Abstract
The protein kinase calcium/calmodulin-dependent kinase II (CaMKII) predominantly consists of the α and β isoforms in the brain. Although CaMKIIα functions have been elucidated, the unique catalytic functions of CaMKIIβ have remained unknown. Using knockdown analyses in primary neurons and in the rat cerebellar cortex in vivo, we report that CaMKIIβ operates at the centrosome in a CaMKIIα-independent manner to drive dendrite retraction and pruning. We also find that the targeting protein PCM1 localizes CaMKIIβ at the centrosome. Finally, we uncover the E3 ubiquitin ligase Cdc20-APC as a novel centrosomal substrate of CaMKIIβ. CaMKIIβ phosphorylates Cdc20 at Ser51, which induces Cdc20 dispersion from the centrosome, thereby inhibiting centrosomal Cdc20-APC activity and triggering the transition from growth to retraction of dendrites. Our findings define a novel isoform-specific function for CaMKIIβ that regulates ubiquitin signaling at the centrosome and thereby orchestrates dendrite patterning, with important implications for neuronal connectivity in the brain.
The proper formation and morphogenesis of dendrites is fundamental to the establishment of neural circuits in the brain. Following exit from the cell cycle and migration to the appropriate location, post-mitotic neurons undergo carefully orchestrated steps in dendrite morphogenesis, including the generation and elaboration of extensive dendrite arbors followed by dendrite retraction and pruning1–2. These steps in dendrite patterning are necessary for the accurate formation of neuronal circuitry. Defects in dendrite patterning may result in severe neurodevelopmental disorders3–4. Although the molecular basis of dendrite growth has been the subject of intense scrutiny5–8, the signaling mechanisms governing the transition from dendrite elaboration to pruning and retraction have remained poorly understood.
The calcium/calmodulin-dependent protein kinases (CaMKs) represent a critical link between the external environment and cellular responses in neurons. CaMKII is one of the major CaMKs in the brain, representing 1–2% of total brain protein9–11. CaMKII exists as a holoenzyme composed of multiple subunits, which form a complex via their association domains12–15. Brain CaMKII predominantly consists of the α and β isoforms, which form heteromeric or homomeric complexes16–18. Previous studies have focused on the functions of CaMKIIα19–22. However, a specific role for CaMKIIβ as a protein kinase in the mammalian brain has remained unknown.
In this study, we report that CaMKIIβ has a distinct and specific function in the regulation of dendrite patterning in the mammalian brain. We identify a unique centrosomal targeting sequence (CTS) within the variable region of CaMKIIβ but not CaMKIIα. The CTS mediates the specific interaction of CaMKIIβ with the centrosomal targeting protein pericentriolar material 1 (PCM1). Consequently, PCM1 localizes CaMKIIβ to the centrosome, where CaMKIIβ drives dendrite retraction and pruning independently of CaMKIIα. Importantly, we uncover the ubiquitin ligase Cdc20-APC as the centrosomal substrate of CaMKIIβ. CaMKIIβ phosphorylates the APC coactivator Cdc20 at Ser51 in neurons, which induces Cdc20 dispersion from the centrosome, thereby inhibiting centrosomal Cdc20-APC activity and triggering a switch from growth to retraction of dendrites. Our findings define a unique CaMKIIβ pathway that regulates ubiquitin signaling at the centrosome to orchestrate dendrite patterning and hence the establishment of neuronal connectivity in the mammalian brain.
CaMKIIβ regulates dendrite patterning in mammalian neurons
To characterize the role of CaMKIIβ in neurons, we used granule neurons of the developing rodent cerebellar cortex. Granule neurons are the most abundant neurons in the brain and follow a highly typified spatial and temporal program of differentiation characteristic of neurons in the central nervous system, providing a robust system for studies of neuronal morphogenesis and connectivity23–25. Using an antibody that specifically recognizes CaMKIIβ, we first confirmed that the distinct isoforms of CaMKIIβ (β, β′, βε, β′ε) are expressed in primary granule neurons, and protein levels increased with maturation (Fig. 1a).
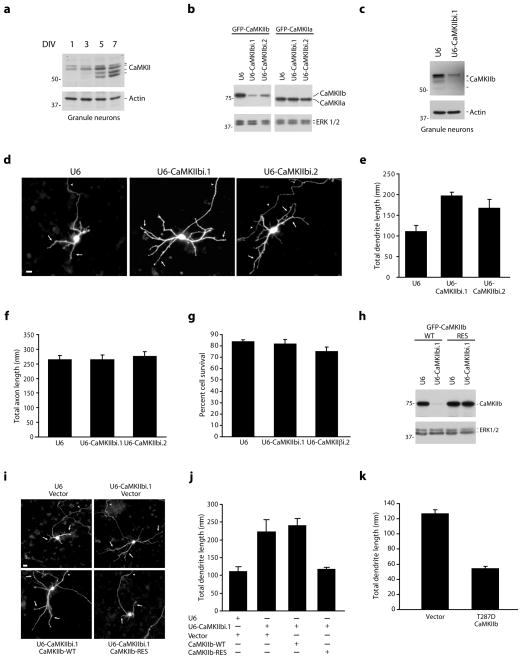
Figure 1. CaMKIIβ restricts the elaboration of dendrites.
a, Lysates of granule neurons were immunoblotted with the indicated antibodies. DIV = days in vitro. Full-length blots for all immunoblotting analyses are presented in Supplementary Fig. 11. b, Lysates of COS cells transfected with GFP-CaMKIIβ or GFP-CaMKIIα together with the CaMKIIβ RNAi or control U6 plasmid were immunoblotted with the indicated antibodies. c, Lysates of granule neurons electroporated with the CaMKIIβ RNAi or control U6 plasmid were immunoblotted with the indicated antibodies. d, Granule neurons transfected with a CaMKIIβ RNAi or control U6 plasmid together with GFP were subjected to immunocytochemistry using the GFP antibody. In all images of neuronal morphology, arrows and arrowheads indicate dendrites and axons, respectively. Scale bar = 10 μm. e, Total dendrite length for granule neurons treated as in (d) was quantified and is presented as mean + SEM. Total dendrite length was significantly increased in CaMKIIβ knockdown neurons compared to control U6-transfected neurons (ANOVA; p < 0.005). 270 neurons were measured. f, Granule neurons were analyzed as in (d). Total axon length was not significantly different in CaMKIIβ knockdown neurons and control U6-transfected neurons. g, Granule neurons were transfected with a CaMKIIβ RNAi or control U6 plasmid and analyzed for cell survival. Cell survival was not significantly different in CaMKIIβ knockdown neurons and control U6-transfected neurons. h, Lysates of COS cells transfected with GFP-CaMKIIβ-WT or GFP-CaMKIIβ-RES together with the CaMKIIβ RNAi or control U6 plasmid were immunoblotted with the indicated antibodies. i, Granule neurons transfected with the CaMKIIβ RNAi or control U6 plasmid together with the expression plasmid encoding CaMKIIβ-WT, CaMKIIβ-RES, or control vector and GFP were analyzed as in (d). Scale bar = 10 μm. j, Total dendrite length for granule neurons treated as in (i) was quantified. CaMKIIβ-RES, but not CaMKIIβ-WT, significantly reduced total dendrite length compared to control vector in the background of CaMKIIβ RNAi (ANOVA; p < 0.005). 360 neurons were measured. k, Granule neurons transfected with constitutively active T287D CaMKIIβ or control vector were analyzed as in (d). Total dendrite length was significantly reduced in T287D CaMKIIβ-expressing neurons compared to control vector-transfected neurons (t-test, p < 0.0005). 180 neurons were measured.
To determine the function of CaMKIIβ in neurons, we employed a plasmid-based method of RNA interference (RNAi) to acutely knockdown CaMKIIβ expression26. Expression of two shRNAs targeting distinct regions of CaMKIIβ reduced the expression of exogenous CaMKIIβ but not CaMKIIα in COS cells (Fig. 1b), and robustly decreased the expression of endogenous CaMKIIβ but not CaMKIIα in granule neurons as determined by immunoblotting and immunocytochemistry (Fig. 1c and Supplementary Fig. 1a–c). To assess the role of CaMKIIβ in neuronal morphogenesis, we induced the knockdown of CaMKIIβ in granule neurons and examined axons and dendrites, which are easily identified based on their morphology and immunocytochemical markers27–28. Remarkably, CaMKIIβ knockdown in granule neurons stimulated the elaboration of dendrites, which were characterized by longer primary dendrites and increased secondary and tertiary dendrite branching (Fig. 1d and Supplementary Fig. 1d). Accordingly, CaMKIIβ knockdown increased total dendrite length by up to 76% in granule neurons (Fig. 1e). In contrast, CaMKIIβ RNAi had little or no effect on axon length (Fig. 1f). In other experiments, CaMKIIβ RNAi had little or no effect on cell survival in granule neurons (Fig. 1g). Together, these results suggest that CaMKIIβ knockdown specifically stimulates the elaboration of dendrites in granule neurons.
To confirm that the CaMKIIβ RNAi-induced dendrite phenotype is the result of specific knockdown of CaMKIIβ, we performed a rescue experiment. We generated an expression plasmid encoding CaMKIIβ that is resistant to RNAi (CaMKIIβ-RES) (Fig. 1h). Expression of CaMKIIβ-RES, but not CaMKIIβ encoded by wild type cDNA (CaMKIIβ-WT), restored the typical appearance of dendrite arbors and reduced dendrite length and branching in the background of CaMKIIβ RNAi to that of control transfected neurons (Fig. 1i,j and Supplementary Fig. 1e). These data suggest that the CaMKIIβ RNAi-induced dendrite phenotype results from specific knockdown of CaMKIIβ rather than off-target effects of CaMKIIβ RNAi. In other experiments, expression of a constitutively active form of CaMKIIβ, in which the site of autophosphorylation Thr287 is replaced with the phosphomimetic residue aspartate (T287D CaMKIIβ), simplified dendrite arbors and profoundly reduced dendrite length and branching in granule neurons (Fig. 1k and data not shown). Thus, based on both inhibition of CaMKIIβ by rigorously-controlled RNAi and gain-of-function analyses, we conclude that CaMKIIβ restricts the extent of dendrite arborization and growth in granule neurons.
