Abstract
Purpose
Somatic mutations in the epidermal growth factor receptor (EGFR) gene occur in a subset of non – small-cell lung cancer (NSCLC) and are highly predictive of the clinical response to selective EGFR kinase inhibitors.The prevalence of EGFR-mutant NSCLC is appreciably higher in females than in males and in East Asian than in Caucasian populations. We hypothesized that these disparate frequencies may be attributable to underlying genetic modifiers. Given the coincident differences in sex and ethnic origin, we tested allozymatic variants of enzymes involved in estrogen biosynthesis and metabolism, encoded by polymorphic alleles known to differ in frequency between Caucasian and Asian populations, as modifying alleles.
Experimental Design
We genotyped nine polymorphisms in the CYP1A1, CYP17A1, CYP19, HSD17B1, COMT, GSTM1, and GSTT1 genes, in a series of 100 Japanese NSCLCs, selected for equal representation of EGFR wild-type (wt) and EGFR-mutant cases, as well as male and female cases. Associations between polymorphic variants and the EGFR genotype and sex of NSCLC cases were examined using Fisher’s exact test of significance.
Results
Only CYP1A1*2C showed a difference in allele frequency that approached statistical significance. Heterozygotes were underrepresented among EGFR-mutant cases compared with EGFR-wt cases (27% versus 47%, P = 0.08), with a concurrent trend toward overrepresentation of CYP1A1*2CIle/Ile homozygotes among EGFR-mutant cases as compared with EGFR-wt cases (69% versus 51%, P = 0.13).
Conclusion
Within the power of this study, our findings suggest that the selected polymorphic variants in the estrogen biosynthesis and metabolism pathways are unlikely to be major genetic modifiers of the prevalence of EGFR-mutant NSCLC.
The clinical response of non–small-cell lung cancer (NSCLC) patients to small-molecule inhibitors of the epidermal growth factor receptor (EGFR), such as gefitinib or erlotinib, is determined by the presence of specific somatic mutations in the kinase domain of EGFR (1–3). More than 90% of mutations described to date consist of in-frame deletions in exon 19 or an amino acid substitution (L858R) in exon 21, leading to selective activation of the receptor and downstream antiapoptotic pathways (4–6). EGFR kinase domain mutations occur at a significantly higher frequency in tumors from East Asians than from non-Asians (30% versus 8%, P < 0.001), from women than from men (59% versus 26%, P < 0.001), from never smokers than from ever smokers (66% versus 22%, P < 0.001), and in adenocarcinomas compared with other NSCLC histologies (49% versus 2%, P < 0.001; refs. 7, 8). The association of smoking-related NSCLCs with Kras mutations and the mutually exclusive nature of Kras and EGFR mutations provide a plausible explanation for the elevated frequency of EGFR mutations in NSCLCs from nonsmokers (3, 7–9). However, although no satisfactory explanation of the observed ethnic and sex-related differences has been uncovered, it has been noted that longer reproductive life span, associated with higher endogenous estrogen exposure, is a risk factor for EGFR-mutant NSCLC (10).
One possible explanation for interethnic differences in EGFR mutation rates would be differing environmental exposures. Large studies to determine the mutation prevalence among second- or later-generation East Asian NSCLC cases living outside the Pacific Rim are lacking. Nonetheless, based on the relatively small number of cases studied to date, the prevalence of EGFR mutations remains higher among Asian cases residing outside the Pacific Rim than among their non-Asian counter-parts. Genetic variation may therefore contribute to interethnic differences in mutation prevalence (8, 11, 12).
To date, the search for germ-line variants that may act as genetic modifiers of susceptibility to EGFR-mutant NSCLC has been limited to naturally occurring variants in the receptor itself. Three polymorphic variants in EGFR, a (CA)n repeat in intron 1 (CA-SSR1) and two SNPs (−216G/T and −191C/A) in the promoter region, reportedly influence EGFR expression (13, 14), and interethnic differences in allele frequencies at all three positions have been noted between East Asian and other populations (14, 15). These polymorphisms may affect the expression levels of mutant EGFR alleles (16, 17). However, no association has been noted between specific EGFR haplotypes and intragenic mutations in the EGFR kinase domain (12).
