Abstract
Avoidance of apoptosis is one of the hallmarks of cancer development and progression. Chemotherapeutic agents aim to initiate an apoptotic response, but often fail due to dysregulation. MSH proteins are capable of recognizing cisplatin damage in DNA and participate in the initiation of cell death. We have exploited this recognition and computationally simulated a MutS homolog (MSH) “death conformation”. Screening and docking experiments based on this model determined that the MSH2-dependent cell-death pathway can be induced by a small molecule without DNA damage, reserpine. Reserpine was identified via virtual screening on structures obtained from molecular dynamics as a small molecule that selectively binds a protein “death” conformation. The virtual screening predicts that this small molecule binds in the absence of DNA. Cell biology confirmed that reserpine triggers the MSH2-dependent cell death pathway. This result supports the hypothesis that the MSH2-dependent pathway is initiated by specific protein conformational changes triggered by binding to either DNA damage or small compound molecules. These findings have multiple implications for drug discovery and cell biology. Computational modeling may be used to identify and eventually design small molecules that selectively activate particular pathways through conformational control. Molecular dynamics simulations can be used to model the biologically-relevant conformations and virtual screening can then be used to select for small molecules that bind specific conformations. The ability of a small molecule to induce the cell death pathway suggests a broader role for MMR proteins in cellular events, such as cell death pathways, than previously suspected.
Keywords: MutS homolog, computational modeling, cell death, small molecule, death conformation
1. Introduction
Mismatch repair (MMR) is a highly conserved process in both prokaryotes and eukaryotes that is responsible for recognizing and repairing replication errors involving insertions, deletions and misincorporation of bases. This function is critical in maintaining the genetic stability of the cell; defects in MMR proteins cause an accumulation of mutations, most evident in the appearance of microsatellite instabilities. The MutS protein in prokaryotes, and its homologs (MSH2/MSH6/MSH3) in eukaryotes, are responsible for the first step in this process; DNA mismatch recognition and the formation of the initial protein/DNA complex. The recognition proteins then recruit additional proteins to complete repair [1, 2]. Recently, it was discovered that these same proteins, MutS and its eukaryotic homologs (MSH2/MSH6/MSH3), also recognize DNA damage and contribute to the initiation of cell death in response to DNA damage [3-7].
The precise role of these proteins in cell death has been subject of considerable debate. Two main hypotheses are entertained for the involvement of MMR proteins in this response; a “futile” repair cycle hypothesis and a direct-signaling hypothesis [4]. The “futile” repair cycles hypothesis suggests repeated attempts to repair damage-containing mismatches in which the damage persists, rather than being repaired, resulting in “futile repair” attempts. These repeated cycles are thought to lead to DNA strand breaks as the actual initiators of cell death [6,9]. The “damage-signaling” or “direct signaling” hypothesis suggests a direct involvement of the MMR proteins in signaling by initiating a pro-cell-death signaling cascade [6]. This hypothesis suggests that there are two functions for at least one MMR protein, MSH2. It further suggests that the choice between repair and cell death pathway is determined by differing conformational changes induced by binding to either damaged DNA, for the cell death function, or to mismatched DNA, for the DNA repair function [3,7]. The direct-signaling process does not necessarily involve the repair function of the MMR proteins, and the repair function does not necessarily require the cell-death function.
Which hypothesis is correct is still an open question, and both processes could, and likely do, occur under different circumstances. However, previous work [7,8] leads credence to the direct signaling model by demonstrating that repair-deficient MMR mutants are functional in MMR-dependent cisplatin cytotoxicity. This suggests that the two functions are distinct, as the cytotoxic response has different functional requirements from DNA repair and vice versa.
Our previous work [7], indicated that the “trigger” for activation of the cell-death pathway is a set of distinct, both local and distant, conformational changes induced in response to DNA damage in form of a 1,2-GpG cisplatin intrastrand crosslink, changes which should then allow for binding of different proteins to the MSH2/MSH6 complex in order to activate the cell-death pathway. The existence of such a trigger was identified via molecular dynamics simulations and confirmed in cell biological experiments [7].
We hypothesized that the existence of a conformational trigger suggests that it might be possible to combine the molecular dynamics results with in silico virtual screening to identify ligands that would bind to the pro-cell death conformation by imitating the structural effects of DNA damage. This conformational change, induced by a small molecule rather than by DNA damage, would then trigger a cell-death pathway. If such a molecule were foundit would imply that the long-range conformational changes which determine the choice between the different cellular pathways are robust. Results reported herein suggest that small molecules indeed are capable of mimicking DNA damage and inducing MSH2-dependent cell death.
2. Materials and methods
2.1. Molecular Dynamics Simulations
The simulation protocol is largely the same as described elsewhere [7], differing in the use of a constant pressure algorithm, and differing simulation lengths, and is briefly summarized here. The X-ray structure of Escherichia coli MutS in complex with DNA was the initial starting point [13]; hydrogen atoms were added using the hbuild facility of CHARMM [14] and TIP3P water molecules and counterions were added using the Visual Molecular Dynamics (VMD) package [15]. The CHARMM27 force field [16] was used for the entire complex with additional parameters added based on pre-existing cisplatin parameters [17]. The platinum adduct crosslinks two adjacent guanines. The simulation was performed in NAMD [18] using standard parameters: a 2.0 fs timestep using SHAKE on all bonds to hydrogen atoms, a 12 Å cutoff, Particle Mesh Ewald with a 128 grid points on a side, Berendsen’s constant pressure algorithm with a target pressure of 1.01325 bar, a compressibility of 45.7 mbar, a relaxation time of 1 ps, and a pressure frequency of 40 fs, and a coordinate save frequency of 200 fs; all as implemented in NAMD [18]. The simulation protocol consisted of 250 ps of thermal equilibration to 300K, followed by 10 ns production simulation. The resulting structures were clustered using CHARMM’s k-means clustering algorithm on alpha-carbon positions, with a 1 Å radius cutoff. The structural models were taken as the centroids of the final equilibrated cluster.