We next determined if the function of CaMKIIβ in dendrite patterning is generalizable in mammalian brain neurons. CaMKIIβ knockdown in hippocampal neurons substantially increased the number of secondary and tertiary dendrite branches and thereby increased dendrite length (Supplementary Fig. 2a–d). CaMKIIβ RNAi-induced dendrite elaboration in hippocampal neurons was reversed by CaMKIIβ-RES but not CaMKIIβ-WT, indicating the specificity of the CaMKIIβ RNAi-induced phenotype (Supplementary Fig. 2e,f). CaMKIIβ knockdown also stimulated dendrite elaboration in primary cerebral cortical neurons (Supplementary Fig. 3). These data suggest that CaMKIIβ inhibits the growth and arborization of dendrites in diverse populations of mammalian brain neurons.
To determine the cellular basis of CaMKIIβ regulation of dendrite patterning, we characterized the temporal dynamics of CaMKIIβ function in dendrite morphogenesis. We subjected individual granule neurons expressing constitutively active T287D CaMKIIβ and control vector-transfected granule neurons to time-lapse analyses. In control vector-transfected granule neurons, dendrites and their branches displayed dynamic elongation and retraction during 48 hours of analysis (Supplementary Fig. 4a,b), with an overall cumulative increase in total dendrite length (Supplementary Fig. 4c–e). In contrast, T287D CaMKIIβ-expressing granule neurons rarely had periods of dendrite growth and almost exclusively displayed retraction of dendrites (Supplementary Fig. 4a,b), which cumulatively reduced total dendrite length (Supplementary Fig. 4c–e). Analyses of individual dendrites yielded similar results, with individual dendrites in T287D CaMKIIβ-expressing neurons also showing an increase in dendrite retraction events compared to individual dendrites in control granule neurons (Supplementary Fig. 4f–h). In a complementary line of experiments, time-lapse analyses of individual neurons and dendrites in control U6-transfected neurons and CaMKIIβ knockdown neurons revealed reduced periods of dendrite retraction and increased periods of dendrite growth, with an overall cumulative increase in total dendrite length, upon CaMKIIβ knockdown (Supplementary Fig. 4i–q). These data suggest that CaMKIIβ restricts the elaboration of dendrite arbors by triggering a switch from growth to active retraction of dendrites.
We next characterized the function of CaMKIIβ in regulating the minute dynamics of dendrite extension and retraction using live environment-controlled imaging analyses. Granule neurons expressing T287D CaMKIIβ or transfected with the control vector were imaged every ten minutes for a one hour period. In control neurons, dendrites had a large number of extension events with few retraction events (Supplementary Fig. 5a,b and Supplementary Video 1). In contrast, T287D CaMKIIβ-expressing neurons had numerous retraction events and few extension events (Supplementary Fig. 5a,b and Supplementary Video 1). In a complementary line of experiments, live imaging analyses revealed a significant increase in extension events and a significant reduction in retraction events in neurons upon CaMKIIβ knockdown (Supplementary Fig. 5c,d and Supplementary Video 2).
CaMKIIβ drives dendrite retraction and pruning in vivo
Having identified a critical function for CaMKIIβ in the control of dendrite patterning in primary neurons, we next determined the role of CaMKIIβ in dendrite morphogenesis in the intact developing cerebellar cortex. We first utilized rat organotypic cerebellar slices in which the architecture of the cerebellar tissue is preserved. Using a biolistics method of transfection, we induced CaMKIIβ knockdown in post-natal day six (P6) cerebellar slices27. CaMKIIβ knockdown robustly stimulated dendrite elaboration in internal granule layer (IGL) granule neurons (Fig. 2a), substantially increasing the number of secondary and tertiary dendrite branches as well as total dendrite length (Fig. 2a,b and Supplementary Fig. 6a).
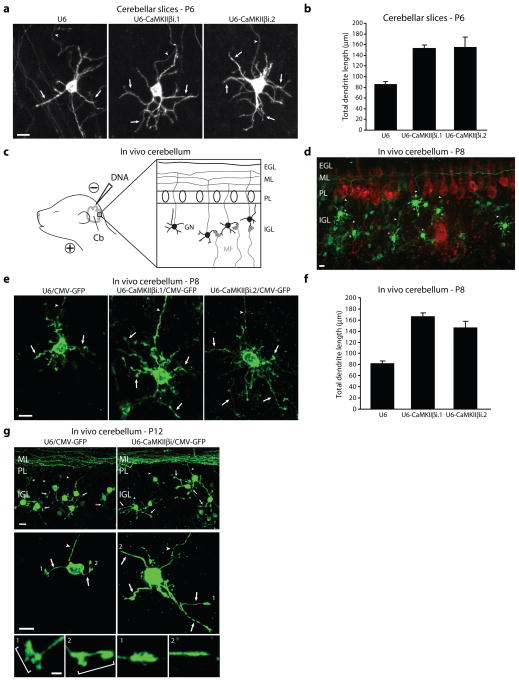
Figure 2. CaMKIIβ regulates dendrite patterning in vivo.
a, Post-natal day six (P6) rat cerebellar slices transfected by a biolistics method with the CaMKIIβ RNAi or control U6 plasmid together with GFP were subjected to immunohistochemistry using the GFP antibody. Scale bar = 10 μm. b, CaMKIIβ knockdown neurons had significantly longer dendrites compared to control U6-transfected neurons (ANOVA, p < 0.0001). 199 neurons were measured. c, Schematic of in vivo electroporation approach. d, Rat pups electroporated in vivo with a U6-CaMKIIβi/CMV-GFP RNAi or control U6/CMV-GFP plasmid were sacrificed five days after electroporation and cerebella were subjected to immunohistochemistry using the GFP and Calbindin antibody. Representative control-transfected granule neurons are shown. Scale bar = 10 μm. e, Rat pups were electroporated as in (d) and representative neurons for each condition are shown. Scale bar = 10 μm. f, IGL granule neurons analyzed as in (d) were subjected to morphometric analysis. Total dendrite length was significantly increased in IGL granule neurons in CaMKIIβ knockdown animals compared to control U6 animals (ANOVA, p < 0.0001). 266 neurons were measured. g, Rat pups electroporated in vivo with the U6-CaMKIIβi/CMV-GFP RNAi or control U6/CMV-GFP plasmid were sacrificed nine days after electroporation and cerebella were analyzed as in (d). (Top) Representative cerebellar sections from each condition are shown. Scale bar = 10 μm. (Bottom) Representative IGL granule neurons for each condition are shown. Scale bar = 10 μm. Inset: Zoomed view of dendritic tips of individual neurons. Scale bar = 2.5 μm. Bracket identifies dendritic claws.
We next assessed CaMKIIβ function in the cerebellar cortex in the organism using an in vivo RNAi approach28. We electroporated P3 rat pups with a CaMKIIβ RNAi plasmid that also expresses GFP (U6-CaMKIIβi/CMV-GFP) or the corresponding control RNAi plasmid (U6/CMV-GFP) (Fig. 2c). Five days after electroporation, animals were sacrificed and cerebella were subjected to immunohistochemical analysis using the GFP antibody (Fig. 2d). At P8, IGL granule neurons in CaMKIIβ knockdown animals had longer dendrites with increased secondary and tertiary dendrite branching than IGL granule neurons in control animals (Fig. 2e). Morphometric analyses revealed a substantial increase in the number of secondary and tertiary dendrite branches and a nearly two-fold increase in total dendrite length in IGL neurons in CaMKIIβ knockdown animals compared to control animals (Fig. 2f and Supplementary Fig. 6b). CaMKIIβ knockdown had little or no effect on parallel fiber patterning or the number of parallel fibers associated with IGL granule neurons (Supplementary Fig. 6c and data not shown). Together, these data reveal a physiological, cell-autonomous function for CaMKIIβ in restricting the elaboration of dendrite arbors in the cerebellar cortex in vivo.
Because CaMKIIβ triggered active retraction of dendrites in primary neurons (Supplementary Figs. 4 and 5), we reasoned that CaMKIIβ might also play a role in pruning of dendrite arbors in vivo, which follows the phase of dendrite growth and arborization1. To test this possibility, we analyzed the effect of CaMKIIβ knockdown on dendrite morphogenesis in P12 rat pups in vivo. In control transfected animals, IGL granule neurons harbored few short dendrites with simplified arbors (Fig. 2g and Supplementary Fig. 6d–g), characteristic of the mature stage of dendrite differentiation in granule neurons. In addition, IGL granule neurons harbored dendritic claws (Fig. 2g and Supplementary Fig. 6h), which house synapses with afferent mossy fiber terminals and Golgi neuron axons1–2,29–32, providing further evidence of dendrite maturation in control P12 animals. In contrast to control animals, IGL granule neurons in CaMKIIβ knockdown animals displayed longer, more branched dendrite arbors (Fig. 2g), with significantly increased total dendrite length (78.3 ± 3.0 μm in CaMKIIβ knockdown animals versus 43.8 ± 1.5 μm in control animals; t-test, p < 0.0001), primary dendrite number (3.53 ± 0.10 in CaMKIIβ knockdown animals versus 3.01 ± 0.08 in control animals; t-test, p < 0.005) and secondary and tertiary dendrite branch number (1.13 ± 0.11 in CaMKIIβ knockdown animals versus 0.63 ± 0.07 in control animals; t-test, p < 0.005) (Supplementary Fig. 6d–g), suggesting that CaMKIIβ knockdown impairs dendrite pruning and blocks the differentiation of dendrites at the stage of exuberant arbors. Consistent with these observations, IGL granule neuron dendrites in CaMKIIβ knockdown pups had a significantly lower percentage of dendrites bearing claws (31.9 ± 2.2% in CaMKIIβ knockdown animals versus 58.9 ± 2.5% in control animals; t-test, p < 0.0001) (Fig. 2g and Supplementary Fig. 6h). Together, these results suggest that CaMKIIβ plays an essential role in dendrite pruning at later stages of dendrite morphogenesis in the cerebellar cortex in vivo. Similar results were obtained in P10 cerebellar slices (Supplementary Fig. 6i,j), corroborating the conclusion that CaMKIIβ promotes dendrite pruning during later stages of dendrite development in the cerebellar cortex. Collectively, our findings suggest that CaMKIIβ regulates the patterning of dendrites throughout development in vivo.