Given that EGFR-mutated lung tumors occur more frequently in both East Asians and in women and are associated with a longer period of fertility, we considered the possibility that estrogen levels might contribute to the differences in EGFR mutation frequency among these two demographic subgroups. Estrogen levels are maintained by a balance in estrogen biosynthesis and metabolism. These complex biochemical processes are regulated by a number of genes, some of which encode allozymatic variants with variable functional activities leading to interindividual variation in estrogen levels. Furthermore, several allozymatic variants are present at significantly higher frequency in East Asian populations compared with Caucasian populations. We hypothesized that polymorphisms that alter enzymatic activity and occur at significantly different frequencies in East Asian and Caucasian populations might contribute to the greater prevalence of EGFR-mutant NSCLC in women and in East Asians. Such an effect could arise through a number of mechanisms: Altered estrogen levels could enhance the development of a form of lung cancer that is marked by the subsequent somatic acquisition of EGFR mutations. Alternatively, estrogen effects could directly modulate the functional properties of mutant EGFRs to enhance their tumorigenic properties. Here, we compare the frequency of functional allelic variants of genes, in the estrogen biosynthesis and metabolism pathways, in EGFR-mutant and EGFR wild-type (wt) NSCLCs from Japan.
Materials and Methods
Clinical material
Tumors were resected from patients undergoing treatment for NSCLC, with an adenocarcinoma histology, at Aichi Cancer Center, Japan. Surgical tissue was snap-frozen in liquid nitrogen at the time of resection and subsequently stored at −80°C, after obtaining written informed consent and Institutional Review Board approval. Tissues were anonymized after recording demographic and clinicopathologic features. Exons 18 to 24 of the EGFR gene were genotyped for recurrent EGFR mutations previously associated with clinical sensitivity to EGFR tyrosine kinase inhibitors. Of the 435 consecutive NSCLC cases, 100 cases of adenocarcinoma were retro-spectively selected for study based on the EGFR genotype of tumors. The study cohort was composed of equal numbers of wt (n = 50) and mutant (n = 50) EGFR cases, and in each genotypic subgroup, cases were further selected for an equal representation of males and females, individually matched for smoking history (ever smokers and never smokers) and age.
Genotyping
DNA was isolated from tumor specimens by standard phenol-chloroform extraction. Exons 18 to 21 of EGFR were PCR amplified and subjected to automated nucleotide sequencing as described previously (7, 10). Tumor DNAs were genotyped for the CYP1A1*2A6235T/C (rs4646903), CYP1A1*2CIle462Val (rs1048942), CYP17A1−34T/C (rs743572), CYP19A1Arg2 64C ys (rs700519), HSD17B1Gly312Ser (rs605059), COMTVal158Met (rs4680), GSTM1, and GSTT1 polymorphic variants. PCR amplification was done using primer pairs (5′-3′) for CYP1A1*2A6235T/C (GCAGTGAAGAGGTG-TAGCCGCTG and GATTAGGAGTCTTGTCTCATGCCTG), CYP1A1* 2CIle462Val (CCAGTGGCAGATCAACCATGACC and CTAAGAGCG-CAGCTGCATTTGGAAG), CYP19A1Arg264Cys (CCTTAACATGAAGTG-TAGGGTCTATG and CTACACAGTCATAACATATGTGGC), HSD17B1Gly312Ser (GGATGCGCCTGGACGACCCCAGC and GCGCTGGTAAAACTGGCTAACGC), COMTVal158Met (GCAAGATCGTG-GACGCCGTGATTC and CTTTAGGGTTCTGGGATGACAAGG), GSTM1 (GAACTCCCTGAAAAGCTAAAGC and GTTGGGCTCAAATA-TACGGTGG), and GSTT1 (TTCCTTACTGGTCCTCACATCTC and TCACCGGATCATGGCCAGCA). Amplification consisted of 40 cycles with annealing temperatures of 52°C (CYP19A1Arg264Cys), 54°C (CYP17A1−34T/C), 58°C (CYP1A1*2A6235T/C, CYP1A1*2CIle462Val), and 61°C (HSD17B1Gly312Ser). Null alleles of GSTM1 and GSTT1 were genotyped by multiplex PCR, with the β-globin gene as a positive control for amplification, according to published conditions (18). Amplicons surrounding the CYP19A1(TTTA)7-13 tetranucleotide repeat were genotyped by size separation on a Wavemaker system (Transgenomic, Inc.).
