Abstract
The highly specialized morphology of a neuron, typically consisting of a long axon and multiple branching dendrites, lies at the core of the principle of dynamic polarization, whereby information flows from dendrites toward the soma and to the axon. For more than a century neuroscientists have been fascinated by how shape is important for neuronal function and how neurons acquire their characteristic morphology. During the past decade, substantial progress has been made in our understanding of the molecular underpinnings of neuronal polarity and morphogenesis. In these studies, transcription factors have emerged as key players governing multiple aspects of neuronal morphogenesis from neuronal polarization and migration to axon growth and pathfinding to dendrite growth and branching to synaptogenesis. In this review, we will highlight the role of transcription factors in shaping neuronal morphology with emphasis on recent literature in mammalian systems.
Introduction: The life of a neuron is a transcriptional smorgasbord
To integrate into neuronal circuits, newly generated neurons engage in a series of stereotypical developmental events. After exit from the cell-cycle, post-mitotic neurons first undergo axo-dendritic polarization, a process that encompasses the initial specification of axons and dendrites and their coordinate growth giving rise to the unique neuronal shape. Concurrently, many neurons undergo extensive migration to reach their final destinations in the brain. Axons grow to their appropriate targets, dendrites arborize and prune to cover the demands of their receptive field, and synapses form and are refined to ensure proper connectivity. How neurons accomplish all these tasks has been the subject of intense scrutiny during the past few decades. A large body of work has established that these fundamental developmental events are regulated by extrinsic cues including secreted polypeptide growth factors, adhesion molecules, extracellular matrix components, and neuronal activity (Dijkhuizen and Ghosh, 2005b; Huber et al., 2003; Katz and Shatz, 1996; Markus et al., 2002a; McAllister, 2002; Tessier-Lavigne and Goodman, 1996). Extrinsic cues are thought to regulate both the overall design of neuronal shape as well as their fine structural elements such as axon branch points and dendritic spines. Growth factors, guidance proteins, and other extrinsic cues act via specific cell surface receptor proteins, which in turn regulate intracellular signaling proteins that directly influence cytoskeletal elements. Members of the Rho GTPase family of proteins and protein kinases have emerged as key signaling intermediaries that couple the effects of extrinsic cues to the control of actin and microtubule dynamics (Dhavan and Tsai, 2001; Dickson, 2002; Govek et al., 2005; Hur and Zhou, 2010; Luo, 2000; O’Donnell et al., 2009; Wayman et al., 2008).
Accumulating evidence also supports the concept that cell-intrinsic mechanisms have major roles in neuronal morphogenesis and connectivity. These mechanisms comprise developmentally inherited pathways that operate largely independently of cellular environments, orchestrate neuronal responses to extrinsic cues, and in turn may be influenced by these cues. Invertebrate model organisms have been invaluable to the study of the cell-intrinsic mechanisms that orchestrate neuronal morphogenesis. Elegant studies in Drosophila have spearheaded the discovery of in vivo functions for transcription factors in diverse aspects of neuronal morphogenesis. In particular, studies of the da sensory neurons in the fly peripheral nervous system have defined roles for different transcription factors in distinct aspects of dendrite development, from growth and branching to tiling (Jan and Jan, 2003, 2010).
Several observations also highlight the importance of cell-intrinsic mechanisms in the control of neuronal morphogenesis and connectivity in mammalian neurons. For example, the in vivo developmental programs of polarization, migration, axon and dendrite growth, and synapse formation are recapitulated in distinct populations of neurons dissociated in primary culture (Banker and Goslin, 1991; Powell et al., 1997). Of course, extrinsic cues and cell-intrinsic mechanisms do not operate in isolation. Isolated primary Purkinje neurons polarize and extend axons, but the proper formation of their dendrites and dendritic spines requires signals from granule neurons (Baptista et al., 1994). Nevertheless, although extrinsic signals influence neuronal morphogenesis, neurons often seem to carry a memory or intrinsic potential that is not altered by a new and different environment. Transplantation studies have suggested that neuronal precursors of the cerebral cortex that give rise to later-born upper layer neurons are restricted in their developmental potential and do not give rise to earlier-born deep-layer neurons when placed in the subventricular zone (SVZ) of younger hosts undergoing deep layer neurogenesis (Desai and McConnell, 2000; Frantz and McConnell, 1996). Likewise, transplantation studies have revealed that dendrite morphology and laminar specificity of granule neurons in the rat olfactory bulb appear to be specified at the time of birth in the SVZ (Kelsch et al., 2007). These studies are consistent with the idea that cell intrinsic mechanisms specify a developmental template for different populations of neurons that is retained in new environments. This intrinsic identity may also influence how neurons respond to extrinsic cues. Application of the same neurotrophic factor to neurons located in distinct cerebral cortical layers elicits differential effects on dendrite morphology (McAllister et al., 1997; McAllister et al., 1995), suggesting that neurons inherit distinct developmental programs that dictate their responses to extrinsic signals. Purified rat embryonic retinal ganglion neurons cultured in a variety of conditions grow axons much faster than ganglion neurons from postnatal animals (Goldberg et al., 2002b). In addition, with maturation retinal granule neurons undergo a switch from preferential axon growth to preferential dendrite growth (Goldberg et al., 2002b). Collectively, these observations suggest that neurons harbor developmentally inherited cell-intrinsic mechanisms that determine in large part neuronal morphogenesis.
Transcriptional control of gene expression represents a major mode of cell-intrinsic regulation of neuronal development. Transcription factors can govern entire developmental programs, directing distinct stages of neuronal development as well as altering the competency and response of cells to extrinsic cues. Accordingly, often the expression of one or a set of transcription factors is sufficient to direct the subtype specification of distinct neuronal populations and thus their morphology and projection patterns (Arlotta et al., 2005; Chen et al., 2005b; Hand et al., 2005; Lai et al., 2008; Liodis et al., 2007; Molyneaux et al., 2005; Molyneaux et al., 2007; Polleux et al., 2007). The current challenge is to understand the extent of intrinsic regulation by identifying the transcription factors responsible in different aspects of neuronal morphogenesis, their direct targets, and the interplay with extrinsic cues.
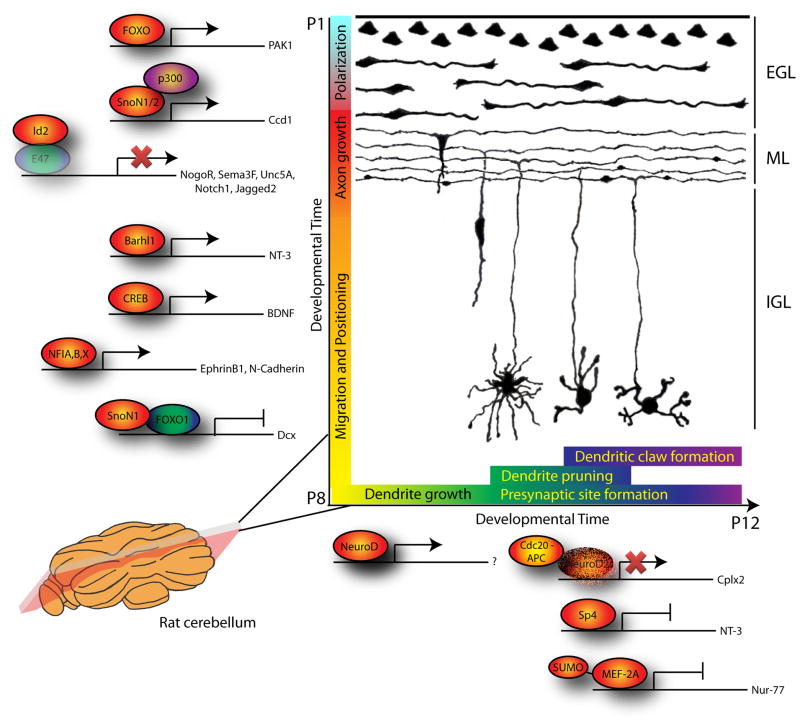
Studies of the mammalian cerebellar cortex have highlighted the importance of transcription factors in distinct aspects of neuronal morphogenesis and connectivity (Figure 1). The rodent cerebellar cortex provides an excellent model system for the study of mechanisms that shape neurons (Altman and Bayer, 1997; Hatten, 1999; Palay and Chan-Palay, 1974; Ramón y Cajal, 1995). Postmitotic granule neurons are generated after division of progenitors located in the external granule layer (EGL). A newly generated granule neuron first extends a single process along the molecular layer (ML). A second process is then generated at the opposite pole of the neuron, giving it a bipolar morphology. A phase of tangential migration follows as the bipolar processes continue to grow before the neuron generates a third leading process perpendicular to the plane of the ML that directs somal migration radially toward the internal granule layer (IGL). As the soma migrates inward in the cerebellar cortex, the two processes in the ML fuse while the neuron continues to extend a trailing process perpendicular to the plane of the ML. The intersection of these orthogonally oriented processes gives rise to the characteristic T-shaped parallel fiber axon of granule neurons. Once granule neurons reach the IGL, they begin to extend dendrites, which following pruning and maturation establish synaptic connections (Altman and Bayer, 1997; Ramón y Cajal, 1995).
Figure 1. Transcription factors orchestrate distinct stages of neuronal morphogenesis in the cerebellar cortex.
The morphogenesis of granule neurons in the cerebellar cortex proceeds in discreet stages governed by distinct transcription factors. After exit from the cell cycle, the FOXO transcription factors trigger granule neuron polarization by regulating the expression of the kinase Pak1. Axon growth is promoted by at least two transcriptional regulators, SnoN1 and SnoN2 acting as transcriptional coactivators in association with p300, and the bHLH inhibitor protein Id2. The transcriptional regulator SnoN2 promotes migration from the IGL into the EGL by inhibiting the transcriptional repressor complex SnoN1/FOXO1. The transcription factors Barhl1, NFIA, B, X and CREB also promote migration into the IGL. The SnoN1/FOXO1 transcriptional complex directly represses DCX and thereby controls the positioning of granule neurons within the IGL. Dendrite morphogenesis includes the stages of growth, pruning and maturation. The transcription factor NeuroD promotes the initiation of dendrite growth and branching, while Sp4 regulates dendrite pruning, and the sumoylated repressor form of the transcription factor MEF2A drives the differentiation of postsynaptic dendritic claws. Concurrent with dendrite pruning and maturation, development of presynaptic structures in parallel fiber axons is regulated by the transcription factor NeuroD2, which is regulated by the ubiquitin ligase Cdc20-APC. Image depicts a coronal or parlobular section of the rat cerebellar cortex at different stages of development as drawn by Ramon y Cajal. See text for details.
The stereotyped sequence of events by which granule neurons acquire a polarized morphology by sequentially generating axons and then dendrites in vivo is closely reproduced when these cells are grown in primary culture or in organotypic cerebellar explants (Kawaji et al., 2004; Powell et al., 1997). These studies suggest that the basic set of instructions to shape granule neurons is intrinsically encoded. In light of the high abundance of granule neurons in the cerebellum, the existence of methods to obtain a highly homogeneous population of granule neurons from the rat or mouse brain (Bilimoria and Bonni, 2008), a relatively simple circuit architecture and accessibility for in vivo studies, the cerebellar cortex has become an excellent system to study intrinsic determinants of neuronal morphogenesis.
