Abstract
Psychologists, with their long-standing tradition of studying mechanistic processes, can make important contributions to further characterizing the risk associated with genes identified as influencing risk for psychiatric disorders. We report one such effort with respect to CHRM2, which codes for the cholinergic muscarinic 2 receptor and was of interest originally for its association with alcohol dependence. We tested for association between CHRM2 and prospectively measured externalizing behavior in a longitudinal, community-based sample of adolescents, as well as for moderation of this association by parental monitoring. We found evidence for an interaction in which the association between the genotype and externalizing behavior was stronger in environments with lower parental monitoring. There was also suggestion of a crossover effect, in which the genotype associated with the highest levels of externalizing behavior under low parental monitoring had the lowest levels of externalizing behavior at the extreme high end of parental monitoring. The difficulties involved in distinguishing mechanisms of gene-environment interaction are discussed.
Keywords: adolescent development, genetics, behavior genetics, drug/substance abuse, antisocial behavior
Gene identification efforts are specifying a growing number of susceptibility genes that influence risk for behavioral disorders. Most of these large-scale gene-mapping efforts focus on psychiatric diagnoses as the primary outcome of interest. Accordingly, the resultant genotype-phenotype associations open the door to many additional research areas, including characterizing the phenotypic spectrum of risk associated with specific genes, mapping the pathways by which genotypic risk eventually contributes to clinical disorders, and studying how specific environmental factors may moderate genetic risk. Psychologists, who have a long-standing tradition of studying mediating and moderating variables, mechanistic processes, and developmental pathways, are well equipped to make important contributions to this work. Many such efforts are underway. For example there are rapidly growing literatures on interactions between a purportedly functional polymorphism in the monoamine oxidose A gene (MAOA) and maltreatment in predicting behavioral problems (Caspi et al., 2002; Ducci et al., 2008; Kim-Cohen et al., 2006) and between the long/short polymorphism in the serotonin transporter gene and stressful life events in predicting depressive outcomes (Caspi et al., 2003; Kaufman et al., 2004; Risch et al., 2009). Now that gene-mapping efforts are producing replicated associations (Edenberg & Foroud, 2006), it will be fruitful for researchers to move beyond mapping risk pathways associated with the “usual suspects” and to expand risk-characterization efforts to novel genes from the psychiatric genetics literature (Dick et al., 2009).
We report analyses from one such effort with respect to CHRM2, a gene first associated with alcohol dependence in the Collaborative Study on the Genetics of Alcohol Dependence (COGA; Wang et al., 2004). CHRM2 codes for the cholinergic muscarinic 2 receptor. Muscarinic acetylcholine receptors (mAchRs) activate a multitude of signaling pathways important for modulating neuronal excitability, synaptic plasticity, and feedback regulation of acetylcholine release (Volpicelli & Levey, 2004). There is evidence that muscarinic receptors are involved in many brain functions, such as learning and memory, so it is biologically plausible that CHRM2 plays a role in psychiatric and behavioral outcomes. The association between CHRM2 and alcohol dependence was replicated in an independent sample (Luo et al., 2005), and subsequent analyses demonstrated that the gene is involved more generally in externalizing behavior, which encompasses a variety of disruptive conditions, including antisocial behavior and conduct problems, in addition to substance use (Dick et al., 2008).
All previously demonstrated associations between the CHRM2 gene and externalizing disorders have been in high-risk clinical samples. We expanded on these analyses by studying the association between CHRM2 and prospectively reported externalizing behavior in community-based samples. In addition, we built on the literature by testing for moderation by parental monitoring. Twin studies have demonstrated that parental monitoring can moderate the importance of genetic influences on substance use: Genetic influences (modeled latently, by comparisons of monozygotic- and dizygotic-twin correlations) become increasingly important with decreasing parental monitoring (Dick, Purcell, et al., 2007). More recently, we have demonstrated such moderation with respect to a specific measured gene, GABRA2 (which encodes the gamma-aminobutyric acid receptor subunit alpha-2), finding that there was a stronger association between GABRA2 and externalizing behavior with lower parental monitoring (Dick et al., 2009). In the study reported here, we tested for main effects of CHRM2 on externalizing behavior across adolescence, and for potential moderation of this effect by parental monitoring, in a longitudinal community-based sample.
Method
Sample
Our sample consisted of participants in the Child Development Project (CDP), who were originally recruited through kindergartens at three cities in the United States in 1987 and 1988. CDP participants represented the broader population demographically and behaviorally, as determined by teacher and sociometric ratings of the entire populations at those sites. The original CDP sample consisted of 585 children (52% male and 48% female; 81% European American, 17% African American, and 2% belonging to other ethnic groups). Data collections began the summer before the participants entered kindergarten (at about 5 years of age); follow-ups have been conducted annually and are ongoing, with a 90% continued-participation rate in the early-adulthood assessments. DNA was collected from CDP participants via saliva sample using Oragene collection kits under the supervision of a specially trained interviewer. Saliva samples were de-identified and mailed to Washington University in St. Louis, Missouri, where DNA extraction and genotyping occurred. The DNA samples were obtained from 452 individuals, representing 93% of the target subset of regular CDP participants. The institutional review boards at all sites approved the study.