Statistical analysis
A two-sided Fisher’s exact test of significance was used to evaluate the associations between polymorphic variants of CYP1A1*2A6235T/C, CYP1A1*2CIle462Val, CYP17A1−34T/C, CYP19A1Arg264Cys, CYP19A1(TTTA)7-13, HSD17B1Gly312Ser, COMTVal158Met, GSTM1, and GSTT1 and the EGFR genotype or sex of NSCLC cases.
Results and Discussion
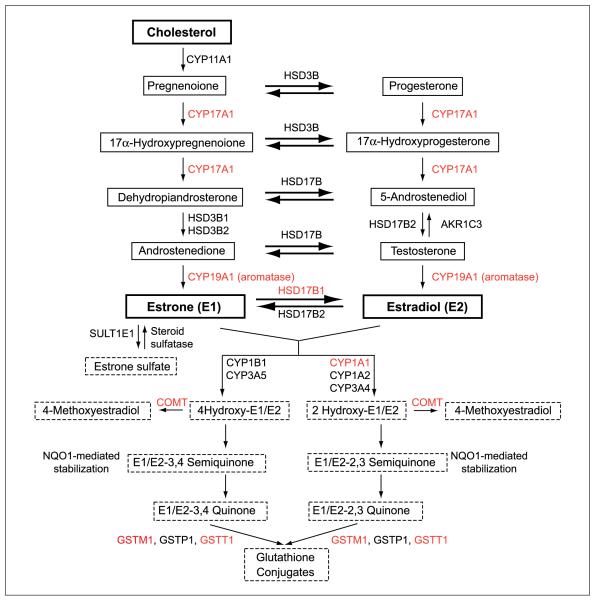
The enzymatic conversion of cholesterol to the estrogens 17β-estradiol and estrone is a multistep process catalyzed by cytochrome P450 (CYP), 3β-hydroxysteroid dehydrogenase (HSD3B), and 17β-hydroxysteroid dehydrogenase (HSD17B) family members (Fig. 1), primarily occurring in the granulosa cells of the ovarian follicles in premenopausal women (19). In contrast, the primary source of estrogen in postmenopausal women is provided by the conversion of androgens into estrogen by aromatase (CYP19) in adipose tissue (Fig. 1).
Fig. 1.
Schematic representation of estrogen biosynthesis and metabolism. The biosynthesis (solid boxes) of estrone and estradiol from cholesterol and their subsequent metabolism (dashed boxes) are sequential processes catalyzed by the action of a number of enzymes including allozymes of the cytochrome P450 (CYP) superfamily, 3β-hydroxysteroid dehydrogenase (HSD3B), HSD17B, COMT, and GSTs. Red, enzymatic variants analyzed in this study.
Estrogen metabolism is a multistep process beginning with CYP1A1-mediated hydroxylation at the 2- or 4-position to produce 2- or 4-hydroxy cathechol estrogens that undergo further oxidation to quinones and semiquinones, respectively (20). Quinones are reportedly mutagenic, being capable of forming stable DNA adducts or depurinating adducts. The inactivation of estrogens, cathechol estrogens, quinones, and semiquinones is achieved by methylation and subsequent sulfation and conjugation, catalyzed by cathecol-O-methyl-transferase (COMT), the glutathione S-transferase (GST) superfamily, and sulfotransferases, respectively (Fig. 1; ref. 19). Among the enzymes participating in estrogen biosynthesis and metabolism, several have naturally occurring allozymatic variants that, in some instances, exhibit differential catalytic activity (21–35). Thus, the genotype of an individual may influence overall tissue estrogen levels. Furthermore, significant interethnic differences in allele frequencies between Caucasian and East Asian populations have been documented for a number of variants (20, 36–41).
