Abstract
Colorectal cancer is a leading cause of cancer deaths in the world. It results from an accumulation of genetic and epigenetic changes in colon epithelial cells that transforms them into adenocarcinomas. There have been major advances in our understanding of cancer epigenetics over the last decade, particularly regarding aberrant DNA methylation. Assessment of the colon cancer epigenome has revealed that virtually all colorectal cancers have aberrantly methylated genes and the average colorectal cancer methylome has hundreds to thousands of abnormally methylated genes. As with gene mutations in the cancer genome, a subset of these methylated genes, called driver genes, is presumed to play a functional role in colorectal cancer. The assessment of methylated genes in colorectal cancers has also revealed a unique molecular subgroup of colorectal cancers called CpG Island Methylator Phenotype (CIMP) cancers; these tumors have a particularly high frequency of methylated genes. The advances in our understanding of aberrant methylation in colorectal cancer has led to epigenetic alterations being developed as clinical biomarkers for diagnostic, prognostic, and therapeutic applications. Progress in the assessment of epigenetic alterations in colorectal cancer and their clinical applications has shown that these alterations will be commonly used in the near future as molecular markers to direct the prevention and treatment of colorectal cancer.
Keywords: Colon cancer, DNA methylation, epigenetics, biomarkers
Introduction
Colorectal cancer results from the accumulation of both acquired genetic and epigenetic changes that transform normal glandular epithelium into invasive adenocarcinomas. The steps involved in this process were first described in the classic adenoma-cancer progression model proposed by Fearon and Vogelstein1. Since this model was originally proposed, our understanding of the molecular pathogenesis of colorectal cancer has advanced considerably, resulting in numerous revisions of the initial model. The original model was based on the premise that tubular and tubulovillous adenomas were the premalignant neoplasms that would progress to invasive adenocarcinoma. However, it is now recognized that there are additional classes of premalignant polyps, such as serrated polyps, that also have significant potential for malignant transformation2. Furthermore, the original model proposed a relatively limited repertoire of genetic alterations that drove the formation of colorectal cancer. It is now appreciated that there are multiple molecular pathways to colon cancer, and that these pathways involve both mutations and epigenetic alterations. For example, serrated polyps are associated with microsatellite instability and aberrant DNA methylation, whereas tubular adenomas more commonly arise via inactivation of the APC tumor suppressor gene and concurrent genetic alterations resulting from chromosomal instability3. At the present time, we appreciate that there are at least three, and likely more, polyp→cancer progression sequences that arise via different molecular events. These changes to the original model highlight that not only are there histologic differences in colorectal cancers but that there are important molecular differences amongst the cancers as well.
We also now understand that there are thousands of molecular alterations found in the average colon cancer genome, and it is widely believed that only a subset of these alterations drive the cellular and clinical behavior of the cancer. These key molecular features of the tumors will ultimately be useful for understanding the environmental and genetic factors that lead to colorectal cancer, for predicting the natural history of the tumors, and for determining a specific cancer’s likelihood of responding to therapy. In fact, the molecular heterogeneity of colorectal cancer is believed to be one of the factors responsible for the significant variability in the treatment response among patients with the same stage of colorectal cancer4. This is a consequence of the fact that, although there are defined molecular subtypes of colorectal cancer, the molecular subgroups cannot be accurately distinguished histologically or clinically at this time. The development of methods that can detect the significant epigenetic and genetic changes in cancers should permit the identification of these molecular alterations, which can then be used as predictive markers. Efforts to characterize the molecular features of colorectal cancers have already led to the development of clinical assays that detect KRAS mutations, which is used to direct therapy with anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer5.
Epigenetic alterations, like gene mutations, contribute to the pathogenesis and molecular heterogeneity of these cancers. This is highlighted by the identification of a CpG island methylator phenotype (CIMP) subtype of colorectal cancer that has a distinct epigenome with a high frequency of methylated genes6. Our understanding of epigenomic alterations in colorectal cancer, however, is less advanced than that of our understanding of gene mutations, but substantial progress has recently been made in this area. In this review, we will provide an overview of the role of epigenetics in colorectal cancer, including the contribution of epigenetics to the molecular heterogeneity of colorectal cancer, and will discuss the clinical applications of these epigenetic alterations as biomarkers for early detection, diagnosis, prognostication and management of patients with colorectal cancer.
The Molecular Pathogenesis of Colorectal Cancer
Gene Mutations and Genomic Instability
The classic view of cancer is that it arises as a consequence of the accumulation of mutations in key tumor-suppressor genes or oncogenes, which deregulate homeostatic functions and cause the transformation of normal cells into cancer cells. Sequencing of colon cancer genomes has revealed that there is only a small number of functionally important gene mutations out of the hundreds of mutations found in the average colon cancer genome. A recent analysis of approximately 13,000 genes revealed mutations in the coding sequence of approximately 67 genes in the average colon cancer genome, of which a subset of 12 genes were proposed to be the genes most likely to be involved with cancer formation in individual cancers7. The current cancer genome sequencing efforts demonstrate that most cancer genomes carry hundreds of mutations, providing a major challenge to determine which of these mutations play a pathogenic role in the formation of the cancer (“driver mutations”) and which are merely a consequence of this process (“passenger mutations”). Furthermore, colorectal cancers can be subgrouped based on the type of genomic instability that they display. They can be grouped into those with chromosomal instability (CIN), which is characterized by aneuploidy and chromosomal gains and losses, and those with microsatellite instability (MSI), which is identified by the presence of frequent insertion and deletion mutation in repetitive DNA sequences8,9. Different patterns of mutant genes can be seen in the MSI vs. the CIN tumors suggesting that the underlying form of genomic instability in the cancer influences the susceptibility to and selection for specific mutations.
The role of gene mutations in cancer formation can be appreciated in even the earliest lesions involved in the polyp→cancer progression sequence. The first identifiable histologic lesion implicated in colon cancer formation is the aberrant crypt focus. Dysplastic aberrant crypt foci can harbor mutations in APC, one of the most frequently mutated tumor suppressor genes in colorectal cancer. Inactivation of APC leads to activation of the Wingless/Wnt pathway, a common mechanism for initiating the polyp→cancer progression sequence10. Subsequent to mutations in APC or other members of the Wnt signaling pathway, mutations in genes such as KRAS or TP53 occur and foster the clonal progression of the polyp cells to cancer10,11. Progression can involve KRAS and TP53 mutations as well as mutations in genes that regulate important cell signaling pathways such as the transforming growth factor –β (TGFB1) signaling pathway12. Mutations in the type II TGF-β receptor (TGFBR2) occur in approximately 30% of colorectal cancers13. The type of mutation that inactivates TGFBR2 depends upon the molecular subtype of the developing cancer cell13,14. In addition, mutations and epigenetic alterations affecting other TGF-β signaling pathway members, including SMAD2, SMAD4, RUNX3, and TSP1, have been identified in colon cancers7,12,15–17. Ultimately, it is believed that although the underlying form of genomic or epigenomic instability determines the types of mutations that occur in colon cancer, the selective pressures that lead to the clonal evolution of the tumors are largely the same across all colorectal cancers. This concept can explain why certain genes are more frequently mutated in colon cancers than in other cancer types (e.g. breast cancer, prostate cancer) and vice versa.
It has been proposed that gene mutations and epigenetic alterations contribute to colon cancer formation through the activation of oncogenic pathways and the inactivation of tumor suppressor genes that regulate hallmark behaviors of cancers18. For example, KRAS is a proto-oncogene that is a downstream effector of the epidermal growth factor receptor. It signals through BRAF to activate the MAPK pathway. Mutations in KRAS or BRAF occur in approximately 55–60% of colorectal cancer, aberrantly activating the MAPK signaling pathway, inducing proliferation and suppressing apoptosis19,20. The most common signaling pathways that carry mutant genes in colorectal cancer include not only the RAS-RAF-MAPK pathway, but also the PI3K pathway, WNT-APC-CTNNB1 pathway, and TGFB1-SMAD pathway21. Insights into the biology of colorectal cancers have resulted from the identification of genes commonly mutated in these particular pathways in colon cancer and have led to paradigms that have informed the study of epigenetic alterations in cancer. These insights are also being currently used to develop new diagnostic and prognostic assays for colorectal cancer as well as therapies for colorectal cancer.
