers were synthesized by eurofins mwg/operon: YinA3 (forward); 5′-ATATATAAGCTTATCTACACSTCSGGCACSACSGGCAAGCCSAAGGG-3′ and YinT (reverse); 5′-GGAAGAAGATCTAWIGAGKSICCICCSRRSIMGAAGAA-3′ (S = G + C; W = A + T; K = T + G; R = G + A; M = A + C; I = inosine; HindIII and BglII sites are bold). The PCR template was S. viridochromogenes genomic DNA. PCR conditions were the same as described previously (Yin et al., 2003) except for substitutions in the forward and reverse primers with YinA3 and YinT as well as the addition of 1 μg of S. viridochromogenes ATCC29814 genomic DNA. PCR products (1.2 kb) were digested with HindIII and BglII, gel-purified and cloned into the E. coli phagemid cloning vector pKS (pBluescript II KS, Short et al, 1988). Individual pKS-derived plasmids from 64 randomly selected clones were analyzed by digestion with HindIII and XbaI to confirm the insert sizes. A total of 54 pKS derivatives were submitted for sequencing with the universal T3 promoter primer. BLAST analysis of these 54 sequences identified 12 unique plasmids, designated pKS-29814PS1, 4, 8, 11, 14, 20, 22, 27, 30, 46, 47 and 60. To amplify the acyl-CoA synthase gene (orf21) and acyl-CoA dehydrogenase gene (orf22), a pair of primers was designed from the sequence obtained by genome scanning (Svacspf: 5′TTCCTTAGATCTCCTCTTCATCTCCGGTTCCTTC -3′ and Svacdpr: 5′-AGAGAGAAGCTTAAGGGCCGCAGGGGCACGTGGGCA-3′, HindIII and BglII sites are in bold). The stop codon of orf21 overlaps the start codon of orf22. An amplicon of the expected size (2 kb) coding for the C-terminal portion of Orf21 and the N-terminal portion of Orf22 was purified and cloned into the pKS vector to obtain plasmid pKS-29814acsd. The insert of plasmid pKS-29814acsd was confirmed by DNA sequencing.
2.4. Genomic scanning of S. viridochromgenes ATCC 29814
High quality genomic DNA isolated from S. viridochromogenes ATCC 29814 was sent out for commercial 454 Sanger sequencing and Illumina next generation sequencing. To identify the putative laspartomycin gene cluster, all contig sequences obtained from the S. viridochromogenes ATCC 29814 genome were scanned against the sequences of the 12 unique peptide synthetase probes identified in Section 2.3 using a BLAST search (Altschul et al., 1990). All contigs harboring the NRPS genes were subjected to the bioinformatics-based substrate specificity analysis of the individual A-domains allowing for identification of the putative laspartomycin (lpm) biosynthetic gene cluster and mapping of the inserts of plasmids pKS-29814PS4, 8, 20, 27 and 60 onto the lpm cluster.
2.5. Development of a genetic system to transform S. viridochromogenes ATCC 29814
To set up a genetic system suitable for transformation of strain ATCC29814, a previously constructed gene disruption plasmid pXY300-sfPS18-AmR (Yin and Zabriskie, 2006) and a new construct pXYUIA-sfPS14 were evaluated. pXY300-sfPS18-AmR is a pXY300-based construct, derived from pGM160 (Muth et al., 1989). sfPS18 encodes the Asp-activating A domain of NRPS EndB-m1. The vector pGM160 and its derivatives are some of the most extensively used Streptomyces gene disruption plasmid vectors. pXY300 is an advanced E. coli-Streptomyces temperature-sensitive conjugal shuttle vector and was developed in our laboratories (Yin et al., 2003). pXYUIA-sfPS14 is a new conjugal integrative construct derived from plasmid pXYUIA created by combination of the integrative and site specific attachment as well as conjugative functions from the commonly used integrative conjugal plasmid pSET152 (Bierman et al., 1992) with the ColE1 ori and the ampicillin resistance gene from pUC18 (Sambrook and Russell, 2001). sfPS14 is a peptide synthetase gene probe which encodes the internal region of the glycine-activating A domain and was obtained by PCR amplification from fosmid pXYF305 (Yin and Zabriskie, 2006). Both plasmids were introduced into the wild-type S. viridochromogenes ATCC 29814 by conjugation.
2.6. Disruption of the NRPS gene lpmC on the S. viridochromogenes ATCC 29814 chromosome
The NRPS substrate specificity sequences extracted from the inserts of plasmids pKS-29814PS4 and pKS-29814PS27 predicted that they activate Ser/Thr and Asp, respectively (Stachelhaus et al., 1999; Challis et al., 2000; Rausch et al., 2005). The DNA sequence from the peptide synthetase probe 29814PS4 is proposed to encode the A domain of the laspartomycin amino acid residue Thr9 while the DNA sequence from the peptide synthetase probe 29814PS27 should encode the A domain of the laspartomycin amino acid residue Asp7. Therefore, these two probes 29814PS27 and 29814PS4 were renamed as 29814PS7 and 29814PS9, respectively, and their corresponding plasmids were accordingly renamed as pKS-29814PS7 and pKS-29814PS9. These two plasmids were used to construct the final gene disruption delivery plasmids pKS-ATR6K-29814PS7 and pKS-ATR6K-29814PS9. pBluescript KS has nonreplicative and nonintegrative functions in Streptomyces. ATR6Kγori is a composite gene disruption cassette designed and constructed in our laboratory. AT stands for an E. coli-Streptomyces bi-functional apramycin resistance gene and oriT element which is required for the intergeneric conjugation. AT was excised from plasmid pIJ773 as an XbaI fragment. R6Kγori carries the plasmid replication function controlled by expression of the pi protein gene (Stalker et al., 1979; Filutowicz et al., 1985) present in the specific E. coli strain EC100D (Epicentre). ATR6Kγori can be used for localizing the disrupted sites by recovery of the plasmids generated by cyclizing the ATR6Kγori-containing fragments and sequencing. To construct the gene disruption plasmids mentioned above, we used two PCR primers (R6Kpf, 5′-CATGCAAGCTTTAAAAGCCTTATA-3′ and R6Kpr, 5′-ATCGGATCGATGCTAGCCCTGAAGCTCTTGT-3′, HindIII and ClaI/NheI sites are in bold) to amplify the R6Kγori fragment from pMod3 (Epicentre). The 0.3 kb R6Kγori fragment was restricted with HindIII and ClaI, and then ligated into the similarly restricted plasmids pXYKS-29814PS7 and pXYKS-29814PS9 to yield plasmids pXYKS-R6Kγori-29814PS7 and pXYKS-R6Kγori-29814PS9. Fragment AT was then ligated into the NheI site to afford the final gene disruption constructs. They were introduced by transformation into E. coli S17-1 and then conjugated with germinated spores of S. viridochromogenes ATCC29814 (Mazodier et al. 1989). Pregermination of strain ATCC 29814 spores was conducted as described previously (Hirsch and Ensign, 1976). The exconjugants showing the apramycin resistance (AmR) phenotype were further purified on ISP2 plates with apramycin (50 μg/mL). Because pKS derivatives have non-plasmid replication function in strain ATCC 29814, and the vector alone does not carry any homologous DNA sequence of strain ATCC 29814, all AmR-resistant exconjugants resulted from single crossover homologous recombination between the copies of peptide synthetase probe 29814PS7 or 29814PS9 on the chromosome and plasmid. The genomic DNA from individual exconjugants was digested with BamHI and XhoI, then analyzed by Southern blot using AmR plus 29814PS7 or 29814PS9 as probe. The correct chromosomal structures for the mutants SvlpmCmtPS7 were confirmed.