To test for an association between polymorphic variants in genes influencing estrogen biosynthesis and metabolism and the prevalence of EGFR-mutant NSCLCs, we genotyped the CYP1A1*2A−6235T/C, CYP1A1*2CIle462Val, CYP17A1−34T/C, CYP19A1 Arg264Cys, CYP19A1 (TTTA)n, HSD17B1 Gly312Ser, COMTVal158Met, GSTM1, and GSTT1 allelic variants within a series of 100 NSCLCs from Japan (Table 1). These specific variants were chosen for analysis because they encode allozymes with reported differential functional activity and/or ethnic distributions among Caucasian and East Asian populations (Table 1). We focused our analysis on NSCLCs from Japan because the higher frequency of EGFR-mutant lung cancer in East Asia would enhance detection of any bias in the prevalence of these polymorphisms between cases with or without EGFR mutations. NSCLC cases were preselected for equal representation of EGFR-wt and EGFR-mutant genotypes and cases arising in males and females.
Table 1.
Frequency and differential activity of allele variants controlling estrogen biosynthesis and metabolism
| Gene | Variant | Observed frequency in NSCLCs (Asian) |
Reported population frequency |
P (Asian vs Caucasian) |
Differential allozymatic activity |
References | |
|---|---|---|---|---|---|---|---|
| Asian | Caucasian | ||||||
|
CYP1A1*2A−6235T/C (rs4646903) |
T | 0.68 | 0.58 | 0.91 | <0.0001 | Minor allele: increased enzymatic activity |
(20, 21) |
| C* | 0.32 | 0.42 | 0.09 | ||||
| n | 200 | 417 | 554 | ||||
|
CYP1A1*2CIle462Val (rs1048943) |
A | 0.78 | 0.74 | 0.95 | <0.0001 | Minor allele: increased inducibility to produce 2-hydroxycatecholestrogens |
(20, 22) |
| G* | 0.22 | 0.26 | 0.05 | ||||
| n | 180 | 417 | 1,920 | ||||
|
CYP17A1−34T/C (rs743572) |
T | 0.56 | 0.49 | 0.50 | Minor allele: elevated expression |
(23, 24, 39) | |
| C* | 0.44 | 0.51 | 0.50 | ||||
| n | 200 | 3,023 | 1,950 | ||||
|
CYP19A1Arg264Cys (rs700519) |
C | 0.68 | 0.87 | 0.96 | None reported | (40, 41) | |
| T | 0.32 | 0.13 | 0.04 | ||||
| n | 200 | 2,052 | 1,614 | ||||
| CYP19A1 (TTTA)n | (TTTA)7 or 7-3 | 0.46 | 0.69 | 0.49 | <0.0001 | None reported | (20, 25, 27–31) |
| (TTTA)8+* | 0.54 | 0.31 | 0.51 | ||||
| n | 186 | 376 | 1,890 | ||||
|
HSD17B1Gly312Ser (rs605059) |
G | 0.54 | 0.46 | 0.47 | None reported | (32, 33, 36) | |
| A* | 0.46 | 0.54 | 0.53 | ||||
| n | 200 | 918 | 6,829 | ||||
|
COMTVal158Met (rs4680) |
G | 0.70 | 0.72 | 0.47 | <0.0001 | Minor allele: decreased enzymatic activity |
(20, 34) |
| A* | 0.30 | 0.28 | 0.53 | ||||
| n | 200 | 453 | 1,379 | ||||
| GSTM1 wt/null | Present | – | – | – | Minor allele: no activity (null allele) |
(35, 37) | |
| Null (−/−)* | 0.46 | 0.53 | 0.50 | ||||
| n | 200 | 2,787 | 6,000 | ||||
| GSTT1 wt/null | Present | – | – | – | Minor allele: no activity (null allele) |
(36, 39) | |
| Null (−/−)* | 0.38 | 0.54 | 0.18 | ||||
| n | 200 | 847 | 1,363 | ||||
Minor allele.
