Abstract
Osteoblasts and osteoclasts are the two major bone cells involved in the bone remodeling process. Osteoblasts are responsible for bone formation while osteoclasts are the bone-resorbing cells. The major event that triggers osteogenesis and bone remodeling is the transition of mesenchymal stem cells into differentiating osteoblast cells and monocyte/macrophage precursors into differentiating osteoclasts. Imbalance in differentiation and function of these two cell types will result in skeletal diseases such as osteoporosis, Paget’s disease, rheumatoid arthritis, osteopetrosis, periodontal disease, and bone cancer metastases. Osteoblast and osteoclast commitment and differentiation are controlled by complex activities involving signal transduction and transcriptional regulation of gene expression. Recent advances in molecular and genetic studies using gene targeting in mice enable a better understanding of the multiple factors and signaling networks that control the differentiation process at a molecular level. This review summarizes recent advances in studies of signaling transduction pathways and transcriptional regulation of osteoblast and osteoclast cell lineage commitment and differentiation. Understanding the signaling networks that control the commitment and differentiation of bone cells will not only expand our basic understanding of the molecular mechanisms of skeletal development but will also aid our ability to develop therapeutic means of intervention in skeletal diseases.
Keywords: osteoblasts, osteoclasts, signaling pathways, transcriptional regulation, skeletal disease, bone genes
I. INTRODUCTION
Bone is an essential mineralized tissue with critical mechanical and metabolic functions. It has the capacity to adapt to its functional environment in such a way that its morphology is “optimized” for the mechanical demand.1 Physiological bone turnover can be divided into two temporal phases: modeling, which occurs during development, and remodeling, a lifelong process involving tissue renewal. Bone integrity and function are maintained by an exquisite balance between the osteoblast and the osteoclast, the two major bone cells involved in the remodeling process. Remodeling starts with removal by osteoclasts of matrix, a mixture of insoluble proteins in which type I collagen is predominant (>90%) and a poorly crystalline, chemically modified hydroxyapatite. Following resorption, osteoblasts are recruited to the site, where they secrete and mineralize new matrix. Bone is continuously remodeled throughout life and an imbalance in this process can result in bone disease. The increased activity of osteoclasts caused by estrogen withdrawal causes bone loss and osteoporosis, a frequent low–bone mass disorder in postmenopausal women leading to structural instability and a high fracture risk. A recent study has shown that estrogen actually induces osteoclast apoptosis.2 Estrogen deficiency is known to play a critical role in the development of osteoporosis, while calcium and vitamin D deficiencies and secondary hyperparathyroidism also contribute.3 Osteoporosis is a factor in more than 1.5 million fractures each year in the United States alone. Costs have been estimated at more than $17 billion a year, particularly from hip fractures. More than 75% of which occur in women. A better understanding of bone quality, coming from biochemical markers and refined imaging techniques, will help predict who is most at risk of debilitating fractures. One of the main approaches to gleaning details about the quality of bones is to measure the activity of osteoclasts and osteoblasts, the cells that remodel bone and thus influence its structural properties.4 The recent discoveries of signal transduction pathways and transcription factors critical for the differentiation and function of osteoblasts and osteoclasts have opened up new approaches to understanding the pathogenesis of osteoporosis. We review what is known about the transcription factors and cytokines that regulate the stages of differentiation in osteoclasts and osteoblasts.
