Abstract
Background
Chronic alcohol feeding of adult Long Evans rats causes major central nervous system abnormalities that link neuronal loss and impaired acetylcholine homeostasis to ethanol inhibition of insulin and insulin-like growth factor (IGF) signaling and increased oxidative stress.
Objectives
We now characterize the integrity of insulin and IGF signaling mechanisms and assess molecular indices of neurodegeneration in the cerebellar vermis and anterior cingulate gyrus of human alcoholics.
Results
Alcoholic cerebella had increased neuronal loss, gliosis, lipid peroxidation, and DNA damage relative to control. Quantitative RT-PCR studies demonstrated reduced expression of insulin, insulin receptor and IGF-II receptor in the anterior cingulate, and reduced expression of insulin, IGF-I, and their corresponding receptors in the vermis. Competitive equilibrium binding assays revealed significantly reduced specific binding to the insulin, IGF-I, and IGF-II receptors in both the anterior cingulate and vermis of alcoholic brains. These effects of chronic alcohol abuse were associated with significantly reduced expression of choline acetyltransferase, which is needed for acetylcholine biosynthesis.
Conclusions
The results suggest that alcoholic neurodegeneration in humans is associated with insulin and IGF resistance with attendant impairment of neuronal survival mechanisms and acetylcholine homeostasis.
Keywords: Alcoholic, Neurodegeneration, Insulin Resistance, Central Nervous System, Receptor-Ligand Binding, Acetylcholine, Human
Alcohol dependence and abuse are among the most costly healthcare problems in the world, and their impact continues to grow due to the rising incidence of heavy alcohol drinking among women and young people in general. Excessive drinking can cause cognitive dysfunction and permanent structural damage to the brain. Although Wernicke-Korsakoff syndrome is one of the most devastating and clinically significant forms of alcohol-associated neurodegeneration, its pathogenesis is largely related to thiamine deficiency (Harper et al., 2003), which is preventable. In contrast, the pathogenesis of the more prevalent alcohol-associated brain lesions, including white matter attrition, ventriculomegaly, cerebellar degeneration, and neuronal loss within the superior frontal association cortex, anterior cingulate region, and hypothalamus, which also cause cognitive and motor deficits (Harper, 1998; Harper et al., 2003; de la Monte, 1988), has not yet been determined.
In the central nervous system (CNS), neuronal survival, energy metabolism, and plasticity, which are critical for maintaining cognitive and motor functions, are regulated through the actions of insulin and insulin-like growth factors (IGF) types I and II. Insulin, IGF-I and IGF-II, and their corresponding receptors are abundantly expressed in various cell types throughout the brain, including neurons (Gammeltoft et al., 1985; Goodyer et al., 1984; Hill et al., 1986; de laMonte and Wands, 2005). In vitro and in vivo experiments demonstrated that insulin and IGF signaling pathways utilized by CNS neurons are virtually identical to those characterized in peripheral organs such as liver (de la Monte and Wands, 2005). The highest levels of insulin and IGF polypeptide and receptor gene expression in the brain are distributed in the hypothalamus, temporal lobe, and cerebellum (de la Monte and Wands, 2005), which represent major targets of ethanol neurotoxicity.
Studies involving immature brains and cultured neuronal cells showed that ethanol inhibition of insulin and IGF signaling (Hallak et al., 2001; de la Monte et al., 2000, 2001; Zhang et al., 1998) downstream through the PI3 kinase-Akt pathway (de la Monte and Wands, 2002; de la Monte et al., 2001; Ramachandran et al., 2001; Zhang et al., 1998) results in increased apoptosis (Ikonomidou et al., 2000; de la Monte et al., 2001; Zhang et al., 1998) and mitochondrial dysfunction (de la Monte and Wands, 2001, 2002; de la Monte et al., 2001; Ramachandran et al., 2001). Recent studies showed that ethanol mediates its adverse effects on insulin signaling in the developing CNS by inhibiting insulin gene expression and impairing insulin receptor binding (de la Monte et al., 2005; Soscia et al., 2006). The resulting insulin resistance is associated with reduced activation of insulin receptor tyrosine kinase, and downstream signaling through critical cell survival pathways. However, little is known about the effects of chronic ethanol abuse on insulin and IGF signaling mechanisms in the adult human brain. The present study shows that in human alcoholics, neurodegeneration is associated with decreased levels of insulin/IGF gene expression, increased insulin/IGF resistance and oxidative stress, and deficits in acetylcholine homeostasis.
METHODS
Human postmortem control and chronic alcoholic brain tissue were received from the NSW Tissue Resource Centre, The University of Sydney, Australia [ethics approval HREC2002/2/3.14(1441), X03-0117]. All cases had documented evidence of chronic ethanol abuse and no evidence of other substance abuse. In addition, 4 of the 6 alcoholics had hepatic macrosteatosis, and 2 of the 6 had alcohol detected in blood at postmortem examination, but none had cirrhosis or hepatocellular carcinoma. Control subjects were matched for age and gender and had documented low levels of ethanol consumption. Two brain regions were studied: the cerebellar cortex (anterior superior cerebellar vermis region) and the anterior cingulate gyrus in the frontal lobe. These regions were selected for study because they represent major targets of ethanol neurotoxicity. Formalin-fixed paraffin-embedded sections of these regions were stained with Luxol Fast Blue, hematoxylin and eosin dyes and examined by light microscopy. Adjacent histological sections were used for immunohistochemical staining to detect gliosis, DNA damage, and lipid peroxidation. Fresh, snap-frozen blocks of tissue from the same regions were used to measure mRNA expression and receptor binding.
Histological Studies
Paraffin sections (8 μM thick) were immunostained with monoclonal antibodies to 8-hydroxy-deoxyguanosine (8-OHdG) (Abcam Corp., Cambridge, MA), 4-hydroxynonenol (HNE) (Chemicon International, Temecula, CA), or glial fibrillary acidic protein (GFAP) (CalBiochem, Carlsbad, CA) to detect DNA damage, lipid peroxidation, or reactive astrocytosis (gliosis), respectively. Prior to immunostaining, the deparaffinized, re-hydrated sections (Xu et al., 2003) were treated with 0.1 mg/ml saponin in phosphate buffered saline (10 mM sodium phosphate; 0.9% NaCl, pH 7.4; PBS) for 20 minutes at room temperature, followed by 3% hydrogen peroxide in methanol for 10 minutes to quench endogenous peroxidase activity, and then a 30-minute incubation SuperBlock-TBS (Pierce Chemical Co., Rockford, IL) to block nonspecific binding sites. The tissue sections were then incubated overnight at 4°C with 0.5 to 1 μg/ml of primary antibody. Immunoreactivity was detected with biotinylated secondary antibody, avidin biotin horseradish peroxidase (HRP) complex (ABC) reagents, and diaminobenzidine as the chromogen (Vector Laboratories, Burlingame, CA) (Chu et al., 2007). The tissue sections were counterstained with hematoxylin, preserved under coverglass, and examined by light microscopy.
Quantitative Reverse Transcriptase Polymerase Chain Reaction Assays
Total RNA was isolated from brain tissue using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA concentrations and purity were determined from the absorbances measured at 260 and 280 nm. RNA (2 μg) was reverse transcribed using the AMV First Strand cDNA synthesis kit (Roche Diagnostics Corporation, Indianapolis, IN) and random oligodeoxynucleotide primers. Quantitative reverse transcriptase polymerase chain reaction (RT-PCR) was used to measure mRNA levels of neuronal (Hu), astrocytic (GFAP), oligodendroglial (myelin-associated glycoprotein-1; MAG-1), microglial (AIF1), and endothelial (endothelin-1; ET-1) cell genes, insulin, IGF-I, and IGF-II growth factors, their corresponding receptors, insulin receptor substrate (IRS) types 1, 2, and 4, acetylcholinesterase (AChE), choline acetyltransferase (ChAT). Ribosomal 18S RNA levels measured in parallel reactions were used to calculate relative abundance of the mRNA transcripts (Chu et al., 2007; Soscia et al., 2006). PCR amplifications were performed in 25 μl reactions containing cDNA generated from 2.5 ng of original RNA template, 300 nM each of gene specific forward and reverse primer (Table 1), and 12.5 μl of 2× QuantiTect SYBR Green PCR Mix (Qiagen Inc, Valencia, CA). The amplified signals were detected continuously with the BIO-RAD iCycler iQ Multi-Color RealTime PCR Detection System (Bio-Rad, Hercules, CA). The amplification protocol used was as follows: initial 15-minutes denaturation and enzyme activation at 95°C, 45 cycles of 95°C × 15 seconds, 55° to 60°C × 30 seconds, and 72°C × 30 seconds. Annealing temperatures were optimized using the temperature gradient program provided with the iCycler software.
Table 1.