Overall, our study cohort seemed to be representative of the Japanese population based on a comparison of the allele frequencies of the variants analyzed here with the reported allele frequencies (Table 1).
A comparison of polymorphic genotype distributions between EGFR-wt and EGFR-mutant NSCLC cases revealed a difference for only one estrogen-related variant, CYP1A1*2C (rs1048943; Table 2). We observed an underrepresentation of CYP1A1*2C heterozygotes (Ile/Val) among EGFR-mutant cases as compared with EGFR-wt cases, although this did not reach statistical significance (27% versus 47%; P = 0.08, two-tailed Fisher’s exact test). We also detected a concurrent overrepresentation of CYP1A1*2C Ile/Ile homozygotes among EGFR-mutant cases compared with EGFR-wt cases, although this, too, did not reach statistical significance (69% versus 51%, P = 0.13). The CYP1A1*2C Val genotype (minor G allele) is more prevalent in the East Asian population compared with the Caucasian population (Table 1) and has been linked to increased production of the 2-hydroxycatecholestrogen metabolite. Thus, whereas interethnic prevalence might favor an association between the CYP1A1*2C Val genotype and EGFR-mutant NSCLC, our analysis of Japanese tumors, in fact, suggests that the CYP1A1*2C Ile genotype was more frequently observed in EGFR-mutant cases. Breakdown of the analysis among male versus female cases did not further support a bias in favor of either CYP1A1*2C variant by sex, with both males and females displaying the trends observed in the combined group (Table 2). We also observed a trend toward an increase (35-57%) in the frequency of CYP19A1(TTTA)7 homozygotes in EGFR-mutant female cases compared with wt female cases, although this did not reach statistical significance (Table 2). A similar trend for CYP19A1(TTTA)7 was not observed in males. The CYP19A1(TTTA)7 allele occurs at higher frequency in Asian than non-Asian populations (0.69 versus 0.49; Table 1); however, to our knowledge, there is currently no known effect of repeat length of this intronic polymorphism on aromatase activity. No other polymorphism tested, by itself suggested a significant difference in prevalence between Japanese EGFR-mutant and EGFR-wt NSCLC. We note, however, that estrogen levels are modulated by the concerted action of a number of enzymes, and that the limited number of samples prevented analysis of combinatorial associations of all genotypes with respect to EGFR status and sex. In theory, an analysis of 50 wt cases and 50 mutant cases has 80% power to detect differences of ≥28%. Accordingly, larger studies will be required to confirm our observations.
Table 2.
Genotype of NSCLCs by EGFR mutation status and sex
| Gene | Variant | % of NSCLC cases (n) |
|||||
|---|---|---|---|---|---|---|---|
|
EGFR wt |
EGFR mutant |
||||||
| Male | Female | Total | Male | Female | Total | ||
| CYP1A1*2A−6235T/C (rs4646903) | T-T | 33 (9) | 43 (10) | 38 (19) | 55 (15) | 35 (8) | 46 (23) |
| T-C | 63 (17) | 48 (11) | 56 (28) | 41 (11) | 52 (12) | 46 (23) | |
| C-C | 4 (1) | 9 (2) | 6 (3) | 4 (1) | 13 (3) | 8 (4) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| CYP1A1*2C Ile462Val (rs1048943) | A-A | 48 (11) | 55 (12) | 51 (23) | 74 (17) | 64 (14) | 69 (31) |