2.2. Virtual Screening
AutoDock 3.0 [19] was used to perform 3D docking into the structural models generated from the simulations. Docking grids form vdW and electrostatic interactions were generated using standard AutoDock charges, vdW and electrostatic parameters on cubic grids, 18.75 Å on a side. The grids were centered on the midpoint of the line connecting the two guanine nitrogens that are crosslinked in the platinated simulation, but the grids were generated in the absence of DNA and the platinum adduct. This preparatory work was performed using AutoDock Tools. The NCI diversity dataset was obtained in 3D mol2 form, converted into autodock form, and screened using default Autodock Lamarckian genetic algorithm parameters [19] except the number of runs was increased to 256. The estimated inhibition constant, Ki, calculated by AutoDock was used as an estimator of the quality of binding. Twenty-fold differential binding was required to consider a structure as a suitable potential lead.
2.3. Cell Survival Assay
HEC59 cells and their isogenic counterpart with chromosome 2 transfer were grown in standard growth media of DMEM-F12 +FBS.
The CellTiter 96® AQueous One Solution Cell Proliferation Assay® is a colorimetric method for determining the number of viable cells in proliferation, cytotoxicity or chemosensitivity assays. The CellTiter 96® AQueous One Solution Reagent contains a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] and an electron coupling reagent (phenazine ethosulfate; PES). PES has enhanced chemical stability, which allows it to be combined with MTS to form a stable solution.
Assays were performed by adding a small amount of the CellTiter 96® AQueous One Solution Reagent directly to culture wells, incubating for 1–4 hours and then recording absorbance at 490nm with a 96-well plate reader. The quantity of formazan product as measured by the amount of 490 nm absorbance is directly proportional to the number of living cells in culture.
Cells were plated in 96 well plates at a concentration of 5000 cells/well in 100 μl media and incubated overnight. Media was replaced with fresh media containing drug at indicated concentrations and allowed to incubate for 24 hrs. Untreated cells received fresh media containing 0.1% DMSO. One solution reagent (CellTiter 96(r) AQueous One Solution Reagent) was added to existing media (20 μl/well) and allowed to incubate 3-4 hrs. A plate reader was used to record the absorbance at 490 nm.
2.4. Western Blot analysis
Protein extracts of HEC59 and HEC59 chr. 2 cells were separated on a 16% SDS polyacrylamide gel, transferred to a PDBV membrane and probed with a monoclonal antibody against cleaved caspase-3 (Cell Signaling Technology). This antibody detects only the large fragment (17/19 kDa) of cleaved caspase-3, and not the full length caspase-3 or other cleaved caspases. The activated caspase-3 results from cleavage at Asp 175.
2.5. Overexpression and purification of S. cerevisiae Msh2/Msh6 complex
The plasmids pET11a-MSH2 and pLANT2/RIL-MSH6 were transformed into BLR(DE3) cells for coexpression of S. cerevisiae MSH2 and MSH6. A fresh transformant was grown in LB media at 37 °C to an OD600 0.500 and induced with 0.5 mM IPTG for 3 hrs. Cells were pressure lysed in Buffer C (25 mM Tris-HCl, pH 8.0, 5% glycerol, and 1 mM EDTA). Cell debris was removed by centrifugation and supernatant was loaded on a SP Sepharose column equilibrated with buffer C + 25 mM NaCl. The column was washed with buffer C + 25 mM NaCl, and then eluted with a linear gradient of buffer C, 0-360 mM NaCl. Fractions containing MSH2-MSH6 were pooled and loaded on a S200 size exclusion column equilibrated with buffer C + 200 mM NaCl. The column was washed with buffer C + 200 mM NaCl until MSH2-MSH6 elution. Fractions with purified MSH2-MSH6 were pooled. The protein was aliquoted and frozen for storage at −80 °C until needed.
2.6. Gel shift experiments with purified protein
Oligonucleotides sequences and reactions were as described [3]. Cisplatination of the non-fluorescent strand was performed as described [3]. Heteroduplex DNA was hybridized to an end concentration of 100 nM, as described previously [3]. The sequence of the oligonucleotides is as previously described [3]. One oligonucleotide in the double-stranded DNA molecule wa fluorescently labeled with TAMRA (Oligos Etc.) for non-radioactive visualization on a TYPHOONTM. 100 nM protein were incubated with 10 nM of unplatinated, mismatched or platinated DNA, , and binding assays were performed as described previously [3]. Samples were separated on 6 % non-denaturing polyacrylamide gels.
2.7. Limited Proteolysis
375 nM purified wild type Msh2/Msh6 protein was incubated with the indicated DNA substrates in binding buffer [3] in a total volume of 25 μl. 6 μl of a 320 ng/ml trypsin solution was added and reactions incubated at room temperature for 15 min. The reactions were stopped by boiling for 10 min, and mixed with 5 μl denaturing gel loading dye (Invitrogen). Protein fragments were separated on a 4-20% Tris-Glycine gel (Invitrogen) under denaturing conditions. Silver staining was performed to visualize banding patterns. ImageQuant TL (Amersham Biosciences) was used to analyze the lanes of the gel. Pixels are shown as peak diagrams.