Human Primer Pairs for Quantitative RT-PCRa
| mRNA | Sequence (5′ → 3′) | Position | Amplicon (bp) |
|---|---|---|---|
| Insulin | TTC TAC ACA CCC AAG TCC CGT C | 189 | 134 |
| ATC CAC AAT GCC ACG CTT CTG C | 322 | ||
| Insulin R | GGT AGA AAC CAT TAC TGG CTT CCT C | 1037 | 125 |
| CGT AGA GAG TGT AGT TCC CAT CCA C | 1161 | ||
| IGF-I | CAC TTC TTT CTA CAC AAC TCG GGC | 1049 | 130 |
| CGA CTT GCT GCT GCT TTT GAG | 1178 | ||
| IGF-I R | TAC TTG CTG CTG TTC CGA GTG G | 295 | 101 |
| AGG GCG TAG TTG TAG AAG AGT TTC C | 395 | ||
| IGF-II | CTG ATT GCT CTA CCC ACC CAA G | 1024 | 76 |
| TTG CTC ACT TCC GAT TGC TGG C | 1099 | ||
| IGF-II R | CAC GAC TTG AAG ACA CGC ACT TAT C | 403 | 132 |
| GCT GCT CTG GAC TCT GTG ATT TG | 534 | ||
| IRS-1 | TGC TGG GGG TTT GGA GAA TG | 4527 | 68 |
| GGC ACT GTT TGA AGT CCT TGA CC | 4594 | ||
| IRS-2 | AAA ATT GGC GGA GCA AGG C | 753 | 64 |
| ATG TTC AGG CAG CAG TCG AGA G | 816 | ||
| IRS-4 | CCG ACA CCT CAT TGC TCT TTT C | 570 | 74 |
| TTT CCT GCT CCG ACT CGT TCT C | 643 | ||
| ChAT | CTG ATG GAA CGG GAA CAG TCA C | 730 | 140 |
| CAC TTG GGA CCA GGA AAC TAT CC | 869 | ||
| AChE | TGC TGC CTC AAG AAA GCG TC | 1095 | 149 |
| ATA CGA GCC CTC ATC CTT CAC C | 1243 | ||
| Hu | ATG GAG CCT CAG GTG TCA AAT G | 246 | 113 |
| TTG CTG TCA TCT GTG GTT GCC | 358 | ||
| GFAP | TAT GAG GCA ATG GCG TCC AG | 738 | 130 |
| AGT CGT TGG CTT CGT GCT TG | 867 | ||
| MAG-1 | TGGAAGCCAACAGTGAACGG | 1128 | 104 |
| TTGAAGATGGTGAGAATAGGGTCC | 1231 | ||
| AIF-1 | TGA AAA CCC TCC AGT CAG CG | 42 | 240 |
| ATC TCT TGC CCA GCA TCA TCC | 281 | ||
| 18S | GGA CAC GGA CAG GAT TGA CA | 1278 | 50 |
| ACC CAC GGA ATC GAG AAA GA | 1327 |
R, receptor; IGF, insulin-like growth factor; IRS, insulin receptor substrate; ChAT, choline acetyltransferase; AChE, acetylcholinesterase; Hu, neuronal RNA binding protein; GFAP, glial fibrillary acidic protein; MAG-1, myelin-associated glycoprotein 1; AIF-1, allograft inflammatory factor-1.
Nucleic acid sequences of gene specific oligodeoxynucleotide primer pairs used for real-time quantitative RT-PCR analysis of human brain tissue. The positions within the cDNAs bound by the most 5′ nucleotide of the primers, and the amplicon sizes (bp) (PCR products) are listed in columns 3 and 4, respectively.
In preliminary studies, SYBR Green-labeled PCR products were evaluated by agarose gel electrophoresis, and the authenticity of each amplicon was verified by nucleic acid sequencing. The complementary (c) DNAs were cloned into the PCRII vector (Invitrogen). Serial dilutions of known quantities of recombinant plasmid DNA containing the specific target sequences were used as standards in the PCR reactions, and the regression lines generated from the Ct values of the standards were used to calculate mRNA abundance. Relative mRNA abundance was determined from the nanogram ratios of specific mRNA to 18S measured in the same samples (Chu et al., 2007; Soscia et al., 2006; Xu et al., 2003). Results were normalized to 18S because 18S is highly abundant and the levels were essentially invariant among the samples, whereas housekeeping genes were modulated with disease state. Inter-group statistical comparisons were made using the calculated mRNA/18S ratios. Control studies included real-time quantitative PCR analysis of the following: (1) template-free reactions, (2) RNA that had not been reverse transcribed, (3) RNA samples that were pretreated with DNAse I, (4) samples treated with RNAse A prior to reverse transcriptase reaction, and (5) genomic DNA.
Receptor Binding Assays
Competitive equilibrium binding assays were used to assess growth factor binding in relation to chronic alcohol abuse exposure (Rivera et al., 2005; Soscia et al., 2006). To extract membrane proteins, fresh frozen tissue (~100 mg) was homogenized in 5 volumes of NP-40 lysis buffer (50 mM Tris–HCl, pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 2 mM EGTA) containing protease (1 mM PMSF, 0.1 mM TPCK, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, 0.5 μg/ml leupeptin, 1 mM NaF, 1 mM Na4P2O7) inhibitors. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Pierce). Exploratory studies determined the amounts of protein and concentrations of radiolabeled ligand required to achieve 20% specific binding. Insulin receptor binding assays were performed using 100 μg protein. IGF-I binding assays required 25 μg protein per sample, and IGF-II receptor binding assays were optimized using 10 μg protein. For total binding, duplicate individual protein samples were incubated in 100-μl reactions containing binding buffer [100 mM HEPES, pH 8.0, 118 mM NaCl, 1.2 mM MgSO4, 8.8 mM dextrose, 5 mM KCl, 1% bovine serum albumin (BSA)] and 100 nCi/ml of [125I] (2,000 Ci/mmol; 50 pM) insulin, IGF-I, or IGF-II. To measure nonspecific binding, replicate samples were identically prepared but with the addition of 0.1 μM unlabeled (cold) ligand.
All reactions were performed in 1.5-ml Eppendorff tubes, and the incubations were performed at 4°C for 16 hours with gentle platform agitation. Bound radiolabeled tracer was then precipitated by adding 500 μl of 0.15% bovine gamma globulin (prepared in 100 mM Tris–HCl, pH 8.0) followed by 400 μl 37.5% polyethylene glycol 8000 (PEG-8000; prepared in 100 mM Tris–HCl, pH 8.0) to each tube. The samples were thoroughly mixed by vortexing, and then incubated on ice for at least 2 hours. The precipitates were collected by centrifuging the samples at 15,000 × g for 5 minutes at room temperature. The supernatant fraction, which contained unbound (free) ligand, was transferred in its entirety to a Gamma counting tube (Sarstedt, Newton, NC). The Eppendorff tube tip containing the pellet was cut and released directly into a separate Gamma counting tube. The samples were counted for 1 minute in an LKB Compu- Gamma CS Gamma counter. Specific binding was calculated by subtracting fmol of nonspecific binding, i.e., amount bound in the presence of cold ligand, from the total fmol bound (absence of unlabeled competitive ligand). The results were analyzed and plotted using the GraphPad Prism 4 software (GraphPad Software, Inc., San Diego, CA).
Enzyme-Linked Immunosorbant Assays
Immunoreactivity for 8-OHdG, HNE, activated Caspase 3, and nitrotyrosine was examined by enzyme linked immunosorbant assays (ELISA). Brain tissue was homogenized in 5 volumes of radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl, pH 7.5, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 2 mM EGTA) containing protease and phosphatase inhibitors (1 mM PMSF, 0.1 mM TPCK, 1 mg/ml aprotinin, 1 mg/ml pepstatin A, 0.5 mg/ml leupeptin, 1 mM NaF, 1 mM Na4P2O7, 2 mM Na3VO4). Protein concentrations were determined using the BCA assay (Pierce). ELISAs were used to measure immunoreactivity corresponding to HNE, activated Caspase 3, and β-actin. ELISAs were performed in 96-well opaque polystyrene plates (Nunc, Rochester, NY). RIPA protein extracts diluted to in TBS (40 ng in 100 μl) were adsorbed to the bottoms of the wells by over night incubation at 4°C. After rinsing in TBS, the wells were blocked for 4 hours with 250 μl/well of 3% BSA in TBS. The proteins were then incubated with primary antibody (0.01 to 0.1 μg/ml) for 1 hour at room temperature. Immunoreactivity was detected with HRP-conjugated secondary antibody (1:10000; Pierce), and either Amplex Red soluble fluorophore (Molecular Probes, Eugene, OR), or enhanced chemiluminescence reagents. Amplex Red fluorescence was measured (Ex 530/Em 590) in a SpectraMax M5 microplate reader (Molecular Devices Corp., Sunnyvale, CA). Luminescence was measured in a TopCount machine (Packard Instrument Company, Meriden, CT). Binding specificity was determined from parallel negative control incubations with nonrelevant antibodies, the primary or secondary antibody omitted, or the protein samples omitted. The calculated ratios of HNE/β-actin and activated Caspase 3/β-actin fluorescence were used to assess relative levels of immunoreactivity.
8-OHdG immunoreactivity was measured by ELISA using purified DNA (Kikuchi et al., 2002). Total DNA was extracted with Qiaquik DNA spin columns (Qiagen) according to the manufacturer’s protocol. DNA was quantified and its purity assessed from the absorbances measured at 260 and 280 nm. Duplicate samples of DNA (10 ng/100 μl PBS) were placed into individual wells of 96-well Universal DNA Bind plates (Corning, Corning, NY) and allowed to adsorb to the bottom surfaces overnight at 4°C. DNA was cross-linked by UV irradiation according to the manufacturer’s protocol. After blocking with 2% BSA in PBS, the samples were incubated with 0.1 μg/ml monoclonal anti-8-OHdG (Abcam Corp., Cambridge, MA) for 1 hour at room temperature, followed by HRP-conjugated secondary antibody as described above. Immunoreactivity was detected with Amplex Red soluble fluorophore, and fluorescence (Ex 530/Em 590) was read in a SpectraMax M5 microplate reader (Molecular Devices Corp.). Then, to correct for differences in the amounts of DNA adsorbed to the well surfaces, bound DNA was labeled with H33342, and fluorescence was measured (Ex 360/Em 460) in the SpectraMax M5. The calculated ratio of Amplex Red/H33342 fluorescence in corresponding wells was used as the index of 8-OHdG immunoreactivity.