Transcription factors play critical roles in all major stages of the life of a granule neuron in the cerebellar cortex (Figure 1). These will be briefly described here and examined in depth in subsequent dedicated sections. Axon growth in granule neurons is controlled by the transcriptional regulators SnoN and Id2, both of which are subject to degradation by the ubiquitin proteasome system (Konishi et al., 2004; Lasorella et al., 2006; Stegmller et al., 2006). Cdh1-anaphase promoting complex (Cdh1-APC), an E3 ubiquitin ligase, targets SnoN and Id2 for degradation and in turn restricts axon growth (Konishi et al., 2004; Lasorella et al., 2006; Stegümller et al., 206). Interestingly, a recent study has revealed that SnoN also regulates in an isoform-specific manner granule neuron migration and positioning by controlling the expression of the microtubule-binding protein doublecortin (Dcx) (Huynh et al., 2011).
Following parallel fiber axon growth, establishment of synaptic connections in the molecular layer occurs through complex interactions between pre-synaptic sites in parallel fiber axons and dendritic spines in Purkinje neurons. The development of parallel fiber presynaptic sites has recently been discovered to be under the purview of transcription factor regulation as well, with the basic helix-loop-helix (bHLH) family member NeuroD2 inhibiting the formation of presynaptic sites in newly generated granule neurons (Yang et al., 2009). Analogous to SnoN-and Id2-control of axon growth, NeuroD2 is also regulated by the ubiquitin-proteasome pathway where the Cdh1-APC-related ligase Cdc20-APC triggers NeuroD2 degradation in mature neurons and thereby promotes presynaptic differentiation (Yang et al., 2009). Thus, different aspects of axon development, growth and presynaptic development are regulated by the APC acting on different transcription factors.
Dendrite development in granule neurons consists of a series of events beginning with the initiation of growth and branching, leading to the formation of an exuberant arbor, followed by pruning, and culminating in the formation of postsynaptic structures termed dendritic claws at the ends of the remaining few dendrites. Dendritic claws house sites of connectivity with mossy fiber terminals and Golgi neuron axons (Altman and Bayer, 1997; Palay and Chan-Palay, 1974; Ramón y Cajal, 1995). As with the axons, dendrite growth and maturation are also under transcriptional control in granule neurons. Intriguingly, transcription factors in these developmental steps are strongly influenced by neuronal activity and calcium signaling. The bHLH transcription factor NeuroD promotes dendrite growth in response to activation of L-type voltage sensitive calcium channels (VSCCs) (Gaudilliére et al., 2004). In a later phase of development, the sumoylated repressor form of the transcription factor myocyte enhancer factor 2A (MEF2A) drives postsynaptic dendritic claw differentiation in a manner that is also regulated by VSCC activation (Shalizi et al., 2006). These studies suggest that activity-dependent calcium signaling regulates dendrite growth and maturation at least in part through changes in gene expression governed by transcription factors.
The rather ubiquitous presence of transcription factor regulation in different aspects of neuronal morphogenesis has been extended to the earliest step of neuronal polarization. Accordingly, the FOXO transcription factors (Forkhead domain type O) have been discovered to trigger neuronal polarization in the mammalian brain (de la Torre-Ubieta et al., 2010). Thus, as soon as neurons are born, transcription factors go to work orchestrating programs of gene expression to shape axons and dendrites and ultimately synapses with other neurons.
FOXO transcription factors regulate neuronal polarization and positioning
The polarization of neurons leading to the generation of axons and dendrites represents an essential step in the establishment of neuronal circuits in the developing brain. Mature axons and dendrites are morphologically, biochemically, and functionally distinct (Craig and Banker, 1994; Falnikar and Baas, 2009). Understanding the mechanisms by which neurons acquire and maintain a polarized morphology is a fundamental question in neurobiology.
The study of the molecular basis of neuronal polarization is a relatively recent endeavor. Within this growing field, the majority of the molecular players regulating neuronal polarity have been characterized in studies of primary hippocampal neurons (Dotti et al., 1988). After plating, dissociated hippocampal neurons first issue several undifferentiated neurites (stage 2). Afterwards, one of the neurites is selected by an apparent stochastic process to become an axon, displaying accelerated growth with concomitant expression of axon markers (stage 3) (Craig and Banker, 1994). Axon specification, which occurs during the transition from stage 2 to stage 3 represents a critical step in neuronal polarization. An array of proteins including molecular scaffolds, Rho-GTPases and their regulators, protein kinases, kinesin motors, and microtubule-associated proteins (MAPs) converge at the nascent axon to regulate cytoskeletal dynamics and promote axon specification and growth (Arimura and Kaibuchi, 2007; Barnes and Polleux, 2009). Other studies using the cerebral cortex as a model system are beginning to implicate extracellular signals in axon formation. The neurotrophin BDNF and the growth factor TGF-β act via the protein kinases SAD-A/B and the Par complex, respectively, to promote axonogenesis (Barnes et al., 2007; Shelly et al., 2007; Yi et al., 2010). Extrinsic cues may also regulate neuronal polarization by preventing axon differentiation in favor of dendrite morphogenesis. The guidance cue Semaphorin 3A (Sema 3A) repels axons and attracts apical dendrites in cortical neurons (Polleux et al., 2000). Two recent studies have expanded upon these findings, suggesting that Sema 3A signaling in diverse populations of neurons suppresses axon specification and instead promotes dendrite formation (Nishiyama et al., 2011; Shelly et al., 2011). Sema 3A suppresses axon differentiation by inducing cGMP/PKG signaling and concomitantly reducing cAMP levels and inhibiting PKA activity, thus leading to decreased activity of the axon-promoting kinases LKB1 and SAD-A/B and increased activity of GSK3β (Shelly et al., 2011). However, Sema 3A knockout as well as BDNF knockout mice do not display overt defects of neuronal polarity, suggesting that alternative compensatory mechanisms are at play (Behar et al., 1996; Ernfors et al., 1994; Jones et al., 1994; Polleux et al., 1998; Polleux et al., 2000). Other studies suggest that the plane of the last mitotic division and the position of the centrosome provide spatial cues that establish the site of axon generation in both primary hippocampal and cortical neurons in vivo (de Anda et al., 2010; de Anda et al., 2005). Although these studies have begun to elucidate the local mechanisms responsible for axon specification and polarization, the cell-intrinsic regulatory mechanisms that might orchestrate neuronal polarization have been largely unexplored.
Recently, the FOXO transcription factors have been identified as key regulators of neuronal polarity (Figure 2). The FOXO proteins are expressed in developing neurons in the brain, including in hippocampal and cerebellar granule neurons at a time when they undergo neuronal polarization and morphogenesis. Knockdown of FOXO1, FOXO3, and FOXO6 by RNA interference (RNAi) in primary granule or hippocampal neurons leads to profound impairment of neuronal polarity (de la Torre-Ubieta et al., 2010). FOXO knockdown neurons extend several unspecified, morphologically similar processes that express both axonal and dendritic markers. This phenotype is recapitulated in the cerebellar cortex in vivo upon induction of FOXO RNAi in postnatal rat pups. FOXO knockdown triggers the formation of aberrant processes in the IGL and the loss of associated parallel fiber axons (de la Torre-Ubieta et al., 2010). Expression of an RNAi-resistant form of FOXO6 in the background of FOXO RNAi reverses the polarity phenotype in primary neurons and in postnatal rat pups. These findings suggest that the FOXO transcription factors, and in particular the brain-enriched protein FOXO6, play a critical role in the regulation of neuronal polarity.
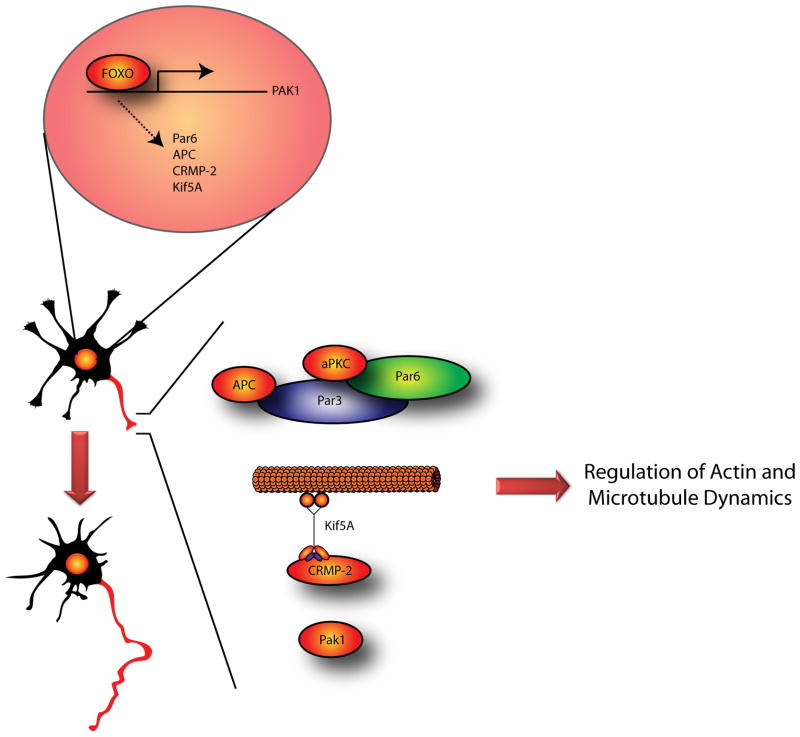
Figure 2. FOXO transcription factors drive neuronal polarization.
FOXO transcription factors occupy the Pak1 gene promoter and induce its expression to promote neuronal polarization. The kinase Pak1 regulates actin and microtubules dynamics by distinct mechanisms at the nascent axon and is required for neuronal polarization. A number of additional FOXO putative transcriptional targets, including mPar6, APC, Kif5A and CRMP-2 regulate polarization by acting locally at the nascent axon. The Par complex protein mPar3 is targeted to the nascent axon by APC. The microtubule-associated protein CRMP-2 is transported to the axon by Kif5A.
How might transcription factors drive neuronal polarization, an event that is specified locally within neuronal processes? A plausible model would be that they trigger the expression of polarity-associated proteins and thereby establish the competency of neurons to undergo polarization. Consistent with this model, analysis of an array of genes implicated in neuronal polarity suggests that the FOXO transcription factors regulate the expression of the polarity complex protein mPar6, the Ras-GTPase R-Ras, the Rac1-GEF STEF, the MAPs adenomatous polyposis coli (APC) and collapsing response mediator protein 2 (CRMP-2), the kinesin family member KIF5A, and the protein kinase Pak1 (Figure 2) (de la Torre-Ubieta et al., 2010). Within this set of genes, Pak1 is the most robustly downregulated gene in FOXO-knockdown neurons. The FOXO proteins occupy the Pak1 gene and thereby directly activate Pak1 transcription in neurons. Knockdown of Pak1 in granule neurons phenocopies the polarity phenotype induced by FOXO knockdown, and expression of Pak1 partly reverses the polarity phenotype triggered by FOXO RNAi (de la Torre-Ubieta et al., 2010). These findings suggest that the protein kinase Pak1 is a direct and physiologically relevant transcriptional target of the FOXO proteins in the control of neuronal polarity, though additional targets mediating FOXO-dependent neuronal polarity remain to be identified.