3. Results
3.1. Computational identification of small molecule inducers of MSH-dependent cell death
The conformational trigger for MMR-induced cell death was identified via a molecular dynamics simulation of MutS, a homolog to MSH2/MSH6, containing a (1,2)GpG-intrastrand crosslink, a major adduct of cisplatin treatment. The resulting model for the MutS response involved local and distant changes in protein-DNA and monomer-monomer interactions [7]. A full discussion of the changes are beyond the scope of this paper, and two of the main differences are as reported previously [7]; local rearrangements in the DNA binding region – which becomes the small molecule binding region in the virtual screening -- and a conformational change in the variable loop-disordered loop interaction at the dimer interface were observed. We simply note that the simulations used to generate the model for the virtual screens show structural differences consistent with our previous work, and that these differences are sufficient to selectively screen small molecules for MSH2-dependent cell death. While there has been some recent work [10,11] combining virtual screening with molecular dynamics, which illustrates the potential utility of such an approach, the approach used hereis novel in that it targets specific conformations to selectively activate an endogenous biological pathway. This raises the possibility of using conformational selectivity [12] in drug discovery, e.g., targeting specific conformations of proteins and inducing pathways directly.
The National Cancer Institute has several databases that can be screened for drug discovery (http://dtp.nci.nih.gov/webdata.html). For our virtual screening, the NCI diversity set was selected. This collection is designed to present a diverse group of pharmacophores for screening and almost all of the structures for the small molecules are readily available for 3D grid docking. The diversity set of the NCI consists of 1990 compounds that were selected from the entire NCI compound library based on small numbers of rotatable bonds and pharmacophore diversity to present good candidates for virtual screening.
As the aim of the virtual screening is to identify small molecules that will selectively target the pro-apoptotic conformation, the members of the diversity set were screened against two representative protein structures obtained from molecular dynamics simulations: one structure of the cisplatin-bound DNA/MutS complex, which is known to initiate MSH2-dependent cell death, the “pro-death conformation” and a second structure representative of the undamaged DNA/MutS complex, the “pro-repair conformation” [7]. These structures are the centroids of the most occupied clusters of the molecular dynamics simulations, and so are the optimal models for the two protein/DNA complexes. Each structure was screened twice for a total of four screens. One screen was performed on each structure with DNA, although without the platinum adduct, as the purpose of the screen was to identify small molecules that would mimic the effect of the platinum adduct. A second screen of each structure without any DNA was performed, to identify small molecules that would bind the pro-death protein conformation even in the absence of DNA.
After screening, the relative predicted binding affinities were used to select small molecules that preferentially bind the structure in the pro-death conformation either in the presence or absence of DNA. Several molecules were found computationally to bind the pro-apoptotic conformation with a predicted Ki at least 20-fold better over the normal pro-repair conformation. This change in Ki results from worse atomic contacts in the pro-repair conformation than in the pro-death conformation. As noted previously, the pro-death conformation of MutS shows distinct conformational changes relative to the pro-repair conformation. These conformational changes are sufficient to allow for significantly better binding of specific small molecules to the pro-death conformation than to the pro-repair conformation, as measured by the predicted Ki’s.
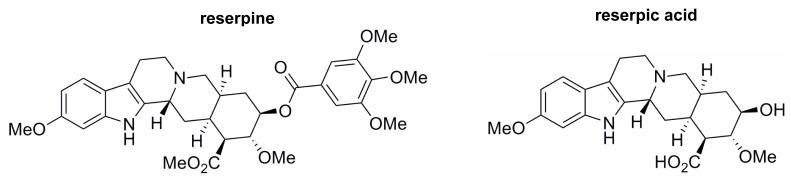
Only one compound, reserpic acid, was readily available commercially for experimental analysis. Reserpic acid also has several commercially available derivatives. Interestingly, one of these derivatives, reserpine (Fig. 1), is an FDA-approved, naturally occurring indole alkaloid previously used to treat hypertension and decrease the heart rate [20]. This compound was chosen for experimental studies along with reserpic acid. Reserpic acid was quickly determined to be a poor compound for cellular studies; the molecule appeared to have poor cellular uptake; not surprising given its low partition coefficient (data not shown). Reserpine, however, was amenable to cellular studies.
Fig. 1.
Structures of reserpic acid and reserpine. Standard two-dimensional chemical drawings of reserpine (left) and reserpic acid (right).
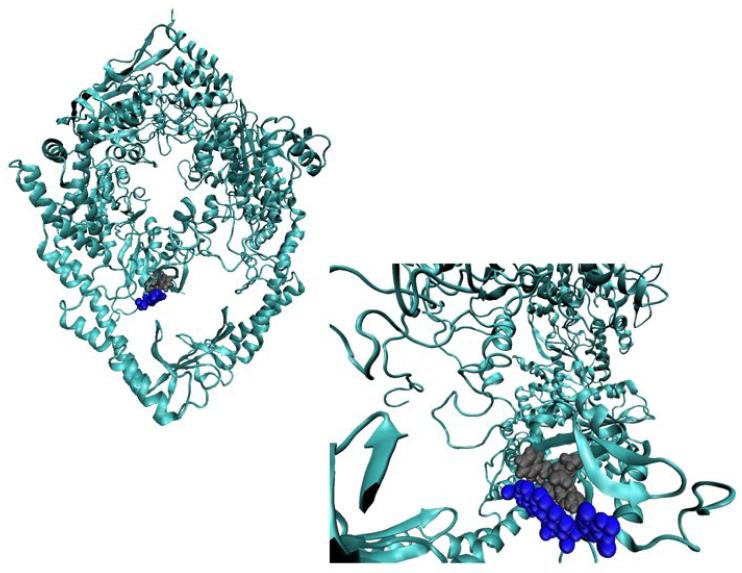
Reserpic acid, and its derivative reserpine (Fig. 1), are predicted to selectively bind to the damaged structure in the same binding pocket as DNA (Fig. 2) The amino acids which are primarily responsible for binding DNA either interact with reserpine, as is the case for Met 33 and Glu 38 in MutS, or are hindered from interactions, as is the case for Phe 36 in MutS, the primary specific contact to mismatched DNA. As a result of these interactions, the binding mode of reserpine overlaps with the binding mode of DNA (Fig. 2, and 6), and the predicted selectivity for the pro-death conformation occurs only in the absence of DNA.