II. BONE CELL ORIGIN AND CELL LINEAGE
A. Osteoblast Lineage
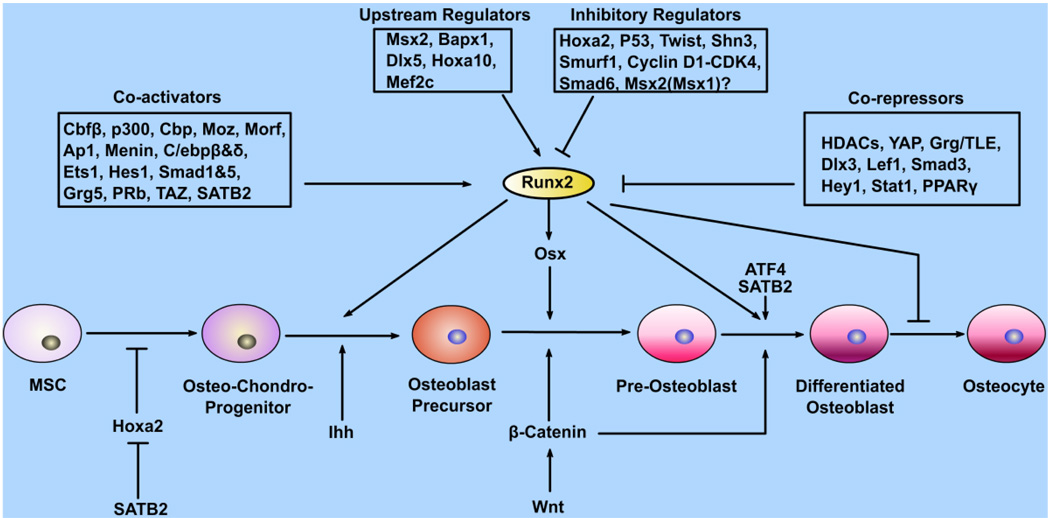
Osteoblasts, which play central roles in bone formation, are derived from undifferentiated mesenchymal cells (Fig. 1), which also have the capacity to differentiate into chondrocytes, adipocytes, and myoblasts.5 There are three major stages of osteoblastogenesis: proliferation, matrix maturation, and mineralization. Osteoblast progenitors can first be identified within the inner perichondrium adjacent to, and coincident with, the first appearance of hypertrophic chondrocytes. This tight linkage reflects a crucial role for Indian hedgehog (Ihh) signaling (Fig. 1), discussed below. Markers of this program in the endochondral-derived skeleton are dependent on an initial Ihh input. During normal development, Ihh signaling appears to act as a switch within a specific population of inner perichondrial mesenchyme to initiate a program of bone formation.6 Failure to activate this switch results in cells adopting an alternative chondrocyte pathway of development. Ihh acts in differentiation of osteoblast progenitors into runt-related transcription factor 2 (Runx2)+ osteoblast precursors. Wnt/β-catenin signaling acts later in the differentiation pathway to osterix+ osteoblast precursors and then to bone-secreting osteoblasts (Fig. 1).6 In addition, alkaline phosphatase (ALP), bone sialoprotein (BSP), and collagen type 1 alpha 1 (Col1a1) are early markers of osteoblast differentiation, while parathyroid hormone/PTH-related peptide (PTH/PTHrP) receptor (PPR) and osteocalcin (OCN) appear late, concomitantly with mineralization. Osteopontin (OPN) peaks twice, during proliferation and then again in the later stages of differentiation.
FIGURE 1.
Transcription factors and signaling involved in the osteoblast differentiation pathway. Osteoblasts and chondrocytes are derived from common mesenchymal stem cell precursors. Runx2 stimulates terminal differentiation. A number of transcription factors are involved in Runx2 regulation and function, either upstream of Runx2 or as coactivators or corepressors. Runx2 functions upstream of Osx, which is required after Runx2 for osteoblast differentiation. Ihh and Wnt/β-catenin are key signaling molecules in osteoblastogenesis.
Following initial lineage commitment, a phase of lineage expansion ensues that culminates normally in permanent cell cycle withdrawal. The initial cell division is asymmetric, giving rise to another stem cell (self-renewal) and a committed osteoprogenitor. Following commitment, the stem cell gives rise to the transit-amplifying compartment.7 This phase is associated with intensive proliferative activity. The preosteoblast is an intermediate stage, which expresses STRO-1, ALP, PPR, and type I collagen, and is committed to the osteoblast lineage with extensive replicative capacity, but no self-renewal capacity.8 In vitro, the use of agents such as retinoic acid can induce further differentiation in the preosteoblast. The mature osteoblast expresses ALP, OPN, BSP, and OCN, and lies adjacent to newly synthesized osteoid. This stage, which is responsible for the laying down of bone, has limited replicative potential.9 The cumulative effect of the recruitment of stem cells and their expansion, and the functional capacity of mature osteoblasts, is measured by rates of bone formation in vivo. The second key step initiates terminal differentiation and permanent cell cycle withdrawal. The terminal stage of the bone lineage is the postmitotic osteocyte, often found isolated within bone, presumably embedded within advancing osteoids. As an alternate fate, a proportion of cells in the transient amplifying compartment may also terminate in apoptosis.