Genotyping
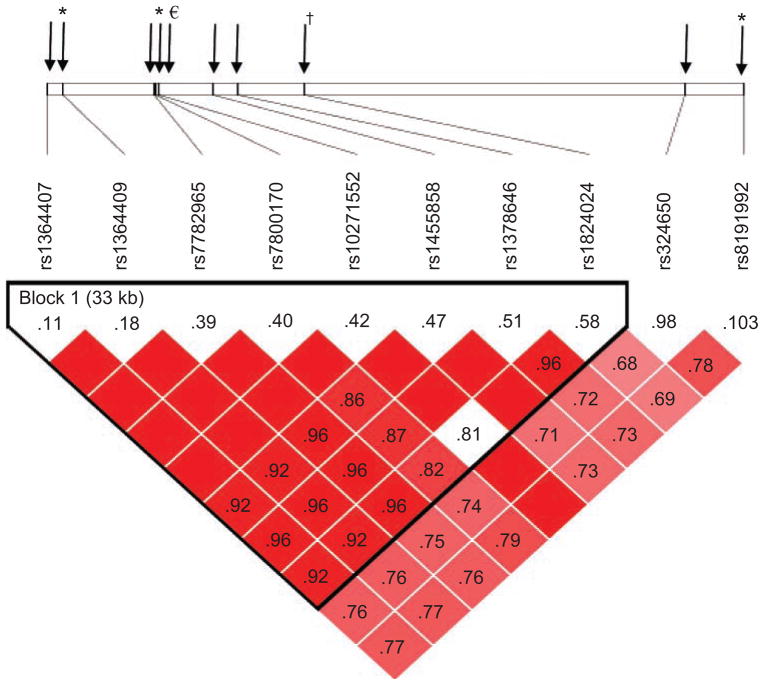
Nine single-nucleotide polymorphisms (SNPs) were selected in CHRM2 for genotyping, on the basis of evidence of association with one or more externalizing disorders in the COGA sample (Dick, Agrawal, et al., 2007; Wang et al., 2004). A SNP is a DNA sequence variation in which a single nucleotide in the genome differs between individuals or between paired chromosomes in an individual. Some SNPs have functional consequences (e.g., they change the protein product or alter the rate at which the protein is produced), but most have no known consequence. SNPs are used as markers for the surrounding DNA sequence. Multiple SNPs were genotyped across the CHRM2 gene because it is possible that variation at multiple locations could alter the function of the gene and contribute to differential susceptibility. Figure 1 shows the location of the genotyped SNPs and the correlation structure across these SNPs using Haploview (Barrett, Fry, Maller, & Daly, 2005). Because the correlation pattern differs across the SNPs across the gene, one would not expect all SNPs across the gene to yield similar results; rather, the observed pattern of significance across SNPs should map broadly onto the correlation pattern.
Fig. 1.
Location of (top), and correlations between (bottom), the single-nucleotide polymorphisms (SNPs) genotyped in the CHRM2 gene in the Child Development Project (CDP) sample. The output was obtained from Haploview (Barrett, Fry, Maller, & Daly, 2005) using the CEPH (Centre d’Etude du Polymorphisme Humain) data from the HapMap database (The International HapMap Consortium, 2003). Shading indicates the degree of correlation as measured by D′ (Hedrick & Kumar, 2001); darker red shading indicates higher correlations, and white shading indicates that markers are unlinked or uncorrelated. The numbers inside the diamonds are R2 values, another measure of correlation between SNPs. R2 is more sensitive to allele frequencies than D′ is and ranges from 0 to 1.0 (as a standard correlation does). The black triangle grouping a subset of SNPs on the figure indicates a block (33 kilobases, or kb) of SNPs that are highly correlated (as defined by criteria detailed in Gabriel et al., 2002). Not all SNPs genotyped in the CDP sample were in the HapMap database; in these cases, proxy SNPs that were the SNPs most highly correlated with the genotyped SNPs are listed: rs1364409 represents rs36210735 (r2 = .7), and rs1364407 represents rs978437 (r2 = .9). The dagger and asterisks indicate SNPs that showed a significant or marginally significant interaction with parental monitoring (†p < .10; *p < .05). In addition to the nine SNPs genotyped in the CDP sample, the figure shows the location of a SNP that showed a significant interaction with parental monitoring in the TRacking Adolescents’ Individual Lives Survey (TRAILS; marked €).