Epigenetic Alterations and Cancer
Overview
The study of gene mutations in colorectal cancer has established our current understanding of the role of molecular alterations in the development of colon cancer, and has led to the intense investigation of epigenetic alterations in cancer. Epigenetics broadly refers to heritable alterations in gene expression that are not mediated by changes in the DNA sequence. The epigenetic regulation of gene expression occurs in normal tissue and plays an important role in embryonic development, imprinting, and tissue differentiation22. In 1982, aberrant epigenetic alterations were first discovered in colorectal cancer. Since that time, epigenetic research has revealed an epigenetic landscape consisting of a complex array of epigenetic regulatory mechanisms that control gene expression in both normal tissue and cancerous tissue23,24 The epigenetic landscape is largely a reflection of factors that determine the condensation state of the chromatin, which determines whether the DNA is accessible to proteins that control gene transcription. A relaxed or “open” chromatin state allows for gene transcription, whereas a condensed or “closed” chromatin state prevents gene transcription22. The epigenetic mechanisms currently believed to play a role in cancer include: 1) DNA methylation of cytosine bases in CG rich sequences, called CpG Islands; 2) post-translational modifications of histones, which are proteins that form the nucleosomes, which regulate the packaging structure of the DNA (called chromatin); 3) microRNAs and noncoding RNAs; and 4) nucleosome positioning22. In this review, we will focus on aberrant DNA methylation as it is the most extensively studied deregulated epigenetic mechanism in colorectal cancer. A number of excellent reviews on other classes of epigenetic alterations, such as histone modifications, have recently been written, and the interested reader is directed to those reviews25–28.
DNA Methylation-Overview
DNA methylation refers to the enzymatic addition of a methyl group to the 5-position of cytosine by DNA methyltransferases (DNMT) to produce 5-methylcytosine, a normal base in DNA29. Generally, the favored substrate for the DNMT’s is a CG dinucleotide sequence, which has been termed CpG. The majority of CpGs are methylated in normal mammalian cells with unmethylated CpGs being typically present only in regions of DNA called CpG Islands. CpG islands are regions in the genome where the percentage of the CpG dinucleotides is higher than would be expected based upon a random distribution of nucleotides. Of interest, CpG’s are generally under-represented in the genome presumably due to the susceptibility of 5-methylcytosines to undergo transition mutations secondary to deamination30. CpG islands are often defined as sequences greater than 200–500 bases in length with greater than 50% GC content and a ratio of observed to expected CpG ratio of greater than 0.631. CpG islands overlap the promoter region of 60–70% of genes and tend to be protected from methylation; however, they can become aberrantly methylated in cancer. Methylation of CpG islands within the promoter region is correlated with transcriptional silencing although it appears that decreased gene expression is only characteristic of a subset of methylated genes in colorectal cancer32,33. (Figure 1) Methylation that occurs in CpG sites outside of promoter regions, termed gene body methylation, does not correlate with transcriptional inactivation. In fact, paradoxically, gene body methylation has been correlated with transcriptional activation34.
Figure 1.
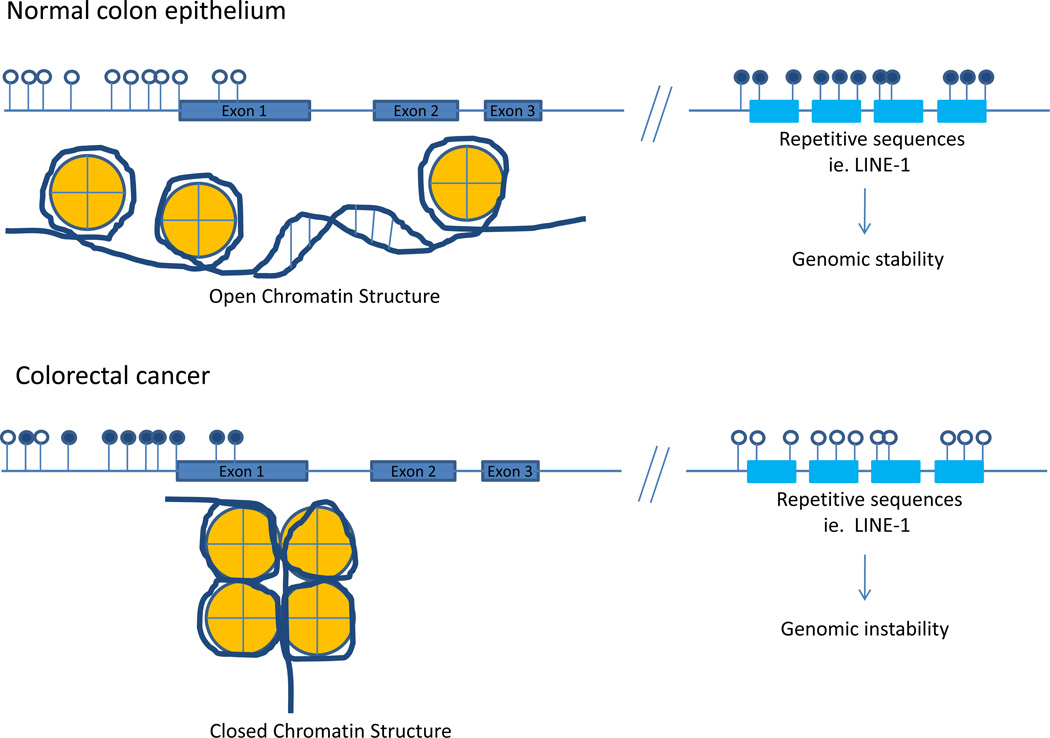
CpG Island DNA hypermethylation and global DNA hypomethylation in colorectal cancer as compared to normal colonic epithelium. Open lollipops represent unmethylated CpG sites in the genome. Closed lollipops represent methylated CpG sites within the genome. Unmethylated CpG islands within the promoter region of genes are correlated with an open chromatin structure (euchromatin) whereas methylated CpG islands are correlated with a condensed, closed chromatin structure (heterochromatin) and transcriptional silencing. Normal colonic epithelium generally has unmethylated CpG islands in the promoter regions of genes, whereas aberrant hypermethylation of promoter associated CpG islands is a hallmark of neoplasms. In addition to the aberrant local hypermethylation seen in colorectal cancers, global hypomethylation at LINE-1 sequences is also observed, which has been shown to associate with genomic instability. Interestingly, there is an inverse association between local CpG island hypermethylation and global LINE-1 hypomethylation as colonic neoplasms progress.
It is important to recognize that DNA methylation is a normal mechanism in the mammalian genome by which cells regulate gene expression, and that the majority of the CpG sites in the human genome, which are located outside of promoter regions, are heavily methylated. For example, DNA methylation is a major mechanism for X chromosome inactivation and genomic imprinting35. As mentioned previously, the methylation state of a gene is regulated by a family of DNA methyltransferases (DNMTs), including, DNMT1, DNMT3a, and DNMT3b, which transfer methyl groups from s-adenosylmethionine to DNA in either a de novo fashion (DNMT3a and DNMT3b) or maintenance fashion (DNMT1)29,36,37. Other important roles for DNA methylation include silencing of repetitive centromeric sequences as well as transposons and other endoparasitic sequences; these later functions appear to be important for maintaining genomic stability38–40. (Figure 1)
Gene methylation patterns are established during development by the de novo DNMTs and are critical for gene regulation. CpG methylation can lead to transcriptional inactivation via multiple mechanism, including directly inhibiting cis-binding elements, including the following transcription factors: AP-2, CREB, E2F, CBF and NF-KB41–45. A prominent mechanism by which DNA methylation is thought to regulate gene expression is through cooperative interactions with enzymes that regulate the chromatin structure, which can induce a compacted chromatin environment that represses gene expression33. One mechanism involved in these cooperative interactions is based on a class of proteins called methyl-binding proteins. Methyl-binding proteins are a family of proteins that bind with high affinity to methylated DNA resulting in a cascade of events that can alter chromatin structure by recruiting proteins that can regulate histone acetylation, histone methylation, and chromatin remodeling, thereby condensing the chromatin and blocking access of transcription factors to the promoter region33. These proteins include the methyl domain binding proteins: MeCP2, MBD1, MBD2, and MBD4; the zinc finger proteins Kaiso, ZBTB4, and ZBTB38; and the SET-and RING-finger associated proteins, UHFR1 and UHFR2 among others28,46. The interaction between DNA methylation, histone modification and chromatin structure is complex, with abundant crosstalk. DNA methylation can impact chromatin structure, but the converse is also true. Because of the epigenetic crosstalk between DNA methylation and histone modification, aberrant DNA methylation can alter chromatin structure and gene expression, and dysregulation of histones and their modifying proteins may cause aberrant DNA methylation. Indeed polycomb group proteins (PcG) have been implicated in predisposing promoters to both normal methylation and cancer related methylation46. PcG protein complexes, PRC1 and PRC2, regulate gene expression during embryonic development through their effects on histone modification and chromatin condensation. Polycomb mediated repression involves proteins commonly overexpressed in cancer, such as EZH2, and cooperates with aberrant DNA methylation to silence gene expression by regulating chromatin compaction46. There is a close association between methylated CpG islands and histones containing repressive post-translational modifications, such as H3K9me3 and H3K27me3, shown to be mediated by polycomb repressive complexes47,48. This association has raised the possibility that abnormal activity of PRC2 may be the mechanism for inducing aberrant DNA methylation in cancer30.