Fig. 2.
Small-Molecule binding mode is in the DNA binding pocket. The predicted binding poise is depicted in two different representations. On the left, the entire protein complex, taken from the MutS/cisplatinated DNA molecular dynamics simulation [7], is shown in a cartoon representation, F36, M33 and E38 are in a gray vdW representation, and reserpine is in a blue vdW representation. On the right is a rotated close-up of the binding pocket with the same color scheme.
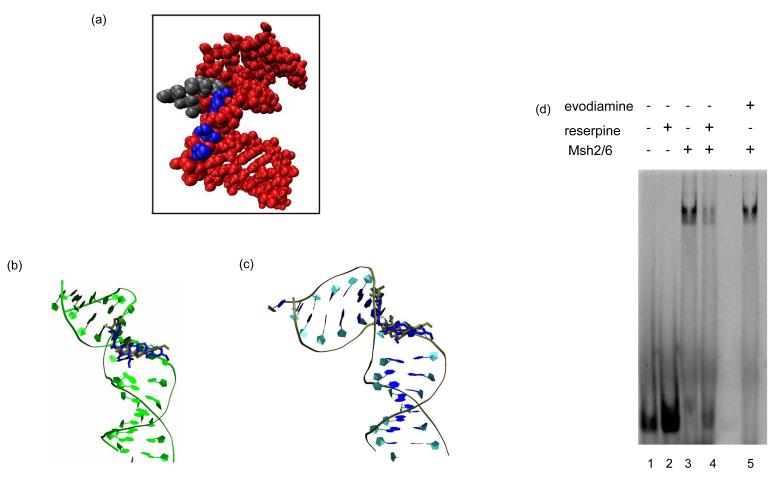
Fig. 6.
(A) Van der Waals image of DNA containing a single 1,2GpG cisplatin adduct (red) as predicted to bind by MutS in the structural model and its overlap with reserpine in its predicted binding mode with MutS (F36, M33 and E38 are in a gray vdW representation, and Reserpine is in a blue vdW representation). Predicted binding of reserpine (blue) and rescinnamine (gold) as it overlaps with (B) mismatched DNA, and (C) cisplatinated DNA, respectively. (D, E) Gel shift assay with yeast Msh2/Msh6 protein in the presence of reserpine and evodiamine, as indicated. 100 nM protein were incubated with 10 nM TAMRA-labeled oligonucleotides (mismatched DNA) in the absence or presence of 200 μM reserpine and evodiamine, respectively (see Materials and Methods).
3.2. Reserpine induces MSH2-dependent cell death
Our prediction from molecular modeling is that reserpine would bind the MutS/MSH proteins in the same conformation found with proteins that are bound to the platinated DNA adduct, i.e., the “pro-death” conformation, but would bind less well when the proteins are in the conformation associated with mismatch recognition, i.e., the pro-repair conformation. Due to our modeling results, we hypothesized that this specific binding to the “pro-death” conformation would induce a similar MMR protein-dependent cell death response as was found with platinated DNA [Topping and Scarpinato; unpublished, 7]. To test this hypothesis, we determined if the cell death response after exposure to reserpine is MSH2-dependent. If so, this would indicate that a small molecule can indeed induce MSH2-dependent cell death.
The specific, MSH2-dependent cell death response was tested in several cell biological experimental assays. We first performed a standard MTS assay, in which cells proficient and deficient in MSH2 were exposed to increasing concentrations of the drug and cell viability was determined. This assay is based on the mitochondrial activity of living cells that reduces the tetrazolium compound (MTS) into a formazan product that can be spectrophotometrically detected at OD490. MSH2-dependent cell-death would be seen if the lack of MSH2, i.e., the msh2-deficient cells, resulted in increased cell survival over those cells with MSH2, i.e., the MSH2-proficient cells. Although these cells are deficient in p53, which may alter sensitivity or resistance to compounds, this internal comparison remains valid.
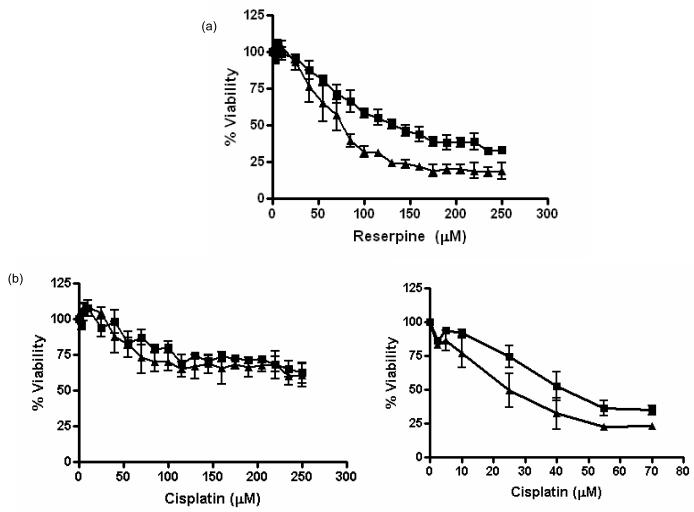
As predicted from the computational modeling, the msh2-deficiency results in a significant increase in resistance to reserpine, as demonstrated by the cell survival results for MSH2-proficient and –deficient cells exposed to reserpine (Fig. 3). The roughly 2-fold difference in IC50 values (93 μM in deficient cells vs. 61 μM in proficient cells) is the reminiscent of the difference observed after treatment with cisplatin [7]. This result suggests that reserpine is as effective in inducing cell death as DNA damage caused by cisplatin. However, cisplatin response differs from reserpine in that it is a slower response. Cisplatin only kills cells 72 to 96 hours after exposure, while reserpine response is already observed 24 hours after exposure (Fig. 3).