The overall genotyping success rate was 98.4%. Allele frequencies were analyzed for deviations from Hardy-Weinberg equilibrium (HWE), which can be an indication of genotyping problems or stratification. No deviations from HWE were detected. Additional details about the genotyping in CDP are available elsewhere (Dick et al., 2009). Table 1 contains additional descriptive information about the SNPs, including their relative location within the gene—as derived from estimates of chromosomal position within the dbSNP (Mailman et al., 2007) database—and marker-specific major and minor nucleotide bases with corresponding allele frequencies. Because allele frequencies and correlation structures between SNPs in a gene often differ across populations, we limited all analyses and data reported in this article to the subsample of Caucasian individuals (N = 374; 189 male, 185 female).
Table 1.
Tested Single-Nucleotide Polymorphisms (SNPs) and Their Effects on Externalizing Behavior in the Child Development Project Sample
| SNP | Chromosomal position | Minor allele frequency | Alleles | Genotype frequency (n)
|
Main effect of CHRM2
|
CHRM2 × Parental Monitoring
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous for minor allele | Heterozygous | Homozygous for major allele | β | p | β | p | ||||
| rs36210735 | 136080901 | .325 | A:T | 78 | 180 | 116 | 0.023 | .673 | −0.820 | .031 |
| rs978437 | 136264717 | .325 | T:C | 39 | 147 | 186 | 0.006 | .907 | −0.617 | .103 |
| rs7782965 | 136274672 | .317 | C:T | 35 | 148 | 191 | −0.003 | .958 | −0.551 | .143 |
| rs7800170 | 136274859 | .433 | A:C | 77 | 180 | 112 | 0.031 | .565 | −0.913 | .017 |
| rs1455858 | 136282242 | .333 | C:T | 38 | 148 | 186 | −0.017 | .748 | 0.602 | .117 |
| rs1378646 | 136285540 | .342 | T:C | 41 | 151 | 183 | 0.011 | .845 | −0.217 | .234 |
| rs1824024 | 136294233 | .342 | A:C | 39 | 150 | 184 | −0.020 | .716 | 0.673 | .081 |
| rs324650 | 136344201 | .467 | A:T | 81 | 171 | 122 | −0.032 | .558 | 0.517 | .192 |
| rs8191992 | 136351848 | .442 | A:T | 82 | 164 | 127 | −0.029 | .592 | −0.828 | .045 |
Note: Chromosomal positions are listed in base pairs, according to dbGaP Release 21 (Mailman et al., 2007). SNPs with significant interactive effects (p < .05) are highlighted in boldface.
Phenotypes
Externalizing behavior
The CDP collected data on externalizing behavior using Achenbach’s (1991, 1997) Child Behavior Checklist (CBCL) and Youth Self-Report (YSR). The Externalizing scale consists of 33 items in the CBCL and 30 items in the YSR and covers both delinquency (e.g., “I cut classes or skip school”) and aggression (e.g., “I am mean to others”). Respondents indicate whether each behavior is “not true,” “somewhat or sometimes true,” or “very or often true.” These measures have been shown to have excellent psycho-metric properties, including high test-retest reliability, content validity, criterion-related validity, and construct validity (Achenbach & Rescorla, 2001, 2003). We created mean externalizing scores by averaging all available parent ratings (CBCL) and youth ratings (YSR) of externalizing behavior across the ages 10 through 17. CBCL ratings (reported by mothers) were obtained for participants at ages 10 through 17, and YSR ratings (reported by participants) were obtained for participants at ages 12, 14, 15, and 16.
Parental monitoring
When participants were 14 years old, the CDP collected data on their perceptions of their parents’ knowledge of their whereabouts, companions, and activities. This measure consisted of five items asking participants how much their mother or father knew about (a) their friends, (b) how they spent their money, (c) how they spent their after-school time, (d) how they spent their free time, and (e) where and when they went out. The 3-point response scale included the options “nothing,” “some,” and “a lot.” A composite score of child-reported parental monitoring was computed as the mean response to the five items (α = .74). This measure likely reflects both parental efforts at obtaining information and participants’ willingness to inform their parents (Kerr & Stattin, 2000). We are unable to tease apart these possibilities. Though we refer to this measure as parental monitoring, as have previous publications using this scale (Dick et al., 2009), it is more generally a measure of parental knowledge.
Statistical analyses
All analyses, including the calculation of the mean externalizing scores and the genetic analyses, were conducted using the statistical program SPSS Statistics 17 (PASW Statistics Release 17.0.2). We ran standard linear regression analyses to test for an association between mean externalizing score and each SNP from CHRM2. Our coding system reflected an additive genetic model, with each of the SNPs coded 0, 1, or 2 according to the number of copies of the major (i.e., most common) allele. We first tested main-effects models separately for each measured polymorphism and for parental monitoring. We then tested for interaction effects between each SNP and parental monitoring. Sex was included as a covariate in all analyses.