Pak1 activity is regulated by the Rho-GTPases Cdc42 and Rac1 (Bokoch, 2003), which interact with the Par polarity complex (Joberty et al., 2000; Lin et al., 2000), suggesting that Pak1 may be activated locally at the nascent axon downstream of the Par complex. Thus, the FOXO transcription factors may control both the expression of Pak1 and its upstream regulators (Figure 2). The FOXO proteins regulate the expression of the microtubule-associated protein APC (de la Torre-Ubieta et al., 2010), which localizes mPar3 to the nascent axon (Shi et al., 2004), and expression of the kinesin KIF5A which is important for the transport of CRMP-2 to the axon (Kimura et al., 2005). Therefore, the FOXO transcription factors may act as critical regulators of polarity by triggering the expression of several components of the local machinery controlling neuronal polarity. The discovery of FOXO proteins as key determinants of polarity should pave the way for future studies aimed at identifying additional potential transcriptional regulators in neuronal polarity.
The FOXO transcription factors are tightly controlled by posttranslational modifications, raising the question of how their function in neuronal polarity might be regulated. Growth factors inhibit FOXO-dependent transcription via the PI3K-Akt signaling pathway (Biggs et al., 1999; Brunet et al., 1999; Gan et al., 2005; Guo et al., 1999; Kops et al., 1999; Nakae et al., 2000; Zheng et al., 2002). Interestingly, IGF-1 signaling and localized PI3K activity at the nascent axon promote axon specification in hippocampal neurons (Jiang et al., 2005; Menager et al., 2004; Shi et al., 2003; Sosa et al., 2006; Yoshimura et al., 2006), raising a potential paradox of how the FOXO transcription factors, which are inhibited by the PI3K-Akt signaling pathway, promote neuronal polarization. It remains unclear, however, whether localized Akt signaling in the axon influences the activity of the FOXO transcription factors in the nucleus. Notably, growth factor inhibition of FOXO proteins can be countered in cellular contexts whereby the protein kinases MST1, JNK, and AMPK promote the nuclear accumulation of FOXO proteins and thereby induce FOXO-dependent transcription (Essers et al., 2004; Greer et al., 2007; Lehtinen et al., 2006). It will be interesting to determine if these or other signals stimulate FOXO-dependent transcription in neuronal polarization.
There has been much interest in the specific biological roles of different FOXO family members. The FOXO proteins are expressed in overlapping patterns in the brain and other tissues and appear to bind to similar sites within responsive genes (Furuyama et al., 2000; Hoekman et al., 2006). Accordingly, the FOXO transcription factors have redundant roles as tumor suppressors in hematopoietic stem cells in vivo (Paik et al., 2007; Tothova et al., 2007). However, genetic ablation of different FOXO family members in mice results in distinct phenotypes in vivo (Castrillon et al., 2003; Furuyama et al., 2004; Hosaka et al., 2004; Kitamura et al., 2002; Lin et al., 2004; Nakae et al., 2002; Polter et al., 2009; Renault et al., 2009), suggesting specific roles for individual family members. The FOXO proteins FOXO1, FOXO3, and FOXO6 appear to operate redundantly in driving neuronal polarization (de la Torre-Ubieta et al., 2010). However, in rescue experiments in the background of FOXO RNAi, expression of FOXO1 or FOXO3 only partially restores polarity, whereas expression of FOXO6 substantially restores polarity. Therefore, FOXO6 may have some non-overlapping functions in neuronal polarity. It will be important in the future to characterize the transcriptional targets of individual FOXO family members to understand the contribution of each FOXO protein to neuronal polarity.
Neuronal polarization temporally overlaps with radial migration in certain populations of neurons in the mammalian brain. In the developing cerebral cortex, cortical neurons undergo a transition from a multipolar to bipolar morphology as they leave the intermediate zone (IZ) and move toward the cortical plate, and this morphological transition is regarded as polarization in cortical neurons (Calderon de Anda et al., 2008; Noctor et al., 2004; Tabata and Nakajima, 2003). Interestingly, genes implicated in axo-dendritic polarization including the mPar complex protein Par6, the kinases LKB1, MARK2, GSK3β, and Pak1 and the Rho GTPases Rac1 and Cdc42 are also required for migration (Asada and Sanada, 2010; Asada et al., 2007; Barnes and Polleux, 2009; Causeret et al., 2009; Konno et al., 2005; Sapir et al., 2008; Solecki et al., 2004). The migration phenotypes associated with misregulation or inhibition of these genes often coincide with a multipolar or aberrant morphology in the stalled neurons. In view of these observations, in some instances it is challenging to determine whether the failure of neurons to polarize precedes the migration defects, or whether the inverse relationship holds. Notably, postmitotic granule neurons of the cerebellum undergo axo-dendritic polarization before the onset of radial migration. In this sense, cerebellar granule neurons provide a simpler system for the study of signaling pathways specific for migration or polarity.
Taking advantage of this experimental system, a recent study has uncovered that FOXO1 and the transcriptional regulator SnoN play key roles in the migration and positioning of granule neurons in the cerebellar cortex (Figure 3) (Huynh et al., 2011). Alternative splicing generates two isoforms of the SnoN protein, SnoN1 and SnoN2, which differ by a 46 amino acid region present only in SnoN1 (Pearson-White and Crittenden, 1997; Pelzer et al., 1996). SnoN1 has an essential function in limiting the extent of migration of granule neurons within the IGL and thus in the correct positioning of granule neurons within the IGL. Specific knockdown of SnoN1 in granule neurons in vivo results in abnormal accumulation of granule neurons within the deep IGL close to the white matter (Huynh et al., 2011). By contrast, SnoN2 promotes the migration of granule neurons from the EGL to the IGL. Accordingly, SnoN2 knockdown impairs migration into the IGL, leading to the accumulation of granule neurons in the EGL (Huynh et al., 2011). Therefore, SnoN1 and SnoN2 have opposing functions in the control of granule neuron migration (Figure 3).
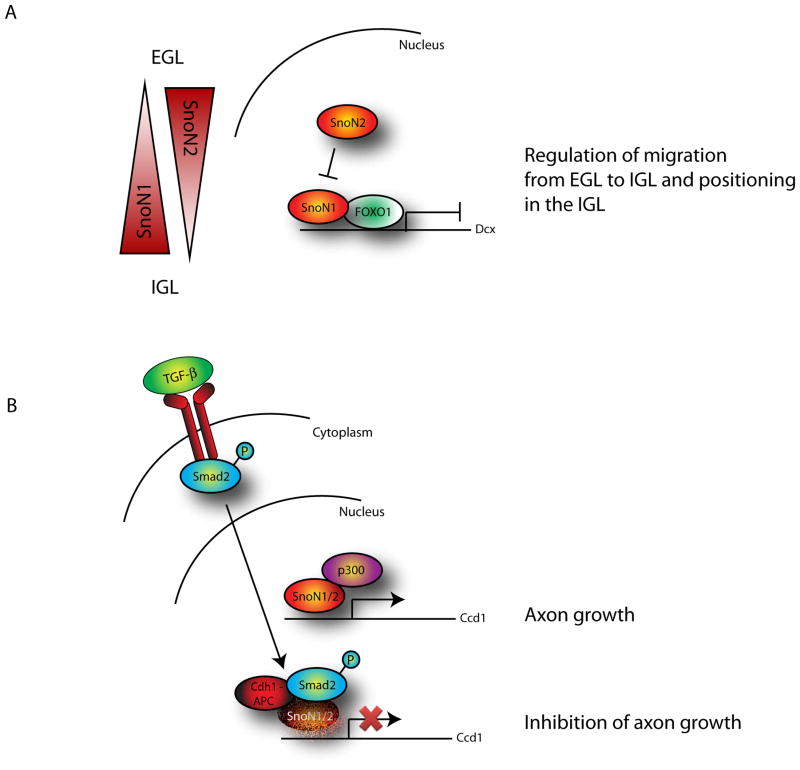
Figure 3. Isoform-specific functions of SnoN in the control of migration and axon growth.
(A) The two spliced isoforms of the Sno gene, SnoN1 and SnoN2, have distinct roles in granule neuron migration. SnoN2 promotes migration from the EGL into the IGL, while SnoN1 regulates positioning within the IGL. SnoN1 associates with the transcription factor FOXO1 and thereby represses Dcx gene expression. SnoN2 inhibits SnoN1 function.
(B) SnoN associates with the transcriptional coactivator p300 and promotes axon growth by inducing the expression of Ccd1. Extrinsic cues impinge on SnoN function during development. TGF-β induces Smad2 phosphorylation and thereby stimulates Cdh1-APC-dependent degradation of SnoN and consequent inhibition of axon growth.
The SnoN isoforms control migration in part by regulating the expression of the X-linked mental retardation and epilepsy gene encoding doublecortin (Dcx). Dcx promotes microtubule stability and polymerization and is thought to be critical for the dynamic coupling between the nucleus and the centrosome during nucleokinesis (Gleeson et al., 1999; Horesh et al., 1999; Koizumi et al., 2006). SnoN1 forms a transcriptional complex with FOXO1 that occupies the Dcx gene and thereby represses its expression in neurons (Figure 3) (Huynh et al., 2011). Consistent with these findings, knockdown of the SnoN1-FOXO1 complex derepresses Dcx expression and hence stimulates excessive migration of granule neurons within the IGL in the cerebellar cortex (Huynh et al., 2011). SnoN2 antagonizes SnoN1 function by directly associating with SnoN1 via a coiled coil domain interaction and inhibiting the ability of SnoN1 to repress FOXO1-dependent transcription (Figure 3) (Huynh et al., 2011). Thus, these findings both define a transcriptional repressor complex for FOXO proteins and illustrate how control of abundance of two splicing isoforms of the same transcriptional regulator leads to opposing cellular responses during development.
The study of neuronal polarization is relevant beyond the context of brain development. Spinal cord injury presents a scenario where neurons have to re-grow axons from an axonal stump. Axon transection can lead to the re-specification of a dendrite into an axon (Bradke and Dotti, 2000; Gomis-Ruth et al., 2008; Goslin and Banker, 1989; Takahashi et al., 2007). In particular, depending on the proximity of the injury to the soma, neurons either extend an axon from the original stump or from a dendrite (Gomis-Ruth et al., 2008). These studies suggest that neuronal polarity is plastic, and conversely the polarized state is actively maintained by dedicated mechanisms (Bisbal et al., 2008; Hedstrom et al., 2008; Jiang et al., 2005; Kobayashi et al., 1992; Nakada et al., 2003; Winckler et al., 1999; Yin et al., 2008). Thus, regulators of neuronal polarity might influence axon regeneration by directing axon re-specification and extension. In this regard, it will be important to determine if FOXO-dependent transcription is required for axon regeneration and in particular whether activators of FOXO proteins can accelerate axon growth after injury. Along these lines, increased SIRT1 activity is associated with protection of dorsal root ganglion (DRG) axons from Wallerian degeneration (Araki et al., 2004). In light of the observation that SIRT1-induced deacetylation of FOXO proteins stimulates FOXO-dependent transcription (Brunet et al., 2004; Daitoku et al., 2004), the FOXO proteins might mediate the protective effect of SIRT1 against axon degeneration. Because the FOXO proteins are regulated by distinct signaling pathways in response to cellular stress, including the protein kinase JNK which stimulates axon regeneration after injury (Lindwall et al., 2004), the FOXO proteins are ideally positioned to promote axon regeneration after injury.
Transcription factors regulate axon growth and guidance from development to regeneration
Several classes of neurons, including projection neurons in the cerebral cortex must extend axons over very long distances in a stereotyped path to innervate specific targets. Beyond the fundamental question of how neurons accomplish this monumental task during development, understanding the mechanisms that promote axon growth may form the basis of treatments aimed at recovery in the central nervous system following injury or disease.