Construction and screening of S. viridochromogenes ATCC 29814 genomic cosmid library
Genomic DNA of strain ATCC 29814 was employed to construct a cosmid library using SuperCos1 (Invitrogen) as the vector. Cosmid library construction followed the manufacturer’s instructions except for the following modification. The genomic DNA was fractionated on 1% low-melting point agarose gel and the approx. 40 kb bands were recovered by following the pCC1Fos fosmid library protocol (Epicentre). Both gel-purified vector and recovered genomic fragments were end-repaired using the End-It DNA End-Repair kit (Epicentre). The end-repaired vector and genomic fragments were then ligated and the ligation mixture was packaged into phage with MaxPlax Lambda Packaging Extracts (Epicentre). Freshly prepared E. coli EPI300 (Epicentre) was infected with pre-incubated phage packaging solution and plated on LB agar plates supplemented with 100 μg/mL ampicillin. The resulting library produced approx. 8000 colony forming units (CFU). The glycerol stocks for 6048 isolated colonies were prepared in plates of 63 × 96 wells and stored at −80 °C. Half of these colonies were replicated on LB plates supplemented with 100 μg/mL ampicillin and 0.2% maltose to evenly distribute the colonies while the same cosmid library was amplified for in situ colony hybridization. Two digoxigenin-labeled probes were used to screen this library. First, the mixture of PCR-amplified peptide synthetase gene probes 29814PS7 and 29814PS9 were employed, and second, a PCR-amplified fragment harboring the portions of the genes lpm21 and lpm22. The library screening with the first probe led to 47 positively hybridizing cosmid clones. After stripping the probes, the same membranes were re-screened with the second probe that yielded 14 positive cosmid clones. A total of 61 cosmid clones were purified and grown in LB liquid culture for cosmid mini-preparation. Restriction analysis with BamHI indicated that some of these recombinant cosmids shared overlapping inserts.
2.8. HPLC analysis of laspartomycin production
Fermentation conditions for the production of laspartomycin from the strain S. viridochromogenes ATCC 29814 and for the disruptant strain SvlpmCmtPS7, and preparation of the laspartomycins from the fermentation broth were the same as previously described (Naganawa et al., 1968; Borders et al., 2007). The purified laspartomycins were dissolved in 50% aqueous methanol. After filtering through a 0.45 μm syringe filter, a 10 μL sample was injected onto a Gemini C18 column (5 μm, 4.6 × 150 mm, Phenomenex, Torrance, CA) attached to a Shimadzu HPLC. Separation was achieved using a 45 min linear gradient from 5% to 95% acetonitrile at a flow rate of 1.0 mL/min. The UV region from 200-300 nm was scanned with a SPD M20A photodiode array detector.
2.9. MALDI-TOF MS analysis of the metabolites from the wild-type S. viridochromogenes ATCC 29814 and disruptant SvlpmCmtPS7 and Bioassay
MALDI-TOF MS analyses of the metabolites isolated from the wild-type strain ATCC 29814 and the crude extract from the disruptant strain and the bioassay of the fractions corresponding to the laspartomycins were conducted as described previously (Yin et al., 2010).
2.10. Southern hybridization
S. viridochromogenes genomic DNA was restricted and electrophoresed in 0.8% agarose gels and transferred onto Hybond-N nylon membranes (Roche). The procedure for colony lifts for in situ hybridization was according to the manufacturer’s protocol (Roche Applied Science). DNA probes were prepared using digoxigenin-labeled dUTP, and hybridization was revealed using a digoxigenin-DNA detection kit (Roche).
2.11. Sequencing and analysis
End sequencing of cosmid inserts, and routine plasmids, PCR product and primer walking sequencing were performed at the Oregon State University Center for Genome Research and Biocomputing (CGRB) using the Amplitaq dye-terminator sequencing system (Perkin Elmer) and Applied Biosystems automated DNA sequencers (models 373 and 377). The nucleotide sequences were determined for both strands. Sequence analysis was carried out using the VectorNTI (Invitrogen) software package. Nucleotide and amino acid sequence similarity comparisons were carried out in public databases using the BLAST program (Altschul et al., 1990).
2.12. GenBank accession numbers
The nucleotide sequence for the region of the S. viridochromogenes ATCC 29814 genome harboring the laspartomycin biosynthetic gene cluster was deposited in GenBank under the Accession No. HM756254. The nucleotide sequence of the S. viridochromogenes ATCC 29814 genomic segment predicted to encode the biosynthesis of L-2,3-diaminopropionate was deposited in GenBank under the Accession No. JF290477.
3. Results and discussion
3.1. Establishment of a genetic system for the transformation of S. viridochromogene ATCC 29814
The ability to genetically transform a streptomycete antibiotic producer is the key prerequisite for in vivo genetic manipulation of its secondary metabolite biosynthetic gene cluster. Prior to our work on strain ATCC 29814, no genetic transformation of this strain was reported, though S. viridochromogenes Tü494, the producer of an important herbicidal phosphinothricin-tripeptide (PTT), was shown to be genetically amenable with pGM-derived constructs (Schwartz et al., 1996). During the development of the genetic tools for the bleomycin producer Streptomyces verticillus ATCC 15003, Galm and co-workers found that the integrative vectors including pSET152 and pRT801 were able to produce true exconjugants while other tested replicative vectors failed (Galm et al., 2008). To set up the suitable genetic transformation system for S. viridochromogenes ATCC 29814 two plasmids, pXY300-sfPS18-AmR and pXYUIA-sfPS14, were respectively introduced into E. coli S17-1 (Simon et al., 1983) and conjugated with heat shock-treated and pre-germinated spores of strain ATCC 29814. The resulting AmR exconjugant colonies were further purified and their BamHI-restricted genomic DNA analyzed by Southern blot using as probes the digoxigenin-labeled apramcyin resistance genes, plasmid vectors and inserts. Southern blot results indicated plasmid pXY300-sfPS18-AmR was present as a free plasmid and no single or double crossovers occurred between the chromosome of strain ATCC 29814 and the plasmid introduced even though the exconjugants passed through the high temperature (40 °C) selection. Plasmids pXYUIA-sfPS14 has non-replicative function in Streptomyces. Its integration into the chromosome of strain ATCC 29814 via a single crossover at the site specific attachment site was confirmed by Southern hybridization and sequencing of the recovered plasmids from the recombinant strain genome (data not shown). These results confirmed that strain ATCC 29814 was genetically amenable.
3.2. Cloning the laspartomycin peptide synthetase (PS) gene probes
PCR amplification of the A-domain-encoding regions from the S. viridochromogenes ATCC 29814 genomic DNA allowed us to identify and clone 12 unique peptide synthetase probes into the pKS vector. The inserts of the resulting plasmids pKS-29814PS1, 4, 8, 11, 14, 20, 22, 27, 30, 46, 47 and 60 were analyzed for their predicted substrate specificities (Stachelhaus et al., 1999; Challis et al., 2000; Rausch et al., 2005). Except for PS1(Cys), PS14 (Orn), PS20 and PS30 (unknown), PS46 and PS47 (multiple), the rest of them, PS4 (Thr/Ser), PS8, 11, 22 and 60 (Gly), and PS27 (Asp) match well with the constituent amino acids found in the laspartomycins (Fig. 1).