Recently, Feinberg and colleagues have extended the concept of CpG islands to “CpG island shores,” which are also abnormally methylated in cancer. CpG island shores refer to areas of less dense CpG dinucleotides within two kilobases upstream of a CpG island49. The methylation of CpG island shores is also associated with transcriptional inactivation and expression of splice variants. The methylation of these CpG island shores tends to be tissue specific, and has recently been shown to be altered in colorectal cancer49,50. This driving role of aberrant methylation of CpG island shores in the development of cancer is still controversial and remains to be proven with additional studies, but if true, it could potentially explain certain phenomenon related to cancer epigenetics that are poorly understood at this time, such as cancer-related alterations in RNA splicing.
DNA methylation and cancer
Although the importance of gene mutations in the pathogenesis of cancer is widely accepted, the role of epigenetic alterations in cancer has been controversial until recently. The first epigenetic alteration in cancer was reported nearly three decades ago by Feinberg and Vogelstein, who showed extensive global loss of 5’-methylcytosine content in colon cancers as compared to the normal colon51. The global loss of DNA methylation predominantly affects CpG dinucleotides found in repetitive sequences of DNA, such as LINE-1 and satellite repeats. This global hypomethylation can be found in the colon in an age-dependent fashion as well as in the early steps of the polyp→cancer progression sequence52–54. Thus, it appears that the global hypomethylation of DNA is an early event in the colorectal cancer formation process. Studies of Dnmt1+/− mice demonstrate that the DNA hypomethylation may contribute to cancer formation by permitting or inducing genomic instability39. It may also lead to loss of imprinting (LOI) which can result in the overexpression of imprinted genes like IGF2 and predispose to an individual to cancer55,56.
Following the discovery of DNA hypomethylation in cancer, the DNA methylation pattern of normally unmethylated CpG islands in gene promoters of many types of neoplasms was shown to be aberrantly hypermethylated, which is yet another distortion of normal methylation patterns52. Genome-wide studies of cancer epigenomes reveal 1–10% of CpG islands are aberrantly methylated, which suggests that thousands of gene promoters may be hypermethylated in the average cancer57,58. The identification of hypermethylation of classic tumor suppressor genes, including CDKN2A, MLH1, CDH1, and VHL, provided indirect support for the functional role of aberrant DNA methylation in the cancer formation process59–62. However, the significance of the global hypomethylation, and aberrant CpG island hypermethylation was not immediately evident leaving open the idea that the epigenetic alterations in cancers, including colorectal cancers are merely bystander phenomenon in the cancer genome. The idea of epigenetic alterations being only passenger events in cancer has since been disproven over the last decade by innovative studies that have led to a richer understanding of the “driver” role of epigenetic alterations in the development of cancer63–65. It is now accepted that both epigenetic alterations as well as gene mutations cooperate to drive the progression of normal cells to cancer cells. The aberrant hypermethylation of genes appears to be a common molecular mechanism for silencing tumor suppressor genes and can contribute to cancer formation through the transcriptional repression of these genes. In fact, it appears that in the average colon cancer genome, many more genes are affected by aberrant methylation than by mutations, and that aberrant methylation is the only mechanism observed for the silencing of many genes in cancer46,66. Thus, while Knudson’s “two-hit hypothesis” initially referred to gene mutations, either germline or somatic67, it is now understood that the “second hit” can be an epigenetic alteration. Both genetic and epigenetic events are comparable in the selective advantage that the event can confer to a cell’s ability to become a cancer cell. Interestingly, the specific genes that are commonly found to be methylated in colon cancer often differ from those commonly found to be methylated in other tumor types, suggesting that there is a selective process driving the occurrence of methylated genes in different tumor types.
Mechanisms causing cancer related aberrant DNA methylation
The processes that mediate aberrant DNA hyper and hypo-methylation are currently under investigation. Although specific mechanisms have yet to be identified, it is now clear that DNA methylation is regulated through reciprocal interactions with histones, and that alterations in the post-translational state of histones are closely associated with cancer related alterations in DNA methylation30. It is currently not clear if the aberrant methylation of genes in colon cancer results primarily from “selective targeting”, reflecting a predisposition of certain genes to become hypermethylated, “selective advantage”, reflecting the clonal outgrowth of cells that have methylated specific genes, or a combination of the two forces30. With regards to mechanisms that could be involved with selective targeting, the two major competing models to explain the aberrant methylation of DNA observed in cancer are: 1) the enzymes that mediate DNA methylation, DNMT1, DNTM3a, and DNMT3b, are over expressed, hyperactive, or misdirected; or 2) the “barrier elements” that normally prevent DNA methylation of specific regions of DNA are impaired. Increased DNMT expression has been proposed as a mechanism for the increased methylation seen in the promoter region of tumors. Both increased expression and increased activity of the DNA methyltransferases has been reported in human cancers, including colon cancer, as compared to normal tissues68–71. Both animal and cell line models have consistently demonstrated that overexpression of DNMT1 and DNMT3b can induce aberrant methylation of normally unmethylated CpG islands, similar to what is seen in human colon cancer30,69,70,72. However, the results of studies in primary cancers are mixed with some studies showing increased DNMT1 expression and others showing no evidence of increased expression, particularly after normalizing for proliferation73. In fact, it may be that the fundamental defect is not increased activity of the DNMTs, but rather misdirected activity. DNMT activity can be directed to specific loci at least in part by histone methyltransferases and demethylases, such as EHMT2, PRMT5, and SUV39H1, which may mediate some aspects of aberrant DNA methylation in certain cancers. Furthermore, polycomb mediated pre-marks, indicated by histone H3K27 methylation, has been shown to recruit DNA methylatransferases, and may, therefore, have a role in the misdirection of methyltransferases in cancer74,75. There is evidence for cis-acting DNA sequences that are susceptible to cancer-specific DNA methylation. In vitro studies have shown 500-fold differences in the methylation rate of preferred sequences (e.g. CTTGCGCAAG) compared to resistant sequences (e.g. TGTTCGGTGG)22,76,77. The deregulation of DNMT activity in cancer can also be shown by assessing the methylation entropy status of the cancer genome, which reveals that cancers cells have decreased DNMT fidelity at Alu repetitive sequences and promoter associated CpG islands, suggesting that the inherent defect in cancer cells is related to decreased enzyme fidelity and not hyperactivity78.
Another major mechanism that may be involved in aberrant methylation of specific loci is the impairment of barrier elements in cancer cells as compared to normal cells. Barrier elements are postulated to be established by specific DNA sequence elements and their associated binding protiens, and are thought to protect genes from inappropriate signals from the surrounding genomic landscape, including DNA methylation. Proposed barrier elements include occupancy of DNA regions by proteins such as PolII, DNA binding by histones with specific modification states, SP1 binding sites, and VEZF1 elements46,79. The concept of methylation spreading from densely methylated areas to surrounding sequences is termed the “DNA neighborhood effect”, and has been proposed as a mechanism for aberrant hypermethylation seen in tumors as a result of impaired barrier function. In support of this concept, it has been shown that epigenetic silencing by DNA methylation can occur across a band of chromosome up to 1Mb in size80. However, there are also many examples of locus specific aberrant DNA methylation with no obvious “neighborhood effect” on adjacent loci46.
In addition to selective targeting, there is also evidence for an element of “selective advantage” driving the aberrant methylation of genes in cancer. Although it is unlikely that aberrant DNA methylation is strictly a random event and that the methylated genes observed in colon cancer merely reflect clonal growth advantage resulting from the abnormal methylation, it is likely that many of the methylated genes observed in colon cancer have been clonally selected. Consistent with the concept of “selective advantage”, cancers have tumor specific methylation patterns and the frequency of specific methylated gene varies widely between tumors40.
There are also other models that have been proposed to explain the aberrant DNA methylation observed in cancers. One model is that the epigenetic alterations observed in colon cancers are reflective of the stem cells from which the colon epithelial cells are derived, and that the alterations observed in the cancers merely indicate the undifferentiated state of the tumor cells22.It has also been proposed that unrepaired inflammation-mediated halogenated DNA damage products can mimic 5-methylcytosine due to their high affinity for methyl-binding proteins and ability to induce methylation of the daughter strand after DNA replication, leading to the establishment and propagation of aberrant methylation81. This model proposes a mechanism then for the model of misdirected rather than hyperactive DNMT activity that leads to aberrant DNA methylation. Dysregulation of noncoding RNA’s, including microRNAs and piwiRNA’s, also contributes to epigenetic deregulation in plants and may have a similar role in the human genome. Emerging data show that microRNAs can induce aberrant DNA methylation by directly targeting DNA methyltransferases82–84. Furthermore, to date there is only one example of this regulatory mechanism in mammalian genomes involving piwi RNA’s [Watanabe, #4849]. Neither of these proposed mechanisms appear to be a generalized mechanism for regulating DNA methylation. However, it does appear that aberrant DNA methylation can silence microRNA expression in cancers, playing an important role in the pathogenesis of colorectal cancer85. In addition to misdirection of DNA methylation and microRNA dysregulation, it is also possible that environmental agents may act as epimutagens and contribute to aberrant DNA methylation in some tumors40.