CaMKIIβ functions independently of CaMKIIα at the centrosome
The identification of a role for CaMKIIβ in the control of dendrite patterning in the mammalian brain led us to determine the molecular basis of CaMKIIβ function in neurons. We took advantage of the ability of CaMKIIβ-RES to rescue the CaMKIIβ RNAi phenotype to perform structure-function analyses of CaMKIIβ in the regulation of dendrite morphogenesis. In contrast to CaMKIIβ-RES, a catalytically inactive mutant of CaMKIIβ-RES in which the ATP binding site was disrupted (K43R CaMKIIβ-RES) failed to restrict the elaboration of dendrites in the background of CaMKIIβ RNAi in granule neurons (Fig. 3a), suggesting that CaMKIIβ requires the catalytic activity of the kinase to control dendrite patterning.
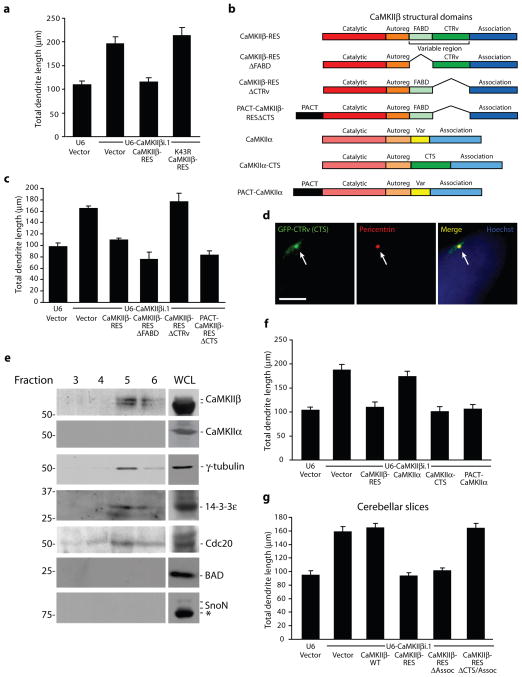
Figure 3. CaMKIIβ operates at the centrosome to specifically control dendrite patterning.
a, Granule neurons transfected with the CaMKIIβ RNAi or control U6 plasmid together with CaMKIIβ-RES, K43R CaMKIIβ-RES, or control vector and GFP were analyzed as in Fig. 1d. CaMKIIβ-RES, but not K43R CaMKIIβ-RES, significantly reduced total dendrite length compared to control vector in the background of CaMKIIβ RNAi (ANOVA; p < 0.0001). 390 neurons were measured. b, Schematic of major structural domains of CaMKIIβ. c, Granule neurons transfected with the CaMKIIβ RNAi or control U6 plasmid together with CaMKIIβ-RES, CaMKIIβ-RESΔFABD, CaMKIIβ-RESΔCTRv, PACT-CaMKIIβ-RESΔCTS, or control vector and GFP were analyzed as in (a). CaMKIIβ-RES, CaMKIIβ-RESΔFABD, and PACT-CaMKIIβ-RESΔCTS, but not CaMKIIβ-RESΔCTRv, significantly reduced total dendrite length compared to control vector in the background of CaMKIIβ RNAi (ANOVA; p < 0.0001). 660 neurons were measured. d, Granule neurons transfected with GFP fused to the C-terminal variable region (GFP-CTRv) were subjected to immunocytochemistry using the GFP and pericentrin antibody. Arrows indicate co-localization of GFP-CTRv with pericentrin. Scale bar = 5 μm. e, Fractions isolated from a granule neuron centrosome preparation were immunoblotted using the indicated antibodies. Asterisk indicates non-specific band. WCL, whole cell lysate. f, CaMKIIβ-RES, CaMKIIα-CTS, and PACT-CaMKIIα, but not CaMKIIα, significantly reduced total dendrite length compared to control vector in the background of CaMKIIβ RNAi in granule neurons (ANOVA; p < 0.0001). 600 neurons were measured. g, CaMKIIβ-RES and CaMKIIβ-RESΔAssoc, but not CaMKIIβ-WT or CaMKIIβ-RESΔAssoc/CTS, significantly reduced granule neuron total dendrite length compared to control vector in the background of CaMKIIβ RNAi in cerebellar slices (ANOVA; p < 0.0001). 479 neurons were measured.
The requirement of the kinase domain in CaMKIIβ to inhibit growth and stimulate retraction of dendrites was intriguing given that CaMKIIα, which harbors a very similar catalytic domain as CaMKIIβ, promotes dendrite growth and arborization27,33. We therefore reasoned that CaMKIIβ but not CaMKIIα might be localized at a subcellular site that endows CaMKIIβ specifically with the ability to phosphorylate local substrates and restrict the elaboration of dendrites. CaMKIIβ harbors an F-actin binding domain within the N-terminal portion of the variable region, but CaMKIIβ bound to F-actin appears to function independently of its catalytic domain as a scaffold protein that recruits CaMKIIα to F-actin34–36. Accordingly, deletion of the F-actin binding domain (ΔFABD) had little or no effect on the ability of CaMKIIβ-RES to restrict dendrite elaboration in the background of CaMKIIβ RNAi in granule neurons (Fig. 3b,c). We next focused on the C-terminal segment of the variable region (CTRv) whose function has remained unknown. Remarkably, a CaMKIIβ-RES mutant in which the CTRv was deleted (CaMKIIβ-RESΔCTRv) failed to restrict dendrite elaboration in granule neurons in the background of CaMKIIβ RNAi (Fig. 3b,c). These results suggest that the CTRv is required for CaMKIIβ-regulation of dendrite patterning.
To characterize the function of the CTRv within CaMKIIβ, we assessed whether the CTRv localizes CaMKIIβ to a distinct subcellular site where CaMKIIβ controls dendrite patterning. A GFP-fusion protein of the CTRv appeared as a perinuclear punctate signal that colocalized with the centrosomal protein pericentrin (Fig. 3d). In immuno-electron microscopy analyses, a pool of endogenous CaMKIIβ was present in the pericentriolar region in granule neurons (Supplementary Fig. 7a). In complementary biochemical fractionation experiments using granule neuron lysates, a pool of endogenous CaMKIIβ cofractionated with the centrosomal proteins γ-tubulin, Cdc20, and 14-3-3ε (Fig. 3e). Importantly, endogenous CaMKIIα did not appear in the centrosomal fraction despite the presence of CaMKIIα in whole cell lysate (Fig. 3e). Together, these data demonstrate that the CTRv represents a centrosomal targeting sequence (CTS) that localizes CaMKIIβ to the centrosome in neurons.
If the principal function of the CTS is to localize CaMKIIβ to the centrosome, forcibly targeting CaMKIIβ lacking the CTS to the centrosome by a different means should restore the ability of the CaMKIIβ mutant to inhibit dendrite growth and stimulate retraction. We therefore generated a chimeric protein in which we fused CaMKIIβ-RESΔCTS at its N-terminus to the PACT domain of AKAP450, which localizes proteins to the centrosome (PACT-CaMKIIβ-RESΔCTS) (Fig. 3b and Supplementary Fig. 7b)37. Notably, addition of the PACT domain restored the ability of CaMKIIβ-RESΔCTS to restrict the elaboration of dendrites in the setting of CaMKIIβ RNAi (Fig. 3c). In complementary experiments, replacing the variable region of CaMKIIα with the CTS (CaMKIIα-CTS) remarkably conferred CaMKIIα with the ability to robustly restrict dendrite elaboration in the background of CaMKIIβ RNAi (Fig. 3b,f). Likewise, in contrast to wild type CaMKIIα, expression of a PACT-CaMKIIα fusion protein restricted dendrite elaboration in the background of CaMKIIβ RNAi (Fig. 3b,f). Collectively, these results suggest that the CTS endows CaMKIIβ with the ability to localize at the centrosome and control dendrite patterning.
The finding that CaMKIIβ regulates dendrite patterning from the centrosome, where CaMKIIβ but not CaMKIIα is localized, suggested that centrosomal CaMKIIβ might function independently of CaMKIIα in the control of dendrite morphogenesis. To test this hypothesis, we assessed the ability of a CaMKIIβ-RES mutant in which we removed the association domain (CaMKIIβ-RESΔAssoc) to restrict dendrite elaboration in the background of CaMKIIβ RNAi. In control biochemical analyses, deletion of the association domain abrogated the interaction of CaMKIIβ with CaMKIIα (Supplementary Fig. 7c). In morphology assays, deletion of the association domain remarkably had little or no effect on the ability of CaMKIIβ-RES to restrict the elaboration of dendrites in the background of CaMKIIβ RNAi in primary granule neurons and in cerebellar slices (Fig. 3g and Supplementary Fig. 7d–f), suggesting that the association domain is dispensable for CaMKIIβ control of dendrite patterning. Importantly, as with full-length CaMKIIβ, deletion of the CTS or mutation of the ATP binding site (K43R) blocked the ability of the CaMKIIβΔAssoc mutant to restrict the elaboration of dendrites (Fig. 3g, Supplementary Fig. 7d–f, and data not shown). In immunocytochemical analyses, although full-length GFP-CaMKIIβ was localized throughout the cell soma and processes (Supplementary Fig. 7g), CaMKIIβΔAssoc was enriched at the centrosome in granule neurons (Supplementary Fig. 7h). Deletion of the CTS blocked centrosomal enrichment of the CaMKIIβΔAssoc mutant (Supplementary Fig. 7h). Together, our data demonstrate that centrosomal CaMKIIβ functions independently of multimerization with CaMKIIα to regulate dendrite patterning.
The targeting protein PCM1 localizes CaMKIIβ at the centrosome
We next characterized the mechanism that localizes CaMKIIβ at the centrosome. In view of the importance of the centrosomal targeting sequence (CTS) in CaMKIIβ’s localization at the centrosome, we reasoned that the CTS might associate with a protein that targets CaMKIIβ to the centrosome. We therefore performed a yeast two hybrid assay using the CTS as bait and a human fetal brain library as prey to identify CTS-interacting proteins. In these assays, we identified the centrosomal targeting protein pericentriolar material 1 (PCM1) as an interactor of the CTS (data not shown). Importantly, in coimmunoprecipitation analyses in cells, PCM1 formed a complex with CaMKIIβ but not CaMKIIα, and CaMKIIβ interacted with PCM1 in a CTS-dependent manner (Fig. 4a,b). In addition, endogenous PCM1 formed a complex with endogenous CaMKIIβ within centrosomal fractions of granule neurons (Fig. 4c).