It was recently found that calcineurin plays a role in osteoblast differentiation. Calcineurin α null mice display severe osteoporosis and reduced Runx2, BSP, and OCN expression.10 A range of cytokines modulate osteoblast differentiation, including bone matrix–derived transforming growth factor beta (TGF-β), bone morphogenic protein 2 (BMP-2), BMP-4, and BMP-7, and their inhibitors noggin, chordin, gremlin, and sclerostin, the last identified by positional cloning of families with increased bone mass. Similarly, numerous hormones impact osteoblast function positively including insulinlike growth factor-1 (IGF-1), PTH, PTHrP, 1,25(OH)2D3, leptin, glucocorticoids, the Notch pathway, and members of the leukemia inhibitory factor/interleukin-6 (IL-6) family. Deletion of the neuropeptide Y2 receptor was shown to increase bone mineralization by increasing the number of mesenchymal progenitor cells and osteoblast activity.11 Y1 null mice also showed elevated bone mass, showing the inhibitory effects of neuropeptide Y on bone cells via the Y receptors.12
B. Osteoclast Lineage
Osteoclast (OC) lineage, phenotype, and gene expression are associated with OC differentiation. Transplantation studies have demonstrated that the OC precursor is a mononuclear cell that is hematopoietic in origin.13 Earlier studies performed in vivo14 and in vitro15–17 suggested that OCs are derived from cells of the mononuclear phagocyte system. However, more recent evidence indicates that although macrophages and OCs share a common precursor, these lineages diverge on further differentiation.18 OCs possess a distinct phenotype and functional capabilities compared to cells of the macrophage series, particularly the ability to resorb bone. OC hematopoietic precursors migrate via vascular pathways to the skeleton. Osteoblasts, chondrocytes, and their mineralized matrices, together with stromal and endothelial cells, provide the microenvironment for homing of these precursor cells. Stromal cells produce cytokines, including macrophage-colony stimulating factor (M-CSF) and IL-6, which induce and modulate growth and differentiation of the precursors to mature OCs.19,20 Annexin II is an OC stimulatory factor that stimulates OC formation by activating T cells to secrete granulocyte macrophage colony-stimulating factor (GM-CSF), which expands the OC precursor pool.21 In addition, the vitamin D metabolite calcitriol and the parathyroid hormone support OC development.22,23 Usui et al. found that hypertrophic chondrocytes regulate osteoclastogenesis through BMP-2– induced receptor activator of NF-κB ligand (RANKL) expression.24 BMP-2 regulation of RANKL activity was abolished by Runx2 mutation. It was recently discovered that follicle-stimulating hormone (FSH) effects osteoclast formation.25 FSH was found to activate several signaling mechanisms in osteoclasts and their precursors, such as Erk kinase/mitogen–activated protein kinase (MEK/Erk), nuclear factor kappa-B (NF-κB), thymoma viral proto-oncogene (Akt), and inhibitory kappa-B alpha (IκBα).
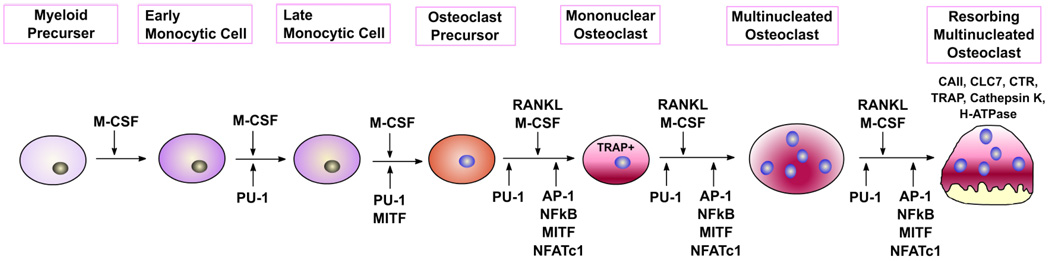
The OC precursor cells have multiple Golgi complexes and abundant mitochondria,14 and are positive for nonspecific esterase26 and possibly type IV collagenase. In a suitable microenvironment, OC precursors differentiate into preosteoclasts (preOCs) or mononuclear osteoclasts (Fig. 2). The term “preOC” has been used to describe a direct mononuclear precursor showing morphological and cytochemical features that are similar to those of the multinucleated OC.27–30 They are tartrate-resistant acid phosphatase (TRAP)– positive cells that express mRNAs for all OC-associated phenotypes [e.g., TRAP, calcitonin receptor (CTR), and cathepsin K], and are poised to fuse into multinucleated OCs on further differentiation.29 OCs possess a highly specialized proton-generating mechanism for the rapid dissolution of mineral, and secrete collagenases,31 cathepsin K,32 and other hydrolases active in the degradation of bone matrix proteins. OC formation in vitro can be studied either in organ culture of murine embryonic metatarsals,19 in a coculture system in which hematopoietic precursor cells (spleen or marrow cells) are cultured with osteoblasts,33 or in a cloned SV40-transformed OC precursor cell line, that is, MOCP-5.34 Soluble RANKL and M-CSF allow spleen or marrow cells to differentiate into OCs without coculture with osteoblasts.