Results
Parental-monitoring scores ranged from 0.0 to 2.0 (M = 1.65, SD = 0.369). The mean externalizing score ranged from 0.62 to 31.36 (M = 9.42, SD = 5.42). Table 1 shows results for the unconditional main effect of each SNP and for the interactions between SNP and parental monitoring. No SNPs had significant main effects of association with externalizing behavior. There was a significant main effect of parental monitoring (p ≤ .001, β = −0.328). Parental monitoring also moderated the effects of CHRM2 on externalizing behavior in the case of three of the nine SNPs (p = .017–.045), and the interaction showed a trend toward significance for one additional SNP (p = .081). It is not straightforward to correct for multiple tests in genetic studies that test multiple genetic variants across a gene because these variants are correlated with one another, which makes straightforward Bonferroni correction inappropriate. The genetic analysis program Plink (Purcell et al., 2007) has implemented a permutation approach that gives a set-based empirical p value indicating the likelihood of observing the pattern of p values across the SNPs, taking into account their correlation structure. This program yielded an empirical p value of .039 for the observed interaction effect, suggesting that it was significant after correction.
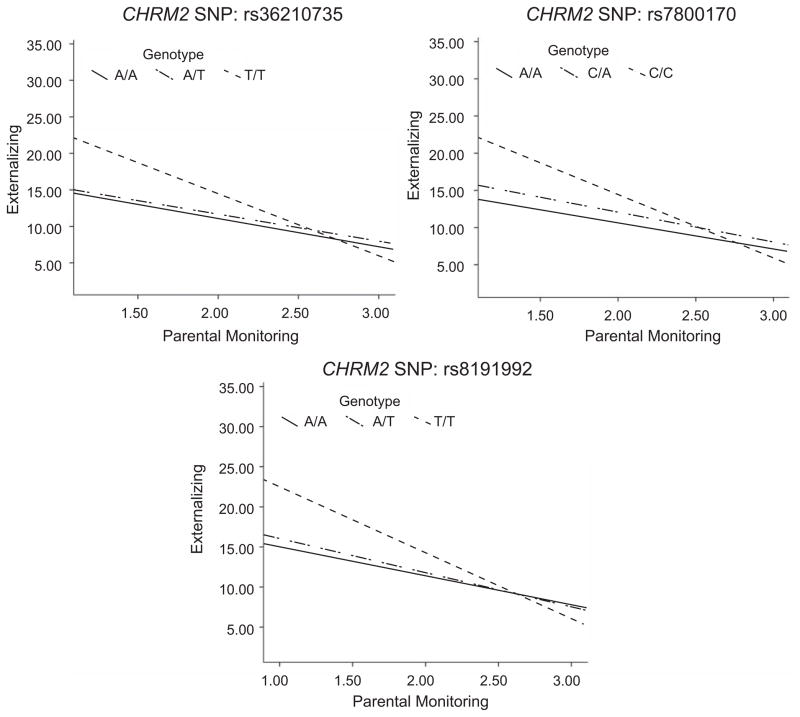
Figure 1 shows the location of the SNPs exhibiting interactions in the CDP sample. The interaction effects are largely clustered in the central block of the gene. Figure 2 plots the regression lines for the interaction effect for the SNPs that interacted significantly with parental monitoring. Across the three SNPs in Figure 2, there was a stronger association between genotype and externalizing behavior with decreased parental monitoring. The regression lines for all three SNPs also cross over: The genotype associated with the highest levels of externalizing behavior at the low end of parental monitoring in each case was associated with the lowest levels of externalizing behavior at the extreme high end of parental monitoring. However, the confidence intervals at the environmental ends of the distribution were large and overlapping; accordingly, the mean differences in externalizing behavior across genotypes were nonsignificant at either end. Thus, there was insufficient power to conclude that the genotypic effects exhibited significant crossing over.
Fig. 2.
Regression lines showing externalizing behavior as a function of parental monitoring and genotype for each of the three CHRM2 single-nucleotide polymorphisms (SNPs) showing a significant gene-environment interaction in the Child Development Project sample. A median split on parental monitoring was used to graph the data to illustrate the shape of the interaction; however, a continuous measure of parental monitoring was used in the statistical analyses reported in the text.