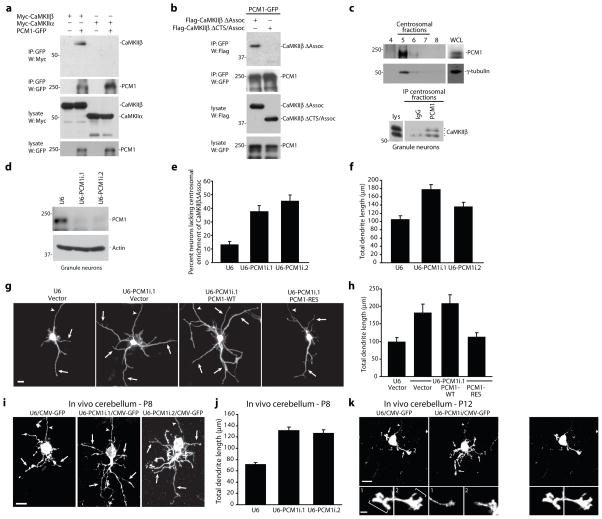
Figure 4. The centrosomal targeting protein PCM1 localizes CaMKIIβ at the centrosome.
a, Lysates of 293T cells transfected with Myc-CaMKIIβ or Myc-CaMKIIα together with PCM1-GFP or control vector were immunoprecipitated using the GFP antibody and immunoblotted with the indicated antibodies. b, Lysates of 293T cells transfected with Flag-CaMKIIβΔAssoc or Flag-CaMKIIβΔCTS/Assoc together with PCM1-GFP were immunoprecipitated using the GFP antibody and immunoblotted with the Flag or GFP antibody. c, Fractions isolated from a granule neuron centrosome preparation were immunoblotted using the PCM1 or γ-tubulin antibody. Pooled centrosomal fractions were immunoprecipitated with the PCM1 or control (IgG) antibody and immunoblotted with the CaMKIIβ antibody. d, Lysates of granule neurons electroporated with the PCM1 RNAi or control U6 plasmid were immunoblotted with the indicated antibodies. e, Granule neurons transfected with a PCM1 RNAi or control U6 plasmid together with GFP-CaMKIIβΔAssoc were analyzed as in Fig. 3d. The percentage of neurons lacking centrosomal enrichment of GFP-CaMKIIβΔAssoc was significantly increased in PCM1 knockdown neurons compared to control U6-transfected neurons (ANOVA, p < 0.005). 394 neurons were analyzed. f, Total dendrite length was significantly increased in PCM1 knockdown neurons compared to control U6-transfected neurons (ANOVA; p < 0.0001). 180 neurons were measured. g, Granule neurons transfected with the PCM1 RNAi or control U6 plasmid together with PCM1-WT, PCM1-RES, or control vector and GFP were analyzed as in (f). Scale bar = 10 μm. h, PCM1-RES, but not PCM1-WT, significantly reduced total dendrite length compared to control vector in the background of PCM1 RNAi (ANOVA; p < 0.0001). 240 neurons were measured. i, Rat pups electroporated in vivo with a U6-PCM1i/CMV-GFP RNAi or control U6/CMV-GFP plasmid were sacrificed at P8 and analyzed as in Fig. 2d. Scale bar = 10 μm. j, Total dendrite length was significantly increased in IGL granule neurons in PCM1 knockdown animals compared to control U6 animals (ANOVA, p < 0.0001). 270 neurons were measured. k, Rat pups electroporated in vivo with the U6-PCM1i/CMV-GFP RNAi or control U6/CMV-GFP plasmid were sacrificed at P12 and analyzed as in Fig. 2g. Scale bar = 10 μm. Inset: Scale bar = 2.5 μm.
To determine PCM1’s role in targeting a pool of CaMKIIβ to the centrosome, we assessed the effect of PCM1 knockdown on the localization of CaMKIIβΔAssoc, which does not interact with CaMKIIα and is enriched at the centrosome (Supplementary Fig. 7h). We found that knockdown of endogenous PCM1, achieved efficiently by two shRNAs targeting distinct regions of PCM1, led to the loss of centrosomal enrichment of CaMKIIβΔAssoc in neurons (Fig. 4d,e). These results suggest that PCM1 specifically localizes CaMKIIβ at the centrosome.
The requirement for PCM1 in localizing CaMKIIβ at the centrosome led us next to determine the functional role of PCM1 in dendrite morphogenesis. We found that PCM1 knockdown substantially increased dendrite arborization and growth in primary granule neurons (Fig. 4f,g and data not shown). Importantly, the PCM1 RNAi-induced dendrite phenotype was reversed by expression of PCM1 encoded by an RNAi-resistant cDNA (PCM1-RES) but not wild type cDNA (PCM1-WT) (Fig. 4g,h and data not shown), indicating the specificity of the PCM1 RNAi-induced phenotype. These results suggest that like CaMKIIβ, PCM1 induces dendrite retraction. We also determined the effect of PCM1 knockdown on dendrite morphogenesis in the cerebellar cortex in vivo. We found that PCM1 knockdown triggered robust elaboration of IGL granule neuron dendrite arbors in P8 rat pups (Fig. 4i,j and Supplementary Fig. 8a). Analyses at later stages of dendrite development in P12 rat pups revealed that granule neuron dendrite arbors in PCM1 knockdown animals remained long and highly branched, containing substantially fewer dendritic claws as compared to control animals (Fig. 4k and Supplementary Fig. 8b–d). Thus, PCM1 knockdown phenocopied the effect of CaMKIIβ knockdown in impairing dendrite pruning and blocking dendrite differentiation at a stage of exuberant arbors in vivo. In other experiments, PCM1 knockdown suppressed the ability of activated CaMKIIβ to stimulate dendrite retraction in granule neurons (Supplementary Fig. 8e). Collectively, these results suggest that PCM1 is required for CaMKIIβ localization at the centrosome and hence the ability of CaMKIIβ to regulate the patterning of dendrites.
The ubiquitin ligase Cdc20-APC is a novel CaMKIIβ substrate
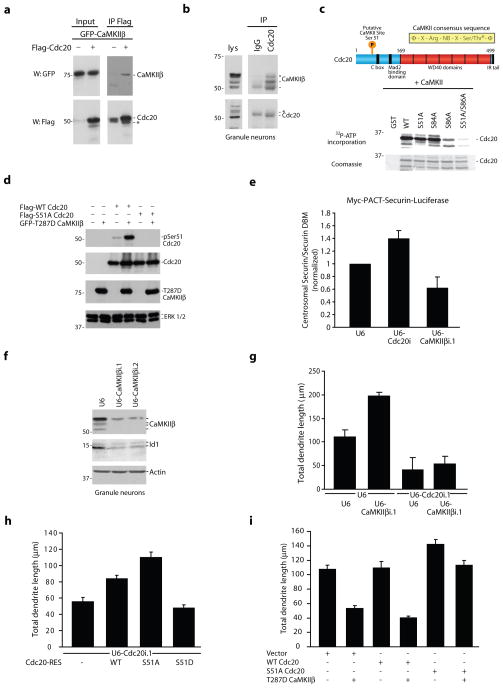
Since the catalytic activity of CaMKIIβ at the centrosome is required for regulating dendrite morphogenesis, we next sought to identify the protein substrates of centrosomal CaMKIIβ in neurons. We considered proteins that have established roles in dendrite development, localize to the centrosome, and contain a consensus sequence for CaMKII phosphorylation. Recently, the major mitotic E3 ubiquitin ligase Cdc20-APC has been identified as a critical mediator of dendrite growth that operates at the centrosome in post-mitotic neurons38. We found that the N-terminus of the APC coactivator Cdc20 has several potential CaMKII sites, including Ser51 and Ser86 based on the CaMKII consensus sequence39. Consistent with the possibility that CaMKIIβ might phosphorylate Cdc20, CaMKIIβ and Cdc20 formed a complex in 293T cells and neurons (Fig. 5a,b and Supplementary Fig. 9a).
Figure 5. CaMKIIβ phosphorylates Cdc20 at Ser51 and thereby inhibits centrosomal Cdc20-APC activity in neurons.
a, Lysates of 293T cells transfected with GFP-CaMKIIβ together with Flag-Cdc20 or control vector were immunoprecipitated using the Flag antibody and immunoblotted with the indicated antibodies. b, Lysates of granule neurons were immunoprecipitated with the Cdc20 or control (IgG) antibody and immunoblotted with the indicated antibodies. Asterisk indicates IgG heavy chain. c, (Top) Schematic of CaMKII consensus sequence. (Bottom) Recombinant WT and S51A, S84A, S86A, and S51A/S86A mutants of an N-terminal region of Cdc20 (1–101) fused to GST were incubated in vitro with purified CaMKII. d, Lysates of 293T cells transfected with Flag-WT Cdc20 or Flag-S51A Cdc20 together with GFP-T287D CaMKIIβ or control vector were immunoblotted with the indicated antibodies. e, Granule neurons were transfected with the Cdc20 RNAi, CaMKIIβ RNAi, or control U6 plasmid together with Myc-PACT-securin-luciferase or Myc-PACT-securin-DBM-luciferase and analyzed by luminometry. The relative amount of the centrosomal securin-luciferase reporter was significantly increased in Cdc20 knockdown neurons and significantly reduced in CaMKIIβ knockdown neurons compared to control U6-transfected neurons (Kruskal-Wallis; p < 0.01, n = 4). f, Lysates of granule neurons electroporated with the CaMKIIβ RNAi or control U6 plasmid were immunoblotted with the indicated antibodies. g, CaMKIIβ knockdown significantly increased total dendrite length compared to control, and Cdc20 RNAi significantly reduced total dendrite length in granule neurons in the presence or absence of CaMKIIβ RNAi (ANOVA, p < 0.0001). 420 neurons were measured. h, Expression of S51A Cdc20-RES, but not S51D Cdc20-RES, significantly increased total dendrite length compared to control vector or WT Cdc20-RES in the background of Cdc20 RNAi in granule neurons (ANOVA, p < 0.0001). 428 neurons were measured. i, Expression of T287D CaMKIIβ significantly reduced total dendrite length compared to control in the background of control vector or WT Cdc20, but not in the background of S51A Cdc20 (ANOVA, p < 0.0001). 540 neurons were measured.