Source of Reagents
Human recombinant [125I] Insulin, IGF-I, and IGF-II were purchased from Amersham Biosciences (Piscataway, NJ). Unlabeled human insulin was purchased from Sigma-Aldrich (St. Louis, MO). Recombinant IGF-I and IGF-II were obtained from Bachem (King of Prussia, PA). Monoclonal antibodies to 8-OHdG, activated caspase 3, and HNE were purchased from Chemicon International (Temecula, CA) or Abcam Corp (Cambridge, MA). All other fine chemicals and reagents were purchased from CalBiochem (Carlsbad, CA) or Sigma-Aldrich (St. Louis, MO).
Statistical Analysis
Data are depicted as mean ± SEM in the graphs. Inter-group comparisons were made using Student t-tests. In addition, we used multivariate analysis of variance (MANOVA) to examine the effects of ethanol abuse on multiple dependent variables, i.e., insulin/IGF receptor, Hu, and ChAT expression. Statistical analyses were performed using the Number Cruncher Statistical System (Kaysville, UT). Significant p-values (<0.05) are indicated over the graphs.
RESULTS
Subject Profiles
This study included 6 male chronic alcoholics who had a mean age of 57.7 ± 7.1 years, and 6 male controls who had a mean age of 57.5 ± 6.7 years at the time of death (Table 2). Blood alcohol levels were significantly higher in the alcoholic (168.3 ± 122.7 g/d) relative to the control (11 ± 7.2 g/d) group (p = 0.01). Primary causes of death included cardiac disease (n = 4), chronic lung disease (n = 1), and complications of chronic liver disease (n = 1) in the alcoholic group, and cardiac disease (mainly ischemic; n = 6) in the control group. Mean postmortem interval was slightly but not significantly longer in the alcoholic group. The livers had hepatic steatosis in 4 of the 6 alcoholics compared with none of the controls. Although none of the brains were judged to be globally atrophic, the mean brain weight in the alcoholic group was significantly lower than in the control group (p = 0.04). The higher brain weight among controls may have been due to acute edema resulting from fatal ischemic cardiac disease.
Table 2.
Population Profile
| Index | Alcoholic | Control | p-Value |
|---|---|---|---|
| Number | 6 | 6 | |
| Age (mean ± SD) | 57.7 ± 7.1 | 57.5 ± 6.7 | |
| % Male | 100% | 100% | |
| Alcohol consumed (g/d) (mean ± SD) | 168.3 ± 51.1 | 11.0 ± 7.2 | 0.01 |
| Postmortem interval (mean ± SD) | 36.2 ± 21.1 | 22.3 ± 8.3 | 0.17 |
| Brain Weight (g) (mean ± SD) | 1335.7 ± 133.2 | 1505.8 ± 115.1 | 0.04 |
Increased Oxidative Stress and DNA Damage With Alcoholic Neurodegeneration
Alcoholics’ brains had no specific histopathologic abnormalities identified in the anterior cingulate region of the frontal lobe. In contrast, alcoholics’ brains had variable degrees of cell loss in the granule and Purkinje cell layers of the cerebellar cortex, and reduced thicknesses of white matter cores within the folia (Fig. 1). Adjacent sections were immunostained to detect gliosis, lipid peroxidation, and DNA damage using antibodies to GFAP, HNE, and 8-OHdG, respectively (Fig. 2). GFAP immunoreactivity was detected in glial cells distributed in the cortex, white matter, and perivascular regions. HNE immunoreactivity was detected in the nucleus and cytoplasm of neurons based on their location, size (12 to 16 micron diameter), and pyramidal shapes, although other cell types including glia and vascular endothelial cells distributed in both gray and white matter structures were also HNE-positive. 8-OHdG immunoreactivity overlapped with that of HNE, suggesting that ethanol-mediated DNA damage occurred in neurons as well as other cell types.
Fig. 1.

Alcoholic cerebellar degeneration. Formalin-fixed, paraffin-embedded sections of anterior superior cerebellar vermis from control and chronic alcoholic patients were stained with Luxol Fast Blue, hematoxylin and eosin, and examined by light microscopy. (A, C, E) Control brains had distinct lamination of the cortex with densely populated granule cell (GC) layers, broad bands of white matter (WM), and abundant large neurons within the Purkinje cell layer (arrows). (B, D, F) In chronic alcoholics, the cerebellar cortex had reduced cell densities within the (B) GC and (F) Purkinje (arrows) cell layers and (D) reduced thickness of WM within the cores of the folia. The background stain is somewhat bluer in control samples due to more intense Luxol Fast Blue (myelin) staining. Original magnifications 100× (A–D) or 650× (E–F).
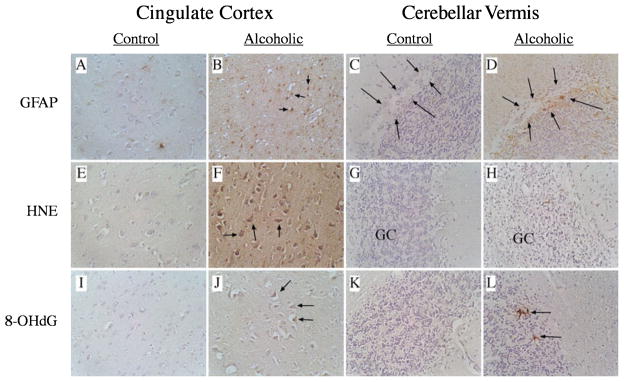
Fig. 2.
Increased gliosis, lipid peroxidation, and DNA damage in alcoholics’ brains. Histological sections of (A, B, E, F, I, J) anterior cingulate cortex and (C, D, G, H, K, L) cerebellar vermis from (A, C, E, G, I, K) control or (B, D, F, H, J, L) alcoholics’ brains were immunostained to detect (A–D) GFAP, (E–H) HNE, or (I–L) 8-OHdG (see Methods). Immunoreactivity was revealed by the ABC method using DAB as the chromogen (brown precipitate). In Panels B, F, and J, arrows point to examples of (B) glial cells or (F, J) neurons with positive immunoreactivity. In Panels C and D, arrows flank the Purkinje cell layer of the cerebellar cortex. GC, granule cell layer. In Panel L, arrows point to examples of 8-OHdG cellular immunoreactivity within the GC layer. The background stain is somewhat bluer in control samples due to increased GFAP, HNE, and 8-OHdG immunoreactivity in alcoholic brains.
In the anterior cingulate gyrus, despite the absence of overt cell loss, prominently increased GFAP and HNE immunoreactivities were observed in both the cortex and subcortical white matter (Fig. 2A–B, E–F). In contrast, 8-OHdG immunoreactivity was only slightly increased and localized in scattered cells within the cortex and white matter (Fig. 2I–J). Alcoholics’ cerebellar vermis had increased GFAP (Fig. 2C–D) immunoreactivity in the granule cell layer and in the white matter cores underlying the cortex, and increased HNE immunoreactivity in the Purkinje and granule cell layers of cortex, and in subcortical white matter (Fig. 2G–H). Increased 8-OHdG immunoreactivity was detected in only scattered cells within the Purkinje and granule layers of the cerebellar cortex and in the subcortical white matter in alcoholics (Fig. 2K–L).
To quantify the effects of chronic alcohol abuse on oxidative stress and DNA damage in the brain, HNE and 8-OHdG immunoreactivities were measured in protein homogenates using direct ELISAs. In addition, we measured the levels of activated Caspase 3 because previous studies showed that chronic ethanol exposure results in increased apoptosis mediated by Caspase 3 activation in experimental animals (Xu et al., 2003). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) immunoreactivity was measured because its expression levels can by reduced by insulin resistance in the context of chronic ethanol exposure (Soscia et al., 2006), and β-actin was measured as a negative control. As β-actin expression was previously observed to be unaffected by chronic ethanol exposure (Soscia et al., 2006), the levels were used to normalize levels of immunoreactivity corresponding to other proteins and thereby correct for small differences in sample loading. The studies demonstrated significantly higher levels of HNE immunoreactivity in both the anterior cingulate cortex and cerebellar vermis of alcoholics relative to control (Fig. 3A–B). In addition, 8-OHdG was significantly increased while GAPDH was significantly reduced in alcoholics’ anterior cingulate gyrus (Fig. 3). Otherwise, there were no significant differences between the groups with respect to the mean levels of activated Caspase 3 or β-actin.
Fig. 3.
Quantification of 4-hydroxynonenol (HNE), 8-hydroxy-deoxyguanosine (8-OHdG), activated caspase 3, glyceraldehydes-3-phosphate dehydrogenase (GAPDH), and β-actin in (A, C, E, G, I) the anterior cingulate cortex or (B, D, F, H, J) cerebellar vermis of control or alcoholic subjects’ brains. Immunoreactivity was measured by enzyme-linked immunosorbant assay (ELISA) using Amplex Red fluorescence detection. To control for differences in sample loading, fluorescence indices were normalized to β-actin measured in parallel reactions. Graphs depict the mean (±SEM) fluorescence ratios corresponding to (A, B) HNE, (C, D) 8-OHdG, (E, F) caspase-3, or (G, H) GAPDH and β-actin. Significant inter-group differences determined with Student t-tests are indicated over the bars.