3.3. Identification of the laspartomycin biosynthetic gene cluster by genome scanning
Sequencing of the S. viridochromogenes ATCC 29814 genome by the commercial 454 Sanger method generated a total of 9,836,847 bp of sequence data consisting of 4,293 large contigs for strain ATCC 29814. BLAST analysis of these contigs against known A-domain-encoding insert sequences from plasmid pKS-29814PS4, 8, 11, 20, 22, 27, 30 and 60, allowed us to identify multiple NRPS genes and clusters. Among them is a contig, designated 29814-64 (ca. 64 kb), which harbors sequences identical to the inserts of plasmids pKS-29814PS4, 8, 20, 27 and 60. The A-domain substrate specificity analysis of the contig 29814-64 revealed the amino acid sequence predicted is consistent with that found in laspartomycin C (Fig. 1). Across the entire genome sequence of strain ATCC 29814, there are no other NRPS clusters with the predicted amino acid composition and sequences similar or identical to those found in laspartomycins. Therefore, contig 29814-64 was believed to carry the putative laspartomycin gene cluster. Contig 29814-64 contains 26 gaps ranging from 6 bp to 2.5 kb. By using a PCR strategy and primers designed to amplify the gap fragments from chromosomal DNA and sequencing of the PCR products, we have been able to close most of the gaps in contig 29814-64. In order to close all gaps, find the up and downstream contigs flanking 29814-64 and further verify the genome sequence data obtained by the 454 Sanger sequencing method, a genomic DNA sample of S. viridochromogenes ATCC 29814 was sequenced by an Illumina next generation sequencing. This time, a total of 9,941,817 bp of genome sequence data from 1583 large contigs was obtained. Comparison of the whole genome sequence data of this strain, obtained by both 454 Sanger and Illumina sequencing methods, allowed us to fill all gaps present in the contig 29814-64, expand its up and downstream sequences and accurately determine a segment of 88912 bp sequence of the strain ATCC 29814 genome. It is worth noting that both Sanger and Illumina sequencing methods produced a similar genome size for strain ATCC 29814 (9.84 vs. 9.94 Mb), and the sequence data from the two methods agree closely. However, there are still numerous gaps to be sequenced before we have the entire genome of strain ATCC 29814. The following considerations have been the major impetus for us to initiate sequencing the genome of wild-type S. viridochromogenes ATCC 29814. First, we wanted to quickly identify the putative laspartomycin biosynthetic gene cluster. Based on the known structure of laspartomycin C and prediction of the A-domain amino acid substrate specificity (Stachelhaus et al., 1999; Challis et al., 2000; Rausch et al., 2005), bioinformatic scanning of the available genome sequence should enable us to recognize a 50-60 kb NRPS cluster specifically coding for the putative laspartomycin biosynthesis pathway. Second, we wanted to facilitate the screening of a S. viridochromogenes genomic cosmid library by selecting and using pathway-specific genes besides the common highly conserved NRPS fragments as in situ colony hybridization probes. Third, the cyclic lipopeptide laspartomycin is a representative member of the amphomycin-group antibiotics that include the friulimicins (Vertesy et al., 2000). The biosynthesis of the friulimicins has been very well studied (Heinzelmann et al., 2005; Muller et al., 2007). The biosynthetic gene cluster for the friulimicins has been cloned, annotated and individual gene functions have been demonstrated. The friulimicins hold great promise as antibiotics because they appear to act via a mechanism which differs from the clinical agent daptomycin (Schneider et al., 2009; Wecke et al., 2009). The general similarities of the friulimicin and laspartomycin will provide an excellent molecular platform for combinatorial biosynthesis. Finally, except for the friulimicin producer, no other amphomycin-group antibiotic producer has been reported to be genetically amenable, and none of their biosynthetic gene clusters has been cloned and sequenced. Sequencing of the S. viridochromogenes genome has not only allowed us to speed up identification and cloning of the lasparotmycin gene cluster, but also provided us comprehensive information useful for genetic manipulation of this antibiotic pathway.
3.4. Inactivation of the 29814PS7- and 29814PS9-containing laspartomycin NRPS gene lpmC
During evaluation of the vector systems for transformation of strain ATCC 29814, we found that pXYUIA-sfPS14 produced five times more exconjugants than pXY300-sfPS18-AmR. This might be due to crossover at the site-specific attachment site of the ATCC29814 chromosome. To eliminate possible crossover via the site-specific attachment site as observed for pXYUIA-sfPS14, we constructed two gene disruption plasmids pXYKS-ATR6Kγori-29814PS7 and pXYKS-ATR6Kγori-29814PS9 on the non-integrative vector pBluescript KS. They were introduced by transformation into E. coli S17-1 and then conjugated with germinated spores of strain ATCC 29814. The exconjugants showing AmR phenotype were further purified on ISP2 plates with apramycin (50 μg/mL). Because pXYKS derivatives have the non-plasmid replication function in strain ATCC 29814, and the vector alone does not carry any homologous DNA sequence of strain ATCC 29814, all AmR-resistant exconjugants resulted from single crossover homologous recombination between the copies of 29814PS7 or 29814PS9 on the chromosome and plasmid. The genomic DNA from individual exconjugant mutant candidates was digested with BamHI and XhoI, and analyzed by Southern blot using AmR plus 29814PS7 as probes. The correct chromosomal structures for the mutant strain SvlpmCmtPS7 was confirmed by Southern hybridization (Fig. 2A).
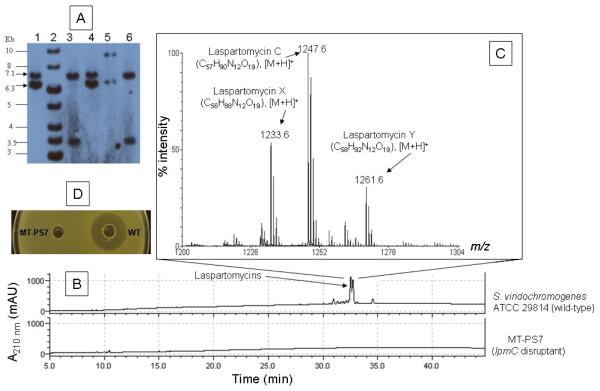
Fig. 2.
A. Southern blot confirming the disruption of the NRPS gene lpmC. The genomic DNA was digested with BamHI and XhoI. The mixture of the peptide synthetase gene segment PS7 amplified by PCR and apramycin-resistance gene AmR was Dig-labeled and used as a Southern blot probe. Lane 2, 1 kb DNA ladder from Fermentas. Lane 5, Dig-labeled molecular marker II from Roche. Lanes 1 & 4: genomic DNA from the wild type strain ATCC 29814. Lanes 3 and 6: genomic DNA from two independent mutant SvlpmCmtPS7 colonies. A 6.3 kb BamHI fragment was present in the wild-type but absent in the mutant because the new XhoI site carried by AmR was introduced into the mutant upon insertional disruption which was also reflected by a 3.5 kb hybridizing-band only present in the mutant. A common 7.1 kb BamHI fragment hybridized with the specific probe mentioned above was observed for both wild-type and mutant. Sequence alignment indicates it has significant identity (92% to 100%) region with the probe. B. HPLC analysis of the metabolites isolated from the wild-type strain ATCC 29814 and the mutant SvlpmCmtPS7. C. MALDI-TOF MS analysis confirming the production of the laspartomycins by strain ATCC 29814 as a mixture of the peptides. D. Bioassay of the metabolites isolated from strain ATCC 29814 and mutant SvlpmCmtPS7. Bioassay was conducted as described previously (Yin et al., 2010)
3.5. Analysis and bioassay of the metabolites from the wild-type strain ATCC 29814 and disruption mutant SvlpmCmtPS7
Figure 2B shows the HPLC chromatographic profiles of the metabolites for the wild-type and mutant strains. Production of the laspartomycins by strain ATCC 29814 and complete abolishment of laspartomycin production in mutant SvlpmCmtPS7 were confirmed by HPLC analysis. MALDI-TOF MS analysis of the purified chromatographic peaks also confirmed the three components of laspartomycins produced by strain ATCC 29814 (Fig. 2C). Three molecular ions detected at m/z 1233.6, 1247.6 and 1261.6 reflected the same peptide core of laspartomycins with different (C14, C15 or C16) fatty acid side chains. MS data also indicated that laspartomycin C is the major component of these naturally occurring peptide mixtures. In addition, the purified laspartomycins from the wild-type culture showed strong inhibitory effects on S. aureus ATCC 29213 while the crude extract from the mutant lacked inhibitory activity against S. aureus (Fig. 2D). The yield of the laspartomycins produced by the wild type strain ATCC 29814 was estimated to be 50 mg/L. The targeted disruption of the NRPS gene lpmC in the region corresponding to the A-domain for Asp7 in the lpm cluster proved that lpmC does participate in laspartomycin biosynthesis. Therefore, the putative laspartomycin gene cluster does encode the laspartomycin biosynthesis pathway in strain ATCC 29814.