Purified CaMKII catalyzed the phosphorylation of full-length and an N-terminal domain of Cdc20 (1–101) in vitro, and mass spectrometric analyses of phosphorylated Cdc20 revealed that CaMKII phosphorylated Cdc20 at Ser51, Ser84, and Ser86 (data not shown). Mutation analysis demonstrated that Ser86 and Ser51 contributed to CaMKII-mediated phosphorylation of Cdc20, while mutation of Ser84 to alanine did not significantly alter 32P-ATP incorporation (Fig. 5c), suggesting that CaMKIIβ does not stoichiometrically phosphorylate Ser84 in vitro. Notably, Ser51 but not Ser86 is evolutionarily conserved from zebrafish to humans. We therefore generated an antibody to specifically recognize Cdc20 when phosphorylated at Ser51. The phospho-Cdc20 antibody recognized wild type Cdc20 but not the Ser51 mutant of Cdc20 when co-expressed with activated CaMKIIβ in cells (Fig. 5d). Immunoblotting experiments using the phospho-Cdc20 antibody revealed that endogenous Cdc20 is phosphorylated at Ser51 in granule neurons, and knockdown of CaMKIIβ reduced the level of endogenous Ser51-phosphorylated Cdc20 in neurons (Supplementary Fig. 9b). Together, these results indicate that endogenous CaMKIIβ phosphorylates Cdc20 at Ser51 in neurons, raising the question of whether CaMKIIβ regulates the activity of Cdc20-APC at the centrosome to control dendrite patterning.
To determine the role of CaMKIIβ in regulating the ubiquitin ligase activity of centrosomal Cdc20-APC in neurons, we first utilized a Myc-PACT-securin-luciferase reporter, which is a validated readout of centrosomal Cdc20-APC activity in neurons38. The reporter encodes the first 88 amino acids of the mitotic Cdc20-APC substrate securin fused to firefly luciferase. This portion of securin harbors the destruction box (D-box), an APC degron40, and the PACT domain localizes the reporter to the centrosome38. A mutant version of the reporter containing a mutation in the securin D-box (securin-DBM) serves as control40. Thus, the ratio of securin to securin-DBM luciferase is inversely proportional to centrosomal Cdc20-APC activity38. As reported, Cdc20 knockdown increased the relative levels of the securin-luciferase reporter in granule neurons (Fig. 5e)38. In contrast, we found that CaMKIIβ knockdown reduced the relative levels of the securin-luciferase reporter in neurons (Fig. 5e), suggesting that CaMKIIβ inhibits centrosomal Cdc20-APC activity in neurons. CaMKIIβ knockdown also reduced levels of the centrosomal HLH protein Id1 (Fig. 5f), which is a physiological substrate of centrosomal Cdc20-APC in neurons38. Collectively, these data suggest that CaMKIIβ inhibits Cdc20-APC activity at the centrosome and the consequent degradation of its substrates in neurons.
In view of Cdc20-APC’s function in dendrite growth and arborization, the finding that CaMKIIβ inhibits the activity of Cdc20-APC at the centrosome in neurons suggested that CaMKIIβ might stimulate dendrite retraction via the regulation of centrosomal Cdc20-APC activity. Consistent with this prediction, in epistasis analyses, Cdc20 knockdown suppressed the effect of CaMKIIβ knockdown on dendrite elaboration in granule neurons (Fig. 5g). In addition, knockdown of the centrosomal Cdc20-APC substrate Id1 suppressed the ability of CaMKIIβ to induce dendrite retraction (data not shown). Together, these results suggest that CaMKIIβ stimulates dendrite retraction via inhibition of centrosomal Cdc20-APC activity in neurons.
Phosphorylation induces Cdc20 dispersion from the centrosome
We next assessed the role of Cdc20 phosphorylation at Ser51 in CaMKIIβ-regulated dendrite patterning. Expression of Cdc20-RES promotes dendrite growth and elaboration in the background of Cdc20 RNAi38. We found that expression of a mutant Cdc20-RES protein in which Ser51 was replaced with the non-phosphorylatable residue alanine (S51A Cdc20-RES) stimulated dendrite growth more effectively than Cdc20-RES (Fig. 5h). In contrast, expression of a mutant Cdc20-RES protein in which Ser51 was replaced with phosphomimetic residue aspartate (S51D Cdc20-RES) failed to stimulate dendrite growth (Fig. 5h). Notably, expression of S51A Cdc20 but not wild type Cdc20 substantially impaired the ability of constitutively active CaMKIIβ to induce dendrite retraction (Fig. 5i). In other experiments, mutation of Ser84 or Ser86 had little or no effect on the ability of Cdc20-RES to drive dendrite elaboration (Supplementary Fig. 9c). Collectively, our results suggest that CaMKIIβ phosphorylates Cdc20 at Ser51 and inhibits centrosomal Cdc20-APC activity in neurons, thereby triggering a switch from dendrite growth and arborization to dendrite retraction.
We next determined the mechanism by which CaMKIIβ-induced phosphorylation of Cdc20 inhibits Cdc20-APC function at the centrosome in neurons. Because the localization of Cdc20 at the centrosome is critical for its function in dendrite growth and arborization38, we asked whether CaMKIIβ phosphorylation of Cdc20 might interfere with the centrosomal localization of Cdc20. Strikingly, expression of wild type or constitutively active CaMKIIβ (T287D), but not the catalytically inactive K43R CaMKIIβ mutant or CaMKIIα, induced dispersion of endogenous Cdc20 away from the centrosome in granule neurons (Fig. 6a,b and Supplementary Fig. 9d,e). Importantly, expression of CaMKIIβ had little or no effect on centrosomal structure in neurons as monitored by endogenous pericentrin or co-transfection with GFP-centrin (Supplementary Fig. 9e and data not shown). Conversely, CaMKIIβ knockdown reduced basal Cdc20 dispersion in granule neurons (Supplementary Fig. 9f), suggesting that endogenous CaMKIIβ inhibits the centrosomal localization of Cdc20 in neurons.
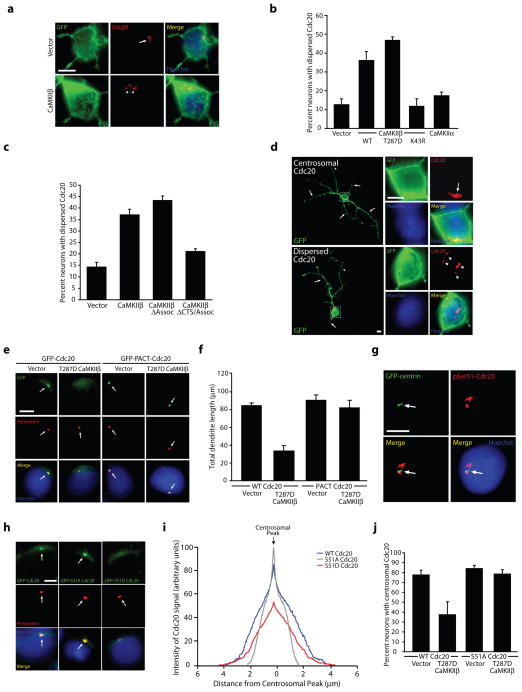
Figure 6. CaMKIIβ-induced phosphorylation of Cdc20 promotes Cdc20 dispersion from the centrosome, leading to the restriction of dendrite elaboration.
a, Granule neurons transfected with CaMKIIβ or control vector together with farnesylated GFP (fGFP) were subjected to immunocytochemistry using the GFP or Cdc20 antibody. Arrows indicate Cdc20 localized to the centrosome, while arrowheads denote dispersed Cdc20. Scale bar = 5 μm. b, The percentage of neurons displaying dispersed endogenous Cdc20 was significantly increased in T287D CaMKIIβ- and WT CaMKIIβ-expressing neurons, but not K43R CaMKIIβ- or CaMKIIα-expressing neurons, as compared to control vector-transfected neurons (ANOVA; p < 0.0001). 622 neurons were analyzed. c, The percentage of neurons displaying dispersed endogenous Cdc20 was significantly increased in CaMKIIβ- and CaMKIIβΔAssoc-expressing neurons, but not CaMKIIβΔCTS/Assoc-expressing neurons, as compared to control vector-transfected neurons (ANOVA; p < 0.0001). 360 neurons were analyzed. d, Granule neurons transfected with T287D CaMKIIβ together with fGFP were analyzed as in (a). Arrows indicate dendrites, while dashed box indicates the magnified region in inset images. Scale bar = 5 μm. e, Granule neurons transfected with GFP-Cdc20 or GFP-PACT-Cdc20 together with T287D CaMKIIβ or control vector were analyzed as in Fig. 3d. Arrows indicate the location of the centrosome, which is labeled with pericentrin. Scale bar = 5 μm. f, Total dendrite length was significantly reduced in T287D CaMKIIβ-expressing neurons compared to control-transfected neurons in the background of Cdc20 (ANOVA, p < 0.0005), but not in the background of PACT-Cdc20. 360 neurons were measured. g, Granule neurons transfected with GFP-centrin were subjected to immunocytochemistry using the GFP or phosphoSer51-Cdc20 antibody. Arrowheads indicate phosphoSer51-Cdc20 immunoreactivity. Scale bar = 5 μm. h, Granule neurons transfected with GFP-Cdc20, GFP-S51A Cdc20, or GFP-S51D Cdc20 were analyzed as in (e). Scale bar = 5 μm. i, Granule neurons treated as in (h) were analyzed by linescan analysis. The location of the centrosome was identified based on pericentrin immunoreactivity. S51A Cdc20 had enhanced signal at the centrosome and reduced cytosolic signal compared to WT Cdc20. In contrast, S51D Cdc20 had reduced centrosomal signal and broader non-centrosomal signal compared to WT Cdc20 (based on peak centrosomal signal intensity, ANOVA, p < 0.0001). 270 neurons were analyzed. j, Expression of T287D CaMKIIβ significantly reduced the percentage of neurons with centrosomally-enriched WT Cdc20 compared to control vector, but had no effect on the centrosomal localization of S51A Cdc20 (ANOVA; p < 0.01). 361 neurons were analyzed.