FIGURE 2.
Stages of osteoclast differentiation from hematopoietic lineage cells. M-CSF and RANKL are essential external stimuli for osteoclastogenesis. PU.1, Mitf, NF-κB, AP-1, and NFATc1 are required for differentiation of mature osteoclasts.
III. BONE CELL SIGNALING
A. Osteoblast Signaling
1. Wnt Signaling Pathway
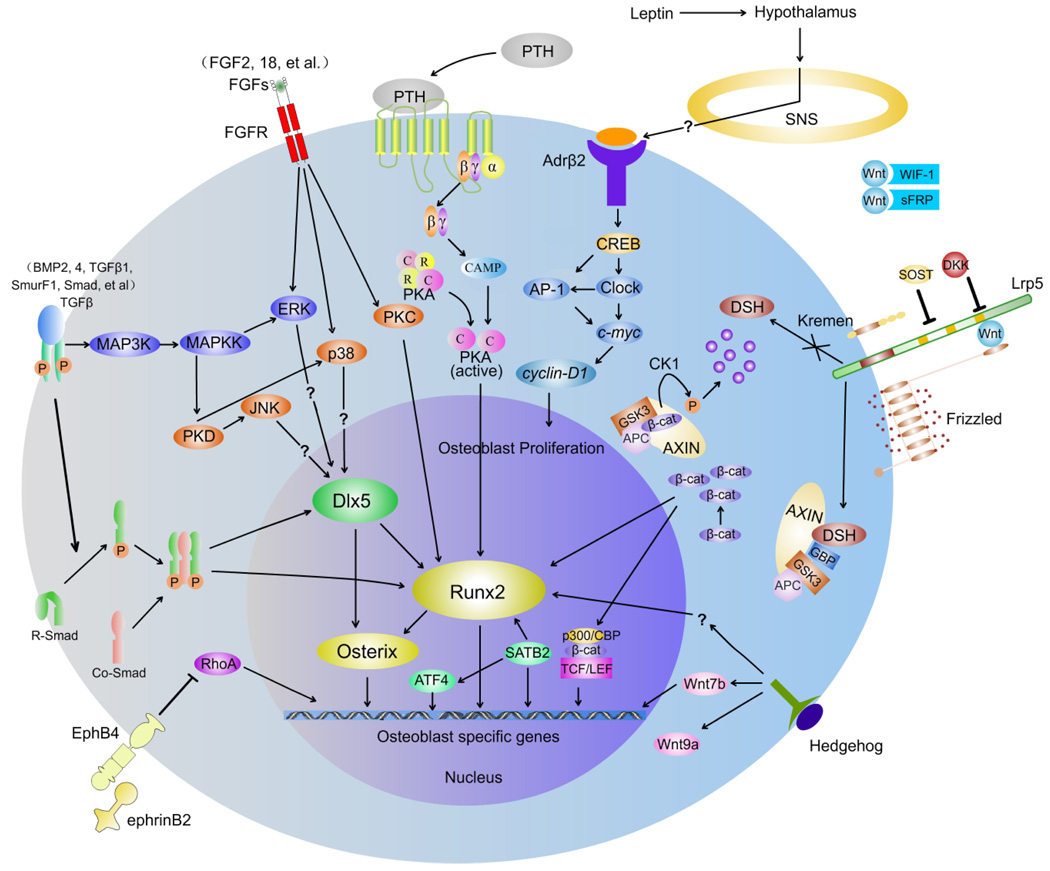
Wnt proteins signal through several pathways to regulate cell growth, differentiation, function, and death. The Wnt/β-catenin, or canonical, pathway appears to be particularly important for bone cell signaling.35–38 Here we outline the current model of Wnt signal transduction, shown in Figure 3. Wnt proteins released from or presented on the surface of signaling cells act on target cells by binding to the frizzled homolog (Drosophila)/low-density lipoprotein receptor– related protein 5 and 6 (FZD/LRP5/6) complex at the cell surface. Signals are generated through the proteins Disheveled, Axin, and Frat-1, which disrupt the protein complex and inhibit the activity of glycogen synthase kinase 3 (GSK3), thus causing hypophosphorylation of its substrate, β-catenin (Fig. 3).39 The on state involves increasing the post-translational stability of β-catenin, through Wnt-dependent inhibition of GSK3 (Fig. 3). Stabilized β-catenin accumulates in the cytosol and translocates to the nucleus to activate target gene transcription. The lymphoid enhancer-binding factor/T-cell factor (Lef/Tcf) transcription factor family members are well-studied nuclear partners of β-catenin.40,41 β-catenin displaces co-repressors of Lef/Tcfs [e.g., Groucho, silencing mediator of retinoid and thyroid receptors and nuclear receptor corepressor (SMRT/NCoR)], and forms heterodimers with the Lef/Tcf proteins to bind DNA and initiate the transcription of target genes.42
FIGURE 3.
Seven important signaling networks of osteoblast differentiation. Binding of Wnt to the FZD receptor induces β-catenin accumulation, which translocates to the nucleus to activate target gene transcription. Several transcription factors have been found crucial for osteoblast differentiation downstream of this signaling pathway, such as Runx2, Osterix, and ATF4. They are essential for differentiation of mesenchymal stem cells into differentiated osteoblasts and also function in the transcription of osteoblast-specific genes.