Thus, there are many proposed mechanisms by which epigenetic gene regulation is thought to be dysregulated in cancer, and each mechanism is supported by evidence from a variety of different lines of experimental evidence. It is likely that no single mechanism is sufficient to alter the cellular process and drive the pathogenesis of aberrant DNA methylation in cancer. It is most likely that all of these mechanisms contribute to the aberrant epigenetic regulation seen in human cancers, depending on the circumstances of the tumor.
Aberrant DNA Methylation in the Pathogenesis of Colorectal Cancer
Age and DNA methylation
Age is an important risk factor in the development of colorectal cancer, and aging colon mucosa shows age-related global hypomethylation as well as locus-specific DNA hypermethylation86. Originally, aberrant methylation of ESR1, IGF2, and TUSC3 was shown to occur in histologically normal colon epithelium in an age dependent fashion, and subsequently other genes were also shown to exhibit age-dependent methylation87–89. Approximately half of the genes that show age-related methylation are the same genes as those involved in the pathogenesis of colon cancer, suggesting a role for these genes in the increased cancer susceptibility that is associated with aging. Interestingly, not only is aberrant hypermethylation of specific gene promoters seen in aging, but there is also evidence that global loss of methylation occurs during the aging process, suggesting that the mechanism(s) responsible for age-related and cancer-related DNA methylation may be shared90–92. However, the cause of age-related aberrant methylation is not known. There is evidence to suggest that age-related hypermethylation occurs as a result of the development of local predisposing factors in DNA that affect sites such as SP1 binding sites or tandem B1 elements, as the result of environmental exposures, or secondary to a genetic predisposition to DNA methylation. It has been shown that SNP variants in MTHFR exist, playing a role in regulating the availability of the methyl donor for DNMT193,94. Transcriptional control of DNMTs is altered in aged cell lines, and it is possible that the global decrease in methylation seen in aging is a result of a progressive decrease in DNMT1 maintenance activity on heterochromatin, whereas upregulation of the de novo methyltransferase DNMT3b could be responsible for the hypermethylation seen in aging68,92,95. The implication of the aberrant DNA methylation observed in the normal tissues of older individuals is that colon cancers arising in older people may have more epigenetic driver events than those seen in younger people96.
DNA Methylation and Field Cancerization
In 1953 Slaughter et al initially introduced the concept of field cancerization in the context of oral squamous cell carcinoma to explain the development of multiple primary tumors, local recurrence, abnormal tissue surrounding the cancer and mutifocal areas of precancerous change97. The field change reflects a susceptibility of the normal tissue to develop cancer, possibly as the result of an environmental exposure. It was conceived based upon histologic observations; however, today it is clear that field cancerization occurs at the molecular level. Pre-neoplastic cells with clonal relationship to the tumor can persist after complete resection of the tumor, and it is not clear how far the cancer field can extend from the primary tumor; it may even involve the entire organ98. There is evidence consistent with field cancerization for numerous types of cancers including colorectal cancer99. Some studies have established that epigenetic alterations may be indicators of field cancerization in the colon100,101. In support of the role of methylated genes as effectors of the earliest steps in cancer formation, studies of primary human mammary epithelial cells have shown that the aberrant methylation of genes associates with immortalization as well as with subsequent steps in the malignant progression of these cells102. It has also been proposed that the aberrant methylation of genes may lock stem cells in an undifferentiated state predisposing them to malignant transformation103. Shen et al showed that 50% of tumor-adjacent histologically normal tissue carried detectable methylated MGMT when the primary tumor had methylated MGMT, whereas only 12% of normal histologic tissue from cancer-free control subjects had MGMT promoter methylation. In cases where the primary tumor did not have methylated MGMT, only 6% of the tumor adjacent normal tissue carried methylated MGMT104. A field effect as assessed by hypomethylated CDH3 (the gene for P-cadherin), methylated EVL, and methylated p14 in normal colon epithelium adjacent to colon cancer has also been shown105–107. In an extension of single gene studies, Belshaw et al assessed age-dependent gene methylation of a panel of genes, APC, AXIN2, DKK1, HPP1, TUSC3, p16, SFRP1, SFRP2 and SFRP4, in normal colon mucosa and found significant differences in gene methylation levels between people with and without colon neoplasms. They employed a multinomial logistic regression model based on the DNA methylation status of APC, TMEFF2 (HPP1), CDKN2A (p16), SFRP4, WIF1, and ESR1 of the normal mucosa of cancer patients as well as of normal patients, and were able to correctly identify 79% of cancer patients and 88% of cancer-free patients108. Other groups have similarly found panels of methylated genes in the normal colon mucosa that correlate with concurrent neoplasm status, although different genes have been used in the different studies101. Further evidence for the role of methylation field defects as effectors in cancerization comes from studies of synchronous tumors. Synchronous colorectal cancers have been shown to have similar epigenetic events through concordant DNA methylation patterns in multiple genes thought to be in important in colorectal carcinogenesis71,109. A major question related to field cancerization is what underlying factor mediates this process. Although some studies have shown a correlation with folate exposure, most studies have not identified an association with folate exposure and the methylation state of genes in the normal colon mucosa110.
Of interest, the methylation state of DNA repair genes, such as MGMT and MLH1, in the adjacent normal mucosa can also be shown to correlate with specific mutations in the tumors, such as KRAS mutations or microsatellite instability, respectively, which are predicted to result from loss of function of these genes. This suggests that epigenetic alterations are creating a predisposition for specific cancer related mutations100,111. Hypermethylation and downregulation of SFRP genes are present in monoclonal aberrant crypt foci lacking APC mutations. This is thought to contribute to constitutive WNT ligand signaling and decreased apoptosis, which may cancerize the field making those cells particularly sensitive to further activating events in the WNT pathway65. The methylation patterns of SFRP genes, which regulate the Wnt signaling pathway, in this case may be useful in predicting the risk of developing colon cancer108. However, the relationship between the aberrant gene methylation in the normal colon mucosa and priming of the mucosa to undergo cancer formation appears to be complex as there is also data suggesting that hypomethylation of genes that undergo age-related DNA methylation correlates with the presence of colorectal neoplasms in the CpG Island Methylator Phenotype (CIMP) molecular class101. At this time, it still remains to be determined whether the methylated genes detected in the normal mucosa are truly indicative of a field effect in the colon or are a marker of an associated phenomenon, such as folate status or tobacco exposure112,113. It is also clear that the methylation state of the right and left colon vary, and that this aspect of normal mucosa sampling needs to be taken into consideration when measuring the methylation state of genes in normal colon mucosa114,115.