To assess whether CaMKIIβ stimulates Cdc20 dispersion independently of CaMKIIα in neurons, we tested the effect of deletion of the association domain on the ability of CaMKIIβ to induce endogenous Cdc20 dispersion in granule neurons. Remarkably, the CaMKIIβΔAssoc mutant protein induced Cdc20 dispersion as effectively as CaMKIIβ (Fig. 6c), suggesting that the association domain is dispensable for CaMKIIβ-induced dispersion of Cdc20 in neurons. Importantly, deletion of the CTS in CaMKIIβΔAssoc diminished its ability to induce Cdc20 dispersion (Fig. 6c). Together, our results suggest that the protein kinase CaMKIIβ functions at the centrosome independently of CaMKIIα to drive Cdc20 dispersion in neurons.
To determine whether the regulation of Cdc20 localization by CaMKIIβ is relevant to dendrite morphogenesis, we first compared dendrite length in granule neurons displaying centrosomal enrichment of Cdc20 with neurons in which Cdc20 was dispersed. Neurons with dispersed endogenous Cdc20 had substantially shorter dendrites compared to neurons with centrosomally localized Cdc20 (35.8 ± 10.2 μm in neurons with dispersed Cdc20 versus 105.5 ± 9.9 μm in neurons with centrosomal Cdc20; t-test, p < 0.01) (Fig. 6d and Supplementary Fig. 9g). In other experiments, dispersion of endogenous Cdc20 in granule neurons increased with maturation (Supplementary Fig. 9h), suggesting that Cdc20 dispersion correlates temporally with CaMKIIβ function in dendrite retraction and pruning. If Cdc20 dispersion from the centrosome is critical for CaMKIIβ-induced dendrite retraction, then forcibly localizing Cdc20 to the centrosome should override the CaMKIIβ response. Consistent with this prediction, we found that a Cdc20 protein fused to the centrosome localizing PACT domain (PACT-Cdc20) suppressed the ability of T287D CaMKIIβ to promote dendrite retraction in granule neurons (Fig. 6f). In control experiments, although expression of T287D CaMKIIβ induced Cdc20 dispersion, T287D CaMKIIβ failed to disperse PACT-Cdc20 (Fig. 6e), suggesting that PACT-Cdc20 is insensitive to CaMKIIβ regulation. These results suggest that Cdc20 dispersion from the centrosome is critical for CaMKIIβ regulation of dendrite patterning.
We next assessed the role of CaMKIIβ-induced Cdc20 phosphorylation at Ser51 in the control of Cdc20 localization at the centrosome. In immunocytochemical analyses using the phospho-Cdc20 antibody, we found that endogenous Ser51-phosphorylated Cdc20 immunoreactivity had a broader distribution beyond the centrosome in granule neurons, suggesting that the dispersed pool of Cdc20 is phosphorylated at Ser51 (Fig. 6g). Importantly, expression of activated CaMKIIβ increased the number of neurons harboring phosphoSer51-Cdc20 immunofluorescence (Supplementary Fig. 9i), whereas knockdown of endogenous CaMKIIβ but not CaMKIIα reduced the number of neurons with Ser51-phosphorylated Cdc20 (Supplementary Fig. 9j). To directly test the importance of Ser51 phosphorylation in Cdc20 dispersion, we characterized the effect of mutations of Ser51 on Cdc20 localization in neurons. We found that the non-phosphorylatable S51A Cdc20 mutant protein appeared to be more enriched at the centrosome than wild type Cdc20, while the phosphomimetic S51D mutant appeared to be less enriched at the centrosome (Fig. 6h). Quantification of the Cdc20 immunofluorescence signal using linescan analyses revealed that the S51A Cdc20 signal peaked at the centrosome and displayed a more limited distribution outside the centrosome (Fig. 6i). By contrast, S51D Cdc20 had a widened centrosomal peak with a broader non-centrosomal signal (Fig. 6h,i). In other experiments, expression of T287D CaMKIIβ robustly induced dispersion of wild type Cdc20 but not S51A Cdc20 from the centrosome in granule neurons (Fig. 6j), suggesting CaMKIIβ induces Cdc20 dispersion in a Ser51 phosphorylation-dependent manner. Importantly, addition of the PACT domain to S51D Cdc20 restored the ability of S51D Cdc20 to stimulate dendrite growth and elaboration, suggesting that the function of Ser51 phosphorylation is to localize Cdc20 away from the centrosome (Supplementary Fig. 9k). These data suggest that CaMKIIβ-induced phosphorylation of Cdc20 at Ser51 triggers its dispersion from the centrosome, leading to the inhibition of centrosomal Cdc20-APC activity and the promotion of dendrite retraction in neurons. Collectively, we have elucidated a centrosomal CaMKIIβ signaling pathway that controls the patterning of dendrite arbors with important consequences on the establishment of neuronal connectivity in the mammalian brain (see model in Supplementary Fig. 10).
DISCUSSION
In this study, we have discovered the first catalytic function of CaMKIIβ in the mammalian brain. Remarkably, CaMKIIβ operates at the centrosome in a CaMKIIα-independent manner to drive dendrite retraction and pruning. CaMKIIβ localizes at the centrosome by forming a complex with the centrosomal trafficking protein PCM1. Accordingly, PCM1 is required for CaMKIIβ-regulation of dendrite morphogenesis and the pruning of dendrites in postnatal rat pups in vivo. We have also identified the ubiquitin ligase Cdc20-APC, which is enriched at the centrosome in neurons, as a novel substrate of CaMKIIβ. CaMKIIβ phosphorylates Cdc20 at Ser51 and thereby triggers the dispersion of Cdc20 from the centrosome, culminating in the inhibition of centrosomal Cdc20-APC activity and consequent dendrite retraction. Collectively, our findings define a novel isoform-specific function of CaMKIIβ that regulates ubiquitin-dependent protein degradation at the centrosome and thereby orchestrates dendrite patterning in mammalian brain neurons.
The identification of an unexpected function for CaMKIIβ in dendrite retraction and pruning bears significant ramifications for our understanding of the major protein kinase CaMKII. As the predominant CaMKII isoforms in the brain, CaMKIIα and CaMKIIβ form holoenzyme complexes in neurons16–18. However, nearly all of the reported functions of CaMKII have focused on CaMKIIα in homomeric or heteromeric complexes19–22. Accordingly, CaMKIIβ has been previously relegated to a largely redundant role within CaMKIIα heteromeric complexes or as a scaffold recruiting CaMKIIα to specific cellular locales such as dendritic spines34–36. Our finding that CaMKIIβ operates at the centrosome in a CaMKIIα-independent manner unveils a biological function for CaMKIIβ homomeric complexes. Thus, rather than simply contributing to CaMKIIα complexes, CaMKIIβ homomers harbor a major biological function at the centrosome as drivers of dendrite retraction and pruning and consequent neuronal connectivity in the mammalian brain. During brain development, CaMKIIβ expression peaks at earlier time points relative to CaMKIIα41. Therefore, it will be interesting to determine in future studies whether centrosomal CaMKIIβ homomeric complexes might also contribute to earlier aspects of neuronal development in which centrosomal signaling is thought to play a critical role, including neuronal polarization and migration42–43. Beyond the nervous system, it will be important to identify the role of CaMKIIβ homomers in cellular homeostasis and development in other organ systems.
Elucidation of the CaMKIIβ/Cdc20 signaling link at the centrosome provides a mechanism by which CaMKIIβ promotes dendrite retraction and pruning. By inducing the phosphorylation of Cdc20 and inhibiting Cdc20-APC ubiquitin ligase activity at the centrosome, CaMKIIβ triggers a switch from dendrite growth and elaboration to dendrite retraction and pruning. These findings suggest that the centrosome may represent a critical signaling platform that integrates diverse cellular signals to determine the phase of dendrite morphogenesis in neurons. In view of the fundamental role of centrosome signaling in diverse systems from the control of cell polarity and migration to ciliary morphogenesis to vesicular transport43, it will be interesting to explore whether centrosomal CaMKIIβ phosphorylation of Cdc20, or of other substrates yet to be identified, might contribute to these diverse biological processes.
The identification of the CaMKIIβ/Cdc20 signaling link also advances our understanding of the regulation of the major ubiquitin ligase Cdc20-APC. CaMKIIβ phosphorylates Cdc20 at Ser51, inducing dispersion of Cdc20 from the centrosome and inhibiting centrosomal Cdc20-APC activity. The phosphorylation-dependent control of the subcellular localization of Cdc20 represents an entirely new mode of regulation of the ubiquitin ligase Cdc20-APC. Interestingly, CaMKIIβ had little or no effect on Cdc20 binding to core APC subunits, the in vitro ubiquitin ligase activity of Cdc20-APC, or Cdc20 polyubiquitination (data not shown), highlighting the significance of Cdc20 dispersion as a critical cellular mechanism for regulating centrosomal Cdc20-APC activity. Future research should explore whether Cdc20 has additional roles outside of the centrosome, bearing in mind the exciting possibility that Cdc20 dispersion might be part of a developmental program that coordinates dendrite patterning with other steps in the establishment of neuronal connectivity.
Although CaMKIIβ and CaMKIIα are similar in their catalytic, autoregulatory, and association domains, they diverge in their variable region41. We have identified a centrosomal targeting sequence (CTS) in the unique variable region of CaMKIIβ, thus providing spatial specificity for CaMKIIβ function. Further, we have found that the CTS mediates the interaction of CaMKIIβ specifically with the centrosomal targeting protein PCM1. Although PCM1 targets structural proteins to the centrosome44, our findings suggest that PCM1 may also drive the centrosomal localization of signaling proteins. Notably, in addition to interacting with CaMKIIβ, endogenous PCM1 formed a complex with endogenous Cdc20 in neurons and stimulated CaMKIIβ phosphorylation of Cdc20 (data not shown). These observations suggest that beyond recruiting CaMKIIβ to the centrosome, PCM1 may also operate as a scaffold protein that organizes the CaMKIIβ/Cdc20 signaling pathway at the centrosome.