The role of extrinsic cues, including neurotrophic factors, in promoting axon elongation is compelling. Exposure of distinct populations of primary neurons, including retinal ganglion cells, DRG neurons, and hippocampal neurons to NGF, BDNF or NT-3 promotes axon growth robustly (Goldberg et al., 2002a; Lentz et al., 1999; Markus et al., 2002b; Shinoda et al., 2007). Importantly, a requirement for neurotrophin signaling in normal axon development has been validated in vivo (Glebova and Ginty, 2004; Kuruvilla et al., 2004; Patel et al., 2000; Patel et al., 2003; Tucker et al., 2001). Neurotrophins act through the distinct Trk receptors activating signaling cascades relayed by the PI3K-Akt and Ras-MAPK signaling pathways, which in turn directly regulate cytoskeletal elements modulating actin and microtubule polymerization at the growth cone (Huber et al., 2003; Zhou and Snider, 2006). However, neurotrophins also induce changes in transcription that are thought to play critical roles in axon growth (Segal and Greenberg, 1996). Accordingly, neurotrophin signaling regulates the transcription factors CREB and NFAT to stimulate axon growth (Graef et al., 2003; Lonze et al., 2002). Conversely, transcription factors regulate the expression of neurotrophin receptors to specify neuronal subtypes and promote axon growth. For example, the transcription factor Runx1 induces the timely expression of TrkA to promote the specification of nociceptive neurons and growth of their axons (Marmigere et al., 2006). These findings suggest that cell-intrinsic mechanisms orchestrate responses to neurotrophins in the control of axon growth.
Several lines of evidence support the concept that the capacity of a neuron to extend axons and project to the appropriate targets is intrinsically encoded. Neurons of the peripheral nervous system (PNS), but not the central nervous system (CNS), have the capacity to regenerate axons after injury (Aguayo et al., 1991). The axon growth-inhibiting environment of the adult CNS, chiefly generated by myelin proteins, contributes to this differential response (Filbin, 2003; He and Koprivica, 2004; Schwab, 2004). However, the observation that embryonic CNS or adult PNS neurons can extend axons on top of adult white matter suggests that an intrinsic property of neurons in the adult CNS contributes to the failure of axon regeneration after injury (Davies et al., 1997; Davies et al., 1999; Schwab and Bartholdi, 1996). Consistently, embryonic RGCs have a higher capacity to extend axons than postnatal RGCs, and this change in the capacity of axon growth requires new gene transcription (Moore et al., 2009). Importantly, emerging evidence suggests that the intrinsic axonal growth capacity is regulated by transcription factors, both during development and in the context of injury.
Evidence for a cell-intrinsic mechanism regulating axon growth has also emerged from studies of granule neurons of the developing cerebellar cortex. The ubiquitin ligase Cdh1-APC plays a critical role in the control of axon growth and patterning in the rodent cerebellar cortex (Konishi et al., 2004). Knockdown of Cdh1 in primary granule neurons stimulates axon growth even in the presence of the growth-inhibiting environment of myelin. Localization of Cdh1 in the nucleus is required for Cdh1-APC-inhibition of axon growth (Stegmüller et al., 2006). As briefly described above, subsequent studies have identified the transcriptional modulator SnoN and the HLH protein Id2 as substrates of neuronal Cdh1-APC in the control of axon growth (Lasorella et al., 2006; Stegmüller et al., 2008; Stegmüller et al., 2006). Expression of SnoN alone can overcome myelin-dependent growth inhibition, suggesting that SnoN drives a genetic program that promotes axon growth under different extrinsic stimuli (Stegmüller et al., 2006). Interestingly, in contrast to the opposing functions of SnoN1 and SnoN2 in the control of granule neuron migration and positioning, the two isoforms of SnoN collaborate to promote axon growth (Huynh et al., 2011; Stegmüller et al., 2006). Although SnoN is widely considered to have transcriptional repressive functions (Luo, 2004), including in the control of neuronal positioning (Huynh et al., 2011), SnoN functions as a transcriptional coactivator in the control of axon growth (Figure 3) (Ikeuchi et al., 2009). In particular, SnoN associates with the histone acetyltrasferase p300 and thereby induces the expression of a large set of genes in neurons (Ikeuchi et al., 2009). These findings support the concept that SnoN acts in a dual transcriptional activating or repressive manner in a cell-or target-specific manner (Pot and Bonni, 2008; Pot et al., 2010). In promoting axon growth, the cytoskeletal scaffold protein Ccd1 represents a critical downstream target of SnoN (Ikeuchi et al., 2009). Ccd1 localizes to the actin cytoskeleton at growth cones and activates the protein kinase c-Jun kinase (JNK) (Ikeuchi et al., 2009), which has been implicated in axon growth (Oliva et al., 2006).
Whereas SnoN drives axon growth by triggering the expression of regulators of the actin cytoskeleton, Id2 is thought to promote axon growth by antagonizing the function of the bHLH transcription factor E47, which induces the expression of a number of genes involved in axon repulsion including NogoR, Sema3F and Unc5A (Lasorella et al., 2006). Thus, Id2 stimulates axon growth by modulating the response of neurons to guidance cues. Interestingly, TGFβ signaling through the protein Smad2 regulates the abundance of SnoN protein and consequently axon growth (Stegmüller et al., 2008), thus highlighting how intrinsic determinants integrate signals from extrinsic cues for proper development.
Although transcriptional regulators such as NFAT, SnoN, and Id2 appear to regulate axon growth in postmitotic neurons, transcription factors that primarily regulate neurogenesis may also coordinate axon growth in differentiated neurons. In studies of retinotectal projection neurons and spinal cord motor neurons, several transcription factors including Vax2, Zic2, Lim1 and Lmx1b have been reported to regulate the timely and cell-specific expression of proteins involved in axon guidance, including Ephrins A and B and their receptors (Barbieri et al., 2002; Dufour et al., 2003; Herrera et al., 2003; Kania and Jessell, 2003; Kania et al., 2000; Mui et al., 2002; Schulte et al., 1999; Williams et al., 2003). Neurons residing in distinct cerebral cortical layers display specific projection patterns, suggesting that subtype specification is linked to aspects of axon morphogenesis. Functional characterization of transcription factors exhibiting layer-specific expression in the cerebral cortex of knockout mice has uncovered a requirement for the transcription factors FEZL and CTIP2 in the generation of proper layer V neuron projections to subcortical targets (Chen et al., 2005a; Molyneaux et al., 2005). The transcription factors Tbr1 and Sox5 are required for the specification of layer VI neurons and their projections to the thalamus (Bedogni et al., 2010; Lai et al., 2008; McKenna et al., 2011), and Sat2b is required for callosal projections (Alcamo et al., 2008; Britanova et al., 2008). Expression of Fezl or Ctip2 in vivo forces neurons to acquire a deep-layer projection pattern while suppressing the expression of Tbr1 and Sat2b (Chen et al., 2008; Molyneaux et al., 2005). Conversely, Tbr1 and Sat2b are thought to directly repress Fezl and Ctip2, respectively (Alcamo et al., 2008; McKenna et al., 2011). Thus, a complex transcriptional network specifies a particular cortical subtype, in part by suppressing the expression of factors that drive alternate subtypes. Identification of the downstream mechanisms mediating the effects of these transcription factors on cortical projection patterns will reveal whether they are directly linked to the machinery controlling axon growth and guidance or if they act primarily in neuron specification.
Considering the evidence indicating that neurons have a developmentally regulated intrinsic axon growth capacity (Blackmore and Letourneau, 2006; Bouslama-Oueghlani et al., 2003; Goldberg et al., 2002b), several groups have sought to characterize the transcriptome of neurons exhibiting different axon growth capabilities in development and in the context of injury (Costigan et al., 2002; Mechaly et al., 2006; Moore et al., 2009; Zou et al., 2009). Transcriptional profiling of retinal ganglion neurons from embryonic to postnatal stages has revealed that several members of the Krupel-like family (KLF) of transcription factors are regulated throughout development (Moore et al., 2009). The expression pattern of a number of KLF proteins from embryonic to postnatal stages correlates with their ability to suppress or promote axon growth. Overexpression of KLF4, which is upregulated postnatally in retinal ganglion neurons, suppresses axon growth. Conversely, retinal ganglion neurons from KLF4 knockout mice exhibit increased axon growth in culture and regenerate after optic nerve crush injury (Moore et al., 2009). These findings support the idea that the regulated expression of transcription factors during development controls the intrinsic potential for axon growth in neurons.
Studies of axon regeneration in dorsal root ganglion (DRG) neurons have also supported an important role for transcription factors in the control of axon growth. Transection of the peripheral branch, but not the central branch, in DRG neurons triggers axon growth and promotes axon regeneration of central branches following a later spinal cord injury (Neumann and Woolf, 1999; Richardson and Issa, 1984; Smith and Skene, 1997). Comparing the transcriptional profiles of DRG neurons with transected central versus peripheral branches reveals that approximately 10% of the genes with altered expression 12 hours after the procedure are transcription factors (Zou et al., 2009). The transcriptional regulator Smad1 represents one of the genes upregulated in DRGs with transected peripheral branches relative to central branches. Smad1 promotes axon growth in DRG neurons following injury, an effect that is potentiated by BMP signaling. Similar studies have identified a role for the transcription factors STAT3, ATF3, CREB, and c-Jun in promoting axon growth after injury (Gao et al., 2004; Lindwall et al., 2004; Qiu et al., 2005; Raivich et al., 2004; Seijffers et al., 2007; Tsujino et al., 2000). Changes in the expression of transcription factors have also been identified in other models of neuronal injury, including stroke. A number of these transcription factors, including STAT3 and KLF7 may play a role in axon sprouting after stroke (Li et al., 2010b). Thus, there might be shared transcriptional responses following stroke with those promoting axon regeneration after neuronal injury. Taken together, these studies highlight the importance of transcriptional responses in axon regeneration and offer the prospect that cell-intrinsic responses might provide a target for development of new therapeutic possibilities in neurological diseases.
A major focus in the study of the role of transcription factors in axon growth and regeneration is the identity of the relevant target genes. Axon guidance molecules including members of the ephrin and semaphorin families of proteins have been identified as key targets (Polleux et al., 2007). Fewer studies have identified direct cytoskeletal regulators that might act at the growth cone or in axon protein transport. The transcription factor COUP-TFI (NR2F1) plays a critical role in neurogenesis, differentiation, migration and formation of commissural projections. Primary hippocampal neurons from COUP-TFI knockout mice initially grow short abnormal axons but later grow to the same extent as wildtype cells (Armentano et al., 2006). The expression of the cytoskeletal regulators MAP1B and RND2 is altered in COUP-TFI knockout brains in microarray analyses (Armentano et al., 2006). The tumor suppressor p53 has also been reported to promote axon growth by regulating the expression of cGKI, a kinase that counteracts growth cone collapse induced by semaphorin 3A signaling (Tedeschi et al., 2009b) or by inducing the expression of cytoskeletal regulators including GAP-43, Coronin1, and the GTPase Rab13 following axonal injury (Di Giovanni et al., 2006; Tedeschi et al., 2009a). The case of p53 is more complex as other studies suggest that p53 can promote axon growth independently of transcription by inhibiting Rho kinase (ROCK) at the axon (Qin et al., 2009; Qin et al., 2010). Collectively, these studies support the idea that transcription factors can independently regulate two different aspects of axon development, growth and guidance, by inducing different target genes according to the developmental requirements of the cell.