3.6. Identification of the overlapping cosmids harboring the entire lpm gene cluster
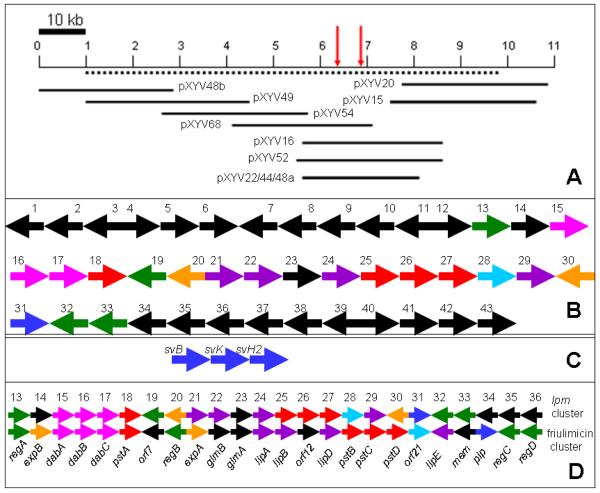
To distinguish the overlapping cosmid sets encoding the laspartomycin biosynthesis pathway from those involved in other secondary metabolite clusters and to correlate them with the genes identified by genome-sequencing and gene-disruption, sequencing was carried out on the ends of the inserts of 61 positive cosmids selected by in situ colony hybridization with either the NRPS probe or the PCR amplified fragment of orf21 and orf22. BLAST alignment of individual sequences with the laspartomycin gene cluster sequence provided the orientation of a total of 11 overlapping fosmids spanning approx. 110 kb of the strain ATCC 29814 genome and harboring the entire lpm cluster and its flanking regions. These overlapping cosmids in order from left to right hand ends were designated as pXYV48b, pXYV49, pXYV54, pXYV68, pXYV16, pXYV52, pXYV22, pXYV44, pXYV48a, pXYV15 and pXYV20 (Fig. 3A).
Fig. 3.
A. Overlapping cosmids harboring the lpm gene cluster and its flanking regions. The dotted line indicates the region covered by the sequence deposited in GenBank. Vertical arrows indicate the insertional disruption sites on lpm gene cluster. B. Genetic organization of the lpm gene cluster and its flanking regions. Each open reading frame is not drawn to scale. The 88912 bp sequences were determined and deposited in GenBank under the Accession No. HM756254. The sequences of the putative biosynthesis genes for diaminopropionate were deposited in the GenBank under the Accession No. JF290477. Color codes: NRPS (red); Fatty acid biosynthesis and attachment (green); Putative regulatory (purple) and resistance genes (orange). The biosynthetic genes for non-constituent amino acid Dab (pink) and for Pip (blue), and MbtH-like gene (light blue). C. The putative biosynthetic genes for the nonproteinogenic constituent amino acid Dap. D. Comparison of the laspartomycin and friulimicin gene clusters. The counterpart genes are indicated with the same colors except for those orfs in black.
3.7. Overall analysis of the laspartomycin (lpm) biosynthetic gene cluster
The lpm gene cluster resides on an 88912 bp segment of the S. viridochromogenes ATCC 29814 chromosome. Using NCBI BLAST search analysis programs, we identified and annotated 43 orfs from the lpm cluster and its flanking regions (Table 1 and Fig. 3B). Genes identified include those required for the formation of the laspartomycin fatty acid side chain and non-proteinogenic amino acid precursors, assembly of the peptide backbone, regulation of laspartomycin production, antibiotic export or self-resistance. Nineteen ORFs shared significant similarity with those of the friulimicin cluster (Table 1 and Fig. 3D) (Muller et al., 2007). The probable boundaries of the lpm cluster were established by comparison with the friulimicin gene cluster and according to the predicted functions of the lpm and flanking gene products.
Table 1.
Summary of ORFs identified in the lpm gene cluster and its flanking regions. Size indicates the number of amino acids in the translated product. Homologues found in the related friulimicin biosynthetic gene cluster are presented separately from the best matching protein identified by BLAST analysis.
| ORF | Gene | Size (aa) |
Friulimicin homologue (% aa identity) |
Best match accession no. (% aa identity) |
Proposed function |
|---|---|---|---|---|---|
| 1* | orf1 | 629 | EEW73624 (82) | ABC transporter related protein | |
| 2* | orf2 | 588 | EDY53507 (79) | Sodium/hydrogen exchanger | |
| 3* | orf3 | 316 | EDY53506 (84) | Integral membrane protein | |
| 4 | orf4 | 605 | EDY53505 (83) | Glycosyl hydrolase | |
| 5 | orf5 | 314 | EDY53504 (83) | Cyclase | |
| 6 | orf6 | 526 | BAC69641 (86) | Putative choline dehydrogenase | |
| 7* | orf7 | 346 | EDY53501 (81) | Cytochrome d ubiquinol oxidase, subunit II |
|
| 8* | orf8 | 482 | EDY53500 (86) | cytochrome bd-I oxidase subunit I | |
| 9* | orf9 | 446 | EDY53499 (83) | NADH dehydrogenase | |
| 10* | orf10 | 280 | EDY53498 (88 ) | 2-Deoxy-D-gluconate 3- dehydrogenase |
|
| 11* | orf11 | 305 | BAI48077(37) | LysR-type transcriptional regulator | |
| 12 | orf12 | 219 | EFL23326(67) | HAD superfamily (subfamily IIIB) phosphatase |
|
| 13 | orf13 | 278 | RegA (CAM56765) (51) |
ADK54894 (52) | Regulatory protein |
| 14 | orf14 | 364 | ACU35079 (59) | Daunorubicin resistance ABC transporter ATPase subunit |
|
| 15 | orf15 | 358 | DabA (69) | CAM56767 | Diaminobutyric acid synthase A |
| 16 | orf16 | 472 | DabB (64) | CAM56768 | Diaminobutyric acid synthase B |
| 17 | orf17 | 424 | DabC (63) | CAM56769 | Diaminobutyric acid synthase C |
| 18 (LpmA) | lpmA | 942 | PstA (59) | CAD32904 | NRPS: CIII-AAsp 1-T |
| 19* | orf19 | 315 | RegB (62) | CAD32906 | Regulatory protein B |
| 20* | orf20 | 280 | ExpA (CAD32907) (67) |
ADK54902 (67) | ABC transporter protein |
| 21 | orf21 | 607 | LipA (58) | CAD32910 | Acyl-CoA synthase |
| 22 | orf22 | 579 | LipB CAJ18234 (60) |
ADK54906 (61) | Acyl-CoA dehydrogenase |
| 23 | orf23 | 268 | Orf12 (CAJ18235) (40) |
ADK54907 (35) | Hypothetical protein |
| 24 | orf24 | 93 | LipD (CAJ18236) (70) |
ADK54908 (71) | Acyl carrier protein |
| 25 (LpmB) | lpmB | 3187 | PstB (60) | CAJ18237 | NRPS: C-ADap 2-T-C-A-C-APip 3-T-E |
| 26 (LpmC) | lpmC | 6771 | PstC (62) | CAM56770 | NRPS: C-AGly
4-T-C-AAsp
5-T-C-AGly
6- T-C-AAsp 7-T-C-AGly 8-T-C-AThr 9-T-E |
| 27 (LpmD) | lpmD | 2405 | PstD (CAM56771) (64) |
ADK54911 (64) | NRPS: C-AIle 10-T-C-APro 11-T-TE |
| 28 | svH1 | 74 | Orf21 (CAM56772) (79) |
MbtH-like protein | |
| 29 | orf29 | 268 | LipE (73) | CAM56773 | LipE homolog |
| 30* | orf30 | 773 | Mem (69) | CAM56774 | Predicted drug exporters of the RND superfamily |
| 31 | orf31 | 342 | Pip (68) | CAM56775 | Lysine cyclodeaminase |
| 32* | orf32 | 214 | RegC (CAM56776) (74) |
ADK54916 (74) | Regulatory protein C |
| 33* | orf33 | 428 | RegD (53) | CAM56777 | Regulatory protein D |
| 34* | orf34 | 267 | EFE79485 (72) | Hypothetical protein | |
| 35* | orf35 | 584 | EFE71562 (69) | VanW family protein | |
| 36* | orf36 | 373 | EDY66402 (83) | Integral membrane Protein | |
| 37* | orf37 | 501 | EAP98312 (81) | Integral membrane protein | |
| 38* | orf38 | 179 | EEP71906 (65) | Integral membrane protein | |
| 39* | orf39 | 318 | EFC61413 (76) | Conserved hypothetic protein | |
| 40 | orf40 | 553 | ZP05535703 (71) |
Two component system histidine kinase |
|
| 41 | orf41 | 227 | EFL36342 (74) | Two-component system response regulator |
|
| 42 | orf42 | 344 | EFL30101(63) | Predicted protein | |
| 43 | orf43 | 225 | ACZ23633(71) | Response regulator with CheY-like receiver domain and winged-helix DNA-binding domain |
indicates the translation from the complementary strand.