Although signaling and cell-intrinsic mechanisms that promote dendrite growth have been the subject of substantial interest5–8, the master regulatory mechanisms that govern the developmental transition from dendrite elaboration to dendrite pruning in the mammalian brain have remained to be elucidated. The identification of centrosomal CaMKIIβ signaling as a mechanism that restricts dendrite elaboration and promotes dendrite pruning suggests that pathways that actively drive dendrite retraction have evolved to sculpt dendrite arbors and thus establish accurate neuronal circuits. Since abnormalities in dendrite development represent prominent pathological features in mental retardation and autism spectrum disorders4,45–46, it will be interesting to determine if deregulation of centrosomal CaMKIIβ signaling contributes to the pathogenesis of neurodevelopmental disorders of cognition.
ONLINE METHODS
Antibodies
Monoclonal CaMKIIβ and CaMKIIα antibodies (Zymed), polyclonal rabbit phosphoThr287-CaMKIIβ antibody (Badrilla), polyclonal rabbit CaMKIIα, CaMKIIβ, and ERK 1/2 antibodies (Cell Signaling), rabbit BAD, mouse Myc, mouse Actin, rabbit Cdc20, rabbit Id1, and rabbit SnoN antibodies (Santa Cruz Biotechnology), mouse Flag, γ-tubulin, Calbindin, and MAP2 antibodies (Sigma), mouse α-tubulin, mouse HA (clone 16B12), and rabbit pericentrin antibodies (Covance), rabbit and mouse GFP antibodies (Invitrogen), mouse beta-galactosidase antibody (Promega), and rat HA (clone 3F10) antibody (Roche) were purchased. Rabbit 14-3-3ε antibody was a generous gift from Dr. Alastair Aitken (University of Edinburgh). Rabbit PCM1 antibody has been described44. Rabbit phosphoSer51-Cdc20 antibody was generated by injection of rabbits with the phospho-peptide CAANRSHpSAGRTPG (amino acids 44–57 in rat Cdc20) and purified by standard affinity purification at Cell Signaling Technology.
Plasmids
Rat CaMKIIβ and CaMKIIα cDNA was a gift from Dr. Tobias Meyer (Stanford) and was cloned into pcDNA3 to produce Myc epitope-tagged and Flag epitope-tagged CaMKIIβ expression plasmids and into pEGFP-C1 (Clontech) to produce an N-terminal GFP-tagged CaMKIIβ expression plasmid. C-terminal GFP-tagged chicken PCM1 in pEGFP-N1 (Clonetech) has been described44. GFP-centrin was a gift from Dr. Karl Munger (Harvard).
shRNA plasmids were produced by insertion of the following oligonucleotides into pBS/U6 or pBS/U6-CMV-GFP: U6-CaMKIIβi.1: 5′-GT CCG ACG CTG TGT CAA GCA CAAGTTAAC AGC TTG ACA CAG CGT CGG ACC TTTTTG; U6-CaMKIIβi.2: 5′-GCA GCT AAG ATC ATT AAC ACG CAAGTTAAC GGT GTT AAT GAT CTT AGC TGC CTTTTTG; U6-PCM1i.1: 5′-CTT GAA GCT CTA ATG GCT GAA CTTTTTG; U6-PCM1i.2: GGA GCA CAT GGA TGA AGT ATG CTTTTTG; U6-Cdc20i and U6-Id1i have been described38.
Mutations in CaMKIIβ and other expression plasmids were performed using standard protocols and confirmed by sequencing. F-actin binding domain and C-terminal variable region deletion constructs were generated by deletion of the F-actin binding domain and C-terminal variable region (consisting of the linker and C-terminal segment) as described36,41. CaMKIIβ-RESΔAssoc and CaMKIIβ-RESΔCTS/Assoc were created by standard PCR subcloning of CaMKIIβ lacking the association domain and CaMKIIβ lacking the association domain and C-terminal variable region36,41 into expression constructs.
Primary neuron cultures and transfection
Primary cerebellar granule neurons were prepared from P6 rat pups and transfected via a modified calcium phosphate protocol as described28. To avoid the possibility that morphological effects of RNAi or protein expression were a result of changes in cell survival, we included the expression plasmid encoding the anti-apoptotic protein Bcl-xl in all neuronal transfections. The CaMKIIβ RNAi-induced dendrite phenotype was identical in the presence or absence of Bcl-xl expression (data not shown). Hippocampal and cerebral cortical neuron cultures were prepared from E18 rat embryos as described47 and transfected using a modified calcium phosphate protocol.
Cerebellar slice cultures and in vivo electroporation
P6 and P10 rat cerebella were prepared as described27. Individual neurons in P6 and P10 slices were transfected after two or four days, respectively, using biolistics (Helios gene gun, Bio-Rad) as described27. Four days after transfection, slices were subjected to immunohistochemical analyses.
All experiments using live animals have been approved by the HMS Standing Committee on Animals and strictly conform to their regulatory standards. In vivo electroporation of P3 Sprague-Dawley rat pups was performed as described28. Five or nine days after electroporation (P8 or P12, respectively), animals were euthanized, and cerebella harvested. Coronal sections of cerebella (40 μm) were prepared and subjected to immunohistochemistry with the GFP and Calbindin antibody and the DNA dye bisbenzimide (Hoechst 33258).
Time-lapse analyses of dendrite morphogenesis
For initial time-lapse analyses, granule neurons were plated on etched coverslips (Bellco) and transfected after one day in vitro (DIV1). Beginning at DIV2, individual neurons were monitored by noting their position on the grid and obtaining sequential images of that position over the 48 hour period of analysis. Neurons were randomly selected. For CaMKIIβ knockdown experiments, granule neurons were similarly plated but transfected on DIV0. Beginning at DIV2, CaMKIIβ knockdown and control U6-transfected neurons were imaged.
For live confocal imaging analyses, granule neurons were plated on glass-bottom multi-well plates (MatTek) and transfected as described for initial time-lapse analyses. Beginning at DIV2, live imaging was performed using a PerkinElmer Life and Analytical Sciences UltraVIEW spinning-disk confocal system with a Nikon Ti-E Perfect-Focus microscope. An environment-controlled chamber maintained the neurons at 37°C, 5% CO2. Images were acquired and analyzed using Volocity software (PerkinElmer Life and Analytical Sciences). A 20x objective was used in combination with an automated stage to capture images over 48 hours. Neurons were chosen randomly and followed by saving their coordinates on the motorized stage. Dendrite extension and retraction events were defined as events where dendrites extended or retracted by greater than 2 μm over the one hour period analyzed.
Immunocytochemistry
For visualization of centrosomal proteins, neurons were fixed in absolute methanol for ten minutes at −20°C and subjected to immunofluorescence analysis after blocking and staining with the indicated antibodies according to standard protocols. For other immunocytochemistry experiments, neurons were fixed in 4% paraformaldehyde for 20 minutes at room temperature and analyzed as described28. Cells were counted as having dispersed Cdc20 if there were multiple discrete Cdc20 puncta (three or greater) or if puncta of Cdc20 immunofluorescence localized away from the centrosome based on GFP-centrin or endogenous pericentrin.
Immuno-electron microscopy analyses
Granule neurons were fixed in 4% paraformaldehyde with 0.025% glutaraldehyde and 5 μg/mL Taxol in Brinkley Buffer 1980 (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA) for ten minutes at 37°C. Neurons were permeabilized in 0.1% Triton X-100 in BRB80 with 5 μg/mL Taxol and immunostained with CaMKIIβ or control (IgG) antibody overnight. Samples were then incubated with 5 nm gold-conjugated Protein A, sectioned using an ultramicrotome (Reichert), and collected on coated copper grids. Sections were visualized using a JEOL 1200EX transmission electron microscope.
Centrosomal fractionation of primary neuronal lysates
Centrosomal fractions from granule neurons or cortical neurons were isolated as described48. Briefly, neurons were treated with 2 μg/mL nocodazole and 1 μg/mL cytochalasin D at 37°C for one hour, washed sequentially in cold 1X PBS, 0.1X PBS + 8% sucrose (w/w), and 8% sucrose, and then lysed in a hypotonic lysis buffer with a protease inhibitor cocktail (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and pepstatin) and NaF. Homogenates were centrifuged at 1500 g for 10 min, and PIPES pH 7.2 (final concentration 10 mM) was added to the post-nuclear supernatant. The post-nuclear fraction was treated with 2 μg/mL DNase I for 30 min at 4°C and then layered on top of a discontinuous sucrose gradient (70%/50%/40% sucrose (w/w) in 10 mM PIPES pH 7.2, 0.1% Triton X-100, and 0.1% β-ME) and subjected to ultracentrifugation at 20,000 rpm in a SW-60 rotor (Beckman) for 1.5 hr. Fractions were collected from the bottom of the tube and analyzed by immunoblotting.
Immunoprecipitation analyses
Cells were lysed in 150 mM NaCl, 20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1% NP40 containing protease inhibitors. Lysates were briefly pre-cleared with a combination of Protein A/G sepharose beads and then incubated with either the appropriate antibody or antibody-conjugated beads overnight. For non-conjugated antibodies, the antibody-protein complexes were immunoprecipitated with Protein A/G beads. Immunoprecipitated proteins bound to beads were washed several times, and lysates were analyzed by SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting analysis.
Purification of GST-Cdc20 fusion proteins and in vitro kinase assays
GST-Cdc20 constructs were expressed in E. coli, and purified via solubilization in buffer containing 500 mM NaCl, 20 mM Tris pH 7.5, 0.2 mM EGTA, 0.2 mM EDTA, and protease inhibitors. Purified GST-Cdc20 proteins were incubated with CaMKII purified from rat forebrain (Calbiochem) for ten minutes at 37°C in the presence of 10 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM Na3VO4, 0.5 mM CaCl2, 2.5 mM dithiothreitol, 10 μg/mL calmodulin, 200 μM ATP (containing trace 32P-γ-ATP). Reactions were terminated by addition of SDS sample buffer, and samples were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. Radioactivity was visualized by Phosporimager analysis.