Epigenetic alterations in the initiation and progression of colorectal cancer
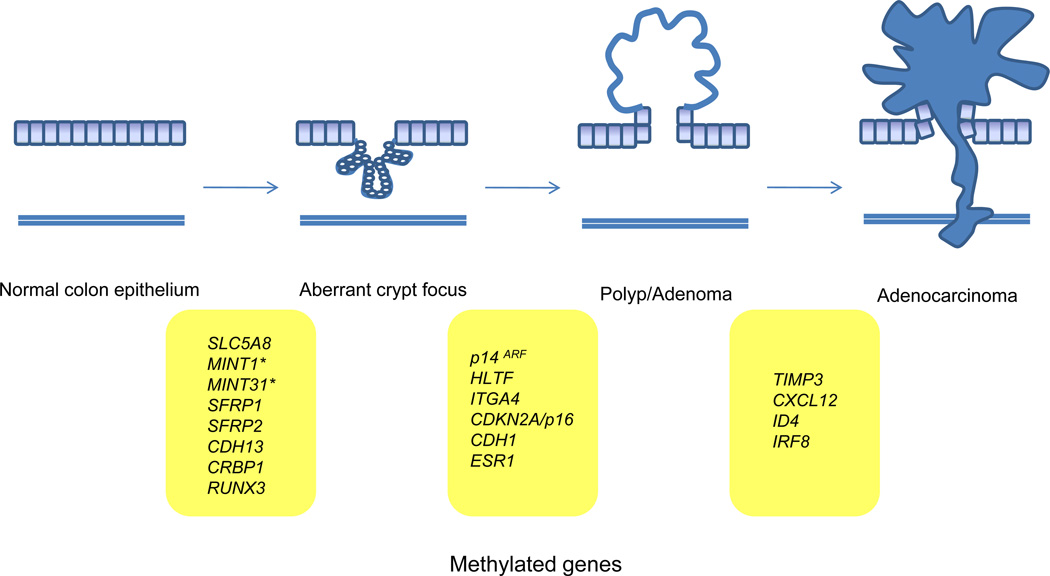
Hundreds to thousands of genes are thought to be aberrantly methylated in the average colorectal cancer genome, and only a subset are likely important in the pathogenesis of these cancers. Of these many methylated genes, there are already numerous genes that have been shown to be commonly hypermethylated as well as silenced in colorectal cancer (Table 1), and our understanding of the role that aberrantly methylated genes play in the initiation and progression of the polyp→cancer sequence continues to evolve as additional methylated genes are identified (Figure 2). As might be expected given the detection of methylated genes in the normal mucosa of people at elevated risk of colorectal cancer, methylated genes can be detected in aberrant crypt foci and adenomas as well as in adenocarcinomas116. Examples of genes methylated early in the adenoma→carcinoma sequence include ITGA4, MGMT, SLC5A8, SFRP2, and MINT1116–121. Notably, the most substantial increase in gene methylation is seen in the normal mucosa to adenoma step of the polyp→cancer progression sequence, with the most substantial increases seen when comparing normal mucosa to early adenomas, and early adenomas to advanced adenomas121. The proportion of neoplasms that carry methylated genes is not significantly different between advanced adenomas and adenocarcinomas but the frequency of methylation of specific genes does vary between early adenomas, advanced adenomas and adenocarcinomas121,122. LINE-1 DNA hypomethylation in the polyp→cancer sequence shares a similar pattern with local promoter hypermethylation in that there is a substantial difference in the levels of LINE-1 hypomethylation and local hypermethylation occurring at the polyp to adenoma step of the sequence, but no difference between levels at the adenomas to cancer step123. There is no clear functional class of genes that appear to be more commonly affected by DNA methylation in the adenoma step of colon cancer development compared to other steps in the progression sequence122. Despite the lack of definitive proof, it has been proposed that aberrant methylation of DNA repair genes, such as MGMT and MLH1, in colon adenomas may promote the progression of adenomas to cancer by creating a permissive state for G->A mutations, the most common mutation seen in KRAS in the case of methylated MGMT and a permissive state for microsatellite instability in the case of methylated MLH1124,125.
Table 1.
Representative Genes Commonly Methylated and Silenced in Colorectal Cancer
| Gene | Protein | Function* | Effect of Loss of Function |
|---|---|---|---|
| APC | Adenomatous polyposis coli | Wnt signaling pathway inhibition | Increased Wnt/β-catenin signaling |
| MLH1 | MutL homolog 1 | DNA Mismatch repair | Microsatellite instability |
| MGMT | O-6-methylguanine-DNA methyltransferase | Repair of alkylation DNA damage | Increased G→A mutation frequency |
| RASSF1A | Ras association domain family 1 (isoform A) | Negative RAS effector, pro-apoptotic, microtubule stablization | Increased RAS/RAF/MAP kinase signaling, death receptor-dependent apoptosis |
| SLC5A8 | Sodium solute symporter family 5 member 8 | Sodium and short chain fatty acid transporter, suppresses colony formation | ? |
| RUNX3 | Runt-related transcription factor 3 | Transcription factor | Decreased TGF-β/BMP signaling |
| MINT1** | Methylated in tumor locus 3 | NA | NA |
| MINT31** | Methylated in tumor locus 31 | NA | NA |
| SFRP1 | Secreted frizzled-related protein 1 | Wnt antagonist | Increased Wnt/β-catenin signaling |
| SFRP2 | Secreted frizzled-related protein 2 | Wnt antagonist | Increased Wnt/β-catenin signaling |
| CDH1 | E-Cadherin | Calcium dependent cell-cell adhesion glycoprotein | Loss of cell adhesion, possible increased Wnt/β-catenin signaling |
| CDH13 | Cadherin 13 | Selective cell recognition and adhesion, anti-apoptotic | Increased PI3K/Akt/mTOR signaling, MAPK signaling |
| CRABP1 | Retinol-binding protein 1 | Carrier protein for transport of retinol, promotes apoptosis | ? |
| CDKN2A/p16 | Cyclin-dependent kinase inhibitor 2A | Regulates cell cycle G1 progression | Increased cell proliferation |
| HLTF | Helicase-like transcription factor | dsDNA translocase, fork remodeling activity, ubiquitin ligase | Impaired DNA repair |
| CDKN2A (P14, ARF) | Protein 14 | Inhibits E3 ubiquitin ligase | Decreased p53 stabilization and activation |
| ESR1 | Estrogen receptor 1 | Ligand-activated transcription factor | Loss of estrogen receptor signaling |
| TIMP3 | Tissue inhibitor of metalloproteinase 3 | Inhibition of MMPs and ADAMs | Increased EGF receptor signaling, TNFalpha signaling |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 | Alpha chemokine | Increased tumor cell metastases |
| ID4 | Inhibitor of DNA binding 4 | Transcription factor | ? |
| IRF8 | Interferon regulatory factor 8 | Transcription factor | Interferon signaling |
| THBS1/TSP1 | Thrombospondin 1 | Cell-to-cell and cell-to-matrix adhesive glycoprotein | Decreased TGF-β1 signaling |
| DAPK | Death associated protein kinase | Induction of cell death | Interferon gamma signaling, TNF alpha signaling, Fas/APO1 signaling |
| VIM | Vimentin | Stablizing cytoskeleton | No known biological effect |
| SEPT9 | Septin 9 | GTPase, formation of filaments | Impaired Cytokinesis and loss of cell cycle control |
Many of these gene products have multiple functions. The listed function in this table is the one most commonly cited as the one responsible for colorectal cancer formation.
MINTs are Methylated IN Tumor loci, and are not specific genes.
Figure 2.
Common methylated genes (and loci) identified at the histologic steps of the colorectal cancer polyp-adenoma-carcinoma sequence. Specific gene and loci hypermethylation has been observed in the different steps of the adenoma-cancer progression sequence implicating these genes in either the initiation of progression of colorectal cancer. The genes listed between normal epithelium and aberrant crypt focus as well as those genes listed between aberrant crypt focus and polyp/adenoma may be involved in the initiation of colorectal cancer. Those genes listed between polyp/adenoma and adenocarcinoma may play a role in the progression and metastasis of colorectal cancer. Note that we have only included genes (and loci) that have consistently been found to be hypermethylated at specific points in the progression sequence. *MINT1 and MINT31 are not genes; rather they are loci that have been found to be “methylated in tumor,” (aka MINT).
In addition to the detection of methylated genes in adenomas, the CIMP phenotype can be detected in advanced tubular adenomas but is not commonly observed in early tubular adenomas. Interestingly, Kwon et al detected CIMP adenomas in a collection of serrated adenomas, but not in tubular adenomas suggesting serrated adenomas may be the precursor polyps for CIMP colorectal cancers126. It is not clear whether the detection of CIMP late in the polyp phase of colorectal cancer formation reflects a technical issue regarding the number of loci assessed in the studies of methylated genes in adenomas, or demonstrates that CIMP occurs late in the polyp→cancer sequence121. This is an issue because it is widely believed that CIMP tumors may arise from serrated adenomas, in which case it would be reasonable to assume that CIMP would be detected in the early adenomas. To date, methylated genes that distinguish sessile serrated adenomas from hyperplastic polyps (CDX2, MLH1, and TLR2) have been identified, but there has yet to be consistent reporting of a subset of early adenomas with CIMP signature of methylated genes127.
Aberrant DNA methylation in the progression and metastasis of colorectal cancer
The progression of colon adenomas to adenocarcinomas is believed to be driven by genetic alterations, such as mutations in TP53, but is also likely a consequence of epigenetic alterations (Figure 2). For example, the aberrant methylation of CXLC12, a chemokine ligand, occurs in human colorectal cancer and can promote the metastatic behavior of colon cancer cell lines128. In addition, the frequency of tumors carrying certain methylated genes, like TIMP3, ID4, and IRF8, is higher in colon cancer and metastases than adenomas suggesting that the inactivation of these genes by methylation provides a clonal growth advantage121,129. In addition to specific aberrantly methylated genes being present in advanced colorectal cancers, hypomethylated LINE-1 elements are found to increase linearly in more progressed colon neoplasms123,130,131(Figure 1). However, in general, it appears that the aberrant DNA methylation of genes is predominantly involved with the early events of colon cancer formation and less with progression events.