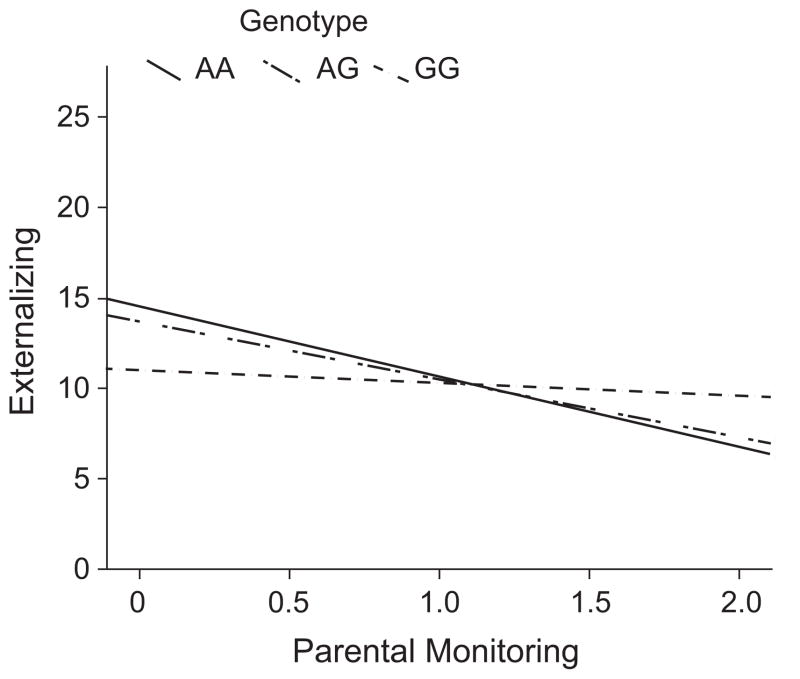
We had available to us data from a second sample, the TRacking Adolescents’ Individual Lives Survey (TRAILS). This sample consists of 1,290 Dutch adolescents with DNA available; at three waves of assessment between the ages 10 and 18, CBCL and YSR data were collected, as was a measure of parental monitoring parallel to the CDP measure. Thirty-five SNPs were genotyped across CHRM2 in that sample, and we conducted analyses parallel to those run in the CDP. One SNP (rs10271552) yielded a p value less than .05 for the interaction effect (p = .035, β = 0.26), and results for 3 additional SNPs were suggestive (ps =.057–.098). However, none of the other SNPs genotyped showed interaction effects. The empirical p value taking into account the multiple SNPs analyzed across the gene was not significant either at the gene level (p = .51) or in a set-based analysis limited to the 6 SNPs genotyped in the linkage disequilibrium block where interaction was detected in the CDP (p = .25). The 1 SNP yielding a significant interaction effect was located in the same block of the gene that yielded interaction effects in the CDP (position indicated on Fig. 1; that particular SNP was not genotyped in the CDP). The shape of the interaction for that SNP also suggested a crossover effect, though (as in the CDP findings for other SNPs) the genotypic means did not differ significantly at either end of the distribution (Fig. 3).
Fig. 3.
Regression lines showing externalizing behavior as a function of parental monitoring and genotype for the single-nucleotide polymorphism (SNP) showing a significant gene-environment interaction in the TRacking Adolescents’ Individual Lives Survey (TRAILS). A median split on parental monitoring was used to graph the data to illustrate the shape of the interaction; however, a continuous measure of parental monitoring was used in the statistical analyses reported in the text.
Discussion
CHRM2 has previously been associated with externalizing symptoms in a sample ascertained through alcohol-dependent individuals (Luo et al., 2005; Wang et al., 2004). We tested for association between this gene and prospectively measured externalizing behavior in a longitudinal, community-based sample of adolescents. No SNPs yielded evidence for a main effect of CHRM2; however, we found significant evidence of gene-environment interaction in the form of an interaction between genotype and parental monitoring. Specifically, the association between the genotype and externalizing behavior was accentuated under conditions of lower parental monitoring. This finding is in line with our previous twin work, which indicated that genetic influences on substance use became more important as parental monitoring decreased (Dick, Viken, et al., 2007). Also, we have previously found evidence for an interactive effect of another gene, GABRA2, and parental monitoring on externalizing behavior in the CDP (Dick et al., 2009). However, the data suggested a crossover effect for all CHRM2 SNPs with significant interaction effects: The genotype associated with the highest levels of externalizing behavior at the low end of parental monitoring was associated with the lowest levels of externalizing behavior at the extreme high end of parental monitoring. Although the overall test of interaction was not significant in the TRAILS sample, the markers with suggestive interaction effects also showed crossover effects.
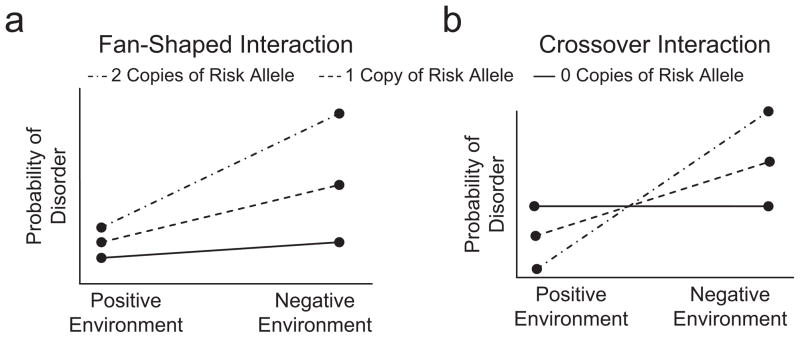
These findings are in line with the differential-susceptibility hypothesis proposed by Belsky et al. (2009). According to this hypothesis, the same individuals who are most adversely affected by negative environments are also those who are most likely to benefit from positive environments. Belsky et al. referred to genetic factors that operate in this way as “plasticity genes,” and contrasted this framework for conceptualizing gene-environment interaction with the diathesis-stress framework, which focuses on “vulnerability genes” and has been the predominant model in psychiatric genetics. Under the diathesis-stress framework, gene-environment interaction is generally conceptualized as certain individuals being more susceptible to psychiatric problems in the context of adverse environmental conditions than other individuals are. A diathesis-stress framework hypothesizes a fan-shaped interaction (as illustrated in Fig. 4a), whereby there is a stronger association between genotype and outcome under adverse environmental conditions than under benign environmental conditions. In contrast, the differential-susceptibility hypothesis predicts a crossover interaction (Fig. 4b), whereby the association between genotype and outcome is actually reversed at environmental extremes, such that the individuals at highest risk under adverse environmental conditions are at lowest risk under positive environmental conditions.