| A-G | 48 (11) | 45 (10) | 47 (21) | 26 (6) | 27 (6) | 27 (12) | |
| G-G | 4 (1) | 0 (0) | 2 (1) | 0 (0) | 9 (2) | 5 (2) | |
| Total | 23 | 22 | 45 | 23 | 22 | 45 | |
| CYP17A1−34T/C (rs743572) | T-T | 33 (9) | 35 (8) | 34 (17) | 30 (8) | 30 (7) | 30 (15) |
| C-T | 52 (12) | 48 (11) | 46 (23) | 40 (11) | 61 (14) | 50 (25) | |
| C-C | 22 (6) | 17 (4) | 20 (10) | 30 (8) | 9 (2) | 20 (10) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| CYP19A1 Arg264Cys (rs700519) | C-C | 44 (12) | 57 (13) | 50 (25) | 40 (11) | 48 (11) | 44 (22) |
| C-T | 44 (12) | 39 (9) | 42 (21) | 48 (13) | 39 (9) | 42 (22) | |
| T-T | 12 (3) | 4 (1) | 8 (4) | 12 (3) | 13 (3) | 12 (6) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| CYP19A1 (TTTA)n | (TTTA)7 | 42 (10) | 35 (8) | 38 (18) | 43 (10) | 57 (13) | 50 (23) |
| (TTTA)7/8+ | 0 (0) | 9 (2) | 4 (2) | 0 (0) | 4 (1) | 2 (1) | |
| (TTTA)8+/8+ | 58 (14) | 56 (13) | 57 (27) | 57 (13) | 39 (9) | 48 (22) | |
| Total | 24 | 23 | 47 | 23 | 23 | 46 | |
| HSD17B1Gly312Ser (rs605059) | G-G | 22 (6) | 30 (7) | 26 (13) | 41 (11) | 35 (8) | 38 (19) |
| G-A | 56 (15) | 40 (9) | 48 (24) | 37 (10) | 48 (11) | 42 (21) | |
| A-A | 22 (6) | 30 (7) | 26 (13) | 22 (6) | 17 (4) | 20 (10) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| COMTVal158Met (rs4680) | G-G | 37 (10) | 61 (14) | 48 (24) | 55 (15) | 52 (12) | 54 (27) |
| G-A | 44 (12) | 30 (7) | 38 (19) | 45 (12) | 35 (8) | 40 (20) | |
| A-A | 19 (5) | 9 (2) | 14 (7) | 0 (0) | 13 (3) | 6 (3) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| GSTM1 wt/null | +/+ or +/− | 59 (16) | 34 (8) | 48 (24) | 67 (18) | 52 (12) | 60 (30) |
| Null allele | 41 (11) | 66 (15) | 52 (26) | 33 (9) | 48 (11) | 40 (20) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
| GSTT1 wt/null | +/+ or +/− | 63 (17) | 61 (14) | 62 (31) | 63 (17) | 61 (14) | 62 (31) |
| Null allele | 37 (10) | 39 (9) | 38 (19) | 37 (10) | 39 (9) | 38 (19) | |
| Total | 27 | 23 | 50 | 27 | 23 | 50 | |
In conclusion, our findings suggest that the selected functionally and/or ethnically distinct variants in the estrogen biosynthesis and metabolism pathways are unlikely to be major genetic determinants of the coincident sex and ethnic bias of EGFR mutations in NSCLC. Our analysis sets the stage for larger population-based studies aimed at defining genome-wide associations that may underlie the genetic contributions toward the strikingly coincident ethnic and sex bias in the prevalence of EGFR-mutant lung cancer (10).
Acknowledgments
We thank Dr. Shyamala Maheswaran for helpful discussions. D.W. Bell acknowledges the Intramural Program of the National Human Genome Research Institute at NIH. Denaturing high-performance liquid chromatography assays were done by Transgenomic, Inc. (Omaha, NE).
Grant support: NIH grants R01CA115830-02 (J. Settleman and D.A. Haber) and P01CA95281 (D.A. Haber and D.W. Bell), the V Foundation Award (J. Settleman), and the Doris Duke Foundation Distinguished Clinical Investigator for Cancer Research Award (D.A. Haber).
Footnotes
Note: Current address for D.W. Bell: Cancer Genetics Branch, National Human Genome Research Institute/NIH, Bethesda, MD 20892. Current address for R. Sordella: Cold Spring Harbor Laboratory, Cold Spring Harbor, NY11724.