DNA Methylation Epigenotypes of Colorectal Cancer
Epigenetic instability in colorectal cancer is manifested in a variety of ways including hypermethylation of gene promoters that contain CpG islands and global DNA hypomethylation. Essentially all colorectal cancers have at least some aberrantly methylated genes, and there appears to be a unimodal distribution of colon cancers when the proportion of methylated genes/cancer genome is assessed. In 1999, Toyota and Issa observed that some colorectal cancers show a high frequency of methylated genes, and they put forth the concept that these tumors have a unique molecular pathogenesis, naming them CpG Island Methylator Phenotype (CIMP) cancers. This concept was met with considerable controversy, but has since been adopted due to the substantial accumulated supporting evidence. Approximately 20% of colorectal cancers are CIMP tumors, and they are currently identified by having >60% of methylated genes in a pre-selected panel of “CIMP marker” genes. The panel of genes for defining CIMP still remain to be standardized, thus the criteria used to identify CIMP tumors can vary across studies, which makes generalizing results across studies difficult6,132–134. A commonly used panel is one defined by Weisenberger et al which includes NEUROG1, SOCS1, RUNX3, IGF2, and CACNA1G, although it remains to be determined which panel will ultimately be selected for determining the CIMP status of colorectal cancers135. Regardless of which panel of CIMP markers is used, CIMP tumors have a high frequency of BRAF c.1799T>A (p.V600E) mutations, tend to occur in the right colon, and are more common in women135,136. The underlying cause of CIMP tumors is not known at this time, but an association with cigarette smoking has been shown113,137. Associations with nutritional status, body size and physical activity have also been shown. Severe energy restriction during childhood has been shown to have a decreased CIMP association, whereas increased BMI is associated with an increase in both CIMP and non-CIMP cancers138,139. Although expected, no correlation between CIMP tumor status and folate status or alcohol consumption, which would contribute to folate deficiency, has been observed94,140 Lastly, it does appear that CIMP tumors arise most often from hyperplastic polyps or sessile serrated polyps suggesting that they are fundamentally different from colorectal cancers arising from tubular adenomas141.
In an extension of the CIMP concept, studies of the epigenomes of colorectal cancers have identified groups of colorectal cancers that segregate by epigenotype32,134,142. The epigenotypes show unique associations with a variety of mutations (ie TP53, BRAF, KRAS) and with the genomic instability status of the cancer (i.e. microsatellite instability, chromosome instability, or neither)142,143. The epigenotypes that have been proposed include different classes of CIMP, including CIMP-low and CIMP-high, which is determined by the proportion of methylated marker loci detected144. Issa and colleagues have suggested the division of CIMP colon cancers into two distinct classes, CIMP1 and CIMP2, based on the results of unsupervised cluster analysis of a large panel of methylation markers143. Based upon a study of 97 primary colorectal cancer cases, CIMP1 tumors are often microsatellite unstable (80%), and have BRAF mutations (53%), whereas CIMP2 tumors have KRAS mutations (92%), but rarely have microsatellite instability or BRAF or TP53 mutations143. Tumors that do not have CIMP have a high frequency of TP53 mutations (71%) and intermediate frequency of KRAS mutations (33%)143. Finally, Hinoue et al performed genome-scale DNA methylation profiling of 125 colorectal cancers and identified four DNA methylation–based subgroups of CRC using model-based cluster analyses. They classified the tumors as CIMP-high (CIMP-H), which exhibits an exceptionally high frequency of cancer-specific DNA hypermethylation, a strong association with methylated MLH1 and mutant BRAF c.1799T>A(p.V600E); CIMP-low (CIMP-L), which is associated with mutant KRAS32,144–147 and has a subset of CIMP-H-associated methylated genes rather than a unique group of CpG islands; and non-CIMP tumors, which are separated into two distinct clusters based on the association with mutant TP53 and location of the tumor32,148.
Although CIMP tumors and the distinct epigenotypes of colorectal cancers described above do appear to represent distinct subsets of colorectal cancers, the clinical utility of these designations is hindered by both the lack of a universally accepted definition of the CIMP phenotype and the lack of validation of the accuracy of the methylated gene panels for designating the epigentoypes. Finally, considerable overlap between CIMP and sporadic MSI tumors adds to the challenge of incorporating CIMP-status into clinical trials and clinical decision making132. Retrospective studies suggest CIMP will ultimately be shown to be a predictive marker for colorectal cancer, however, currently, the data is not robust enough to recommend its clinical use143,149. Thus, the discovery and classification of CIMP tumors has advanced our understanding of the molecular pathology of colorectal cancer but has not yet impacted the clinical care of patients with colorectal cancer.
Mechanisms that give rise to CIMP are not yet clear, although there is a strong association between BRAFc.1799T>A(p.V600E) mutations and CIMP colorectal cancer, suggesting a role for activated BRAF in the pathogenesis of the methylator phenotype and a link between sporadic MSI and CIMP132,144. Recent structural equation modeling by Tanaka et al. show that KRAS and BRAF mutations differentially correlate with the structure of CpG island methylation, suggesting distinct perturbations resulting in CIMP150. However, in vitro studies of mutant BRAF in colorectal cancer cell lines have not demonstrated a direct cause and effect relationship between BRAF and CIMP151.
Clinical Use of Methylated Genes
Biomarkers
Epigenetic biomarkers for cancer for diagnosis, prognosis and prediction of responsiveness to therapy are already in clinical use for colorectal cancer [(Cologuard assay, (LabCorp); Epi proColon assay, (Epigenomics)], lung cancer (Epi proLung, (Epigenomics)) and gliomas (PredictMDX, (MDxHealth)). These are likely the first of many epigenetic biomarkers that will ultimately be used in the clinical setting. In prostate cancer, hypermethylation of the GSTP1 gene is a marker for disease and malignant transformation and may have a role in conjunction with elevated KLK (PSA) for diagnosis and prognosis152. Hypermethylation of the O6-methylguanine methyltransferase gene (MGMT), which is a DNA repair enzyme involved in direct repair of alkylation damage products, was found to be predictive of a favorable response in gliomas to chemotherapy with alkylating agents, carmustine and temozolomide, and is the basis for the PredictMDx assay. Silencing of MGMT secondary to DNA methylation inactivates MGMT leading to an increased susceptibility of the gliomas to chemotherapy-induced alkylation DNA damage; consequently, methylated MGMT has been associated with improved outcomes from therapy with alkylating agents124.
With regards to the use of methylated genes as biomarkers specifically for colorectal cancer, the most advanced uses are as DNA-based colon cancer screening assays. Currently, colonoscopy is still the most accurate test for colorectal cancer screening; however, it is costly and is associated with procedure-related complications as well as poor patient compliance. In contrast, another commonly used colorectal cancer screening test, fecal occult blood testing (FOBT) is inexpensive and simple to perform, but has a relatively low sensitivity and specificity153. Advances in our understanding of the molecular pathology of colorectal cancer, has led to the identification of promising early detection molecular markers for use in non-invasive colorectal cancer screening assays154. Hypermethylation of promoter regions in colorectal cancer occurs early in some genes, and these methylated genes are being used as the basis for early detection markers (Table 2). At this time, stool-based methylated VIMENTIN (mVim) is a clinically validated marker for early detection of colorectal cancer that is now commercially available in the United States under the name ColoGuard assay (LabCorp)155. The test exploits the fact that the vimentin gene (VIM) is aberrantly methylated in a majority of colorectal cancers (53–84%). This early detection test is a PCR-based assay that simultaneously measures methylated VIM as well as DNA integrity and has reported a sensitivity of 83% and a specificity of 82%, with approximately equal sensitivity in Stage I-III colorectal cancer patients156. In addition, in Europe and the Middle East, there is a blood-based assay that detects methylated SEPT9 that is being marketed as a colon cancer screening assay under the name Epi proColon (Epigenomics AG). Currently, methods are under development to enhance the performance of stool- and plasma-based methylation assays for clinical purposes157. The use of molecular assays, such as the fecal-methylated VIM assay, in the clinical care of patients will likely to undergo rapid advances and be adopted into clinical practice over the next decade.
Table 2.