General Comments Regarding Quantitative RT-PCR Studies
The use of real-time quantitative RT-PCR enabled all samples to be analyzed simultaneously and with sufficient replicates to demonstrate consistency of results. With the techniques employed, the quality of the cDNA templates generated from tissue was judged to be excellent based on the similar 18S Ct values and consistent 28S:18S ratios obtained for both the control and alcoholics’ brains. In addition, the 28S:18S ratios were uniformly comparable to the values obtained for RNA isolated from primary neuronal cultures or cell lines. Typically, with cDNA prepared from 10 ng of total RNA, the 18S Ct values ranged from 8 to 10, and the calculated 28S/18S ng ratios ranged from 2.0 to 2.2 as previously reported for brain tissue using Northern blot or slot blot analysis (Payao et al., 1998; da Silva et al., 2000). The use of real-time quantitative RT-PCR was ideally suited for rigorous analysis of gene expression in brain tissue because the amplicons were small (mainly <150 bp), thereby circumventing any potential problems related to partial RNA degradation, e.g., nicking, which may increase with oxidative stress. The specificity of the amplified products was verified by direct nucleic acid sequencing. Control studies in which cDNA templates were excluded, RNA was not reverse transcribed, the RNA samples were pretreated with RNAse A prior to the RT step, or genomic DNA was used in the reactions produced no detectable amplified products by real-time PCR analysis and agarose gel electrophoresis. Treatment of the RNA samples with DNAse I prior to the RT step had no effect on the detection levels of amplified gene products. The mean ribosomal 18S and 28S levels were similar in cerebellum and anterior cingulate gyrus of alcoholic and control human brains.
Alcoholic Neurodegeneration Is Associated With Pathological Shifts in Cell Type
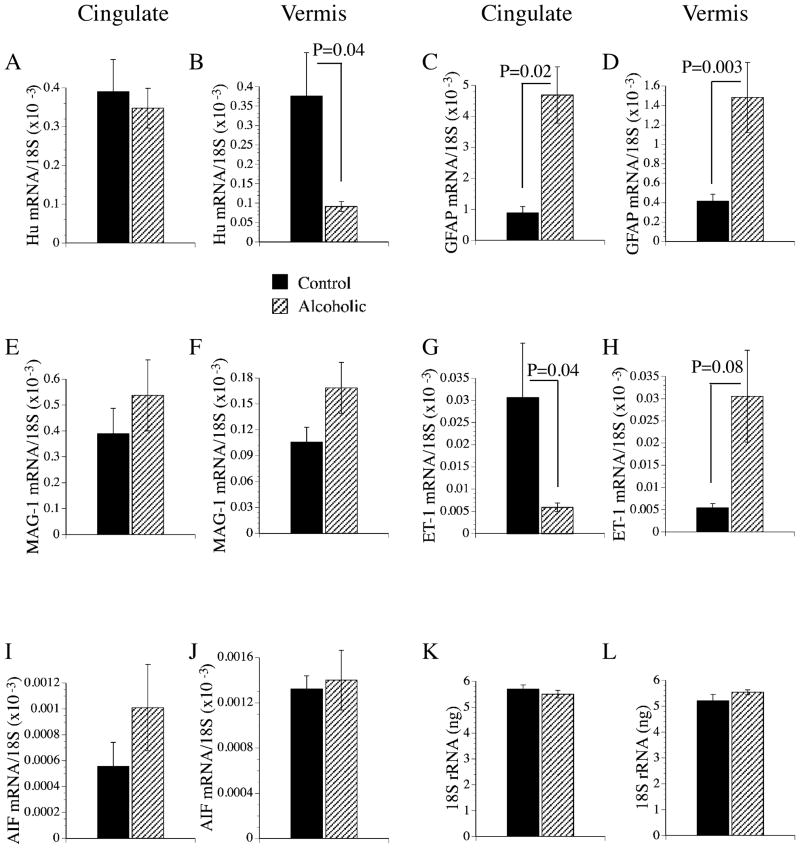
To determine if chronic alcohol abuse caused pathological shifts in the cell populations as observed previously in experimental models of fetal alcohol spectrum disorder (Soscia et al., 2006) and Alzheimer’s disease (AD) (Rivera et al., 2005), quantitative RT-PCR was used to measure mRNA transcripts encoding Hu neuronal ribosomal RNA binding protein (Hu et al., 2004; Kumagai et al., 1999; Szabo et al., 1991), MAG-1 for oligodendroglia, GFAP for astrocytes, ET-1 for endothelial cells, and AIF-1 for microglia (Ito et al., 2001). The nanogram quantities of each specific mRNA transcript detected were normalized to the 18S RNA levels measured in the same samples, and results were analyzed statistically (Fig. 4). The studies demonstrated significant reductions in Hu expression in the cerebellar vermis (Fig. 4B), and ET-1 expression in the anterior cingulate (Fig. 4G), and significantly increased expression of GFAP in both the anterior cingulate and cerebellar vermis (Fig. 4C–D), and ET-1 in the cerebellar vermis (Fig. 4H) of chronic alcoholics. In contrast, there were no significant inter-group differences with respect to in the mean levels of MAG-1, AIF-1 or 18S rRNA (Fig. 4).
Fig. 4.
Pathological shifts in brain cell populations in alcoholics: cell-specific gene expression was measured in the anterior cingulate gyrus (A, C, E, G, I) or cerebellar vermis (B, D, F, H, J) by quantitative RT-PCR with levels normalized to (K, L) 18S rRNA. The mRNA transcript levels corresponding to (A, B) Hu neuronal RNA binding protein, (C, D) astrocytic glial fibrillary acidic protein (GFAP), (E, F) myelin-associated glucoprotein-1 (MAG-1), (G, H) endothelin-1 (ET-1), and (I, J) microglial AIF-1 were used to detect pathological shifts in brain cell types in alcoholics. RNA extracted with Trizol reagent was reverse transcribed using random oligodeoxynucleotide primers. The mRNA transcript levels were measured using gene-specific primers (Table 1) and quantitative RT-PCR. The nanogram quantities of mRNA were normalized to 18S ribosomal RNA measured in the same samples. Graphs depict the mean ± SEM of results obtained from 6 samples per group. Data were analyzed statistically using Student t-tests. Significant p-values are indicated over the bar graphs.
Effects of Chronic Alcohol Abuse on mRNA Expression of Insulin, IGF-I, and IGF-II Polypeptides, and the Insulin, IGF-I, and IGF-II Receptors
Quantitative RT-PCR studies detected mRNA transcripts corresponding to insulin, IGF-I, and IGF-II polypeptides (Fig. 5), and insulin, IGF-I, and IGF-II receptors (Fig. 6). Insulin, IGF-I, and IGF-II were expressed in both control and alcoholics’ brains. In the anterior cingulate, IGF-II was most abundant, followed by IGF-I, and then insulin, whereas in the cerebellar vermis, IGF-I was most abundant followed by IGF-II and then insulin (Fig. 5). Of note is that the mean levels of insulin gene expression were between 5- and 20-fold lower than either IGF-I or IGF-II. Chronic alcohol abuse resulted in significantly reduced levels of insulin gene expression in the anterior cingulate and cerebellar vermis (Fig. 5A–B), and IGF-I expression in the cerebellar vermis (Fig. 5D). In contrast, IGF-II gene expression was significantly increased in alcoholics’ cerebellar vermis (Fig. 5F).
Fig. 5.
Alterations in insulin, IGF-I, and IGF-II gene expression in brains of chronic alcoholics. (A, C, E) Anterior cingulate gyrus and (B, D, F) cerebellar vermis expression levels of (A, B) insulin (INS), (C, D) IGF-I, and (E, F) IGF-II mRNA transcripts were measured by quantitative RT-PCR with levels normalized to 18S rRNA. See Methods and Fig. 4 legend. Graphs depict the mean ± SEM of results obtained from 6 samples per group. Data were analyzed statistically using Student t-tests. Significant p-values are indicated over the bar graphs.
Fig. 6.
Alterations in insulin, IGF-I, and IGF-II receptor gene expression in brains of chronic alcoholics. (A, C, E) Anterior cingulate gyrus and (B, D, F) cerebellar vermis expression levels of (A, B) insulin receptor (INS-R), (C, D) IGF-I receptor (IGF-1R), and (E, F) IGF-II receptor (IGF-IIR) mRNA transcripts were measured by quantitative RT-PCR with levels normalized to 18S rRNA. See Methods and Fig. 4 legend. Graphs depict mean ± SEM of results obtained from 6 samples per group. Data were analyzed statistically using Student t-tests. Significant p-values are indicated over the bar graphs.
Corresponding with the relative abundances of growth factor mRNA transcripts, in the anterior cingulate, IGF-II receptor was most abundantly expressed, followed by IGF-I receptor, and then the insulin receptor, whereas in the cerebellar vermis, IGF-I receptor was most abundant, followed by IGF-II receptor, and then the insulin receptor (Fig. 6). In addition, the mean levels of insulin receptor mRNA were between 5- and 20-fold lower than those corresponding to the IGF-I or IGF-II receptors. Reductions in insulin and IGF-I gene expression were associated with corresponding reductions in the mean levels of insulin receptor mRNA in the anterior cingulate and cerebellar vermis (Fig. 6A–B), and IGF-I receptor expression in the cerebellar vermis (Fig. 6D). IGF-II receptor expression was also significantly reduced in the anterior cingulate region (Fig. 6E). Of note is that the mean level of IGF-II gene expression was also lower in alcoholics’ anterior cingulate (Fig. 5E), but the inter-group differences were not statistically significant due to the large standard deviations. The mean levels of mRNA corresponding to the IGF-I receptor in the anterior cingulate (Fig. 6C) and IGF-II receptor (Fig. 6F) in the cerebellar vermis were similar for the control and alcoholic groups.