Extracellular Wnt ligands can interact with a host of secreted antagonists, including the secreted FZD-related protein (sFRP) family and Wnt inhibitory factor 1 (WIF-1), preventing activation of the pathway. Knockout of sFRP1 in mice leads to a high bone mass phenotype, early induction of collagen type 10a1, and activation of the Runx2 transcription factor leading to increased chondrocyte differentiation.43 Loss of sFRP1 in mice results in reduced osteoblast apoptosis while enhacing proliferation and differentiation of osteoblasts, leading to heightened trabecular bone mass.44 LRP5/6 coreceptor activity is inhibited by members of the SOST (Sclerosteosis gene product)45 and Dickkopf (Dkk) families,46 all of which bind LRP5/6.47 Both Dkk1 and Dkk2 antagonize canonical Wnt signaling by simultaneously binding to LRP5/647 and a single-transmembrane protein called kremen.46 Diarra et al. were able to reverse the bone-destructive pattern of a mouse model of rheumatoid arthritis to the bone-forming pattern of osteoarthritis by inhibiting Dkk1.48 Tumor necrosis factor (TNF)-α was identified as a key inducer of Dkk1 in human rheumatoid arthritis, suggesting that Wnt signaling is a key regulator of joint remodeling. Dkk2 has been shown to be involved in terminal osteoblast differentiation, although the precise mechanisms need further investigation.49
In the absence of Wnt expression, degradation of β-catenin is facilitated via interactions with a protein complex consisting of adenomatous polyposis coli (APC), axin, and GSK3. APC and axin act as scaffold proteins allowing GSK3 to bind and phosphorylate β-catenin, identifying it for degradation by the beta-transducin repeat–containing protein (β-TrCP)–mediated ubiquitin/proteasome pathway. In the nucleus, prospective target genes of the pathway are kept in a repressed state by interacting with TCF and LEF transcription factors, with associated corepressors (Fig. 3). In this off state, cells maintain low cytoplasmic and nuclear levels of β-catenin, although β-catenin is associated with cadherins at the plasma membrane, which spares it from the degradative pathway.41 Despite the prominent role of canonical Wnt signaling in osteoblast biology, the role of the TCF/LEF family of transcription factors is unclear.