3.8. Assembly of the laspartomycin peptide backbone
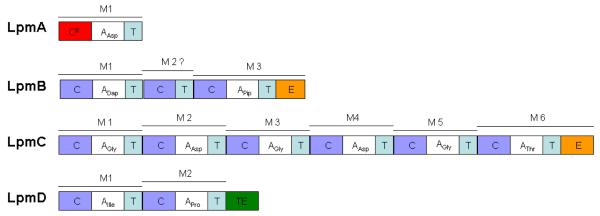
Many bioactive peptides are made on nonribosomal peptide synthetases (NRPSs), which are usually large modular enzymes. Each module typically consists of several discriminative catalytic domains and is responsible for incorporating a specific amino acid residue into a peptide chain. The minimum extension module is comprised of the adenylation domain (A), a peptidyl carrier protein (PCP) or thiolation (T) domain and the condensation (C) domains. The overall process is detailed in several excellent reviews (Marahiel et al., 1997; Walsh and Fischbach, 2010). The process consists of three stages: precursor formation (e.g., dedicated biosynthesis pathways to form the nonproteinogenic amino acid building blocks), peptide backbone assembly involving multiple modules and domains sequentially incorporating amino acids into the growing peptide chain, and post-assembly tailoring that includes α-carbon epimerization, N-methylation, heterocyclization of Cys or Ser/Thr residues to thiazolines and oxazolines, and side chain halogenation or hydroxylation. Other modifications such as oxidation, alkylation, acylation and glycosylation can occur after release of the nascent peptide from the NRPS complex and are often necessary for full biological activity. In the lpm gene cluster, four peptide synthetase genes, designated lpmA, lpmB, lpmC and lpmD were identified (Table 1 and Fig. 3B). Their products LpmA, LpmB, LpmC and LpmD were predicted to be responsible for assembly of the 11 amino acid residue laspartomycin peptide backbone. The number and order of the modules and domains identified in the lpm cluster support the NRPS co-linearity principle (Fig. 1 and 4) (Marahiel at al., 1997). All modules and domains exhibit typical NRPS features. Extra condensation and thiolation or PCP domains immediately following module 1 of LpmB are present (Fig. 4). Nine out of 11 predicted A-domains exactly match the order and sequence of amino acids found in the structure of laspartomycin C (Table 2 and Fig. 1), which is the only structurally defined component of the laspartomycins. However, position 2 in laspartomycin C is expected to have the amino acid residue Dap, but the sequence-based substrate prediction analysis only gave the residue Asp (Table 2). This finding does not seem to be the result of mistaken assembly because the independent genomic sequence results obtained by 454 Sanger and Illumina sequencing methods concluded that no extra A-domain is present in the lpm cluster and no stand-alone A-domain or gene is located near the lpm cluster. It is worth pointing out that the VioF in viomycin cluster was annotated to activate and recognize the nonproteinogenic amino acid Dap, but the predicted substrate for VioF was an Asp instead of Dap (Table 2) (Thomas et al., 2003). Apparently, the substrate specificity sequences for Dap are not included in the NRPS prediction database we used (http://www-ab.informatik.uni-tuebingen.de/toolbox/index.php?view=domainpred) (Rausch et al., 2005). Interestingly, comparison of the substrate specificity sequences for the amino acid Dap between lpm cluster (LTKIGAVN) and vio cluster (AQDLAIVD) found they shared almost no similarity. In contrast, the substrate specificity sequences (LTKIGAVN) predicted for laspartomycin residues Dap2, Asp5 and Asp7 are identical to one another but they shared no similarity with those for L-Asp1 (IWQSTADD). These findings suggest the presence of the multiple versions of the substrate specificity sequences for the same amino acid in the same or different nonribosomally generated peptides. Another subtle substrate deviation is found at position 10 which should be the residue Ile but the prediction gave the best match with Val. It is worth noting that this position in Nature could have either Ile or Val residue as observed in many lipopeptide members of the amphomycin-group antibiotics. No other unusual features were found for the NRPS of the lpm cluster.
Fig. 4.
Module and domain organization of the laspartomycin NRPS. M1, module 1; CIII, starter condensation domain; C, condensation domain; A, adenylation domain; T, thiolation domain or peptidyl carrier protein; E, epimerization domain; TE, thioesterase domain. The extra module with absence of the A domain is indicated with a question mark.
Table 2.
Derived substrate signature sequences for NRPS adenylation domains identified in the laspartomycin (lpm) cluster.
| Module | Substrate recognition sequence |
Corresponding laspartomycins |
amino acid in Predicted amino acid |
|---|---|---|---|
| LpmA-m1 | IWQSTADD | L-Asp1 | Asp |
| LpmB-m1 | LTKIGAVN (AQDLAIVD)a |
L-Dap2 | Asp |
| LpmB-m3 | FQFFGIAA (FQFFGVAV)b | Pip3 | None |
| LpmC-m1 | ILQLGLIW | Gly4 | Gly |
| LpmC-m2 | LTKIGAVN | Asp5 | Asp |
| LpmC-m3 | ILQLGLIW | Gly6 | Gly |
| LpmC-m4 | LTKIGAVN | Asp7 | Asp |
| LpmC-m5 | ILQLGLIW | Gly8 | Gly |
| LpmC-m6 | FWNVGMVH | D-Thr9 | Thr |
| LpmD-m1 | AYFWGVCF | Val10 | Ile |
| LpmD-m2 | VQYIAHVV | Pro11 | Pro |
the substrate specificity sequence in bold for the Dap residue found in the viomycin cluster (Thomas et al., 2003).
the substrate specificity sequence in bold for the Pip residue found in the friulimicin cluster (Muller et al., 2007).
The laspartomycin enzymatic assembly line begins with LpmA, a single module peptide synthetase predicted to activate and incorporate the first amino acid residue L-Asp1 into the laspartomycin peptide backbone. A condensation domain (LpmA-C1) is located immediately upstream of the adenylation domain at the N-terminus of LpmA. Unique C domains have been found in a number of lipopeptide biosynthetic gene clusters (Strieker and Marahiel, 2009). These C domains have been referred to as type CIII or C’ domains (Miao et al., 2005; Miao et al., 2006) and they catalyze the transfer of CoA-activated fatty acids with high substrate specificity to the PCP-bound amino acid (Chooi and Tang, ; Kraas et al.). LpmA-C1 shares 43% amino acid identity with the corresponding PstA in the friulimicin cluster (Muller et al., 2007). Like all other CIII domains, LpmA-CIII is believed to specifically catalyze the acylation of the starting NRPS-bound Asp1 residue prior to peptide assembly.