Is axon growth regulated by epigenetic mechanisms? Compelling evidence on epigenetic mechanisms selectively regulating axon growth in the mammalian brain is scarce. Epigenetic regulators including the histone acetyltransferase CBP and the chromatin modifier Sat2b influence cortical and motor neuron projection patterns, but this is also linked to a role in neuronal subtype specification (Alcamo et al., 2008; Britanova et al., 2008; Lee et al., 2009). Loss of function of the methyl-CpG-binding transcriptional repressor MeCP2 has been associated with several abnormalities in neuronal morphogenesis including disrupted axon projections (Belichenko et al., 2009; Degano et al., 2009). Axonal targeting defects observed in MeCP2 knockout mice are attributed to changes in the expression of the guidance factor Semaphorin3F, albeit in a non cell-autonomous fashion (Degano et al., 2009). Among the genes identified in a screen for axonal sprouting after stroke is ATRX (α-thalassemia/mental retardation syndrome X-linked) (Li et al., 2010b), a chromatin remodeling enzyme linked to mental retardation that has also been implicated in dendrite development and neuronal survival (Berube et al., 2005; Shioda et al., 2011). ATRX appears to be upregulated in sprouting neurons relative to non-sprouting neurons. Knockdown of ATRX by RNAi reduces basal axon growth of cultured DRG neurons and prevents axonal sprouting after stroke in vivo (Li et al., 2010b). Interestingly, ATRX and MeCP2 can interact in vitro and in cells, and in MeCP2 knockout cells ATRX fails to localize to heterochromatin, displaying instead a diffuse expression pattern (Nan et al., 2007). Thus, some of the neuronal defects observed in MeCP2 mutants might be due to abnormal ATRX activity. Future studies will be needed to understand the extent of epigenetic mechanisms in axon growth.
Transcription factors direct distinct stages of dendrite morphogenesis
As the receptive limbs of neurotransmission in the brain, dendrites have evolved to display immense variety of shape and size. Dendrite architecture strongly influences the processing of information (Spruston, 2008), suggesting that the morphogenesis of dendrite arbors directly impacts the flow of information across the brain. Although we will focus on the role of transcription factors on dendrite morphology in mammalian systems, significant contributions in this field have also come from studies in the fly nervous system. We refer the reader to excellent reviews on this topic (Corty et al., 2009; Jan and Jan, 2003, 2010).
Dendrites come in greater variety of shapes and size than axons, and the morphogenesis of dendrites in different populations of mammalian neurons proceeds in distinct stereotypical phases. Just as with axon specification and neuron migration, granule neurons of the rodent cerebellar cortex provide a robust model system for the study of dendrite development including their distinct stages of growth, pruning, and postsynaptic maturation (Figure 1). In recent years, a number of transcription factors have been discovered to regulate distinct stages of dendrite development in granule neurons. As part of the process of establishing neuronal polarity, the FOXO transcription factors, and in particular the brain-enriched protein FOXO6, inhibit the growth of dendrites while simultaneously promoting the growth of axons (de la Torre-Ubieta et al., 2010). Thus, even as neurons migrate and their axons grow, transcriptional mechanisms are at play to inhibit the formation of dendrites. In this capacity, the FOXO proteins may inhibit a cell-intrinsic switch from axon to dendrite growth in the brain. The bHLH protein NeuroD plays a critical role in the initiation of dendrite growth as well as the branching of granule neuron dendrite arbors in the cerebellar cortex (Gaudillière et al., 2004). While NeuroD promotes the initiation of dendrite growth and elaboration, the zinc-finger transcription factor Sp4 promotes the pruning of the granule neuron dendrite arbor (Ramos et al., 2007; Ramos et al., 2009), and the MADS domain transcription factor MEF2A triggers the morphogenesis of the postsynaptic dendritic claws (Shalizi et al., 2007; Shalizi et al., 2006). Collectively, these studies support the concept that different transcription factors are dedicated to distinct aspects of dendrite development (Figure 1). Whether and how these transcription factors might regulate each other in the control of dendrite morphogenesis is an unanswered question.
An interesting feature of the role of transcription factors in the regulation of dendrite development is that they are robustly influenced by calcium signaling and consequently neuronal activity (Figure 4). Membrane depolarization is critical for the development of dendrite growth and branching, including in granule neurons of the cerebellar cortex (Gaudillière et al., 2004; Okazawa et al., 2009). Calcium influx via L-type calcium channels triggers the activation of the protein kinase CaMKIIα (Hudmon and Schulman, 2002; Wayman et al., 2008). Once activated, CaMKIIα induces the phosphorylation of NeuroD at Serine 336 (Gaudillière et al., 2004). Structure-function analyses of NeuroD in the background of NeuroD RNAi indicate that the CaMKIIα-induced phosphorylation of NeuroD, including at Serine 336, is essential for the ability of NeuroD to mediate membrane depolarization-dependent dendrite growth (Gaudillière et al., 2004). How the CaMKIIα-induced phosphorylation activates the transcriptional function of NeuroD remains to be determined. Since depolarization-induced NeuroD-dependent transcription in transient reporter assays is not affected by mutation of Serine 336 (Gaudillière et al., 2004), the possibility that the CaMKIIα-induced phosphorylation triggers changes in NeuroD-recruitment of chromatin remodeling enzymes is an intriguing possibility that remains to be tested. The NeuroD target genes that couple calcium signaling to the growth of dendrites also remain unknown. Interestingly, the role of NeuroD in dendrite morphogenesis seems to extend beyond early postnatal development into the regulation of dendrites in adult-born neurons. Adult-born granule neurons of the hippocampus in NeuroD null mice display shorter dendrites as compared to wild type neurons (Gao et al., 2009). Whether calcium signaling is relevant to NeuroD-dependent dendrite morphogenesis in adult-born neurons remains an open question.
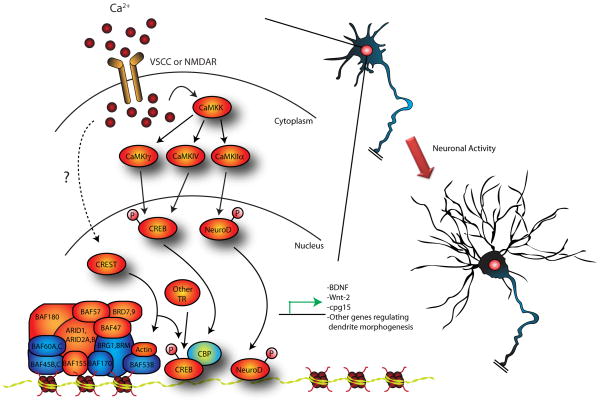
Figure 4. Neuronal activity regulates transcription-dependent dendrite growth.
Activity-dependent gene expression is regulated at multiple levels. Neuronal activity leading to calcium influx via voltage-sensitive calcium channels or NMDA receptors leads to the activation of CaMKK and downstream kinases CaMKIγ CaMKIV, or CaMKIIα to regulate transcription factors that control dendrite growth. Activation of CaMKIγ or CaMKIV leads to phosphorylation and activation of CREB, whereas CaMKIIα phosphorylates NeuroD and induces NeuroD-dependent transcription. A number of transcriptional regulators (TR) including CBP, CREST, TORC1 and CRTC1 associate and regulate CREB-dependent transcription. In another layer of regulation, epigenetic mechanisms, including chromatin modification by the chromatin remodeling complex nBAF have a critical role in activity-dependent dendrite growth. CREST associates with the nBAF complex and regulates the expression of a number of genes important for dendrite growth. The complex interplay between transcription factors, transcriptional regulators and chromatin modifying enzymes that regulate gene expression in response to neuronal activity remains to be elucidated.
Calcium signaling also regulates the function of the transcription factor MEF2A in postsynaptic dendritic differentiation. A calcium-regulated sumoylated transcriptionally repressive form of MEF2A drives the differentiation of dendritic claws in the cerebellar cortex (Shalizi et al., 2007; Shalizi et al., 2006). Sumoylation of MEF2A at Lysine 408, which converts MEF2A into a transcriptional repressor, is dependent on the status of phosphorylation of a nearby site, Serine 403, which in turn is regulated by the calcium-regulated phosphatase calcineurin (Shalizi et al., 2006). The phosphorylation of MEF2A at Serine 403 is required for the sumoylation of MEF2A at Lysine 408, owing to increasing the catalytic efficiency of the SUMO E2 enzyme Ubc9 acting on MEF2A as a substrate (Mohideen et al., 2009; Shalizi et al., 2006). Strikingly, calcineurin-induced dephosphorylation of MEF2A at Serine 403 triggers a switch in the modification of MEF2A Lysine 408 from sumoylation to acetylation, thereby converting MEF2A from a transcriptional repressor form to an activator, and leading to the inhibition postsynaptic of dendritic claw differentiation (Shalizi et al., 2006). Consistent with these findings, activation of MEF2-dependent transcription triggers elimination of postsynaptic sites in other populations of brain neurons (Barbosa et al., 2008; Flavell et al., 2006; Flavell et al., 2008; Pfeiffer et al., 2010; Pulipparacharuvil et al., 2008). What might be the purpose of calcium influx through L-type VSCCs inhibiting the function of sumoylated MEF2A in postsynaptic dendritic claw differentiation? A plausible explanation is that calcium influx in membrane depolarized granule neurons during earlier phases of dendrite development might coordinately promote dendrite growth and branching via NeuroD and concomitantly inhibit the premature formation of postsynaptic dendrite sites. Alternatively, with neuronal maturation, calcium influx induced by trans-synaptic signaling might induce the refinement of postsynaptic dendritic structures.
The idea that calcium signaling and VSCCs induce the early growth of dendrites and simultaneously inhibit the later steps of dendrite development has been further evaluated in recent studies. Interestingly, the resting membrane potential of newly generated granule neurons in the EGL is depolarized, and it is hyperpolarized with maturation in the IGL (Okazawa et al., 2009). Hyperpolarization of granule neurons in cerebellar slices triggers dendritic pruning and differentiation, including the formation of dendritic claws (Okazawa et al., 2009). Switching between these stages of dendrite morphogenesis coincides with changes in the expression of a large number of genes, including the transcription factors Etv1, Math2, Tle1, and Hey1 suggesting that these proteins might regulate dendrite maturation (Okazawa et al., 2009; Sato et al., 2005). Collectively, studies of dendrite morphogenesis in the cerebellar cortex support the idea that both the early phases of dendrite growth and activity-dependent remodeling are under the purview of transcription factor regulation.