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers”and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 5.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations inlung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Greulich H, Jänne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–74. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–23. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 9.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–53. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo K, Ito H, Yatabe Y, et al. Risk factors differ for non-small-cell lung cancers with and without EGFR mutation: assessment of smoking and sex by a case-control study in Japanese. Cancer Sci. 2007;98:96–101. doi: 10.1111/j.1349-7006.2006.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequist LV, Joshi VA, Janne PA, et al. Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing. Oncologist. 2007;12:90–8. doi: 10.1634/theoncologist.12-1-90. [DOI] [PubMed] [Google Scholar]
- 12.Sellers WR, Meyerson M. EGFR gene mutations: a call for global × global views of cancer. J Natl Cancer Inst. 2005;97:326–8. doi: 10.1093/jnci/dji079. [DOI] [PubMed] [Google Scholar]
- 13.Gebhardt F, Bürger H, Brandt B. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations-a link between genetics and epigenetics. Histol Histopathol. 2000;15:929–36. doi: 10.14670/HH-15.929. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Innocenti F, Wu MH, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 15.Liu W, Innocenti F, Chen P, et al. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–12. [PubMed] [Google Scholar]
- 16.Ichihara S, Toyooka S, Fujiwara Y, et al. The impact of epidermal growth factor receptor gene status on gefitinib-treated Japanese patients with non-small-cell lung cancer. Int J Cancer. 2007;120:1239–47. doi: 10.1002/ijc.22513. [DOI] [PubMed] [Google Scholar]
- 17.Nie Q, Wang Z, Zhang GC, et al. The epidermal growth factor receptor intron1 (CA)n microsatellite polymorphism is a potential predictor of treatment outcome in patients with advanced lung cancer treated with Gefitinib. Eur J Pharmacol. 2007;570:175–81. doi: 10.1016/j.ejphar.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Chen CL, Liu Q, Pui CH, et al. Higher frequency of glutathione S-transferase deletions in black children with acute lymphoblastic leukemia. Blood. 1997;89:1701–7. [PubMed] [Google Scholar]
- 19.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi Y, Noguchi S. Polymorphisms of estrogen synthesizing and metabolizing genes and breast cancer risk in Japanese women. Biomed Pharmacother. 2003;57:471–81. doi: 10.1016/j.biopha.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Petersen DD, McKinney CE, Ikeya K, et al. Human CYP1A1gene: cosegregation of the enzyme inducibility phenotype and an RFLP. Am J Hum Genet. 1991;48:720–5. [PMC free article] [PubMed] [Google Scholar]
- 22.Cosma G, Crofts F, Taioli E, Toniolo P, Garte S. Relationship between genotype and function of the human CYP1A1 gene. J Toxicol Environ Health. 1993;40:309–16. doi: 10.1080/15287399309531796. [DOI] [PubMed] [Google Scholar]
- 23.Carey AH, Waterworth D, Patel K, et al. Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet. 1994;3:1873–6. doi: 10.1093/hmg/3.10.1873. [DOI] [PubMed] [Google Scholar]
- 24.Nedelcheva Kristensen V, Haraldsen EK, Anderson KB, et al. CYP17 and breast cancer risk: the polymorphism in the 5′ flanking area of the gene does not influence binding to Sp-1. Cancer Res. 1999;59:2825–8. [PubMed] [Google Scholar]
- 25.Garcia-Closas M, Herbstman J, Schiffman M, Glass A, Dorgan JF. Relationship between serum hormone concentrations, reproductive history, alcohol consumption and genetic polymorphisms in pre-menopausal women. Int J Cancer. 2002;102:172–8. doi: 10.1002/ijc.10651. [DOI] [PubMed] [Google Scholar]