IRS Expression in Chronic Alcoholics’ Brains
Major responses to growth factor-stimulated signaling through IRS molecules include increased cell growth and survival, and inhibition of apoptosis (Burgering and Coffer, 1995; Delcommenne et al., 1998; Dudek et al., 1997; Kulik et al., 1997; Lam et al., 1994; Myers and White, 1993; Puro and Agardh, 1984; Sun et al., 1993). To examine the integrity of signaling pathways that are activated by insulin/IGF-I, IRS-1, IRS-2, and IRS-4 mRNA transcript levels were measured. IRS-3 was not examined because it is only expressed in rodent adipose tissue. Quantitative RT-PCR detected expression of IRS-1, IRS-2, and IRS-4 mRNA transcripts in both control and chronic alcoholics’ brains (Fig. 7). In control anterior cingulate region, IRS-1 mRNA transcripts were most abundant, followed by IRS-2 then IRS-4 (Fig. 7A,C,E), whereas in the cerebellar vermis, IRS-2 mRNA transcripts were slightly more abundant than IRS-I (Fig. 7B,D), and both were 4- or 5-fold higher than the mean level of IRS-4 (Fig. 7F). With the exception of IRS-4 in the anterior cingulate, the mean levels of IRS genes were higher in alcoholics’ anterior cingulate and cerebellar vermis. However, the inter-group differences were not statistically significant due to the large standard deviations.
Fig. 7.
Effects of chronic alcohol abuse on insulin receptor substrate (IRS) gene expression. (A, C, E) Anterior cingulate gyrus and (B, D, F) cerebellar vermis expression levels of (A, B) IRS-1, (C, D) IRS-2, and (E, F) IRS-4 mRNA transcripts were measured by quantitative RT-PCR with levels normalized to 18S rRNA. See Methods and Fig. 4 legend. Graphs depict mean ± SEM of results obtained from 6 samples per group. Data were analyzed statistically using Student t-tests.
Impaired Insulin and IGF Receptor Binding in Chronic Alcoholics’ Brains
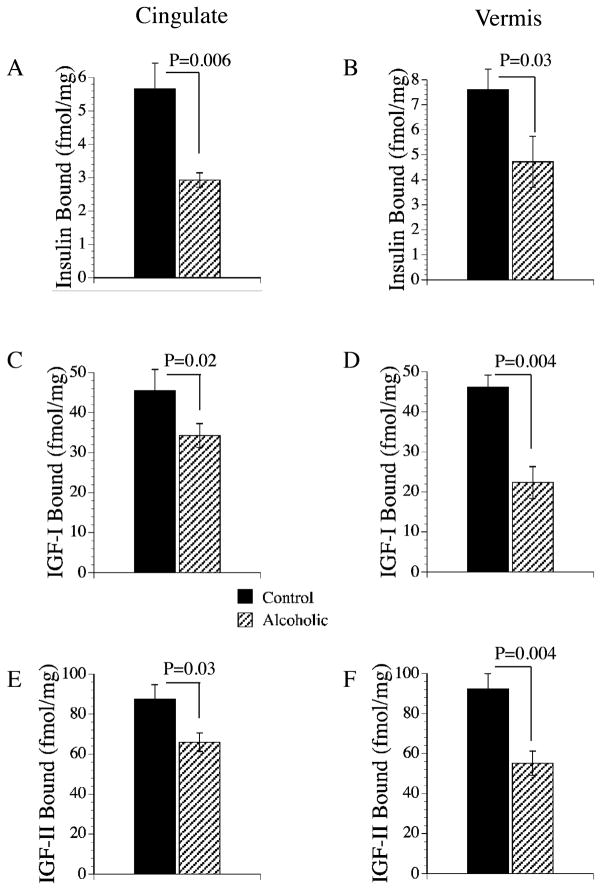
Previous studies in experimental animal models (Lester-Coll et al., 2006; de la Monte et al., 2006; Soscia et al., 2006) and in human neurodegenerative diseases (Rivera et al., 2005; Steen et al., 2005) demonstrated that inhibition of insulin and IGF signaling in the CNS can be mediated by growth factor depletion, reduced expression of the corresponding receptors, or impaired binding to receptors. Effective ligand binding is critical to the insulin and IGF signaling cascades, and many of the downstream effects of impaired insulin signaling that have been reported in ethanol- exposed rat brains, including reduced neuronal survival, were mediated by reduced insulin or IGF-I binding in the CNS (Soscia et al., 2006). In the present study, equilibrium binding assays were performed by incubating anterior cingulate or cerebellar vermis membrane protein extracts with [125I]-labeled insulin, IGF-I, or IGF-II as tracer, in the presence or absence of excess cold ligand. In both brain regions, IGF-II receptor binding (fmol/mg membrane protein) was highest, followed by IGF-I, and then insulin (Fig. 8). These relative differences in the mean levels of receptor binding roughly correspond to the relative abundances of each receptor as measured by quantitative RT-PCR. In chronic alcoholics, the mean levels of insulin, IGF-I, and IGF-II receptor binding were significantly reduced relative to control, in both the anterior cingulate and cerebellar vermis (Fig. 8).
Fig. 8.
Chronic alcohol abuse impairs insulin, IGF-I, and IGF-II receptor binding in the human brain. Equilibrium binding assays were performed by incubating membrane protein extracts of (A, C, E) anterior cingulate gyrus or (B, D, F) cerebellar vermis overnight at 4°C with 50 pM [125I]-labeled insulin, IGF-I, or IGF-II as tracer, in the presence or absence of 100 nM cold ligand. Membrane bound tracer was precipitated by adding bovine gamma globulin and PEG-8000 to the reactions and centrifuging the samples (14,000 × g). Radioactivity present in the supernatant fractions (containing unbound/free ligand) and the pellets (containing bound ligand) was measured in a gamma counter. Specific binding (fmol/mg) was calculated using the GraphPad Prism 4 software. Graphs depict mean ± SEM of results obtained for (A, B) insulin, (C, D) IGF-I, and (E, F) IGF-II specific binding with 6 cases per group. Data were analyzed statistically using Student t-tests. Significant p-values are indicated over the bar graphs.
Impaired Acetylcholine Homeostasis in Chronic Alcoholics’ Brains
Acetylcholine has major roles in cognitive and motor functions. Acetylcholine production requires adequate supplies of choline and acetyl-Co-A. Acetyl-Co-A is generated by energy metabolism, which in turn is driven by insulin and IGF-I stimulation. Recent studies demonstrated that ChAT expression is regulated by insulin and IGF-I stimulation (de la Monte et al., 2005; Rivera et al., 2005; Soscia et al., 2006). Therefore, it was of interest to determine whether the inhibition of insulin and IGF signaling mechanisms observed in alcoholics’ brains was associated with deficits in ChAT gene expression. As the steady-state levels of acetylcholine are negatively regulated by AChE, it was also of interest to measure AChE mRNA levels. Quantitative RT-PCR studies demonstrated significantly reduced mean levels of ChAT mRNA expression in the anterior cingulate (Fig. 9A) and cerebellar vermis (Fig. 9B) in alcoholics. In addition, the mean level of AChE mRNA was reduced in the anterior cingulate (Fig. 9C), but increased in the cerebellar vermis (Fig. 9D) of alcoholics, although the inter-group differences were not statistically significant due to large standard deviations.
Fig. 9.
Effects of chronic alcohol abuse on acetylcholine homeostasis. (A, C) Anterior cingulate gyrus and (B, D) cerebellar vermis expression levels of (A, B) choline acetyltransferase (ChAT) and (C, D) acetyl cholinesterase (AChE) mRNA transcripts were measured by quantitative RT-PCR with levels normalized to 18S rRNA. See Methods and Fig. 4 legend. Graphs depict mean ± SEM of results obtained from 6 cases per group. Data were analyzed statistically using Student t-tests. Significant p-values are indicated over the bar graphs.
The multivariate ANOVA (MANOVA) test with chronic alcohol abuse as the independent factor and Hu, insulin receptor, IGF-I receptor, and IGF-II receptor expression in the cerebellum as dependent variables was highly statistically significant (Hotelling–Lawley Trace T: p < 0.0001). Within-correlation covariance analysis demonstrated significant positive correlations between Hu and insulin receptor (p = 0.0003) and IGF-I receptor (p < 0.0001), and negative correlation between Hu and IGF-IIR (p = 0.02) mRNA expression. Separate ANOVA tests further demonstrated significantly reduced expression of Hu (p = 0.039), insulin receptor (p = 0.019), and IGF-I receptor (p = 0.005), but not IGF-II receptor (p > 0.10) in alcoholic cerebella. MANOVA tests also revealed a significant negative correlation between insulin receptor binding and ChAT expression, and significantly reduced binding to the insulin (p < 0.03), IGF-I (p = 0.005), and IGF-II (p = 0.005) receptors in alcoholic versus control cerebellar vermis. MANOVA tests using data generated from cingulate gyrus samples also demonstrated significantly reduced insulin receptor (p = 0.006), IGF-I receptor (p = 0.02), and IGF-II receptor (p = 0.03) binding, and ChAT (p = 0.016), IGF-II receptor (p = 0.034), and insulin receptor (p = 0.037) mRNA expression in the chronic alcoholics. These results confirm the above results obtained by independent T-test analysis.