Fig. 4.
Different models of gene-environment interactions. The diathesis-stress framework (a) predicts that there is a stronger association between genotype and outcome under adverse environmental conditions than under benign environmental conditions (a fan-shaped interaction). In contrast, the differential-susceptibility hypothesis (b) predicts that the individuals at highest risk under adverse environmental conditions are at lowest risk under positive environmental conditions (a crossover interaction).
In principle, these different types of gene-environment interaction effects should be easily distinguishable, but in practice they are not. One can imagine many variations on these two clear-cut models, including variations in whether there are main effects of genotype at each end of the environmental continuum, and the extent to which the slope for each genotype differs as a function of the environment (see Kendler, in press, for further elaboration of these models and Belsky, Bakermans-Kranenburg, & van IJzendoorn, 2007, for steps to formally test for differential susceptibility). Our data are illustrative of these complexities: Although the shape of the interactions clearly suggests a crossover effect, the confidence intervals at the environmental ends of the distributions are large and overlapping; accordingly, the mean differences in externalizing behavior across genotypes are nonsignificant at either end. Thus, there is insufficient power to conclude that the genotypic effects exhibited significant crossing. This is a limitation frequently encountered in research at the intersection of genetics and psychology: Exploring nuanced questions about the effects associated with identified genes requires samples with detailed phenotypic and environmental information. But the trade-off for depth of measurement is that sample sizes are necessarily decreased. More limited power hampers one’s ability to draw strong conclusions about the nature of the interactions observed.
Absent definitive statistical evidence to support strong conclusions about the mechanism of the observed interaction between CHRM2 and parental monitoring, are there theoretical reasons to believe that CHRM2 could be involved in differential susceptibility? If CHRM2 were a plasticity gene, rather than a vulnerability gene, this could explain many puzzling features of the extant literature on CHRM2. We have focused here on the role of CHRM2 in substance use and externalizing outcomes; however, CHRM2 has also been associated with major depression in three independent samples (Comings et al., 2002; Luo et al., 2005; Wang et al., 2004) and with IQ in three independent samples (Comings et al., 2003; Dick, Aliev, et al., 2007; Gosso et al., 2006). In the studies that explicitly controlled for covarying conditions, it did not appear that the association across multiple outcomes (depression, alcohol dependence, IQ outcomes) was due to correlation among the phenotypes (Dick, Aliev, et al., 2007; Luo et al., 2005). However, these remarkably consistent early findings have more recently been joined by failures to replicate, with a new study finding no evidence of association between CHRM2 and cognitive ability across three independent samples (Lind et al., 2009). Further, we did not find strong evidence for main effects of CHRM2 on externalizing behavior in the CDP.
There are currently no published studies on environmental moderation of effects associated with CHRM2, and in this first effort to examine such moderation, we found suggestions of a crossover effect in two independent samples. The possibility that CHRM2 is a plasticity gene, rather than a vulnerability gene, provides a potential explanation for the observed results involving this gene. CHRM2 may not be a gene “for” any one disorder; rather, if it is a gene involved in biological sensitivity to context, it would be expected to have effects on a range of outcomes, which is the pattern that has been observed for this gene. Further, most of the genetic studies of CHRM2 have not incorporated environmental information, and this could help account for the variability in the findings concerning possible main effects associated with CHRM2: If a sample has an average mean level on the relevant environmental dimension, then no main effect of CHRM2 would be observed, as sampling is occurring essentially where the lines cross on Figure 4b. However, deviation from mean levels on the relevant environmental dimension (in either direction) would lead to the ability to detect significant main effects of CHRM2. This could explain the notable early replications of CHRM2’s effects across multiple independent samples, as many samples were clinical samples, and perhaps more likely than community samples to deviate from mean normative environmental conditions. (However, we note that an equally plausible explanation of mixed findings is normal sampling variability across studies trying to detect small genetic effects.)
It is of interest that in the CDP, we genotyped the same SNPs as were genotyped in the COGA project. The main effect detected in the COGA project mapped onto the ordering of genotypes observed at the low end of parental monitoring in the CDP sample. The COGA families are densely affected with alcohol dependence, and alcohol dependence in the family is known to be adversely related to family relations (Zhou, King, & Chassin, 2006). Accordingly, one might expect that these families would disproportionately be characterized by low parental monitoring, and the genetic effects observed for the low-monitoring families from the community-based CDP sample exactly map onto the genetic main effects observed in the COGA.