- 26.Travis RC, Churchman M, Edwards SA, et al. No association of polymorphisms in CYP17, CYP19, and HSD17-1 with plasma estradiol concentrations in 1,090 British women. Cancer Epidemiol Biomarkers Prev. 2004;13:2282–4. [PubMed] [Google Scholar]
- 27.Salmen T, Heikkinen AM, Mahonen A, et al. Relation of aromatase gene polymorphism and hormone replacement therapy to serum estradiol levels, bone mineral density, and fracture risk in early postmenopausal women. Ann Med. 2003;35:282–8. doi: 10.1080/07853890310006370. [DOI] [PubMed] [Google Scholar]
- 28.Haiman CA, Hankinson SE, Spiegelman D, et al. A tetranucleotide repeat polymorphism in CYP19 and breast cancer risk. Int J Cancer. 2000;87:204–10. [PubMed] [Google Scholar]
- 29.Tworoger SS, Chubak J, Aiello EJ, et al. Association of CYP17, CYP19, CYP1B1, and COMT polymorphisms with serum and urinary sex hormone concentrations in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2004;13:94–101. doi: 10.1158/1055-9965.epi-03-0026. [DOI] [PubMed] [Google Scholar]
- 30.Dick IM, Devine A, Prince RL. Association of an aromataseTTTA repeat polymorphism with circulating estrogen, bone structure, and biochemistry in older women. Am J Physiol Endocrinol Metab. 2005;288:E989–95. doi: 10.1152/ajpendo.00550.2004. [DOI] [PubMed] [Google Scholar]
- 31.Puranen TJ, Poutanen MH, Peltoketo HE, Vihko PT, Vihko RK. Site-directed mutagenesis of the putative active site of human 17β-hydroxysteroid dehydrogenase type 1. Biochem J. 1994;304:289–93. doi: 10.1042/bj3040289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setiawan VW, Hankinson SE, Colditz GA, Hunter DJ, DeVivo I. HSD17B1gene polymorphisms and risk of endometrial and breast cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:213–9. doi: 10.1158/1055-9965.epi-03-0241. [DOI] [PubMed] [Google Scholar]
- 33.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyl-transferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–50. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Brockmöller J, Gross D, Kerb R, Drakoulis N, Roots I. Correlation between trans-stilbene oxide-glutathione conjugation activity and the deletion mutation in the glutathione S-transferase class α gene detected by polymerase chain reaction. Biochem Pharmacol. 1992;43:647–50. doi: 10.1016/0006-2952(92)90591-6. [DOI] [PubMed] [Google Scholar]
- 35.Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase θ (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271–6. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraft P, Pharoah P, Chanock SJ, et al. Genetic variation in the HSD17B1gene and risk of prostate cancer. PLoS Genet. 2005;1:e68. doi: 10.1371/journal.pgen.0010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benhamou S, Lee WJ, Alexandrie AK, et al. Meta- and pooled analyses of the effects of glutathione S-transferase M1 polymorphisms and smoking on lung cancer risk. Carcinogenesis. 2002;23:1343–50. doi: 10.1093/carcin/23.8.1343. [DOI] [PubMed] [Google Scholar]
- 38.Raimondi S, Paracchini V, Autrup H, et al. Meta- and pooled analysis of and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2006;164:1027–42. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 39.Habuchi T, Liqing Z, Suzuki T, et al. Increased risk of prostate cancer and benign prostatic hyperplasia associated with a CYP17 gene polymorphism with a gene dosage effect. Cancer Res. 2000;60:5710–3. [PubMed] [Google Scholar]
- 40.Tao MH, Cai Q, Zhang ZF, et al. Polymorphisms in the CYP19A1 (aromatase) gene and endometrial cancer risk in Chinese women. Cancer Epidemiol Biomarkers Prev. 2007;16:943–9. doi: 10.1158/1055-9965.EPI-06-1012. [DOI] [PubMed] [Google Scholar]
- 41.Lo JC, Zhao X, Scuteri A, Brockwell S, Sowers MR. The association of genetic polymorphisms in sex hormone biosynthesis and action with insulin sensitivity and diabetes mellitus in women at midlife. Am J Med. 2006;119:S69–78. doi: 10.1016/j.amjmed.2006.07.009. [DOI] [PubMed] [Google Scholar]