Fig. 3.
MTS cell survival assay shows MSH2-dependent cell viability after concentration-dependent (a) reserpine and (b) cisplatin treatment. A representative MTS cell survival assay of MSH2-proficient (triangle) and –deficient (square) cells in the presence of increasing concentrations of reserpine (as indicated) and cisplatin is shown. Percent cell survival is plotted in reference to untreated cells. Mean values and standard deviation of three independent experiments are shown. Results are shown 24h ((a) and (b) left panel) and 72h ((b) right panel) after exposure.
These data confirm our computational prediction and show that computational modeling can be used to identify compounds that specifically target the MSH2-dependent cell death pathway.
3.3. Two different Reserpine analogs affect MSH2-dependent cell viability differentially
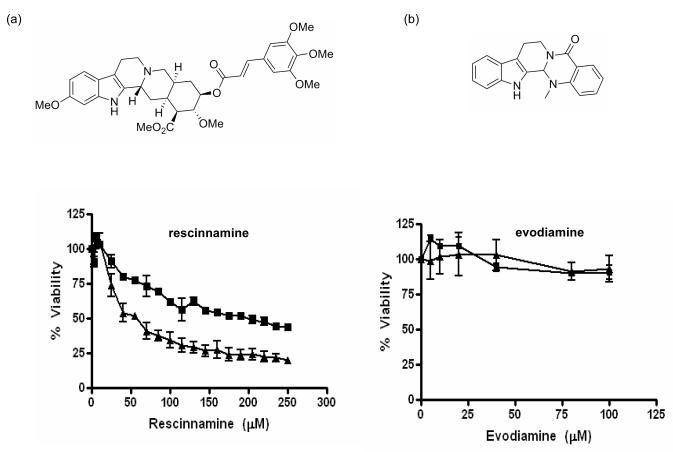
We next determined if the MSH2-dependent cell death response observed with reserpine is limited to this compound or can be re-capitulated by other reserpine-like compounds. Rescinnamine, another FDA-approved compound used for hypertension, was analyzed for its effect on MSH2-dependent cell viability in MTS assays (Fig. 4). This compound contains a longer hydrophobic bond between rings 5 and 6, increasing the length of the molecule without changing the functional groups. MSH2-deficient cells show a significant increase in resistance to increasing concentrations of the compound, again reminiscent of the response to cisplatin. Rescinnamine is hence another example for the induction of MSH2-dependent cell death. Evodiamine, a drug suggested in diet-induced obesity, contains only the basic ring system found in reserpine and rescinnamine, but lacks the additional ring 6 and several functional groups. Exposure of both MSH2-proficient and –deficient cells to evodiamine does not show any cell killing, or a difference between both cell types. With their differential use in hypertension and obesity, reserpine, rescinnamine and evodiamine clearly have multiple effects on the cell. The induction of MSH2-dependent cell death appears to require very distinct structural features of the compound, which has been specified with an array of other additional compounds [Vasilyeva, King, Salsbury and Scarpinato; unpublished].
Fig. 4.
MTS cell survival assay with two reserpine analogs: (A) Structure and MTS assay of MSH2-proficient (triangle) and –deficient (square) cells after exposure to increasing concentrations (as indicated) of rescinnamine. (B) Structure and MTS assay of MSH2-proficient (triangle) and –deficient (square) cells after exposure to increasing concentrations (as indicated) of evodiamine. Shown is cell viability in percent compared to untreated cells. Mean values and standard deviation of three independent experiments are shown.
Our data demonstrate that the induction of MSH2-dependent cell death by a small molecule is not limited to reserpine, but can be re-capitulated by other molecules.
3.4. Reserpine induces MSH2-dependent activation of caspase-3
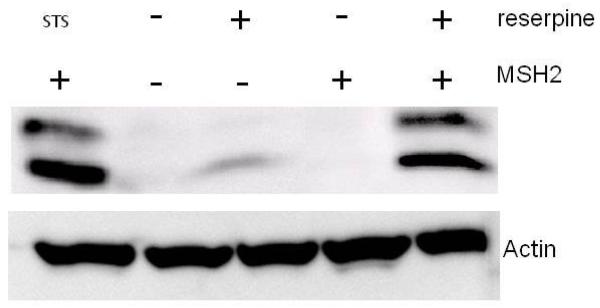
We next determined if we can detect the activation of a protein known to function in apoptosis in response to reserpine. Cells proficient and deficient in MSH2 were exposed to reserpine and cell extracts analyzed via Western blot for cleaved caspase-3. An antibody that recognizes only the large fragment of caspase-3 (17/19 kDa), after cleavage at Asp175 was used in this assay, while uncleaved caspase-3 is not recognized. Fig. 5 shows a representative figure of this experiment. Proficient cells were treated with staurosporine (STS) as a control for caspase-3 cleavage. Neither cell line shows significant levels of cleaved caspase-3 in the absence of insult. Addition of reserpine to MSH2-deficient cells results in a weak induction of the protein, primarily in the appearance of the 17kDa fragment. MSH2-proficient cells exposed to reserpine show a strong induction of caspase-3 cleavage. These results are consistent with a MSH2-dependent apoptotic response to reserpine.