DISCUSSION
Molecular Indices of Neurodegeneration in Chronic Alcoholics
In human chronic alcoholics, CNS degeneration is characterized by cerebellar atrophy, cerebral white matter atrophy, and either loss or impaired function of neurons within the hypothalamus, thalamus, hippocampus, and frontal cortex. These abnormalities cause variable degrees of cognitive and motor deficits, and in severe cases, dementia (Harper, 1998; Harper et al., 2003). In experimental models of chronic ethanol feeding, CNS neurodegeneration is associated with overt cell loss with increased immunoreactivity for HNE, 8-OHdG, and activated Caspase 3 in the cerebellar cortex (Chu et al., 2007; Cohen et al., 2007; Soscia et al., 2006; Xu et al., 2003). In the present study, we demonstrated that the chronic alcoholics had histopathologic and molecular evidence of neuronal loss in the cerebellar vermis. The neuronal loss was associated with gliosis (GFAP expression) as demonstrated by immunohistochemical staining and quantitative RT-PCR, and increased immunoreactivity for HNE and 8-OHdG, reflecting increased lipid peroxidation and DNA damage. Therefore, these findings in human chronic alcoholics’ cerebella correspond with previous observations in both adult and developing experimental animal models (Chu et al., 2007; Cohen et al., 2007; Soscia et al., 2006; Xu et al., 2003). The significance of the results is that the sustained oxidative stress caused by chronic alcohol abuse may render CNS neurons more vulnerable to “second hits” such as hypoxia or ischemia which, in the otherwise normal brain would not necessarily cause permanent injury or neurodegeneration.
In alcoholics’ anterior cingulate, where there was no overt histopathologic evidence of neuronal loss or neurodegeneration, immunohistochemical staining studies demonstrated increased immunoreactivity for GFAP, HNE, and 8-OHdG. These effects of chronic alcohol abuse were confirmed by ELISA or quantitative RT-PCR. These results are of interest because, in contrast to the cerebellar vermis, chronic oxidative stress mediated by lipid peroxidation and DNA damage did not cause significant neuronal death in the anterior cingulate cerebral cortex in chronic alcoholics. Therefore, the neurodegenerative consequences of oxidative stress caused by chronic alcohol abuse vary with brain region. In the cerebellar vermis, a major effect is neuronal loss, whereas in the anterior cingulate, the major effect may be loss of function as evidenced by the significant reductions in ChAT gene expression. The latter may be mediated by alcohol-induced degeneration of neuritis with down-regulation of neurotransmitter gene expression or function, which could result in cognitive impairment (Diamond and Gordon, 1997; Guerri, 1998).
Pathological Shifts in Brain Cell Populations in Alcoholics
In the alcoholic brains, histopathological and/or immunohistochemical indices of neurodegeneration were associated with pathological shifts in brain cell populations within anterior cingulate gyrus and cerebellar vermis. We utilized a novel approach for estimating the proportions of neurons, oligodendroglia, astrocytes, endothelial cells, and microglia by comparing the relative mRNA expression levels of Hu, MAG-1, GFAP, ET-1, and AIF-1, respectively, in the same tissue samples. Molecular profiling of genes that report relative abundance of different cell types within the brain was previously used to characterize neurodegeneration in AD (Rivera et al., 2005) and experimental models of fetal alcohol spectrum disorder (Soscia et al., 2006), adult chronic alcohol feeding (Cohen et al., 2007), and Alzheimer neurodegeneration (Lester-Coll et al., 2006; de laMonte et al., 2006, 2007).
Although both brain regions studied are targets of alcoholic neurotoxicity, the adverse effects of chronic alcohol abuse were inhomogeneous with respect to cell loss. In the anterior cingulate gyrus, the alcoholic group had significantly reduced levels of ET-1, suggesting loss of vascular endothelial cells, and increased GFAP expression, consistent with the gliosis observed by immunohistochemical staining. In the cerebellar vermis, the reduced levels of Hu and increased GFAP mRNA levels corresponded with the neuronal loss and gliosis observed in histological sections. In addition, a significant trend was detected with respect to ET-1 expression in alcoholics’ cerebellar vermis, suggesting modest relative increases in either proliferation and/or activation of endothelial cells. The significance of this result is not certain as it could reflect either a response to or consequence of neuronal loss, or nonspecific response to unrelated pathology such as minor head trauma. The prominent neuronal loss detected in the cerebellar vermis using histopathologic and molecular techniques would account for the motor deficits (ataxias) that occur in alcoholics.
Given the frequent occurrence of white matter atrophy in chronic alcoholics (Harper, 1998; Harper et al., 2003; de la Monte, 1988) and previous demonstration of reduced myelin-associated gene expression in alcoholic brains (Lewohl et al., 2000; Liu et al., 2006), it was anticipated that MAG-1 gene expression would be significantly reduced in alcoholics’ brains. However, the molecular profiling studies did not demonstrate reduced MAG-1 mRNA levels in alcoholics, despite histopathologic evidence of white matter atrophy in the cerebellar vermis. One probable explanation for this discordant result is that the tissue samples analyzed from anterior cingulate gyrus and cerebellar vermis contained predominantly gray matter. This point is corroborated by the higher levels of Hu compared with MAG-1 mRNA in the cerebellum, and similar levels of Hu and MAG-1 mRNA in the anterior cingulate samples. Therefore, additional studies are needed to further characterize the effects of chronic alcohol abuse on cellular gene expression anterior cingulate and cerebellar vermis white matter.
Effects of Chronic Alcohol Abuse on the Expression of Genes Required for Insulin and IGF Signaling
Quantitative RT-PCR studies demonstrated mRNA transcripts corresponding to insulin, IGF-I, IGF-II, their corresponding receptors, and IRS-1, IRS-2, and IRS-4 in the anterior cingulate gyrus and cerebellar vermis, indicating that the genes required for insulin and IGF signaling are expressed in adult human brains. In both brain regions, IGF-1 and IGF-II polypeptide genes were expressed at higher levels than insulin. In the anterior cingulate, IGF-II was the dominant (most abundant) growth factor, whereas in the cerebellum, IGF-I was most abundant of the 3 mRNA transcripts. Chronic alcoholics had markedly reduced levels of insulin gene expression in both the anterior cingulate and cerebellar vermis, and reduced IGF-I expression in the cerebellum. The finding of reduced IGF-I expression in the cerebellar vermis and not in the anterior cingulate correlates with the selective distribution of neuronal loss detected by histopathological examination and quantitative RT-PCR analysis, and suggests that neuronal viability in the adult human cerebellum is largely regulated by IGF-I mediated signaling. Correspondingly, previous studies demonstrated that ethanol-impaired neuronal survival could be prevented by IGF-I treatment (de la Monte et al., 2000; Patel, 2004). These findings together suggest that the differential inhibitory effects of ethanol on insulin and IGF gene expression may account for region-specific differences in neuronal loss that occur in chronic alcoholics.
Although IGF-II signaling mechanisms have not been thoroughly investigated in the CNS, recent studies in other tissues and cell types demonstrated that IGF-II can mediate cell survival by interacting with its own receptor and activating PI3 kinase-Akt via G-coupled protein signaling (de la Monte and Wands, 2005). Alternatively, IGF-II can bind to insulin and IGF-I receptors, and activate growth and survival signaling pathways through IRS-dependent mechanisms (Herr et al., 2003; Zygmunt et al., 2005). Therefore, ethanol-mediated increases in IGF-II expression in the cerebellum could represent a positive compensatory response that would help promote neuronal survival in the setting of insulin and IGF-I withdrawal. Another potential interpretation of the results is that IGF-II gene expression was increased in alcoholics’ cerebellar vermis because of the relatively increased populations of glial and vascular cells because IGF-II is abundantly expressed in CNS cells of mesenchymal origin (de la Monte and Wands, 2005).
Insulin and IGF-I receptors were more abundantly expressed in the cerebellum than in the anterior cingulate gyrus, whereas IGF-II receptors were more abundant in the anterior cingulate gyrus than in the cerebellum. The major effects of chronic alcohol abuse were to reduce the levels of the insulin and IGF-II receptor expression in the anterior cingulate, and insulin and IGF-I receptor expression in the cerebellar vermis. Reduced expression of the insulin, IGF-I, and/or IGF-II receptors could reflect either loss of cells that bear these receptors, or down-regulation of receptors as a feature of neurodegeneration. Loss of insulin receptor-expressing cells in the brain could contribute to insulin resistance and result in decreased expression of insulin-responsive genes. Loss of IGF-I and IGF-II receptor-expressing cells could adversely affect neuronal survival and plasticity. For example, signaling through IGF-I and IGF-II receptors can activate PI3 kinase-Akt via IRS pathways or G-coupled protein signaling mechanisms (de la Monte and Wands, 2005). The PI3 kinase-Akt pathway has a critical role in stimulating neuronal survival and neurite outgrowth, and neuritic sprouting and reorganization are needed for plasticity associated with learning and memory (Dudek et al., 1997; Eves et al., 1998; Jackson et al., 1996; Kurihara et al., 2000; Markus et al., 2002; Virdee et al., 1999).
The studies demonstrated no significant effects of chronic alcohol abuse on the expression levels of IRS-1, IRS-2, or IRS-4, which have critical roles in transmitting growth, survival, and metabolic signals downstream from the insulin and IGF-I receptors. This result contrasts with the findings in experimental models of fetal alcohol spectrum disorders in which prominent reductions in IRS-1, IRS-2, and/or IRS-4 were detected in developing cerebella (de la Monte et al., 2005; Soscia et al., 2006). Therefore, in contrast to the effects of chronic gestational exposure to ethanol in which impairments in insulin and IGF signaling occurs at multiple levels within the cascade, in adult human alcoholics, the major abnormalities in insulin and IGF signaling are mediated by growth factor deficiency and reduced receptor expression and function (see below), i.e., proximal points within the signal transduction cascade. Similar observations were made in adult chronic ethanol fed rats (Cohen et al., 2007).