Mass Spectrometry Analyses
The Coomasie stained band corresponding to the putative Cdc20 protein was excised from the gel and destained using 50% CH3CN/50 mM NH4HCO3. Gel was dehydrated with 100% CH3CN, followed by rehydration with 2 M Urea/25 mM Tris buffer, pH 8.8, containing 25 ng/μl of the endopeptidase Lys-C. In gel digestion was performed overnight at 37°C. LC-MS/MS data were obtained using an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher). The sample was loaded onto a pulled fused silica microcapillary column (125 μm, 15 cm) packed with C18 reverse-phase resin (Magic C18AQ; Michrom Bioresources). Peptides were separated using an Agilent 1200 series binary pump across a 50 min linear gradient of 4–25% CH3CN in 0.125% HCOOH at a flow rate of 800 nL/min. In each data collection cycle, one full MS scan (300–1500 m/z) was acquired in the Orbitrap (6 × 104 resolution, automatic gain control (AGC) target of 106), followed by 10 data-dependent MS/MS scans in the LTQ (AGC target, 104; threshold 5 × 103) using the 10 most abundant ions and electron transfer dissociation (ETD) for fragmentation. We employed a reaction time of 69 ms and an anion target value of 2 × 105 reagent ions. SEQUEST was used to search MS/MS spectra for rat Cdc20 sequence to identify Cdc20 phosphopeptides. The search parameters used for post-translational modification included 79.96633 Da on serine, threonine, and tyrosine for phosphorylation.
Luciferase reporter assays
The Myc-PACT-securin-luciferase or Myc-PACT-securin DBM-luciferase plasmids were transfected with the indicated RNAi plasmid or control U6 vector in granule neurons as described38. Four days later, neurons were treated with cycloheximide (100 μg/mL) for four hours, and lysates were subjected to luciferase measurements using luminometry (Promega). Luminescence values obtained with securin-luciferase transfection were divided by those of securin DBM-luciferase (Centrosomal Securin/Securin DBM), yielding a value inversely proportional to centrosomal Cdc20-APC activity.
Analysis of neuronal morphology and imaging
To analyze the axonal and dendritic morphology of primary neurons in culture, in slices, and in vivo, images of individual neurons were captured randomly in a blinded manner on a Nikon Eclipse TE2000 epifluorescence microscope using a digital CCD camera (Diagnostic Instruments). SPOT software was used to measure individual process length by tracing. Axons and dendrites were identified in transfected neurons based on morphology and selective expression of MAP2 and Tau1 in dendrites and axons, respectively (data not shown). Total dendrite length was determined by summing the lengths of all dendrite processes measured from a single neuron. To analyze dendrite morphology in vivo, granule neurons residing in the IGL were selected for morphometry. Percent parallel fiber association was determined by counting the number of parallel fibers and cell bodies present in a specific region of a section in consecutive sections in a blinded manner as described49. To analyze cell survival, neurons were transfected with the β-galactosidase expression plasmid and subjected to immunocytochemistry using the β-galactosidase antibody and the DNA dye bisbenzimide (Hoechst 33258). Cell survival was scored by assessment of process fragmentation and nuclear condensation.
Confocal images were collected using an Olympus IX81 microscope with FluoView1000 scanning confocal unit (taken with a 40X/0.90 NA Olympus UPlanSApo or 60X/1.42 NA Oil Olympus PlanApoN objective). Labeled neurons were excited at 405 nm, 488 nm and 559 nm, and emission was collected at 425–475 nm, 500–545 nm, and 575–675 nm for Hoechst, Alexa Fluor-488, and Cy3, respectively.
Linescan analyses
Images taken from transfected neuron cohorts using identical acquisition parameters were subjected to linescan analyses. The immunofluorescent signal was analyzed using Olympus Fluoview-1000 image analysis software by taking pericentrin signal as the location of the centrosome and tracing GFP-Cdc20 signal and quantifying immunofluorescence intensity along the line. Linescan intensity plots from individual neurons were aligned at the centrosomal peak (defined as zero on the x-axis) and were combined to generate an average for the cohort. Plots were thresholded at the level of background signal. Intensity values along the traced line are reported as a percentage of the maximum centrosomal peak signal.
Statistics
All analyses were completed from a minimum of three independent experiments. The number of cells contributing to each condition was equally distributed across independent experiments. Statistical analyses were performed with GraphPad Prism 4.0. All histograms are presented as mean + SEM unless otherwise noted. The Student’s t-test was utilized for comparisons in experiments with two sample groups. In experiments with more than two sample groups, analysis of variance (ANOVA) was performed followed by Bonferroni’s post-hoc test. For non-parametric data with more than two sample groups, the Kruskal-Wallis test was used.
Supplementary Material
Acknowledgments
We thank Dr. Maria Ericsson for assistance with electron microscopy experiments and Dr. John Blenis and members of the Bonni laboratory for helpful discussions and critical reading of the manuscript. This work was supported by the National Institutes of Health grant NS051255 (A.B.), a Ruth L. Kirschstein National Research Service Award (National Cancer Institute) and a Brain Science Foundation grant (A.H.K.), and a Human Frontier Science Program Long-term Fellowship (Y.I.).
Footnotes
AUTHOR CONTRIBUTIONS
A.B. directed and coordinated the project. S.V.P., A.H.K., and Y.I. designed and performed in vivo experiments, biochemical assays, and morphological analyses. J.T.W.-G. prepared mass spectrometry samples and completed analyses of Cdc20 phosphorylation in S.P.G.’s laboratory. A.M. contributed molecular reagents. The manuscript was written by S.V.P. and A.B. and critically reviewed and commented on by all authors.
References
- 1.Altman J, Bayer S. Development of the Cerebellar System: In Relation to Its Evolution, Structure, and Functions. CRC Press; New York: 1997. [Google Scholar]
- 2.Palay S, Chan-Palay V. Cerebellar Cortex: Cytology and Organization. Springer-Verlag; New York: 1974. [Google Scholar]
- 3.Lee A, et al. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- 4.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 5.Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller FD, Kaplan DR. Signaling mechanisms underlying dendrite formation. Curr Opin Neurobiol. 2003;13:391–398. doi: 10.1016/s0959-4388(03)00072-2. [DOI] [PubMed] [Google Scholar]
- 7.Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- 8.Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J. 2002;364:593–611. doi: 10.1042/BJ20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erondu NE, Kennedy MB. Regional distribution of type II Ca2+/calmodulin-dependent protein kinase in rat brain. J Neurosci. 1985;5:3270–3277. doi: 10.1523/JNEUROSCI.05-12-03270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg OS, et al. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006;273:682–694. doi: 10.1111/j.1742-4658.2005.05088.x. [DOI] [PubMed] [Google Scholar]
- 14.Kolb SJ, Hudmon A, Ginsberg TR, Waxham MN. Identification of domains essential for the assembly of calcium/calmodulin-dependent protein kinase II holoenzymes. J Biol Chem. 1998;273:31555–31564. doi: 10.1074/jbc.273.47.31555. [DOI] [PubMed] [Google Scholar]
- 15.Griffith LC. Calcium/calmodulin-dependent protein kinase II: an unforgettable kinase. J Neurosci. 2004;24:8391–8393. doi: 10.1523/JNEUROSCI.2888-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanaseki T, Ikeuchi Y, Sugiura H, Yamauchi T. Structural features of Ca2+/calmodulin-dependent protein kinase II revealed by electron microscopy. J Cell Biol. 1991;115:1049–1060. doi: 10.1083/jcb.115.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallano ML. Separation of isozymic forms of type II calcium/calmodulin-dependent protein kinase using cation-exchange chromatography. J Neurosci Methods. 1989;30:1–9. doi: 10.1016/0165-0270(89)90067-8. [DOI] [PubMed] [Google Scholar]
- 18.Miller SG, Kennedy MB. Distinct forebrain and cerebellar isozymes of type II Ca2+/calmodulin-dependent protein kinase associate differently with the postsynaptic density fraction. J Biol Chem. 1985;260:9039–9046. [PubMed] [Google Scholar]
- 19.Colbran RJ. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem J. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicoll RA, Malenka RC. Contrasting properties of two forms of long-term potentiation in the hippocampus. Nature. 1995;377:115–118. doi: 10.1038/377115a0. [DOI] [PubMed] [Google Scholar]
- 21.Zou DJ, Cline HT. Postsynaptic calcium/calmodulin-dependent protein kinase II is required to limit elaboration of presynaptic and postsynaptic neuronal arbors. J Neurosci. 1999;19:8909–8918. doi: 10.1523/JNEUROSCI.19-20-08909.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 23.Powell SK, Rivas RJ, Rodriguez-Boulan E, Hatten ME. Development of polarity in cerebellar granule neurons. J Neurobiol. 1997;32:223–236. doi: 10.1002/(sici)1097-4695(199702)32:2<223::aid-neu7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 24.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 25.Mason CA, Morrison ME, Ward MS, Zhang Q, Baird DH. Axon-target interactions in the developing cerebellum. Perspect Dev Neurobiol. 1997;5:69–82. [PubMed] [Google Scholar]
- 26.Gaudilliere B, Shi Y, Bonni A. RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J Biol Chem. 2002;277:46442–46446. doi: 10.1074/jbc.M206653200. [DOI] [PubMed] [Google Scholar]
- 27.Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- 28.Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 29.Ramon y Cajal S. The cerebellum. In: Swanson N, Swanson L, editors. Histology of the nervous system. Oxford University Press; New York: 1995. [Google Scholar]
- 30.de la Torre-Ubieta L, et al. A FOXO-Pak1 transcriptional pathway controls neuronal polarity. Genes Dev. 2010;24:799–813. doi: 10.1101/gad.1880510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalizi A, et al. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 33.Vaillant AR, et al. Signaling mechanisms underlying reversible, activity-dependent dendrite formation. Neuron. 2002;34:985–998. doi: 10.1016/s0896-6273(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fink CC, et al. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 36.O’Leary H, Lasda E, Bayer KU. CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Mol Biol Cell. 2006;17:4656–4665. doi: 10.1091/mbc.E06-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Songyang Z, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brocke L, Srinivasan M, Schulman H. Developmental and regional expression of multifunctional Ca2+/calmodulin-dependent protein kinase isoforms in rat brain. J Neurosci. 1995;15:6797–6808. doi: 10.1523/JNEUROSCI.15-10-06797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat Rev Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- 44.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5 (Suppl 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 46.Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007;17:434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goslin K, Asmussen H, Banker GA. Rat Hippocampal Neurons in Low-Density Culture. MIT Press; Cambridge, MA: 1998. [Google Scholar]
- 48.Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- 49.Stegmuller J, et al. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.