Belsky et al. (2009) have also pointed out that only with rare exceptions (Bakermans-Kranenburg & van IJzendoorn, 2006; Taylor et al., 2006) has the environment of a study sample included both positive and negative ends of the spectrum. Rather, the absence of environmental stressors has usually constituted the “low” end of the environment—for example, the absence of life stressors (Caspi et al., 2003) or the absence of maltreatment (Caspi et al., 2002). Parental monitoring, the environmental variable studied in the present work, reflects levels of parents’ knowledge about their children’s activities. Low levels of parental monitoring can be considered a negative, high-risk environment for externalizing behavior, whereas high levels of parental monitoring can be considered a positive, low-risk environment (Pettit, Laird, Dodge, Bates, & Criss, 2001). That said, although we studied parental monitoring because of the previous literature indicating the central importance of this construct for externalizing behavior and because of the availability of data on parental monitoring for both samples, we do not necessarily believe that the environmental moderation associated with CHRM2 is limited to parental monitoring. Rather, we believe that parental monitoring, although an important variable in itself (Latendresse et al., 2008), is likely to be a proxy for many correlated parenting variables and provides general information about the nature and quality of the adolescent’s environment. For example, in the CDP sample, parental monitoring is significantly correlated with measures of peer deviance and neighborhood safety. Future work will be devoted to exploring the nature of the environmental variables that might be relevant to the interaction effect observed in the present study.
In summary, our data suggest a gene-environment interaction effect in which the effect of CHRM2 on adolescent externalizing behavior is moderated by parental monitoring. Specifically, the association between the genotype and externalizing behavior is accentuated under conditions of lower parental monitoring. Additionally, there is suggestion of a crossover effect: The genotype associated with the highest levels of externalizing behavior at the low end of parental monitoring was associated with the lowest levels of externalizing behavior at the extreme high end of parental monitoring. These findings may suggest that CHRM2 is a plasticity gene, a gene involved in biological sensitivity to context (Ellis & Boyce, 2008). This would account for the patterns of results observed across studies of CHRM2. We hope our study will act as a springboard for future studies aimed at testing this hypothesis and further characterizing how CHRM2 is associated with a variety of psychological outcomes of interest.
Footnotes
Reprints and permission: sagepub.com/journalsPermissions.nav
Declaration of Conflicting Interests
The authors declared that they had no conflicts of interest with respect to their authorship or the publication of this article.
References
- Achenbach TM. Manual for the Youth Self-Report and 1991 profile. Burlington: University of Vermont, Department of Psychiatry; 1991. [Google Scholar]
- Achenbach TM. Manual for the Young Adult Self-Report and Young Adult Behavior Checklist. Burlington: University of Vermont, Department of Psychiatry; 1997. [Google Scholar]
- Achenbach TM, Rescorla LA. Manual for the ASEBA school-age forms & profiles. Burlington: University of Vermont, Research Center for Children, Youth, & Families; 2001. [Google Scholar]
- Achenbach TM, Rescorla LA. Manual for the ASEBA adult forms & profiles. Burlington: University of Vermont, Research Center for Children, Youth, & Families; 2003. [Google Scholar]
- Bakermans-Kranenburg MJ, van IJzendoorn MH. Gene-environment interaction of the dopamine D4 receptor (DRD4) and observed maternal insensitivity predicting externalizing behavior in preschoolers. Developmental Psychobiology. 2006;48:406–409. doi: 10.1002/dev.20152. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinfor-matics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Belsky J, Bakermans-Kranenburg MJ, van IJzendoorn MH. For better and for worse: Differential susceptibility to environmental influences. Current Directions in Psychological Science. 2007;16:300–304. [Google Scholar]
- Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Molecular Psychiatry. 2009;14:746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Comings DE, Wu S, Rostamkhani M, McGue M, Iacono WG, Cheng LS, MacMurray JP. Role of the cholinergic muscarinic 2 receptor (CHRM2) gene in cognition. Molecular Psychiatry. 2003;8:10–13. doi: 10.1038/sj.mp.4001095. [DOI] [PubMed] [Google Scholar]
- Comings DE, Wu S, Rostamkhani M, McGue M, Iacono WG, MacMurray JP. Association of the muscarinic cholin-ergic 2 receptor (CHRM2) gene with major depression in women. American Journal of Medical Genetics. 2002;114:527–529. doi: 10.1002/ajmg.10406. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Wang JC, Hinrichs AL, Bertelsen S, Bucholz K, et al. Alcohol dependence with comorbid drug dependence: Genetic and phenotypic associations suggest a more severe form of the disorder with stronger genetic contribution to risk. Addiction. 2007;102:1131–1139. doi: 10.1111/j.1360-0443.2007.01871.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Kramer J, Wang JC, Hinrichs AL, Bertelsen S, et al. Association of CHRM2 with IQ: Converging evidence for a gene influencing intelligence. Behavior Genetics. 2007;37:265–272. doi: 10.1007/s10519-006-9131-2. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Grucza RA, Schuckit M, Kuperman S, et al. Using dimensional models of externalizing psychopathology to aid in gene identification. Archives of General Psychiatry. 2008;65:310–318. doi: 10.1001/archpsyc.65.3.310. [DOI] [PubMed] [Google Scholar]