Although studies in the cerebellar cortex have provided compelling evidence for cell-intrinsic regulation of stage-dependent dendrite morphogenesis that is widely relevant to diverse populations of neurons in the brain, transcription factors can also shape the development of dendritic arbors characteristic of a particular neuronal subtype. Transcription factors set up complex dendrite morphologies in a neuron-specific manner in Drosophila (Corty et al., 2009; Jan and Jan, 2003, 2010). Transcriptional mechanisms specifying dendrite arbors in the mammalian brain are also beginning to be described. Temporally-specific or layer-specific expression of transcription factors in the cerebral cortex may define the morphological identity of neurons (Arlotta et al., 2005; Molyneaux et al., 2009; Molyneaux et al., 2007). The zinc finger transcription factor Fezf2 is required for dendritic arbor complexity in layer V/VI neurons specifically (Chen et al., 2005b). The mammalian homologues of the Drosophila transcription factor Cut, Cux1 and Cux2, have been implicated in layer II/III pyramidal neuron dendrite development by two different groups, though with seemingly conflicting conclusions (Cubelos et al., 2010; Li et al., 2010a). Using a combination of knockout mice and in vivo RNAi to generate Cux1-and Cux 2-deficient cortical neurons in the intact cerebral cortex, Cubelos and colleagues have found that Cux1 and Cux2 additively promote dendrite growth and branching as well as dendritic spine formation. Cux1 and Cux2 directly repress the putative chromatin modifying proteins Xlr3b and Xlr4b, which couple Cux1 and Cux2 to regulation of dendritic spine morphogenesis, while the transcriptional targets involved in dendrite arbor formation remain to be identified (Cubelos et al., 2010). In contrast, using cortical cultures Li and colleagues have found that overexpression of Cux1, but not Cux2, decreases dendrite complexity, and conversely that knockdown of Cux1 leads to excessive dendritic arbor size in cortical neurons. Li and colleagues have also reported that Cux1 directly represses the cell-cycle regulator p27kip1 and thereby inhibits dendrite growth through RhoA (Li et al., 2010a). The findings from Cubelos and colleagues whereby Cux1 promotes dendritic complexity are consistent with the function of the fly homologue Cut, suggesting functional evolutionary conservation of this transcription factor.
Just as in the cerebellar cortex, studies of dendrite morphogenesis in the cerebral cortex and hippocampus have highlighted the regulation of transcription factors by neuronal activity and calcium influx (Figure 4). Prominent among these is the transcription factor cAMP-responsive element binding protein (CREB), which is modulated by a variety of extrinsic cues and regulates neuronal survival, dendrite growth, and synaptic function (Flavell and Greenberg, 2008; Lonze and Ginty, 2002; Shaywitz and Greenberg, 1999). Neuronal activity stimulates CaMKIV-dependent phosphorylation and activation of CREB in cortical neurons and thereby induces dendrite growth and arborization (Redmond et al., 2002). In more recent studies, CaMKIγ has been found to mediate neuronal activity-dependent phosphorylation and activation of CREB in hippocampal neurons, leading to increased dendritic arborization (Wayman et al., 2006). The CREB co-activator CBP also participates in neuronal activity-induced dendrite morphogenesis (Redmond et al., 2002). Another calcium-regulated transcriptional coactivator termed CREST, which also associates with CBP, is required for activity-dependent dendrite growth development in the cerebral cortex (Aizawa et al., 2004). Recent studies have identified additional CREB binding partners that act as co-activators required for CREB-dependent dendrite growth, including TORC1 (transducer of regulated CREB activity) and CRTC1 (CREB-regulated transcription co-activator), which operate downstream of activity-dependent signaling and BDNF, respectively (Finsterwald et al., 2010; Li et al., 2009). These studies highlight the complexity of CREB-dependent transcription. It will be important to elucidate the context and signaling mechanisms controlling the association of CREB with different coregulators and the consequences on CREB-dependent transcription.
Although a role for these transcriptional regulators in dendrite development is compelling, the downstream mechanisms are incompletely understood. BDNF represents a potential relevant target of CREB and associated proteins in the control of dendrite development and branching (Cheung et al., 2007; Dijkhuizen and Ghosh, 2005a; Horch and Katz, 2002; McAllister et al., 1997; Tao et al., 1998). The secreted signaling protein Wnt-2, which promotes dendritic arborization, is also induced by CREB downstream of neuronal activity (Wayman et al., 2006). Likewise, the cpg15 gene, which encodes a GPI-anchored protein, is induced by CREB downstream of calcium signaling and promotes dendrite arbor elaboration in Xenopus tectal neurons (Fujino et al., 2003; Nedivi et al., 1998). Taking into account that CREB mediates several aspects of neuronal development including neuronal survival (Bonni et al., 1999; Lonze et al., 2002; Riccio et al., 1999), identifying the specific direct targets involved in dendrite growth will clarify the role of CREB in neuronal morphogenesis.
The complexity of transcriptional regulation in activity-dependent dendrite growth is further highlighted by evidence demonstrating that the nBAF chromatin remodeling complex is required for dendrite development (Figure 4) (Wu et al., 2007). The multimeric nBAF complex is assembled from several homologous proteins in a developmental-specific manner. In the context of this combinatorial assembly, the BAF53a subunit, which is present in neuronal progenitors, is replaced by the BAF53b subunit, which is specific for differentiated neurons (Lessard et al., 2007). Genetic ablation of BAF53b in mice leads to abnormalities in basal and activity-dependent dendrite growth. Interestingly, the nBAF complex associates with CREST and modulates the expression of a large number of genes involved in neurite growth (Wu et al., 2007). This is of particular interest in light of the observation that at least two other epigenetic regulators, the histone demethylase SMCX and the DNA methyl-binding transcriptional repressor MeCP2, which are mutated in cases of X-linked mental retardation (XLMR) and Rett syndrome also control dendrite growth (Iwase et al., 2007; Zhou et al., 2006). These studies suggest that epigenetic mechanisms altering chromatin structure, which can drive longer lasting transcriptional changes or provide additional levels of regulation, contribute to dendrite development. Elucidation of the interplay between these epigenetic regulators and transcription factors in the context of dendrite development should advance our understanding of these disorders.
The few transcription factors that have been characterized in dendrite development in the mammalian brain to date likely only represent the tip of the iceberg. Further, the targets of many of these transcription factors are largely unknown. Regulators of cytoskeletal components, including Rho-GTPases and microtubule-binding proteins, have been identified as targets of transcription factor regulation in the context of dendrite development (Cobos et al., 2007; Hand et al., 2005; Li et al., 2010a; Wu et al., 2007). It will be interesting to determine whether additional mechanisms, including contact-mediated signaling and secretory function through the Golgi apparatus, also operate downstream of specific transcriptional regulators in the control of dendrite morphogenesis.
Concluding remarks
Transcription factors play a prominent role in all facets of neuronal development from neuronal polarization and migration to axon and dendrite morphogenesis to synapse differentiation. A flurry of studies during the past decade has unraveled many functions of transcription factors and regulators in neuronal development in the mammalian brain. Prior to these studies, transcription factors were generally considered to govern the transition from precursor cells to postmitotic neurons, and this transition was thought to unleash a differentiation program, resulting in the mature morphology of neurons. A major conclusion of studies of the past decade is that transcription factors continue to play key regulatory roles in postmitotic neurons to specify and regulate the development of distinct morphological compartments. Another related key conclusion is the concept that different transcription factors are dedicated to distinct phases of neuronal morphogenesis and connectivity. This, however, is an oversimplification. Although some transcription factors have a restricted expression pattern and orchestrate specific aspects of development, others operate in a pleiotropic manner to regulate several steps of development. In some cases, transcription factors operate as nodes to coordinate two different aspects of neuronal development, such as neuronal branching and migration or dendrite growth and synapse formation. In addition, the functions of different transcription factors may overlap temporally to control a specific feature of neuronal morphology and connectivity.
An important goal of future research in the study of transcriptional regulation of neuronal morphogenesis will be to define the relationship between different transcription factors regulating distinct phases of neuronal development. For example, it will be interesting to determine whether and how the functions of FOXO6, NeuroD, Sp4, and sumoylated MEF2A intersect in the course of orchestrating granule neuron dendrite arbor development in the cerebellar cortex. Do any of these transcriptional factors regulate the expression of another factor acting in a subsequent or preceding step of dendrite development? Do any of these factors interact with other transcription factors and thereby regulate their activity? Finally, do upstream signals impinging on a specific transcription factor, such as CaMKIIα or calcineurin that control NeuroD and MEF2A activity respectively, influence the activity of another transcription factor acting on a different stage of dendrite development?
Another important goal of future studies will be to determine the extent of programs of gene expression regulated by different transcription factors acting at distinct stages of neuronal development. Advances in genomic technologies will facilitate these studies and yield large datasets for analysis of transcription factor-dependent networks of genes at distinct developmental stages. Comparisons of these datasets as well as analyses of transcription factor occupancies at target genes should also provide novel insights into cooperative mechanisms between different transcriptional regulators in neuronal morphogenesis and connectivity. Several transcription factors that direct neuronal morphogenesis in postmitotic neurons also have roles in neuron specification. Although dissociating such distinct roles may not always be a simple task, transcriptional profiling coupled with ChIP-Seq analyses may allow for the characterization of targetomes associated with specific developmental programs.
The complexity of transcriptional regulation is vast. Transcription factors are controlled by posttranslational modifications, which lead to changes in protein stability, localization, activity, or interaction partners. These modifications may not simply stimulate or inhibit transcriptional activity of the factor, but may induce a switch in the mode of a transcription factor’s function between activator and repressor. Additionally, association with epigenetic regulators, including chromatin remodeling complexes, may induce longer lasting or widespread changes in gene expression. Finally, transcription factors often regulate the expression of other transcription factors creating complex cascades. How and to what extent these cascades may be involved in other aspects of neuronal morphogenesis is a task for future studies.
Finally, studies of transcriptional regulation offer the basis for elucidation of key mechanisms of brain development as well as serve the foundation for a better understanding of the molecular basis of developmental disorders of the brain in which deregulation of neuronal morphogenesis and connectivity plays a prominent role (Kaufmann and Moser, 2000; McManus and Golden, 2005; Penzes et al., 2011; Schwartzkroin and Walsh, 2000; Sisodiya, 2004). Mutations in several transcriptional regulators have been implicated in diverse array of neurodevelopmental disorders from mental retardation and autism spectrum disorders to inherited ataxias to epilepsy syndromes (Grinberg and Millen, 2005; Gutierrez-Delicado and Serratosa, 2004; Helmlinger et al., 2006; Hong et al., 2005; Orr, 2010). Understanding the normal functions of these transcriptional regulators in neuronal morphogenesis and connectivity will be a major first step toward understanding the pathogenesis of these disorders.