LpmB consists of two complete modules (LpmB-m1 and LpmB-m3) and an extra incomplete module (LpmB-m2), which is missing an A domain. The presence of LpmB-m2 was not expected. The first module of LpmB was predicted to couple N-lipo-L-Asp1 and L-Dap2 to form a dipeptide that is directly passed onto the third module of LpmB. Whether LpmB-m2 plays an additional role in discriminating between Dap and Asp remains to be determined. After completing the formation of the tripeptide N-lipo-L-Asp1-L-Dap2-D-Pip3 by joint action of LpmA and LpmB, the successive peptidyl chain elongation from amino acid residues D-Pip3 to D-Thr9, and D-Thr9 to L-Pro11 would be catalyzed by LpmC (6 modules) and LpmD (2 modules), respectively. The second module of the final NRPS LpmD is immediately followed by a thioesterase (TE) domain that terminates the peptidyl chain elongation and promotes the cyclization and release of the mature active peptide.
3.9. Biosynthesis of the non-proteinogenic amino acids
Two nonproteinogenic amino acid residues L-2,3-diaminopropionate (L-Dap2) and pipecolic acid (D-Pip3) are found in the structure of laspartomycin C (Borders et al., 2007). Inspection of the lpm cluster did not reveal any gene predicted to be involved in the biosynthesis of Dap. The peptide antibiotic viomycin also contains Dap (Thomas et al., 2003). Previous studies on the viomycin cluster predicted that the vioB and vioK gene products were jointly responsible for the formation of L-Dap (Thomas et al., 2003). VioB, a pyridoxal-5′-phosphate-dependent enzyme, is a homolog of cysteine synthase. It was proposed to use L-serine as substrate and ammonia as the nucleophile to catalyze a β-replacement reaction. VioK is a homolog of ornithine cyclodeaminases and was proposed to use L-ornithine as substrate to supply the ammonia for VioB. We used the sequences of VioB and VioK in BLAST searches against the available genomic sequence data from strain ATCC29814. Two genes, designated svB and svK, were identified in an approx. 21 kb contig 29814-84 (Fig. 3C). SvB and SvK shared 38% and 31% amino acid identities with their counterparts VioB and VioK, respectively. Further BLAST analysis indicated that SvB shared strong aa identities with pyridoxal-5′-phosphate-dependent cysteine synthases (GenBank accession Nos.: EFB78112 (76%)) while SvK exhibited strong aa identities with ornithine cyclodeaminases (GenBank accession Nos. EFB78112 (80%)). Therefore, SvB and SvK may direct the biosynthesis of Dap of laspartomycin C, though the relative distances of SvB and SvK to lpm cluster were not determined on the chromosome. The other possibility is that the Dab genes within the lpm cluster may actually mediate Dap formation. This hypothesis is supported by the following observations: (1) the predicted substrate specificity codons for Dap from the laspartomycin cluster and for Dab from the friulimicin cluster are highly similar (LTKVGDVN vs. LTKIGAVN). This similarity may be the key to the substrate flexibility; (2) the presence of an extra incomplete module (M2) in lpmB may be involved in the discrimination between Dap and Dab resulting in exclusive or preferential acceptance of Dap instead of Dab as the substrate amino acid at position 2 of larspartomycin; (3) unlike the genes svB and svK which are distal to the laspartomycin cluster, the predicted biosynthetic genes for Dab are located within the laspartomycin cluster. The 11 amino acid lipopeptide antibiotics laspartomycin and friulimicin are structurally and functionally closely related. Muller and co-workers demonstrated by gene inactivation, heterologous expression and complementation that the pip gene, predicted to code for a lysine cyclodeaminase, was required for the biosynthesis of friulimicin in Actinoplanes friuliensis (Muller et al., 2007). The Pip counterpart Orf31 was identified in the lpm gene cluster. Orf31 shared 68% aa identity with the friulimicin Pip protein. Therefore, Orf31 was predicted to catalyze the deaminative cyclization of lysine to pipecolic acid (Molnar et al., 1996; Muller et al., 2007).
3.10. Formation and attachment of the fatty acid chain
Laspartomycin C possesses a 2, 3-unsaturated acyl side chain attached to the starter Asp1 unit (Borders et al., 2007). The two minor structurally undefined components of laspartomycins are believed to differ from laspartomycin C only in the acyl side chains (Borders et al., 2002; Borders et al., 2007). Four genes (orf21, orf22, orf24 and orf29) are predicted to activate and modify a precursor lipid chain and transfer it to the amine of Asp1 on LpmA. These four gene products have their respective counterparts LipA, LipB, LipD and LipE in the friulimicin cluster. Homologs of Orf21 (an acyl-CoA ligase) and Orf24 (an acyl carrier protein) are commonly present in biosynthetic pathways for lipopeptide antibiotics such as surfactin, CDA, daptomycin and the friulimicins (Strieker and Marahiel, 2009; Kraas et al., 2010). Orf22, an acyl-CoA dehydrogenase, is predicted to introduce the double bond in laspartomycin. Its counterpart LipB was demonstrated to introduce the Δcis3 double bond in the friulimicin lipid tail (Heinzelmann et al., 2005). Orf29 was predicted to be a member of the α,β-hydrolase superfamily. Homologs of Orf28 were found in several lipopeptide gene clusters including LipE (friulimicin cluster) (Muller et al., 2007) and LptH (A54145 cluster) (Miao et al., 2006). The role of Orf29 is uncertain and might be involved in either an acyltransferase reaction or a thioesterase type II function (Muller et al., 2007).
3.11. Genes for self-resistance, regulation and export
The mechanism of antibiotic action of the laspartomycins is still unknown. Some studies on the mode of action of the closely related lipopeptides amphomycin and friulimicin have been reported. Schneider and co-workers recently found friulimicin have a unique mode of action by interrupting the cell wall precursor cycle through the formation of a Ca2+-dependent complex with the bactoprenol phosphate carrier C55-P, which is not targeted by any other antibiotic in use (Schneider et al, 2009). Wecke and co-workers compared the action of daptomycin and friulimicin B on Bacillus subtilis. They found the cell envelope stress-sensing two-component system LiaRS is exclusively and strongly induced by daptomycin, indicating a different mechanism of action in these two lipopeptide antibiotics.
Self-resistance may involve export of the lipopeptides. In the friulimicin cluster, the expA gene encodes the transmembrane component of an ABC transporter. Expression of ExpA in S. lividans TK23, a friulimicin-sensitive strain, conferred increased resistance to friulimicin for this recombinant strain. Therefore, ExpA may be partially or fully responsible for the self-resistance of friulimicin by serving as an efflux pump (Muller et al., 2007). The ExpA counterpart Orf20 was identified in the lpm cluster. It shares 67% amino acid identity with ExpA and may play a similar role in the self-resistance of laspartomycin. Two other laspartomycin genes, Orf14 and Orf30, may be also involved in laspartomycin self-resistance. Orf14 encodes an additional ABC transporter. It does not have a counterpart in the friulimicin cluster. Orf30 encodes a drug export integral membrane protein. Its homolog Mem is present in the friulimicin cluster (Muller et al., 2007). Four gene products Orf13, Orf19, Orf32 and Orf33 are predicted to regulate laspartomycin production. Their corresponding counterparts RegA, RegB, RegC and RegD were identified in the friulimicin cluster (Muller et al., 2007). RegA was demonstrated as a pathway-specific regulator of friulimicin biosynthesis in Actinoplanes friuliensis (Nolden et al., 2009). Inactivation of RegA resulted in abolishment of friulimicin production in the mutant. RegA was also shown to positively regulate the majority of the friulimicin biosynthetic genes. Orf13 shared 51% amino acid identity with RegA. Therefore, we predict for Orf13 a pathway-specific activator role in laspartomycin biosynthesis in strain ATCC 29814. To date, regulatory involvement of RegB, RegC and RegD in friulimicin biosynthesis has not been experimentally documented. Likewise, the regulatory roles of Orf19, Orf32 and Orf33 in laspartomycin production remain to be determined.