- Dick DM, Latendresse SJ, Lansford JE, Budde J, Goate A, Dodge KA, et al. Role of GABRA2 in trajectories of externalizing behavior across development and evidence of moderation by parental monitoring. Archives of General Psychiatry. 2009;66:649–657. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Purcell S, Viken R, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. Journal of Abnormal Psychology. 2007;116:213–218. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. Journal of Abnormal Psychology. 2007;116:213–218. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Enoch MA, Hodgkinson C, Xu K, Catena M, Robin RW, Goldman D. Interaction between a functional MAOA locus and childhood sexual abuse predicts alcoholism and antisocial personality disorder in adult women. Molecular Psychiatry. 2008;13:334–347. doi: 10.1038/sj.mp.4002034. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: Identifying specific genes through family studies. Addiction Biology. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Ellis BJ, Boyce WT. Biological sensitivity to context. Current Directions in Psychological Science. 2008;17:183–187. [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gosso MF, van Belzen M, de Geus EJ, Polderman JC, Heutink P, Boomsma DI, Posthuma D. Association between the CHRM2 gene and intelligence in a sample of 304 Dutch families. Genes, Brain and Behavior. 2006;5:577–584. doi: 10.1111/j.1601-183X.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- Hedrick P, Kumar S. Mutation and linkage disequilibrium in human mtDNA. European Journal of Human Genetics. 2001;9:969–972. doi: 10.1038/sj.ejhg.5200735. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Yang BZ, Douglas-Palumberi H, Houshyar S, Lipschitz D, Krystal JH, Gelernter J. Social supports and serotonin transporter gene moderate depression in maltreated children. Proceedings of the National Academy of Sciences, USA. 2004;101:17316–17321. doi: 10.1073/pnas.0404376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS. A conceptual overview of gene-environment interaction and correlation in a developmental context. In: Kendler KS, Jaffe S, Romer D, editors. The dynamic genome and mental health: The role of genes and environments in youth development. New York, NY: Oxford University Press; (in press) [Google Scholar]
- Kerr M, Stattin H. What parents know, how they know it, and several forms of adolescent adjustment: Further support for a reinterpretation of monitoring. Developmental Psychology. 2000;36:366–380. [PubMed] [Google Scholar]
- Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, Craig IW, Moffitt TE. MAOA, maltreatment, and gene-environment interaction predicting children’s mental health: New evidence and a meta-analysis. Molecular Psychiatry. 2006;11:903–913. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- Latendresse SJ, Rose RJ, Viken RJ, Pulkkinen L, Kaprio J, Dick DM. Parenting mechanisms in links between parents’ and adolescents’ alcohol use behaviors. Alcoholism: Clinical and Experimental Research. 2008;32:322–330. doi: 10.1111/j.1530-0277.2007.00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Luciano M, Horan MA, Marioni RE, Wright MJ, Bates TC, et al. No association between cholinergic muscarinic receptor 2 (CHRM2) genetic variation and cognitive abilities in three independent samples. Behavior Genetics. 2009;39:513–523. doi: 10.1007/s10519-009-9274-z. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence, and affective disorders: Results from an extended case-control structured association study. Human Molecular Genetics. 2005;14:2421–2432. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature Genetics. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit GS, Laird RD, Dodge KA, Bates JE, Criss MM. Antecedents and behavior-problem outcomes of parental monitoring and psychological control in early adolescence. Child Development. 2001;72:583–598. doi: 10.1111/1467-8624.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale BM, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: A meta-analysis. Journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SE, Way BM, Welch WT, Hilmert CJ, Lehman BJ, Eisenberger NI. Early family environment, current adversity, the serotonin transporter promoter polymorphism, and depressive symptomatology. Biological Psychiatry. 2006;60:671–676. doi: 10.1016/j.biopsych.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Volpicelli LA, Levey AI. Muscarinic acetylcholine receptor subtypes in cerebral cortex and hippocampus. Progress in Brain Research. 2004;145:59–66. doi: 10.1016/S0079-6123(03)45003-6. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, et al. Evidence of common and specific genetic effects: Association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Human Molecular Genetics. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Zhou Q, King KM, Chassin L. The roles of familial alcoholism and adolescent family harmony in young adults’ substance dependence disorders: Mediated and moderated relations. Journal of Abnormal Psychology. 2006;115:320–331. doi: 10.1037/0021-843X.115.2.320. [DOI] [PubMed] [Google Scholar]