Current and Potential Methylated Gene Biomarkers for Colon Neoplasms
| Gene | Abnormality | Biomarker function |
|---|---|---|
| MLH1 | Hypermethylation | Sporadic MSI tumor marker (The likelihood of Lynch syndrome decreases if MLH1 is methylated.) |
| VIM | Hypermethylation | Early detection of CRC |
| SEPT9 | Hypermethylation | Early detection of CRC |
Methylated genes may also be useful prognostic or predictive molecular marker for sporadic colon cancer. As noted above, aberrantly methylated genes affect a number of genes in colon cancer, including CDKN2A/p16, MGMT, THBS1, TIMP3, CDKN2A (p14ARF) and MLH1, some of which occur early on in the polyp-cancer sequence making them especially promising as early detection markers. Methylated MLH1 is currently being used as a marker for sporadic as opposed to hereditary (ie Lynch syndrome) MSI cancers (Table 2). The vast majority of sporadic MSI tumors carry methylated MLH1, whereas it is infrequent in Lynch syndrome tumors158. Thus, methylated MLH1 status can be used to identify individuals who should be considered for genetic testing for Lynch syndrome. CIMP status of the cancers has also been assessed as a predictive marker for 5-FU responsiveness, however there is conflicting data surrounding the correlation between CIMP status and responsiveness to adjuvant 5FU therapy159–161. At this time, no single hypermethylated gene has been identified that can predict the responsiveness of colorectal cancer to any one specific treatment with sufficient specificity and sensitivity to be used in a clinical theranostic assay.
Although efforts to find epigenetic markers for clinical use with colorectal cancer have mostly been focused on methylated genes, due to the inverse correlation between CpG island methylation and global DNA hypomethylation130,131, the role of LINE-1 hypomethylation as a biomarker has also been investigated. Recently, LINE-1 hypomethylation has shown promise as a prognostic marker for shorter disease free survival in proximal colorectal cancer162,163. Also, LINE-1 methylation was lower in patients whose cancer recurred as compared to those whose cancer did not recur163.
Ultimately, it is expected that a clinically useful assay will be developed that will incorporate a panel of methylated genes and mutant genes, however, the data to date is not robust enough to support the use of any methylated genes as predictive biomarkers at the time of this publication.
Conclusions
The role of epigenetics in the pathogenesis of cancer has come to the forefront over the last decade. It is now well established that epigenetic events can be driver events in the pathogenesis of colorectal cancer, and that these epigenetic events cooperate with gene mutations in the progression of normal colon mucosa to colon cancer, with more genes in the colon cancer genome affected by altered DNA methylation than by gene mutations. Aberrant DNA methylation has been the most studied aspect of epigenetic alterations in colorectal cancer, and it is clear that global hypomethylation and local hypermethylation occur in the majority of colorectal cancers. It has been shown that aberrant DNA methylation generally occurs early in the polyp-cancer sequence and that these changes contribute to the initiation of colorectal cancers; however, there are also some methylation changes that occur later in the sequence that may contribute to the progression of cancers. These alterations in DNA methylation contribute to the molecular heterogeneity of colorectal cancers, as illustrated by the identification of molecular subtype of colorectal cancers that can be identified by their unique methylated gene signatures. Given the role of altered DNA methylation in directing the pathogenesis of colorectal cancer, studying DNA methylation signatures and developing them as biomarkers for diagnosis, prognosis and direction of therapy is likely to yield clinically useful assays that will be used to direct patient care. Currently, the role of DNA methylation as clinical biomarkers is only beginning to emerge. There are recently developed epigenetic biomarkers for the early detection of colorectal cancer and efforts are in progress to develop epigenetic markers for prognostic and predictive markers relevant for therapy. Finally, although the molecular mechanisms by which aberrant DNA methylation occurs is not yet fully understood, there are multiple models for cancer-related aberrant methylation, involving mechanisms such as altered DNMT function and impaired barrier elements. It is likely that these mechanisms cooperate to alter DNA methylation and other aspects of the epigenome in colorectal cancer. Continued efforts to investigate these molecular mechanisms will allow for a better understanding of the role of epigenetic alterations in colorectal cancer and will lead to the translation of these insights into the clinical arena.
Key Points.
The average colon cancer epigenome has hundreds to thousands of abnormally methylated genes, a subset of which is thought to drive colorectal cancer pathogenesis and clinical behavior.
Alterations in DNA methylation patterns in colorectal cancer contribute to the molecular heterogeneity of colorectal cancer as illustrated by the identification of a CpG island methylator phenotype (CIMP) subtype of colorectal cancer that has a distinct epigenome.
Clinical markers for diagnosis, prognosis and therapeutic applications are being developed using the epigenetic alterations and DNA methylation signatures in colorectal cancer.
Although the mechanisms by which aberrant DNA methylation occurs are not yet fully understood, there may be a mechanistic connection between the altered methylation seen in colorectal cancer and the phenomena of age related methylation and field cancerization.
Box 1.
Epigenetics: Heritable alterations in gene expression that are not mediated by changes in the DNA sequence
DNA Methylation: Enzymatic addition of a methyl group to the 5-position of cytosine by either maintenance or de novo methyltransferases. DNA methylation is critical in orchestrating gene expression and is deregulated in many cancer types including colon cancers.
CpG Island: DNA sequences greater than 200–500 bases in length with greater than 50% GC content and a ratio of observed to expected CpG ratio of greater than 0.6. CpG islands overlap the promoter region of 60–70% genes and tend to be protected from methylation; however, they can become aberrantly methylated in cancer. Methylation of CpG islands is associated with transcriptional inactivation.
CpG Island Shores: Areas of less dense CpG dinucleotides in the genome within two kilobases 5’ of a CpG island. Methylation of CpG island shores has been shown to be associated with transcriptional inactivation and expression of splice variants in some studies.
Polyp: A benign neoplasm resulting from overgrowth of the colonic epithelium. Polyps include tubular adenomas and serrated adenomas, as well as other histologic types of benign neoplasms. Tubular adenomas more commonly arise via inactivation of the APC tumor suppressor gene and via genetic alterations resulting from chromosomal instability. Serrated polyps are associated with microsatellite instability and aberrant DNA methylation.
CpG Island Methylator Phenotype (CIMP): A subgroup of colorectal cancers showing a high frequency of methylated genes, identified by having >60% of methylated genes in a pre-selected panel of “CIMP marker” genes. The panel of genes that defines CIMP remains to be standardized. Approximately 20% of colorectal cancers can be classified as CIMP.
Chromosomal instability (CIN): A subgroup of colorectal cancers showing aneuploidy, and chromosomal losses or gains
Microsatellite instability (MSI): A subgroup of colorectal cancers showing frequent insertion and deletions in repetitive DNA sequences. MSI is a consequence of inactivation of the DNA Mismatch Repair enzymes.