3.12. Boundaries and remaining genes
The boundaries of the lpm cluster were proposed according to the predicted functions of the lpm and flanking gene products and by comparison with the friulimicin gene cluster. The regA and regD were proposed to be the first and last genes of the friulimicin cluster, respectively (Muller et al., 2007). Likewise, orf13 and orf33, corresponding to regA and regD, were assigned to be the first and last gene of the lpm cluster, respectively. The approx. 16.5 kb region sequenced upstream of Orf13 does not harbor any genes with predicted products necessary for laspartomycin production. However, Orf33 may not be the right-hand boundary of the lpm cluster. orf35 encodes an uncharacterized vancomycin resistance protein VanW (Depardieu et al., 2003). orf40 and orf41 encode two component system-type regulators. They may regulate the expression of the laspartomycin self-resistance genes. If Orf35, Orf40 and Orf41 contributed to the laspartomycin self-resistance and/or regulation of laspartomycin production, the right-hand boundary of the lpm cluster would be extended to include orf41.
The biosynthesis genes for diaminobutyric acid (Dab) naturally occur in the lpm gene cluster even though the Dab residue was not found in laspartomycin C. It is unlikely that the Dab residue would be present in the other two structurally unknown minor laspartomycin components because (1) all substrate specificity predictions for the laspartomycin constituent amino acids revealed no such option; (2) all laspartomycin components have the same 11 amino acid peptide and differ in their acyl side chains (Borders et al., 2002; Borders et al., 2007). Additionally, it was noted that the very early amino acid composition analysis of the laspartomycins did show the presence of Dab (Naganawa et al., 1968). Therefore, we can not completely eliminate the possibility that Dab is a residue in one or more unidentified minor laspartomycin components.
Orf28 encodes a MbtH-like protein. MbtH protein has been found in numerous NRPS gene clusters. It was recently demonstrated that the small MbtH-like protein VbsG was required for adenylation activity of the NRPS Vbs (Heemstra et al., 2009), and another MbtH-like protein GlbE was also required for obtaining active and soluble NRPS protein GlbF (Imker et al., 2010).
Finally, the function of Orf23 in the lpm cluster is unknown. Whether Orf23 and any of the genes in the flanking regions that encode products of unknown function are involved in laspartomycin self-immunity or regulation of the peptide production in strain ATCC 29814 remains to be determined.
4. Conclusions
The laspartomycins and friulimicins are closely related lipopeptides of the amphomycin-group antibiotics with strong activity against life-threatening Gram-positive pathogens including MRSA and VRE. Like friulimicin B, the unknown mechanism of the laspartomycins might be unique and different from the clinical antibiotic daptomycin (Cubicin). Cloning, sequencing, annotation and disruption of the lpm biosynthetic gene cluster in S. viridochromogenes ATCC 29814 lay the groundwork for further genetic engineering of the laspartomycin biosynthetic machinery to generate new analogues with improved properties.
Interesting features of the lpm cluster include the presence of apparently unnecessary biosynthetic genes for the non-component amino acid Dab coupled with the absence of the necessary biosynthetic genes for the constituent amino acid Dap, which are located elsewhere on the chromosome. The counterpart genes found in the laspartomycin and friulimicin clusters are highly conserved with respect to their number and local genetic organization. The comparative annotation and the molecular blueprints available for these lipopeptides provide us important information for targeted mutation and combinatorial biosynthesis. Characterization of the resulting mutants and their metabolites will allow us to decipher the mechanism of laspartomycin self-resistance and the means of regulation of peptide production.
Supplementary Material
Acknowledgements
This work was in part supported by NIH Grants R01AI073784 (X.Y.) and AI073784-03S1 (X.Y.), and the OSU-NAU collaborative grant 20101757 (X.Y.). Profs. T. Mark Zabriskie and Philip J. Proteau are thanked for their helpful discussions and critical reading of the manuscript. We thank Prof. K.F. Chater (JIC, Norwich, England) for providing plasmids pSET152 and pIJ773. Yang Wang is supported by the State Scholarship Fund of the People’s Republic of China. The Mass Spectrometry Facility at OSU is supported by NIEHS Grant P30 ES00210.
Abbreviations
- Dab
diaminobutyric acid
- Dap
diaminopropionic acid
- HPLC
high performance liquid chromatography
- bp
base pair
- MIC
minimal inhibitory concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- MS
mass spectrometry
- NRPS
nonribosomal peptide synthetase
- orf
open reading frame
- PCR
polymerase chain reaction
- VISA
vancomycin-intermediate Staphylococcus aureus
- VRE
vancomycin-resistant enterococci
- VRSA
vancomycin-resistant Staphylococcus aureus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon B, Lacal JC, Fernandez-Sousa JM, Carrasco L. Screening for new compounds with antiherpes activity. Antiviral Res. 1984;4:231–44. doi: 10.1016/0166-3542(84)90029-9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aretz W, Meiwes J, Seibert G, Vobis G, Wink J. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. I. Taxonomic studies of the producing microorganism and fermentation. J Antibiot (Tokyo) 2000;53:807–15. doi: 10.7164/antibiotics.53.807. [DOI] [PubMed] [Google Scholar]
- Banerjee DK. Amphomycin inhibits mannosylphosphoryldolichol synthesis by forming a complex with dolichylmonophosphate. J Biol Chem. 1989;264:2024–8. [PubMed] [Google Scholar]
- Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–9. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Borchert S, Patil SS, Marahiel MA. Identification of putative multifunctional peptide synthetase genes using highly conserved oligonucleotide sequences derived from known synthetases. FEMS Microbiol Lett. 1992;71:175–80. doi: 10.1016/0378-1097(92)90508-l. [DOI] [PubMed] [Google Scholar]
- Borders DB, Curran WV, Fantini AA, Francis ND, Jarolmen H, Leese RA. Derivatives of laspartomycin and preparation and use thereof. Pub. No.: WO/2002/005838 U.S. Pat. 2002
- Borders DB, Leese RA, Jarolmen H, Francis ND, Fantini AA, Falla T, Fiddes JC, Aumelas A. Laspartomycin, an acidic lipopeptide antibiotic with a unique peptide core. J Nat Prod. 2007;70:443–6. doi: 10.1021/np068056f. [DOI] [PubMed] [Google Scholar]
- Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–24. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Chooi YH, Tang Y. Adding the lipo to lipopeptides: do more with less. Chem Biol. 17:791–3. doi: 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Cronk GA, Neumann DE. The topical use of amphomycin and amphomycin-neomycin ointments. Antibiotic Med Clin Ther. 1956;3:142–5. [PubMed] [Google Scholar]
- Curran WV, Leese RA, Jarolmen H, Borders DB, Dugourd D, Chen Y, Cameron DR. Semisynthetic approaches to laspartomycin analogues. J Nat Prod. 2007;70:447–50. doi: 10.1021/np068062b. [DOI] [PubMed] [Google Scholar]
- Depardieu F, Bonora MG, Reynolds PE, Courvalin P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol. 2003;50:931–48. doi: 10.1046/j.1365-2958.2003.03737.x. [DOI] [PubMed] [Google Scholar]
- Dini C. MraY Inhibitors as Novel Antibacterial Agents. Curr Top Med Chem. 2005;5:1221–36. doi: 10.2174/156802605774463042. [DOI] [PubMed] [Google Scholar]
- Elbein AD. The effect of tsushimycin on the synthesis of lipid-linked saccharides in aorta. Biochem J. 1981;193:477–84. doi: 10.1042/bj1930477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M, Davis G, Greener A, Helinski DR. Autorepressor properties of the pi-initiation protein encoded by plasmid R6K. Nucleic Acids Res. 1985;13:103–14. doi: 10.1093/nar/13.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori DG, Barr EW, Matthews ML, Koch GM, Yonce JR, Walsh CT, Bollinger JM, Jr., Krebs C, Riggs-Gelasco PJ. Spectroscopic evidence for a high-spin Br-Fe(IV)-oxo intermediate in the alpha-ketoglutarate-dependent halogenase CytC3 from Streptomyces. J Am Chem Soc. 2007;129:13408–9. doi: 10.1021/ja076454e. [DOI] [PubMed] [Google Scholar]