Fig. 5.
Western blot analysis of caspase-3 cleavage in MSH2-proficient (Hec59(2)) and – deficient (Hec59) cells after exposure to reserpine and staurosporine (STS, control), as indicated. The 17 and 19 kDa cleavage products of caspase-3 are shown. Actin was added as a loading control.
3.5. Reserpine can compete with protein-DNA interactions
Computational modeling predicts that the binding of reserpine (and rescinnamine) in the binding pocket of MutS overlaps with bound DNA (Fig. 6A, B, C). The molecules are suggested to bind in the area occupied by the major groove at the site of a mismatch (Fig. 6B) or a 1,2-GpG cisplatin adduct (Fig. 6C). This predicted binding mode would significantly interfere with the binding to both mismatched and damaged DNA. We tested this prediction in a competition assay for DNA binding. Gel shift assays in the absence and presence of reserpine and evodiamine (control) were performed (Fig. 6D). Reserpine itself has no measurable effect on the mobility of the heteroduplex in this assay in the absence of protein (lane 2). Msh2/Msh6 shows the characteristic shift in mobility, indicating binding of the protein to the single G/T mismatch contained in the oligonucleotides (lane 3). In the presence of reserpine, a decrease in the protein-DNA complex is observed (Fig. 6, lane 4), which is not observed in the presence of evodiamine (lane 5), consistent with the differential effects of the drugs in the cell (Fig. 4). This decrease in DNA binding is only observed when a high excess of reserpine over protein and DNA (2,000-fold excess over protein, data not shown) is present, suggesting a kinetic effect in the competition for binding of DNA and small molecule to the protein.
3.6. Reserpine induces conformational changes much like cisplatin
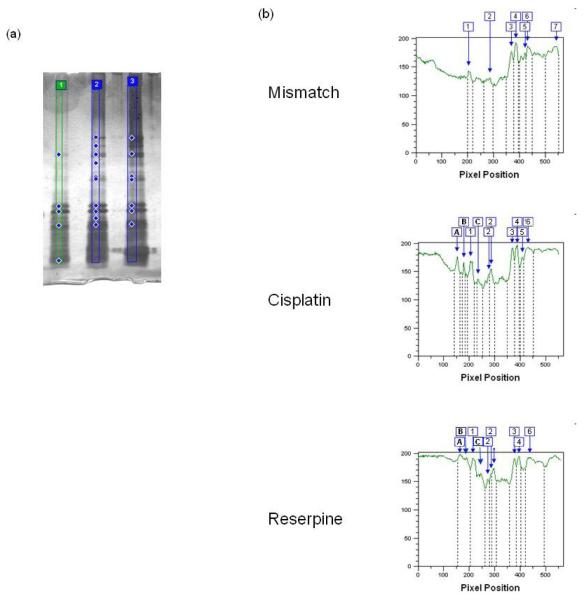
We next performed a preliminary analysis to determine the relative conformational changes induced by reserpine in comparison to mismatched DNA and cisplatinated DNA (Fig. 7.). Limited trypsin proteolysis was performed on yeast Msh2/Msh6 protein in the absence of DNA (not shown) or the presence of mismatched DNA, cisplatinated DNA, reserpine (100 μM), respectively. The silver-stained gel was analyzed using ImageQuant software to detect the banding patterns of samples. Several differences are noted between the presence of mismatched and DNA damage/small molecule (Fig. 7). Fragments present in the presence of mismatched DNA are labeled 1-7. Additional fragments that appear in the presence of cisplatinated DNA, or reserpine are labeled A-C. In addition to the appearance of larger fragments A-C, fragment 2 is more pronounced in the presence of cisplatin (lane 2) or reserpine (lane 3), while fragment 7 is not easily detectable in these experiments. Comparing the pattern obtained with cisplatinated DNA and reserpine, a remarkable similarity is observed (lanes 2 and 3). This is consistent with the induction of distinct conformational changes in the presence of DNA damage or small molecule in comparison to the presence of mismatched DNA, while such differences are less pronounced when comparing the patterns observed with DNA damage or small molecule themselves (lanes 2 and 3).
Fig. 7.
(A) Silver-stained SDS polyacrylamide gel of trypsin proteolysis of MSH2/MSH6 in the presence of mismatched (lane 1), cisplatinated DNA (lane 2) and reserpine (lane 3), respectively. Diamonds indicate fragments detected by the imaging software (B). ImageQuant TL analysis of pixel intensity of individual fragments. Banding was detected automatically after setting the slope to 300. Fragments present with mismatched DNA are labeled 1-7. Additional fragments observed with DNA damage or small molecule are labeled by letters. See Materials and Methods for details.