Acknowledgments
We thank members of the Bonni laboratory, in particular Luis Mejía, Yoshiho Ikeuchi and Chi Zhang, for helpful discussions and critical reading of the manuscript. This work was supported by NIH grant NS041021 (A.B.) and the Albert J. Ryan Foundation (L.T.U.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguayo AJ, Rasminsky M, Bray GM, Carbonetto S, McKerracher L, Villegas-Perez MP, Vidal-Sanz M, Carter DA. Degenerative and regenerative responses of injured neurons in the central nervous system of adult mammals. Philos Trans R Soc Lond B Biol Sci. 1991;331:337–343. doi: 10.1098/rstb.1991.0025. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science (New York, NY. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure, and functions. Boca Raton: CRC Press; 1997. [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science (New York, NY. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Armentano M, Filosa A, Andolfi G, Studer M. COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development. 2006;133:4151–4162. doi: 10.1242/dev.02600. [DOI] [PubMed] [Google Scholar]
- Asada N, Sanada K. LKB1-mediated spatial control of GSK3beta and adenomatous polyposis coli contributes to centrosomal forward movement and neuronal migration in the developing neocortex. J Neurosci. 2010;30:8852–8865. doi: 10.1523/JNEUROSCI.6140-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada N, Sanada K, Fukada Y. LKB1 regulates neuronal migration and neuronal differentiation in the developing neocortex through centrosomal positioning. J Neurosci. 2007;27:11769–11775. doi: 10.1523/JNEUROSCI.1938-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K. Culturing nerve cells. Cambridge, Mass: MIT Press; 1991. [Google Scholar]
- Baptista CA, Hatten ME, Blazeski R, Mason CA. Cell-cell interactions influence survival and differentiation of purified Purkinje cells in vitro. Neuron. 1994;12:243–260. doi: 10.1016/0896-6273(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Barbieri AM, Broccoli V, Bovolenta P, Alfano G, Marchitiello A, Mocchetti C, Crippa L, Bulfone A, Marigo V, Ballabio A, Banfi S. Vax2 inactivation in mouse determines alteration of the eye dorsal-ventral axis, misrouting of the optic fibres and eye coloboma. Development. 2002;129:805–813. doi: 10.1242/dev.129.3.805. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383:525–528. doi: 10.1038/383525a0. [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. The Journal of comparative neurology. 2009;514:240–258. doi: 10.1002/cne.22009. [DOI] [PubMed] [Google Scholar]
- Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. The Journal of clinical investigation. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilimoria PM, Bonni A. Cultures of Cerebellar Granule Neurons. Cold Spring Harbor Protocols. 2008 doi: 10.1101/pdb.prot5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal M, Conde C, Donoso M, Bollati F, Sesma J, Quiroga S, Diaz Anel A, Malhotra V, Marzolo MP, Caceres A. Protein kinase d regulates trafficking of dendritic membrane proteins in developing neurons. J Neurosci. 2008;28:9297–9308. doi: 10.1523/JNEUROSCI.1879-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore M, Letourneau PC. Changes within maturing neurons limit axonal regeneration in the developing spinal cord. J Neurobiol. 2006;66:348–360. doi: 10.1002/neu.20224. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annual review of biochemistry. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science (New York, NY. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Bouslama-Oueghlani L, Wehrle R, Sotelo C, Dusart I. The developmental loss of the ability of Purkinje cells to regenerate their axons occurs in the absence of myelin: an in vitro model to prevent myelination. J Neurosci. 2003;23:8318–8329. doi: 10.1523/JNEUROSCI.23-23-08318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Differentiated neurons retain the capacity to generate axons from dendrites. Curr Biol. 2000;10:1467–1470. doi: 10.1016/s0960-9822(00)00807-1. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, NY. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Calderon de Anda F, Gartner A, Tsai LH, Dotti CG. Pyramidal neuron polarity axis is defined at the bipolar stage. J Cell Sci. 2008;121:178–185. doi: 10.1242/jcs.023143. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science (New York, NY. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Causeret F, Terao M, Jacobs T, Nishimura YV, Yanagawa Y, Obata K, Hoshino M, Nikolic M. The p21-activated kinase is required for neuronal migration in the cerebral cortex. Cereb Cortex. 2009;19:861–875. doi: 10.1093/cercor/bhn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Chin WH, Chen Y, Ng YP, Ip NY. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biol. 2007;5:e63. doi: 10.1371/journal.pbio.0050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SJ, Fitch MT, Memberg SP, Hall AK, Raisman G, Silver J. Regeneration of adult axons in white matter tracts of the central nervous system. Nature. 1997;390:680–683. doi: 10.1038/37776. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Goucher DR, Doller C, Silver J. Robust regeneration of adult sensory axons in degenerating white matter of the adult rat spinal cord. J Neurosci. 1999;19:5810–5822. doi: 10.1523/JNEUROSCI.19-14-05810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci. 2010;30:10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Pollarolo G, Da Silva JS, Camoletto PG, Feiguin F, Dotti CG. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Gaudillière B, Yang Y, Ikeuchi Y, Yamada T, DiBacco S, Stegmüller J, Schüller U, Salih DA, Rowitch D, et al. A FOXO-Pak1 transcriptional pathway controls neuronal polarity. Genes Dev. 2010;24:799–813. doi: 10.1101/gad.1880510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degano AL, Pasterkamp RJ, Ronnett GV. MeCP2 deficiency disrupts axonal guidance, fasciculation, and targeting by altering Semaphorin 3F function. Mol Cell Neurosci. 2009;42:243–254. doi: 10.1016/j.mcn.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–287. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Knights CD, Rao M, Yakovlev A, Beers J, Catania J, Avantaggiati ML, Faden AI. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. The EMBO journal. 2006;25:4084–40. doi: 10.1038/sj.emboj.7601292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science (New York, NY. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen PA, Ghosh A. BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J Neurobiol. 2005a;62:278–288. doi: 10.1002/neu.20100. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen PA, Ghosh A. Regulation of dendritic growth by calcium and neurotrophin signaling. Prog Brain Res. 2005b;147:17–27. doi: 10.1016/S0079-6123(04)47002-2. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–146. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Seibt J, Passante L, Depaepe V, Ciossek T, Frisen J, Kullander K, Flanagan JG, Polleux F, Vanderhaeghen P. Area specificity and topography of thalamocortical projections are controlled by ephrin/Eph genes. Neuron. 2003;39:453–465. doi: 10.1016/s0896-6273(03)00440-9. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. The EMBO journal. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A, Baas PW. Critical roles for microtubules in axonal development and disease. Results Probl Cell Differ. 2009;48:47–64. doi: 10.1007/400_2009_2. [DOI] [PubMed] [Google Scholar]
- Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat Rev Neurosci. 2003;4:703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- Finsterwald C, Fiumelli H, Cardinaux JR, Martin JL. Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. J Biol Chem. 2010;285:28587–28595. doi: 10.1074/jbc.M110.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science (New York, NY. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Fujino T, Lee WC, Nedivi E. Regulation of cpg15 by signaling pathways that mediate synaptic plasticity. Mol Cell Neurosci. 2003;24:538–554. doi: 10.1016/s1044-7431(03)00230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Nakayama K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. The Biochemical journal. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Zheng W, Chabot JG, Unterman TG, Quirion R. Nuclear/cytoplasmic shuttling of the transcription factor FoxO1 is regulated by neurotrophic factors. J Neurochem. 2005;93:1209–1219. doi: 10.1111/j.1471-4159.2005.03108.x. [DOI] [PubMed] [Google Scholar]
- Gao Y, Deng K, Hou J, Bryson JB, Barco A, Nikulina E, Spencer T, Mellado W, Kandel ER, Filbin MT. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron. 2004;44:609–621. doi: 10.1016/j.neuron.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudillière B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Glebova NO, Ginty DD. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J Neurosci. 2004;24:743–751. doi: 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron. 2002a;33:689–702. doi: 10.1016/s0896-6273(02)00602-5. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science (New York, NY. 2002b;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth S, Wierenga CJ, Bradke F. Plasticity of polarization: changing dendrites into axons in neurons integrated in neuronal circuits. Curr Biol. 2008;18:992–1000. doi: 10.1016/j.cub.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Goslin K, Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J Cell Biol. 1989;108:1507–1516. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Millen KJ. The ZIC gene family in development and disease. Clin Genet. 2005;67:290–296. doi: 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Delicado E, Serratosa JM. Genetics of the epilepsies. Curr Opin Neurol. 2004;17:147–153. doi: 10.1097/00019052-200404000-00011. [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- He Z, Koprivica V. The Nogo signaling pathway for regeneration block. Annu Rev Neurosci. 2004;27:341–368. doi: 10.1146/annurev.neuro.27.070203.144340. [DOI] [PubMed] [Google Scholar]
- Hedstrom KL, Ogawa Y, Rasband MN. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol. 2008;183:635–640. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Tora L, Devys D. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 2006;22:562–570. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, Brown S, Mason CA. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell. 2003;114:545–557. doi: 10.1016/s0092-8674(03)00684-6. [DOI] [PubMed] [Google Scholar]
- Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hong EJ, West AE, Greenberg ME. Transcriptional control of cognitive development. Curr Opin Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nat Neurosci. 2002;5:1177–1184. doi: 10.1038/nn927. [DOI] [PubMed] [Google Scholar]
- Horesh D, Sapir T, Francis F, Wolf SG, Caspi M, Elbaum M, Chelly J, Reiner O. Doublecortin, a stabilizer of microtubules. Hum Mol Genet. 1999;8:1599–1610. doi: 10.1093/hmg/8.9.1599. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annual review of biochemistry. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MA, Ikeuchi Y, Netherton S, de la Torre-Ubieta L, Kanadia R, Stegmuller J, Cepko C, Bonni S, Bonni A. An Isoform-Specific SnoN1-FOXO1 Repressor Complex Controls Neuronal Morphogenesis and Positioning in the Mammalian Brain. Neuron. 2011;69:930–944. doi: 10.1016/j.neuron.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Stegmuller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29:4312–4321. doi: 10.1523/JNEUROSCI.0126-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120:123–135. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nature cell biology. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Jones KR, Farinas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science (New York, NY. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kawaji K, Umeshima H, Eiraku M, Hirano T, Kengaku M. Dual phases of migration of cerebellar granule cells guided by axonal and dendritic leading processes. Mol Cell Neurosci. 2004;25:228–240. doi: 10.1016/j.mcn.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Kelsch W, Mosley CP, Lin CW, Lois C. Distinct mammalian precursors are committed to generate neurons with defined dendritic projection patterns. PLoS Biol. 2007;5:e300. doi: 10.1371/journal.pbio.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Nakae J, Kitamura Y, Kido Y, Biggs WH, 3rd, Wright CV, White MF, Arden KC, Accili D. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. The Journal of clinical investigation. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Storrie B, Simons K, Dotti CG. A functional barrier to movement of lipids in polarized neurons. Nature. 1992;359:647–650. doi: 10.1038/359647a0. [DOI] [PubMed] [Google Scholar]