- Galm U, Wang L, Wendt-Pienkowski E, Yang R, Liu W, Tao M, Coughlin JM, Shen B. In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC15003 revealing new insights into its biosynthetic pathway. J Biol Chem. 2008;283:28236–45. doi: 10.1074/jbc.M804971200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemstra JR, Jr., Walsh CT, Sattely ES. Enzymatic tailoring of ornithine in the biosynthesis of the Rhizobium cyclic trihydroxamate siderophore vicibactin. J Am Chem Soc. 2009;131:15317–29. doi: 10.1021/ja9056008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann B, Kaplan MA, Muir RD, Hooper IR. Amphomycin - a new antibiotic. antib. Chemother. 1953;3:239. [PubMed] [Google Scholar]
- Heinzelmann E, Berger S, Muller C, Hartner T, Poralla K, Wohlleben W, Schwartz D. An acyl-CoA dehydrogenase is involved in the formation of the Delta cis3 double bond in the acyl residue of the lipopeptide antibiotic friulimicin in Actinoplanes friuliensis. Microbiology. 2005;151:1963–74. doi: 10.1099/mic.0.27844-0. [DOI] [PubMed] [Google Scholar]
- Hinuma Y. Zaomycin, a new antibiotic from a Streptomyces sp. J Antibiot (Tokyo) 1954;7:134–6. [PubMed] [Google Scholar]
- Hirsch CF, Ensign JC. Heat activation of Streptomyces viridochromogenes spores. J Bacteriol. 1976;126:24–30. doi: 10.1128/jb.126.1.24-30.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imker HJ, Krahn D, Clerc J, Kaiser M, Walsh CT. N-acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem Biol. 2010;17:1077–83. doi: 10.1016/j.chembiol.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M. On Glumamycin, a New Antibiotic. II*. Isolation and identification of Amino Acids Constituting Glumamycin. J Am Chem Soc. 1962;35:1249–1254. [Google Scholar]
- Kong F, Carter GT. Structure determination of glycinocins A to D, further evidence for the cyclic structure of the amphomycin antibiotics. J Antibiot (Tokyo) 2003;56:557–64. doi: 10.7164/antibiotics.56.557. [DOI] [PubMed] [Google Scholar]
- Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol. 2010;17:872–80. doi: 10.1016/j.chembiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Lomakina NN, Brazhnikova MG. Chemical composition of crystallomycin. Biokhimiia. 1959;24:425–31. [PubMed] [Google Scholar]
- Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- Miao V, Brost R, Chapple J, She K, Gal MF, Baltz RH. The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae. J Ind Microbiol Biotechnol. 2006;33:129–40. doi: 10.1007/s10295-005-0028-5. [DOI] [PubMed] [Google Scholar]
- Miao V, Coeffet-Legal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology. 2005;151:1507–23. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- Molnar I, Aparicio JF, Haydock SF, Khaw LE, Schwecke T, Konig A, Staunton J, Leadlay PF. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- Muller C, Nolden S, Gebhardt P, Heinzelmann E, Lange C, Puk O, Welzel K, Wohlleben W, Schwartz D. Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic Friulimicin in Actinoplanes friuliensis. Antimicrob Agents Chemother. 2007;51:1028–37. doi: 10.1128/AAC.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth G, Nußbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol. Gen. Genet. 1989;219:341–348. [Google Scholar]
- Naganawa H, Hamada M, Maeda K, Okami Y, Takeushi T. Laspartomycin, a new anti-staphylococcal peptide. J Antibiot (Tokyo) 1968;21:55–62. doi: 10.7164/antibiotics.21.55. [DOI] [PubMed] [Google Scholar]
- Naganawa H, Takita T, Maeda K, Umezawa H. A novel fatty acid from laspartomycin. J Antibiot (Tokyo) 1970;23:423–4. doi: 10.7164/antibiotics.23.423. [DOI] [PubMed] [Google Scholar]
- Nolden S, Wagner N, Biener R, Schwartz D. Analysis of RegA, a pathway-specific regulator of the friulimicin biosynthesis in Actinoplanes friuliensis. J Biotechnol. 2009;140:99–106. doi: 10.1016/j.jbiotec.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DV. Molecular cloning A laboratory manual. Third edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Schneider T, Gries K, Josten M, Wiedemann I, Pelzer S, Labischinski H, Sahl HG. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother. 2009;53:1610–8. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Alijah R, Nussbaumer B, Pelzer S, Wohlleben W. The peptide synthetase gene phsA from Streptomyces viridochromogenes is not juxtaposed with other genes involved in nonribosomal biosynthesis of peptides. Appl Environ Microbiol. 1996;62:570–7. doi: 10.1128/aem.62.2.570-577.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay AJ, Adam J, Martin JH, Hausmann WK, Shu P, Bohonos N. Aspartocin. I. Production, isolation, and characteristics. Antibiot Annu. 1959;7:194–8. [PubMed] [Google Scholar]
- Shibata M, Kanzaki T, Nakazawa K, Inoue M, Hitomi H, Mizuno K, Fujino M, Akira M. On glumamycin, a new antibiotic. J Antibiot (Tokyo) 1962;15:1–6. [PubMed] [Google Scholar]
- Shoji JI, Kozuki S, Okamoto S, Sakazaki R, Otsuka H. Studies on tsushimycin. I. Isolation and characterization of an acidic acylpeptide containing a new fatty acid. J Antibiot (Tokyo) 1968;21:439–43. [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnology. 1983;1:784–791. [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Stalker DM, Kolter R, Helinski DR. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979;76:1150–4. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieker M, Marahiel MA. The structural diversity of acidic lipopeptide antibiotics. Chembiochem. 2009;10:607–16. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iwai Y, Oiwa R, Shinohara S, Shimizu S, Oka T, Omura S. Studies on bacterial cell wall inhibitors. II. Inhibition of peptidoglycan synthesis in vivo and in vitro by amphomycin. Biochim Biophys Acta. 1977;497:633–40. doi: 10.1016/0304-4165(77)90283-5. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Oiwa R, Matsukura S, Inokoshi J, Omura S. Studies on bacterial cell wall inhibitors. X. Properties of phosph-N-acetylmuramoyl-pentapeptide-transferase in peptidoglycan synthesis of Bacillus megaterium and its inhibition by amphomycin. J Antibiot (Tokyo) 1982;35:1216–21. doi: 10.7164/antibiotics.35.1216. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Oiwa R, Matsukura S, Omura S. Amphomycin inhibits phospho-N-acetylmuramyl-pentapeptide translocase in peptidoglycan synthesis of Bacillus. Biochem Biophys Res Commun. 1979;86:902–8. doi: 10.1016/0006-291x(79)91797-2. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Chan YA, Ozanick SG. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob Agents Chemother. 2003;47:2823–30. doi: 10.1128/AAC.47.9.2823-2830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertesy L, Ehlers E, Kogler H, Kurz M, Meiwes J, Seibert G, Vogel M, Hammann P. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. II. Isolation and structural characterization. J Antibiot (Tokyo) 2000;53:816–27. doi: 10.7164/antibiotics.53.816. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–93. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecke T, Zuhlke D, Mader U, Jordan S, Voigt B, Pelzer S, Labischinski H, Homuth G, Hecker M, Mascher T. Daptomycin versus Friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob Agents Chemother. 2009;53:1619–23. doi: 10.1128/AAC.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Chen Y, Zhang L, Wang Y, Zabriskie TM. Enduracidin analogues with altered halogenation patterns produced by genetically engineered strains of Streptomyces fungicidicus. J Nat Prod. 2010;73:583–9. doi: 10.1021/np900710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, O’Hare T, Gould SJ, Zabriskie TM. Identification and cloning of genes encoding viomycin biosynthesis from Streptomyces vinaceus and evidence for involvement of a rare oxygenase. Gene. 2003;312:215–24. doi: 10.1016/s0378-1119(03)00617-6. [DOI] [PubMed] [Google Scholar]
- Yin X, Zabriskie TM. The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus. Microbiology. 2006;152:2969–83. doi: 10.1099/mic.0.29043-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.