4. Discussion
Our evidence, suggests that a small molecule is capable of initiating MSH2-dependent cell death, independent of DNA damage (Fig. 3). We show that this response is specific for certain compounds (Fig. 4) and induces caspase-dependent apoptosis (Fig. 5). Computational modeling predicted the binding of the small molecules in the same binding pocket as DNA, which would interfere with DNA binding. This competition can be observed in in vitro assays (Fig. 6), however, a high excess of reserpine is required to observe this effect. Likely, this is due to a kinetic effect. Interactions of MSH2/MSH6 and protein involve a multitude of individual interactions [22], which are mostly lost in the interaction with reserpine, due to the size difference between DNA and the small molecule (Fig. 5b, c). Hence, a higher concentration of reserpine is required to actively compete with the DNA. In vivo, several other factors may contribute to the successful induction of reserpine-induced cell death, such as cellular compartmentalization and accessibility of the compound (unpublished).
Reserpine, a Rauwolfia alkaloid, is an FDA-approved drug that causes depletion of catecholamines and had been in clinical use for hypertension. Due to the high concentration used in patients, severe side effects stopped the clinical use of the compound. An early-described carcinogenic effect of the drug was later disputed [23]. Reserpine was shown to not induce any genotoxic or chromosome destabilizing effects, suggesting a lack of direct interactions with DNA [24,25]. Based on these previously discovered consequences of reserpine treatment, and conflicting reports on its effects, this compound is clearly not a choice chemotherapeutic agent. Rather, our data are a proof-of-principle for the induction of MSH2-dependent cell death by small molecules that appear to mimic DNA damage. This result can be exploited for the generation of novel agents that specifically induce mismatch repair-dependent cell death.
The observation that reserpine and some of its analogs can induce MSH2-dependent cell death without DNA damage is unexpected, since MSH2/MSH6 is typically described as a DNA mismatch and damage recognition protein complex. This result is not consistent with a “futile” repair cycle hypothesis for cisplatin induced cell-death. The computational and experimental results together suggest that a small molecule can stimulate the conformational changes induced by DNA damage and initiate the same cellular response as DNA damage. This result further emphasizes the idea that different conformational changes are likely involved in the initiation of cell death. That a small molecule, reserpine, which is predicted to bind in the same region as DNA, can produce the same cellular outcome as cisplatin suggests that there are two general responses of MutS/MSH proteins to interactions with molecules in the DNA-binding domain: one that triggers DNA repair and one that triggers cell death.
This is the first evidence that MMR proteins do not require specific interactions with DNA, but that inducers of certain conformational changes are sufficient to trigger a response. The finding that this response can be triggered by a small molecule, independent of specific DNA binding indicates that previous hypotheses of futile repair cycles and the requirement for a mismatch to initiate MMR-dependent cell death do not apply to all types of damage. Warren et al. [22] recently described that MSH2/MSH6 interacts with methylation damage identical to a mismatch and suggested that the MSH2/MSH6 proteins are not directly involved in the initiation of different cellular responses. This description is not immediately reconcilable with our data, which suggest a direct involvement of the mismatch repair proteins in cell death. The immediate implication of these studies, when taken together, is that not all binding events to MMR proteins trigger the same responses. Also, the authors only describe the methylation damage in conjunction with a mismatch, which will have overlapping effects, that of the damage and that of the mismatch, which cannot easily be distinguished. We suggest that methylation damage and DNA platinum adducts may trigger cell death differently, and that the latter can be mimicked by small molecule binding.
Cisplatin was chosen as a model compound to predict the “death conformation” of MSH proteins, since it appears to be the only compound described today that may function in a futile cycles-independent, MMR-dependent mechanism (Topping, Wilkinson, and Scarpinato; unpublished). As mentioned above, the well-described response to O6-methylguanine primarily functions via futile cycles of repair, a mechanism that we are proposing to be different from the cisplatin response. This is substantiated by the fact that the methylation damage is recognized in the same manner as a mismatch [22], and appears to require the formation of a mismatch as a prerequisite for MMR-dependent cell death initiation. Based on this reasoning, cisplatin adducts were used to model the MSH protein conformation initiating cell death response.
Though resulting in a relatively weak difference in sensitivity between MSH2-proficient and deficient cells, this difference is sufficient to have clinical implications that are observed in increased cisplatin resistance of patients with MMR defects [26].
Interestingly, the MSH2-dependent response to reserpine differs from that to cisplatin in its time-dependence (Fig. 3). The response to cisplatin requires 72 to 96 hours of exposure to show an MSH2-dependent cell death response, which was previously interpreted as a requirement for replication to occur that would provide mismatches and then induce futile repair cycles. In contrast, the response to reserpine can be observed after only 24 hours.
The cultured cells used in this experiment carry a dysfunctional p53 gene [21]. p53 deficiency may alter sensitivity or resistance to DNA damage. However, we here compared cells that should only differ in mismatch repair status. The comparison of results obtained with these two cell lines remains valid. The lack of p53 in the cells used herein leads to the conclusion that the cell-death pathway is both dependent on MSH2 and independent of p53. This is an important discovery for both potential drug development, and for understanding the cell biology of the cell-death response. First, the other actors in the MSH2-dependent cell death are unknown; knowing that it is p53-independent suggests that some conventional apoptotic pathways are not involved. Second, about 50% of tumors carry mutant p53 alleles, and many cancer therapeutics depend on pathways that require active p53. A therapeutic that activates the MSH2-dependent cell-death pathway might be advantageous as it would eliminate the requirement for functional p53.
Though compounds that induce mismatch repair-specific cell death will likely kill benign and cancerous cells, a certain specificity for cancer cells will be achieved by the fact that mismatch repair proteins are more abundant and active in highly proliferating cells. One downside is that such compounds are not suitable for tumors with mismatch repair defects, such as patients with HNPCC.