Field cancerization: Locoregional molecular changes occurring within tissue adjacent to cancer tissue that reflects the susceptibility of normal tissue to develop cancer. Epigenetic alterations may be indicators of field cancerization in the colon
Acknowledgements
This work was supported by funds from the FHCRC CCSG (5P30CA015704), Burroughs Wellcome Fund, NCI/EDRN (UO1CA152756), NCI (U54CA143862, PO1CA077852) to WMG and ASCRS, NIH T32 (T32DK007742-14) to VVL.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein NS. Serrated pathway and APC (conventional)-type colorectal polyps: molecular-morphologic correlations, genetic pathways, and implications for classification. Am J Clin Pathol. 2006;125:146–153. [PubMed] [Google Scholar]
- 3.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343–364. doi: 10.1146/annurev.pathol.4.110807.092317. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 5.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 8.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 9.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 12.Grady W, Markowitz SD. TGF-β Signaling Pathway and Tumor Suppression. In: Derynck R, Miyazano K, editors. The TGF-β Family. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2008. pp. 889–938. [Google Scholar]
- 13.Grady W, et al. Mutation of the type II transforming growth factor-β receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 14.Markowitz S, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 15.Eppert K, et al. Madr2 maps to 18q21 and encodes a TGF-β regulated Mad-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 16.Takaku K, et al. Intestinal Tumorigenesis in Compound Mutant Mice of both Dpc4(Smad4) and Apc Genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 17.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 18.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 19.Samowitz WS, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Nosho K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 24.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 25.Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- 26.Ballestar E, Esteller M. Epigenetic gene regulation in cancer. Adv Genet. 2008;61:247–267. doi: 10.1016/S0065-2660(07)00009-0. [DOI] [PubMed] [Google Scholar]
- 27.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 28.van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol. 2011;29:1382–1391. doi: 10.1200/JCO.2010.28.2319. [DOI] [PubMed] [Google Scholar]
- 29.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 30.McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–291. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 32.Hinoue T, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2011 doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 34.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 35.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 36.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano M, Bell D, Haber D, Li E. DNA methylatransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;29:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 38.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 39.Laird P, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 40.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inamdar NM, Ehrlich KC, Ehrlich M. CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol Biol. 1991;17:111–123. doi: 10.1007/BF00036811. [DOI] [PubMed] [Google Scholar]
- 43.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng G, Chen A, Pong E, Kim YS. Methylation in hMLH1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncogene. 2001;20:7120–7127. doi: 10.1038/sj.onc.1204891. [DOI] [PubMed] [Google Scholar]
- 45.Bednarik DP, et al. DNA CpG methylation inhibits binding of NF-kappa B proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- 46.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21:502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 49.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feinberg A, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. 1983;301:89, 92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 52.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki K, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Sakatani T, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 56.Cui H, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 57.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez-Zulueta M, et al. Methylation of the 5' CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 60.Graff J, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 61.Herman J, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cunningham J, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 63.Grady W, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nature Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 64.Baylin SB, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence. Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 66.Schuebel KE, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knudson AG, Jr, Strong LC. Mutation and cancer: a model for Wilms' tumor of the kidney. J Natl Cancer Inst. 1972;48:313–324. [PubMed] [Google Scholar]
- 68.Issa JP, et al. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst. 1993;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 69.Nosho K, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt WM, et al. Progressive up-regulation of genes encoding DNA methyltransferases in the colorectal adenoma-carcinoma sequence. Mol Carcinog. 2007;46:766–772. doi: 10.1002/mc.20307. [DOI] [PubMed] [Google Scholar]
- 71.Ibrahim AE, et al. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60:499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 72.Steine EJ, et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J Clin Invest. 2011;121:1748–1752. doi: 10.1172/JCI43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehrlich M, et al. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 75.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 77.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;87:572–579. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 78.Xie H, et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011;39:4099–4108. doi: 10.1093/nar/gkr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dickson J, et al. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000804. e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 81.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: a mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67:5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 82.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 83.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 86.Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15:835–842. doi: 10.14670/HH-15.835. [DOI] [PubMed] [Google Scholar]
- 87.Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 88.Issa J-P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nature Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 89.Fink D, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 90.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 91.Fuke C, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 92.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 93.Hazra A, et al. Germline polymorphisms in the one-carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes Control. 2010;21:331–345. doi: 10.1007/s10552-009-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Curtin K, et al. Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28:1672–1679. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 96.Alemayehu A, Sebova K, Fridrichova I. Redundant DNA methylation in colorectal cancers of Lynch-syndrome patients. Genes Chromosomes Cancer. 2008;47:906–914. doi: 10.1002/gcc.20586. [DOI] [PubMed] [Google Scholar]
- 97.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 98.Polley AC, et al. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006;66:6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- 99.Jothy S, et al. Field effect of human colon carcinoma on normal mucosa: relevance of carcinoembryonic antigen expression. Tumour Biol. 1996;17:58–64. doi: 10.1159/000217967. [DOI] [PubMed] [Google Scholar]
- 100.Ramirez N, et al. Epigenetic events in normal colonic mucosa surrounding colorectal cancer lesions. Eur J Cancer. 2008;44:2689–2695. doi: 10.1016/j.ejca.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 101.Worthley DL, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–1662. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- 102.Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009;69:5251–5258. doi: 10.1158/0008-5472.CAN-08-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 104.Shen L, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 105.Milicic A, et al. Ectopic expression of P-cadherin correlates with promoter hypomethylation early in colorectal carcinogenesis and enhanced intestinal crypt fission in vivo. Cancer Res. 2008;68:7760–7768. doi: 10.1158/0008-5472.CAN-08-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen L, Kondo Y, Hamilton SR, Rashid A, Issa JP. P14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology. 2003;124:626–633. doi: 10.1053/gast.2003.50102. [DOI] [PubMed] [Google Scholar]
- 107.Grady WM, et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- 108.Belshaw NJ, et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136–142. doi: 10.1038/sj.bjc.6604432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Konishi K, et al. Concordant DNA methylation in synchronous colorectal carcinomas. Cancer Prev Res (Phila) 2009;2:814–822. doi: 10.1158/1940-6207.CAPR-09-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol. 2008;46:1346–1359. doi: 10.1016/j.fct.2007.09.101. [DOI] [PubMed] [Google Scholar]
- 111.Hiraoka S, et al. Methylation status of normal background mucosa is correlated with occurrence and development of neoplasia in the distal colon. Hum Pathol. 2010;41:38–47. doi: 10.1016/j.humpath.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 112.Wallace K, et al. Effect of calcium supplementation on the risk of large bowel polyps. J Natl Cancer Inst. 2004;96:921–925. doi: 10.1093/jnci/djh165. [DOI] [PubMed] [Google Scholar]
- 113.Limsui D, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst. 2010;102:1012–1022. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Worthley DL, et al. DNA methylation in the rectal mucosa is associated with crypt proliferation and fecal short-chain fatty acids. Dig Dis Sci. 2011;56:387–396. doi: 10.1007/s10620-010-1312-4. [DOI] [PubMed] [Google Scholar]
- 115.Shannon B, Gnanasampanthan S, Beilby J, Iacopetta B. A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut. 2002;50:520–524. doi: 10.1136/gut.50.4.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chan AO, et al. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A. 2003;100:8412–8417. doi: 10.1073/pnas.1430846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7117. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ausch C, et al. Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clin Chem. 2009;55:1559–1563. doi: 10.1373/clinchem.2008.122937. [DOI] [PubMed] [Google Scholar]
- 121.Kim YH, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–789. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- 122.Oster B, et al. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int J Cancer. 2011 doi: 10.1002/ijc.25951. [DOI] [PubMed] [Google Scholar]
- 123.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Esteller M, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 125.Nosho K, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yano Y, et al. Clinicopathological and molecular features of colorectal serrated neoplasias with different mucosal crypt patterns. Am J Gastroenterol. 2011;106:1351–1358. doi: 10.1038/ajg.2011.76. [DOI] [PubMed] [Google Scholar]
- 127.Dhir M, et al. Sessile serrated adenomas and classical adenomas: An epigenetic perspective on premalignant neoplastic lesions of the gastrointestinal tract. Int J Cancer. 2010 doi: 10.1002/ijc.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wendt MK, et al. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene. 2006;25:4986–4997. doi: 10.1038/sj.onc.1209505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 130.Ogino S, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Estecio MR, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 133.Ogino S, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaneda A, Yagi K. Two groups of DNA methylation markers to classify colorectal cancer into three epigenotypes. Cancer Sci. 2011;102:18–24. doi: 10.1111/j.1349-7006.2010.01712.x. [DOI] [PubMed] [Google Scholar]
- 135.Weisenberger DJ, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res. 2008;36:4689–4698. doi: 10.1093/nar/gkn455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int. 2011;2011:902674. doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Samowitz WS, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- 138.Hughes LA, et al. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One. 2009;4:e7951. doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hughes LA, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP) PLoS One. 2011;6:e18571. doi: 10.1371/journal.pone.0018571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schernhammer ES, et al. Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut. 2010;59:794–799. doi: 10.1136/gut.2009.183707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Torlakovic EE, et al. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA) Am J Surg Pathol. 2008;32:21–29. doi: 10.1097/PAS.0b013e318157f002. [DOI] [PubMed] [Google Scholar]
- 142.Silver A, et al. A distinct DNA methylation profile associated with microsatellite and chromosomal stable sporadic colorectal cancers. Int J Cancer. 2011 doi: 10.1002/ijc.26104. [DOI] [PubMed] [Google Scholar]
- 143.Shen L, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barault L, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 145.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 147.Yagi K, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 148.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int J Clin Oncol. 2008;13:498–503. doi: 10.1007/s10147-008-0854-3. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka N, et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. Am J Pathol. 2010;177:2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hinoue T, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bastian PJ, et al. GSTP1 hypermethylation as a molecular marker in the diagnosis of prostatic cancer: is there a correlation with clinical stage, Gleason grade, PSA value or age? Eur J Med Res. 2004;9:523–527. [PubMed] [Google Scholar]
- 153.Smith RA, Cokkinides V, Brooks D, Saslow D, Brawley OW. Cancer screening in the United States: 2010: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin. 2010;60:99–119. doi: 10.3322/caac.20063. [DOI] [PubMed] [Google Scholar]
- 154.Ahlquist DA, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–450. W481. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Itzkowitz SH, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 156.Itzkowitz S, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 157.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bouzourene H, Hutter P, Losi L, Martin P, Benhattar J. Selection of patients with germline MLH1 mutated Lynch syndrome by determination of MLH1 methylation and BRAF mutation. Fam Cancer. 2010;9:167–172. doi: 10.1007/s10689-009-9302-4. [DOI] [PubMed] [Google Scholar]
- 159.Ide T, et al. Expression of the hMLH1 gene is a possible predictor for the clinical response to 5-fluorouracil after a surgical resection in colorectal cancer. Oncol Rep. 2008;19:1571–1576. [PubMed] [Google Scholar]
- 160.Jover R, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ogino S, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 162.Ogino S, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ahn JB, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]