- Koizumi H, Higginbotham H, Poon T, Tanaka T, Brinkman BC, Gleeson JG. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science (New York, NY. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Konno D, Yoshimura S, Hori K, Maruoka H, Sobue K. Involvement of the phosphatidylinositol 3-kinase/rac1 and cdc42 pathways in radial migration of cortical neurons. J Biol Chem. 2005;280:5082–5088. doi: 10.1074/jbc.M408251200. [DOI] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye H, Ginty DD. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmüller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Lentz SI, Knudson CM, Korsmeyer SJ, Snider WD. Neurotrophins support the development of diverse sensory axon morphologies. J Neurosci. 1999;19:1038–1048. doi: 10.1523/JNEUROSCI.19-03-01038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhao CT, Wang Y, Yuan XB. The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons. PLoS One. 2010a;5:e10596. doi: 10.1371/journal.pone.0010596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Overman JJ, Katsman D, Kozlov SV, Donnelly CJ, Twiss JL, Giger RJ, Coppola G, Geschwind DH, Carmichael ST. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat Neurosci. 2010b;13:1496–1504. doi: 10.1038/nn.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nature cell biology. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Lindwall C, Dahlin L, Lundborg G, Kanje M. Inhibition of c-Jun phosphorylation reduces axonal outgrowth of adult rat nodose ganglia and dorsal root ganglia sensory neurons. Mol Cell Neurosci. 2004;27:267–279. doi: 10.1016/j.mcn.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Markus A, Patel TD, Snider WD. Neurotrophic factors and axonal growth. Curr Opin Neurobiol. 2002a;12:523–531. doi: 10.1016/s0959-4388(02)00372-0. [DOI] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002b;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Montelius A, Wegner M, Groner Y, Reichardt LF, Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK. Neurotrophins and cortical development. Results Probl Cell Differ. 2002;39:89–112. doi: 10.1007/978-3-540-46006-0_5. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MF, Golden JA. Neuronal migration in developmental disorders. J Child Neurol. 2005;20:280–286. doi: 10.1177/08830738050200040301. [DOI] [PubMed] [Google Scholar]
- Mechaly I, Bourane S, Piquemal D, Al-Jumaily M, Venteo S, Puech S, Scamps F, Valmier J, Carroll P. Gene profiling during development and after a peripheral nerve traumatism reveals genes specifically induced by injury in dorsal root ganglia. Mol Cell Neurosci. 2006;32:217–229. doi: 10.1016/j.mcn.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Menager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. J Neurochem. 2004;89:109–118. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- Mohideen F, Capili AD, Bilimoria PM, Yamada T, Bonni A, Lima CD. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat Struct Mol Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science (New York, NY. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui SH, Hindges R, O’Leary DD, Lemke G, Bertuzzi S. The homeodomain protein Vax2 patterns the dorsoventral and nasotemporal axes of the eye. Development. 2002;129:797–804. doi: 10.1242/dev.129.3.797. [DOI] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nature cell biology. 2003;5:626–632. doi: 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. The EMBO journal. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Biggs WH, 3rd, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity induced signaling molecule. Science (New York, NY. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Woolf CJ. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Togashi K, von Schimmelmann MJ, Lim CS, Maeda S, Yamashita N, Goshima Y, Ishii S, Hong K. Semaphorin 3A induces CaV2.3 channel-dependent conversion of axons to dendrites. Nature cell biology. 2011;13:676–685. doi: 10.1038/ncb2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- O’Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa M, Abe H, Katsukawa M, Iijima K, Kiwada T, Nakanishi S. Role of calcineurin signaling in membrane potential-regulated maturation of cerebellar granule cells. J Neurosci. 2009;29:2938–2947. doi: 10.1523/JNEUROSCI.5932-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Jr, Atkins CM, Copenagle L, Banker GA. Activated c-Jun N-terminal kinase is required for axon formation. J Neurosci. 2006;26:9462–9470. doi: 10.1523/JNEUROSCI.2625-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT. Nuclear ataxias. Cold Spring Harb Perspect Biol. 2010;2:a000786. doi: 10.1101/cshperspect.a000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V. Cerebellar cortex: cytology and organization. Berlin, Heidelberg, New York: Springer; 1974. [Google Scholar]
- Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–357. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Patel TD, Kramer I, Kucera J, Niederkofler V, Jessell TM, Arber S, Snider WD. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003;38:403–416. doi: 10.1016/s0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- Pearson-White S, Crittenden R. Proto-oncogene Sno expression, alternative isoforms and immediate early serum response. Nucleic Acids Res. 1997;25:2930–2937. doi: 10.1093/nar/25.14.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer T, Lyons GE, Kim S, Moreadith RW. Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn. 1996;205:114–125. doi: 10.1002/(SICI)1097-0177(199602)205:2<114::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Giger RJ, Ginty DD, Kolodkin AL, Ghosh A. Patterning of cortical efferent projections by semaphorin-neuropilin interactions. Science (New York, NY. 1998;282:1904–1906. doi: 10.1126/science.282.5395.1904. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ince-Dunn G, Ghosh A. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat Rev Neurosci. 2007;8:331–340. doi: 10.1038/nrn2118. [DOI] [PubMed] [Google Scholar]
- Polleux F, Morrow T, Ghosh A. Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature. 2000;404:567–573. doi: 10.1038/35007001. [DOI] [PubMed] [Google Scholar]
- Polter A, Yang S, Zmijewska AA, van Groen T, Paik JH, Depinho RA, Peng SL, Jope RS, Li X. Forkhead box, class O transcription factors in brain: regulation and behavioral manifestation. Biol Psychiatry. 2009;65:150–159. doi: 10.1016/j.biopsych.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot I, Bonni S. SnoN in TGF-beta signaling and cancer biology. Curr Mol Med. 2008;8:319–328. doi: 10.2174/156652408784533797. [DOI] [PubMed] [Google Scholar]
- Pot I, Ikeuchi Y, Bonni A, Bonni S. SnoN: bridging neurobiology and cancer biology. Curr Mol Med. 2010;10:667–673. doi: 10.2174/156652410792630616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, Rivas RJ, Rodriguez-Boulan E, Hatten ME. Development of polarity in cerebellar granule neurons. J Neurobiol. 1997;32:223–236. doi: 10.1002/(sici)1097-4695(199702)32:2<223::aid-neu7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Baudry M, Liao G, Noniyev A, Galeano J, Bi X. A novel function for p53: regulation of growth cone motility through interaction with Rho kinase. J Neurosci. 2009;29:5183–5192. doi: 10.1523/JNEUROSCI.0420-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Liao G, Baudry M, Bi X. Role of calpain-mediated p53 truncation in semaphorin 3A-induced axonal growth regulation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13883–13887. doi: 10.1073/pnas.1008652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cafferty WB, McMahon SB, Thompson SW. Conditioning injury-induced spinal axon regeneration requires signal transducer and activator of transcription 3 activation. J Neurosci. 2005;25:1645–1653. doi: 10.1523/JNEUROSCI.3269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal S. Histology of the nervous system of man and vertebrates. New York: Oxford University Press; 1995. [Google Scholar]
- Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Valin A, Sun X, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Mol Cell Neurosci. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science (New York, NY. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Issa VM. Peripheral injury enhances central regeneration of primary sensory neurones. Nature. 1984;309:791–793. doi: 10.1038/309791a0. [DOI] [PubMed] [Google Scholar]
- Sapir T, Sapoznik S, Levy T, Finkelshtein D, Shmueli A, Timm T, Mandelkow EM, Reiner O. Accurate balance of the polarity kinase MARK2/Par-1 is required for proper cortical neuronal migration. J Neurosci. 2008;28:5710–5720. doi: 10.1523/JNEUROSCI.0911-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suzuki K, Yamazaki H, Nakanishi S. A pivotal role of calcineurin signaling in development and maturation of postnatal cerebellar granule cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5874–5879. doi: 10.1073/pnas.0501972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Furukawa T, Peters MA, Kozak CA, Cepko CL. Misexpression of the Emx-related homeobox genes cVax and mVax2 ventralizes the retina and perturbs the retinotectal map. Neuron. 1999;24:541–553. doi: 10.1016/s0896-6273(00)81111-3. [DOI] [PubMed] [Google Scholar]
- Schwab ME. Nogo and axon regeneration. Curr Opin Neurobiol. 2004;14:118–124. doi: 10.1016/j.conb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Bartholdi D. Degeneration and regeneration of axons in the lesioned spinal cord. Physiol Rev. 1996;76:319–370. doi: 10.1152/physrev.1996.76.2.319. [DOI] [PubMed] [Google Scholar]
- Schwartzkroin PA, Walsh CA. Cortical malformations and epilepsy. Ment Retard Dev Disabil Res Rev. 2000;6:268–280. doi: 10.1002/1098-2779(2000)6:4<268::AID-MRDD6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Segal RA, Greenberg ME. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Bilimoria PM, Stegmüller J, Gaudillière B, Yang Y, Shuai K, Bonni A. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science (New York, NY. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annual review of biochemistry. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Shelly M, Cancedda L, Heilshorn S, Sumbre G, Poo MM. LKB1/STRAD promotes axon initiation during neuronal polarization. Cell. 2007;129:565–577. doi: 10.1016/j.cell.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Shelly M, Cancedda L, Lim BK, Popescu AT, Cheng PL, Gao H, Poo MM. Semaphorin3A Regulates Neuronal Polarization by Suppressing Axon Formation and Promoting Dendrite Growth. Neuron. 2011;71:433–446. doi: 10.1016/j.neuron.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Cheng T, Jan LY, Jan YN. APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol. 2004;14:2025–2032. doi: 10.1016/j.cub.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S, Iwamatsu A, Kaibuchi K. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci. 2007;27:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda N, Beppu H, Fukuda T, Li E, Kitajima I, Fukunaga K. Aberrant calcium/calmodulin-dependent protein kinase II (CaMKII) activity is associated with abnormal dendritic spine morphology in the ATRX mutant mouse brain. J Neurosci. 2011;31:346–358. doi: 10.1523/JNEUROSCI.4816-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodiya SM. Malformations of cortical development: burdens and insights from important causes of human epilepsy. Lancet Neurol. 2004;3:29–38. doi: 10.1016/s1474-4422(03)00620-3. [DOI] [PubMed] [Google Scholar]
- Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Sosa L, Dupraz S, Laurino L, Bollati F, Bisbal M, Caceres A, Pfenninger KH, Quiroga S. IGF-1 receptor is essential for the establishment of hippocampal neuronal polarity. Nat Neurosci. 2006;9:993–995. doi: 10.1038/nn1742. [DOI] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Huynh MA, Yuan Z, Konishi Y, Bonni A. TGFbeta-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J Neurosci. 2008;28:1961–1969. doi: 10.1523/JNEUROSCI.3061-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmüller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci. 2003;23:9996–10001. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D, Yu W, Baas PW, Kawai-Hirai R, Hayashi K. Rearrangement of microtubule polarity orientation during conversion of dendrites to axons in cultured pyramidal neurons. Cell Motil Cytoskeleton. 2007;64:347–359. doi: 10.1002/cm.20188. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Nguyen T, Puttagunta R, Gaub P, Di Giovanni S. A p53-CBP/p300 transcription module is required for GAP-43 expression, axon outgrowth, and regeneration. Cell Death Differ. 2009a;16:543–554. doi: 10.1038/cdd.2008.175. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Nguyen T, Steele SU, Feil S, Naumann U, Feil R, Di Giovanni S. The tumor suppressor p53 transcriptionally regulates cGKI expression during neuronal maturation and is required for cGMP-dependent growth cone collapse. J Neurosci. 2009b;29:15155–15160. doi: 10.1523/JNEUROSCI.4416-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science (New York, NY. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Meyer M, Barde YA. Neurotrophins are required for nerve growth during development. Nat Neurosci. 2001;4:29–37. doi: 10.1038/82868. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Mann F, Erskine L, Sakurai T, Wei S, Rossi DJ, Gale NW, Holt CE, Mason CA, Henkemeyer M. Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron. 2003;39:919–935. doi: 10.1016/j.neuron.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Winckler B, Forscher P, Mellman I. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 1999;397:698–701. doi: 10.1038/17806. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science (New York, NY. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin DM, Huang YH, Zhu YB, Wang Y. Both the establishment and maintenance of neuronal polarity require the activity of protein kinase D in the Golgi apparatus. J Neurosci. 2008;28:8832–8843. doi: 10.1523/JNEUROSCI.1291-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Arimura N, Kawano Y, Kawabata S, Wang S, Kaibuchi K. Ras regulates neuronal polarity via the PI3-kinase/Akt/GSK-3beta/CRMP-2 pathway. Biochemical and biophysical research communications. 2006;340:62–68. doi: 10.1016/j.bbrc.2005.11.147. [DOI] [PubMed] [Google Scholar]
- Zheng WH, Kar S, Quirion R. FKHRL1 and its homologs are new targets of nerve growth factor Trk receptor signaling. J Neurochem. 2002;80:1049–1061. doi: 10.1046/j.0022-3042.2002.00783.x. [DOI] [PubMed] [Google Scholar]
- Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Ho C, Wong K, Tessier-Lavigne M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. J Neurosci. 2009;29:7116–7123. doi: 10.1523/JNEUROSCI.5397-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]