Our ability to use computational modeling to predict that a small molecule will selectively interact with a specific conformation, and thereby induce a specific pathway, has implications for drug discovery. First, the predicted cellular outcome, i.e., MSH2-dependent cell death, was confirmed experimentally, which suggests that molecular simulations have achieved a level of accuracy sufficient to model conformational changes relevant for drug discovery, at least in this particular system. Second, these results suggest that future research in structure-based drug discovery should, where relevant, aim specifically at perturbing conformational ensembles.
Our findings change the current assumptions made on the involvement of MMR proteins in cytotoxic response to endogenous and exogenous cytotoxic compounds. They suggest that futile repair cycles of DNA damage are not necessarily required for the participation of MMR proteins in the cell death response, as no requirement for DNA damage exists. This finding should direct future research analyzing the MMR-dependent cell death pathway and those molecules that initiate a MMR-dependent response. Most significantly, our results suggest a much larger role for MMR proteins in major cellular activities that go beyond the mere recognition of DNA damage.
Acknowledgements
We thank Drs. J. Fetrow, G. Kucera, J. Wilkinson and D. Kim-Shapiro for their helpful discussions. This work was sponsored in part by NCI CA101829, the Comprehensive Cancer Center at WFUSM (to KDS), and the Golfers Against Cancer grant (to FRS, KDS and TH). The computations were performed on the WFU DEAC cluster (www.deac.wfu.edu) supported by Wake Forest University Information Systems and the Associate Provost for Research. Disk storage was provided by IBM through a SUR grant to WFU. We thank IBM and WFU for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- [2].Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- [3].Drotschmann K, Topping RP, Clodfelter JE, Salsbury FR. Mutations in the nucleotide-binding motif of MutS homologs uncouple cell death from cell survival. DNA Repair. 2004;3:729–742. doi: 10.1016/j.dnarep.2004.02.011. [DOI] [PubMed] [Google Scholar]
- [4].Irving JA, Hall AG. Mismatch repair defects as a cause of resistance to cytotoxic drugs. Expert Rev. Anticancer Ther. 2001;1:149–158. doi: 10.1586/14737140.1.1.149. [DOI] [PubMed] [Google Scholar]
- [5].Fedier A, Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity. Int. J. Oncol. 2004;24:1039–104. [PubMed] [Google Scholar]
- [6].Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- [7].Salsbury FR, Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Res. 2006;34:2173–2185. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, Umar A, Kucherlapati R, Lipkin M, Kunkel TA, Edelmann W. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;15:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- [9].Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- [10].Lerner MG, Bowman AL, Carlson HA. Incorporating dynamics in E. coli dihydrofolate reductase enhances structure-based drug discovery. J. Chem. Info. Model. 2007;47:2358–2365. doi: 10.1021/ci700167n. [DOI] [PubMed] [Google Scholar]
- [11].Bowman AL, Lerner MG, Carlson HA. Protein Flexibility and Species Specificity in Structure-Based Drug Discovery: Dihydrofolate Reductase as a Test System. J. Am. Chem. Soc. 2007;129:3634–3640. doi: 10.1021/ja068256d. [DOI] [PubMed] [Google Scholar]
- [12].Gunasekaran K, Ma BY, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins: Struct, Funct., Bioinf. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- [13].Lamers MH, Perrakis A, Enzlin JH, Winterwerp HHK, de Wind NM, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a GT mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- [14].Brooks BR, Bruccoleri RD, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- [15].Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [16].MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins and nucleic acids. J. Phys. Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [17].Scheef ED, Briggs JM, Howell SB. Molecular Modeling of the Intrastrand Guanine-Guanine DNA Adducts Produced by Cisplatin and Oxaliplatin. Mol. Pharm. 1999;56:633–643. [PubMed] [Google Scholar]
- [18].Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
- [19].Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- [20].Koshiura R, Miyamoto K, Sanae F. Combination antitumor effect with central nervous system depressants on rat ascites haptomas. Gann. 1980;71:45–51. [PubMed] [Google Scholar]
- [21].Yan T, Schupp JE, Hwang HS, Wagner MW, Berry SE, Strickfaden S, Veigl ML, Sedwick WD, Boothman DA, Kinsella TJ. Loss of DNA mismatch repair imparts defective cdc2 signaling and G(2) arrest responses without altering survival after ionizing radiation. Cancer Res. 2001;61:8290–8297. [PubMed] [Google Scholar]
- [22].Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutS alpha DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- [23].Lina BAR, Woutersen RA, Bruijntjes JP, van Benthem J, van den Berg JAH, Monbaliu J, Thoolen BJJM, Beems RB, van Kreijl CF. Evaluation of the Xpa-deficient transgenic mouse model for short-term carcinogenicity testing: 9-month studies with Haloperidol, Reserpine, Phenacetin and D-Mannitol. Toxicol. Pathol. 2004;32:192–201. doi: 10.1080/01926230490274344. [DOI] [PubMed] [Google Scholar]
- [24].von Poser G, Andrade HH, da Silva KV, Henriques AT, Henriques JA. Genotoxic, mutagenic and recombinogenic effects of rauwolfia alkaloids. Mutat. Res. 1990;232:37–43. doi: 10.1016/0027-5107(90)90107-f. [DOI] [PubMed] [Google Scholar]
- [25].Kevekordes S, Mersch-Sundermann V, Burghaus CM, Spielberger J, Schmeiser HH, Arit VM, Dunkelberg H. SOS induction of selected naturally occurring substances in Escherichia coli (SOS chromotest) Mutat. Res. 1999;445:81–91. doi: 10.1016/s1383-5718(99)00141-2. [DOI] [PubMed] [Google Scholar]
- [26].Martin LP, Hamilton TC, Schilder RJ. Platinum Resistance: The Role of DNA Repair Pathways. Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]