Ethanol-Impairs Insulin and IGF Receptor Binding
Effective ligand binding is critical for transmitting intracellular signals, and many of the previously reported downstream adverse effects of ethanol on insulin signaling including reduced neuronal survival were found to be mediated by impaired insulin or IGF-I binding in the CNS. Equilibrium binding assays demonstrated that chronic alcoholics have significantly reduced levels of insulin, IGF-I and IGF-II receptor binding in both the anterior cingulate gyrus and cerebellar vermis. This suggests that chronic alcohol abuse impairs insulin, IGF-1, and IGF-II signaling mechanisms in 2 different brain regions that represent important targets of ethanol neurotoxicity. The mediators of insulin signaling, i.e., insulin gene expression, insulin receptor expression, and insulin receptor binding were relatively more impaired in the anterior cingulate compared with the cerebellar vermis, and correspondingly, the expression of GAPDH, which is an insulin- regulated gene (Alexander et al., 1992), was also more prominently reduced in the anterior cingulate gyrus. The inhibition of insulin, IGF-I, and IGF-II receptor binding in the cerebellum was associated with histopathological evidence of neuronal loss, whereas in the anterior cingulate gyrus, the tissue architecture was relatively preserved despite evidence of chronic oxidative injury. These differences may have been due to the smaller reductions in IGF-I and IGF-II receptor binding in the anterior cingulate gyrus compared with the cerebellar vermis. These findings and results from previous studies suggest that ethanol inhibition of insulin and IGF signaling, which are required for cell survival and energy metabolism, is mediated at the level of receptor binding, i.e., the most proximal point within the signal transduction cascade. Moreover, the aggregate results highlight the importance of insulin as well as IGF resistance as mediators of impaired neuronal survival and function in chronic alcoholic brain disease.
Consequences of Ethanol Impaired Insulin and IGF Signaling in Relation to Brain Function
As ChAT expression is regulated by insulin and IGF-I stimulation (Rivera et al., 2005; Soscia et al., 2006), and acetylcholine is a major neurotransmitter that mediates CNS cognitive and motor functions, it was of interest to determine if the inhibitory effects of chronic alcohol abuse on insulin and IGF signaling impaired acetylcholine homeostasis in the brain. The quantitative RT-PCR studies demonstrated reduced ChAT expression in alcoholic cingulate gyrus and cerebellar vermis. Reductions in ChAT expression could result in deficits in acetylcholine biosynthesis, and without compensatory reductions in AChE expression, acetylcholine homeostasis would be adversely perturbed. In this regard, it is noteworthy that in the anterior cingulate gyrus, AChE expression was slightly reduced, whereas in the cerebellar vermis, AChE expression was somewhat increased in the alcoholics. Reduced AChE expression in the anterior cingulate gyrus may have helped preserve acetylcholine homeostasis. However, with regard to the cerebellar vermis, the increased levels of AChE, vis-à-vis reduced ChAT expression, would have further impaired acetylcholine homeostasis and thereby worsened motor deficits associated with alcoholic cerebellar degeneration.
CONCLUSIONS
The aggregate results demonstrate that chronic alcohol abuse causes neurodegeneration characterized by neuronal loss, impaired neuronal function, and increased oxidative stress/lipid peroxidation in adult human brains. Neuronal oxidative stress was more pronounced than cell loss, suggesting that many of the remaining neurons in the brain, although histologically intact, had major deficits in function as demonstrated by the significantly reduced levels of ChAT gene expression. The deficits in acetylcholine homeostasis produced by chronic alcohol abuse could impair cognitive and/or motor functions and thereby contribute to the impairments in CNS function commonly observed in chronic alcoholics. One potential limitation of this study is that it included only male alcoholics, and therefore any potential modifying effects of estrogens on IGF signaling in the brain could not be assessed. Nonetheless, the results from this study suggest that alcoholic neurodegeneration is mediated by 2 distinct but overlapping mechanisms: (1) insulin/IGF-1 resistance, which is effectuated in part by impaired receptor binding and (2) increased oxidative stress mediated by lipid peroxidation and DNA damage. It is noteworthy that nearly identical mediators of neuronal loss and impaired neuronal function were identified in association with cerebellar hypoplasia in experimental models of fetal alcohol spectrum disorders (de la Monte et al., 2005; Soscia et al., 2006) and Alzheimer-type neurodegeneration (Lester-Coll et al., 2006; de laMonte et al., 2007), as well as in human cases of AD (de la Monte and Wands, 2006; Rivera et al., 2005; Steen et al., 2005). What distinguishes alcoholic brain disease from AD is that in Alzheimer’s, the fundamental problem centers around CNS insulin and IGF withdrawal followed by degeneration and loss of cells that respond to these trophic factors. In alcoholic brain disease, insulin/IGF resistance mediated by impaired binding to the corresponding receptors results in decreased signaling through growth, survival, and metabolic cascades.
Acknowledgments
Tissues were received from the Australian Brain Donor Programs NSW Tissue Resource Centre, which is supported by The University of Sydney, National Health and Medical Research Council of Australia, Schizophrenia Research Institute, National Institute of Alcohol Abuse and Alcoholism and NSW Department of Health. This study was supported by AA-02666, AA-02169, AA-11431, AA-12908 and K24-AA-16126 from the National Institute of Alcohol Abuse and Alcoholism at the National Institutes of Health, the NSW Department of Health, and the NHMRC.
References
- Alexander BM, Dugast I, Ercolani L, Kong XF, Giere L, Nasrin N. Multiple insulin-responsive elements regulate transcription of the GAPDH gene. Adv Enzyme Regul. 1992;32:149–159. doi: 10.1016/0065-2571(92)90014-q. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Chu J, Tong M, de la Monte SM. Chronic ethanol exposure causes mitochondrial dysfunction and oxidative stress in immature central nervous system neurons. Acta Neuropathol (Berl) 2007;113:659–673. doi: 10.1007/s00401-007-0199-4. [DOI] [PubMed] [Google Scholar]
- Cohen AC, Tong M, Wands JR, de la Monte SM. Insulin and insulin-like growth factor resistance with neurodegeneration in an adult chronic ethanol exposure model. Alcohol Clin Exp Res. 2007;31:1558–1573. doi: 10.1111/j.1530-0277.2007.00450.x. [DOI] [PubMed] [Google Scholar]
- Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci U S A. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond I, Gordon AS. Cellular and molecular neuroscience of alcoholism. Physiol Rev. 1997;77:1–20. doi: 10.1152/physrev.1997.77.1.1. [DOI] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine-threonine protein kinase Akt [see comments] Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- Eves EM, Xiong W, Bellacosa A, Kennedy SG, Tsichlis PN, Rosner MR, Hay N. Akt, a target of phosphatidylinositol 3-kinase, inhibits apoptosis in a differentiating neuronal cell line. Mol Cell Biol. 1998;18:2143–2152. doi: 10.1128/mcb.18.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammeltoft S, Fehlmann M, Van OE. Insulin receptors in the mammalian central nervous system: binding characteristics and subunit structure. Biochimie. 1985;67:1147–1153. doi: 10.1016/s0300-9084(85)80113-9. [DOI] [PubMed] [Google Scholar]
- Goodyer CG, De SL, Lai WH, Guyda HJ, Posner BI. Characterization of insulin-like growth factor receptors in rat anterior pituitary, hypothalamus, and brain. Endocrinology. 1984;114:1187–1195. doi: 10.1210/endo-114-4-1187. [DOI] [PubMed] [Google Scholar]
- Guerri C. Neuroanatomical and neurophysiological mechanisms involved in central nervous system dysfunctions induced by prenatal alcohol exposure. Alcohol Clin Exp Res. 1998;22:304–312. doi: 10.1111/j.1530-0277.1998.tb03653.x. [DOI] [PubMed] [Google Scholar]
- Hallak H, Seiler AE, Green JS, Henderson A, Ross BN, Rubin R. Inhibition of insulin-like growth factor-I signaling by ethanol in neuronal cells. Alcohol Clin Exp Res. 2001;25:1058–1064. [PubMed] [Google Scholar]
- Harper C. The neuropathology of alcohol-specific brain damage, or does alcohol damage the brain? J Neuropathol Exp Neurol. 1998;57:101–110. doi: 10.1097/00005072-199802000-00001. [DOI] [PubMed] [Google Scholar]
- Harper C, Dixon G, Sheedy D, Garrick T. Neuropathological alterations in alcoholic brains. Studies arising from the New South Wales Tissue Resource Centre. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:951–961. doi: 10.1016/S0278-5846(03)00155-6. [DOI] [PubMed] [Google Scholar]
- Herr F, Liang OD, Herrero J, Lang U, Preissner KT, Han VK, Zygmunt M. Possible angiogenic roles of insulin-like growth factor II and its receptors in uterine vascular adaptation to pregnancy. J Clin Endocrinol Metab. 2003;88:4811–4817. doi: 10.1210/jc.2003-030243. [DOI] [PubMed] [Google Scholar]
- Hill JM, Lesniak MA, Pert CB, Roth J. Autoradiographic localization of insulin receptors in rat brain: prominence in olfactory and limbic areas. Neuroscience. 1986;17:1127–1138. doi: 10.1016/0306-4522(86)90082-5. [DOI] [PubMed] [Google Scholar]
- Hu JG, Fu SL, Zhang KH, Li Y, Yin L, Lu PH, Xu XM. Differential gene expression in neural stem cells and oligodendrocyte precursor cells: a cDNA microarray analysis. J Neurosci Res. 2004;78:637–646. doi: 10.1002/jnr.20317. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Bittigau P, Ishimaru MJ, Wozniak DF, Koch C, Genz K, Price MT, Stefovska V, Horster F, Tenkova T, Dikranian K, Olney JW. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science. 2000;287:1056–1060. doi: 10.1126/science.287.5455.1056. [DOI] [PubMed] [Google Scholar]
- Ito D, Tanaka K, Suzuki S, Dembo T, Fukuuchi Y. Enhanced expression of Iba1, ionized calcium-binding adapter molecule 1, after transient focal cerebral ischemia in rat brain. Stroke. 2001;32:1208–1215. doi: 10.1161/01.str.32.5.1208. [DOI] [PubMed] [Google Scholar]
- Jackson TR, Blader IJ, Hammonds OL, Burga CR, Cooke F, Hawkins PT, Wolf AG, Heldman KA, Theibert AB. Initiation and maintenance of NGF-stimulated neurite outgrowth requires activation of a phosphoinositide 3-kinase. J Cell Sci. 1996;109(Pt 2):289–300. doi: 10.1242/jcs.109.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Takeda A, Onodera H, Kimpara T, Hisanaga K, Sato N, Nunomura A, Castellani RJ, Perry G, Smith MA, Itoyama Y. Systemic increase of oxidative nucleic acid damage in Parkinson’s disease and multiple system atrophy. Neurobiol Dis. 2002;9:244–248. doi: 10.1006/nbdi.2002.0466. [DOI] [PubMed] [Google Scholar]
- Kulik G, Klippel A, Weber MJ. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Kitagawa Y, Hirose G, Sakai K. Antibody recognition and RNA binding of a neuronal nuclear autoantigen associated with paraneoplastic neurological syndromes and small cell lung carcinoma. J Neuroimmunol. 1999;93:37–44. doi: 10.1016/s0165-5728(98)00184-2. [DOI] [PubMed] [Google Scholar]
- Kurihara S, Hakuno F, Takahashi S. Insulin-like growth factor-I-dependent signal transduction pathways leading to the induction of cell growth and differentiation of human neuroblastoma cell line SH-SY5Y: the roles of MAP kinase pathway and PI 3-kinase pathway. Endocr J. 2000;47:739– 751. doi: 10.1507/endocrj.47.739. [DOI] [PubMed] [Google Scholar]
- Lam K, Carpenter CL, Ruderman NB, Friel JC, Kelly KL. The phosphatidylinositol 3-kinase serine kinase phosphorylates IRS-1. Stimulation by insulin and inhibition by Wortmannin. J Biol Chem. 1994;269:20648–20652. [PubMed] [Google Scholar]
- Lester-Coll N, Rivera EJ, Soscia SJ, Doiron K, Wands JR, de la Monte SM. Intracerebral streptozotocin model of type 3 diabetes: relevance to sporadic Alzheimer’s disease. J Alzheimers Dis. 2006;9:13–33. doi: 10.3233/jad-2006-9102. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574– 1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- de la Monte SM. Disproportionate atrophy of cerebral white matter in chronic alcoholics. Arch Neurol. 1988;45:990–992. doi: 10.1001/archneur.1988.00520330076013. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Ganju N, Banerjee K, Brown NV, Luong T, Wands JR. Partial rescue of ethanol-induced neuronal apoptosis by growth factor activation of phosphoinositol-3-kinase. Alcohol Clin Exp Res. 2000;24:716–726. [PubMed] [Google Scholar]
- de la Monte SM, Jhaveri A, Maron BA, Wands JR. Nitric oxide synthase 3-mediated neurodegeneration after intracerebral gene delivery. J Neuropathol Exp Neurol. 2007;66:272–283. doi: 10.1097/nen.0b013e318040cfa2. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Neely TR, Cannon J, Wands JR. Ethanol impairs insulin-stimulated mitochondrial function in cerebellar granule neurons. Cell Mol Life Sci. 2001;58:1950–1960. doi: 10.1007/PL00000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte SM, Tong M, Lester-Coll N, Plater M, Jr, Wands JR. Therapeutic rescue of neurodegeneration in experimental type 3 diabetes: Relevance to Alzheimer’s disease. J Alzheimers Dis. 2006;10:89–109. doi: 10.3233/jad-2006-10113. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Wands JR. Mitochondrial DNA damage and impaired mitochondrial function contribute to apoptosis of insulin-stimulated ethanol-exposed neuronal cells. Alcohol Clin Exp Res. 2001;25:898–906. [PubMed] [Google Scholar]
- de la Monte SM, Wands JR. Chronic gestational exposure to ethanol impairs insulin-stimulated survival and mitochondrial function in cerebellar neurons. Cell Mol Life Sci. 2002;59:882–893. doi: 10.1007/s00018-002-8475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte SM, Wands JR. Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer’s disease. J Alzheimers Dis. 2005;7:45–61. doi: 10.3233/jad-2005-7106. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Wands JR. Molecular indices of oxidative stress and mitochondrial dysfunction occur early and often progress with severity of Alzheimer’s disease. J Alzheimers Dis. 2006;9:167–181. doi: 10.3233/jad-2006-9209. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Xu XJ, Wands JR. Ethanol inhibits insulin expression and actions in the developing brain. Cell Mol Life Sci. 2005;62:1131–1145. doi: 10.1007/s00018-005-4571-z. [DOI] [PubMed] [Google Scholar]
- Myers MG, White MF. The new elements of insulin signaling. Insulin receptor substrate-1 and proteins with SH2 domains. Diabetes. 1993;42:643–650. doi: 10.2337/diab.42.5.643. [DOI] [PubMed] [Google Scholar]
- Patel TB. Single transmembrane spanning heterotrimeric g protein-coupled receptors and their signaling cascades. Pharmacol Rev. 2004;56:371–385. doi: 10.1124/pr.56.3.4. [DOI] [PubMed] [Google Scholar]
- Payao SL, Smith MA, Winter LM, Bertolucci PH. Ribosomal RNA in Alzheimer’s disease and aging. Mech Ageing Dev. 1998;105:265–272. doi: 10.1016/s0047-6374(98)00095-5. [DOI] [PubMed] [Google Scholar]
- Puro DG, Agardh E. Insulin-mediated regulation of neuronal maturation. Science. 1984;225:1170–1172. doi: 10.1126/science.6089343. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Perez A, Chen J, Senthil D, Schenker S, Henderson GI. In utero ethanol exposure causes mitochondrial dysfunction, which can result in apoptotic cell death in fetal brain: a potential role for 4- hydroxynonenal. Alcohol Clin Exp Res. 2001;25:862–871. [PubMed] [Google Scholar]
- Rivera EJ, Goldin A, Fulmer N, Tavares R, Wands JR, de la Monte SM. Insulin and insulin-like growth factor expression and function deteriorate with progression of Alzheimer’s disease: link to brain reductions in acetylcholine. J Alzheimers Dis. 2005;8:247–268. doi: 10.3233/jad-2005-8304. [DOI] [PubMed] [Google Scholar]
- da Silva AM, Payao SL, Borsatto B, Bertolucci PH, Smith MA. Quantitative evaluation of the rRNA in Alzheimer’s disease. Mech Ageing Dev. 2000;120:57–64. doi: 10.1016/s0047-6374(00)00180-9. [DOI] [PubMed] [Google Scholar]
- Soscia SJ, Tong M, Xu XJ, Cohen AC, Chu J, Wands JR, de la Monte SM. Chronic gestational exposure to ethanol causes insulin and IGF resistance and impairs acetylcholine homeostasis in the brain. Cell Mol Life Sci. 2006;63:2039–2056. doi: 10.1007/s00018-006-6208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, de la Monte SM. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease—is this type 3 diabetes? J Alzheimers Dis. 2005;7:63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Crimmins DL, Myers MJ, Miralpeix M, White MF. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol. 1993;13:7418–7428. doi: 10.1128/mcb.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- Virdee K, Xue L, Hemmings BA, Goemans C, Heumann R, Tolkovsky AM. Nerve growth factor-induced PKB/Akt activity is sustained by phosphoinositide 3-kinase dependent and independent signals in sympathetic neurons. Brain Res. 1999;837:127–142. doi: 10.1016/s0006-8993(99)01643-1. [DOI] [PubMed] [Google Scholar]
- Xu J, Yeon JE, Chang H, Tison G, Chen GJ, Wands J, de la Monte S. Ethanol impairs insulin-stimulated neuronal survival in the developing brain: role of PTEN phosphatase. J Biol Chem. 2003;278:26929–26937. doi: 10.1074/jbc.M300401200. [DOI] [PubMed] [Google Scholar]
- Zhang FX, Rubin R, Rooney TA. Ethanol induces apoptosis in cerebellar granule neurons by inhibiting insulin-like growth factor 1 signaling. J Neurochem. 1998;71:196–204. doi: 10.1046/j.1471-4159.1998.71010196.x. [DOI] [PubMed] [Google Scholar]
- Zygmunt M, McKinnon T, Herr F, Lala PK, Han VK. HCG increases trophoblast migration in vitro via the insulin-like growth factor-II/mannose-6 phosphate receptor. Mol Hum Reprod. 2005;11:261–267. doi: 10.1093/molehr/gah160. [DOI] [PubMed] [Google Scholar]