Indisputably, Wnts are involved in embryonic skeletal patterning, fetal skeletal development, and adult skeletal remodeling.50–52 Unraveling the function(s) of Wnt proteins in the regulation of skeletogenesis has been a complex problem, however, confounded by questions of functional redundancy, multiple times and sites of action, and the presence of other molecules that compete with Wnt function. The first indication that Wnt signaling plays a critical role in bone formation came from human studies where mutations in the Wnt coreceptor LRP5 are causally linked to alterations in human bone mass.53–59 Eight LRP5 missense mutations (D111Y, G171R, A214T, A214V, A242T, T253I, G171V, M282V) have been found to cause high bone mass,53,54,56,58,59 while several homozygous or heterozygous nonsense, frameshift, and missense mutations have been identified in osteoporosis-pseudoglioma patients leading to low bone density.55,57 LRP5−/− mice also have low bone mass.60 One of the mechanisms whereby Wnt signaling increases bone formation is via stimulation of the development of osteoblasts, and there is considerable in vitro evidence supporting a role for canonical Wnt/β-catenin signaling in this process.61–63 Higher levels of β-catenin enhance bone formation with associated increases in expression of osteoblast-specific genes,61,64 whereas conditional knockdown of the β-catenin gene at an early developmental stage causes ectopic chondrogenesis and abnormal osteoblast differentiation.64–66 Most recently, Hill et al.,64 Day et al.,65 Glass et al.,67 and Hu et al.68 provide compelling evidence that Wnt signaling represents both a cell-autonomous mechanism for inducing osteoblastic and suppressing chondrocytic differentiation in early osteochondroprogenitors, and a mechanism in fully differentiated osteoblasts for stimulating the production of osteoprotegerin (OPG), an inhibitor of osteoclast formation.64,65,67,68
Recent experiments examining the conditional inactivation of β-catenin in skeletal progenitors and using different Cre lines revealed that β-catenin activity is essential for the differentiation of mature osteoblasts and, consequently, for bone formation in endrochondral bones (the long bones of the limbs) and membranous bones (in the skull).64,65,68 These variable results likely arise because Wnt/β-catenin signaling regulates bone development and accrual through different mechanisms at different stages of life.69 This concept is supported by the results of studies using mouse models in which targeted deletion of β-catenin occurs early or late in osteoblastogenesis. It is likely that β-catenin activity is required in a bipotential precursor of the osteoblast lineage, the so-called osteochondroprogenitor, and indeed its absence steers the fate of mesenchymal precursors toward chondrogenesis.64,65 Because Runx2, but not osterix, is expressed in β-catenin−/− mesenchymal cells,65,68 β-catenin seems to be required for osteoblast differentiation at the preosteoblast stage. Furthermore, β-catenin/TCF1 enhances Runx2 expression and Runx2 promoter activity.70 By contrast, for differentiation into the chondrocyte lineage, β-catenin levels must be low.64,65,68 A recent study of the mechanism of Wnt/β-catenin–induced osteoblastogenesis revealed that Wnt/β-catenin signaling allows activation of transcription factors important in osteoblastogenesis by suppressing CCAAT/enhancer binding protein alpha (C/EBPa) and peroxisome proliferator activated receptor gamma (PPARγ).71 Knockdown of C/EBPa or PPARγ expression in ST2 cells and mouse embryonic fibroblasts reduced adipogenic potential and caused spontaneous formation of osteoblasts.
Recently, inactivation of β-catenin function in more mature osteoblasts using a Col1a1- and an OCN-Cre line revealed a novel role for canonical Wnt signaling in postnatal bone homeostasis.66,67 Mice deficient in β-catenin develop osteopenia, while activation of β-catenin function in osteoblasts resulted in increased bone mass and an osteopetrotic phenotype.66,67 However, no change in osteoblast activity or histomorphometric evidence of bone formation was observed. The altered bone resorption was caused by deregulation of OPG, a major inhibitor of osteoclast differentiation.67 OPG is a direct target gene of the β-catenin-TCF complex in osteoblasts and Tcf1 is probably the relevant transcription factor required for OPG regulation; nevertheless, a possible role for Tcf4 cannot be excluded.62,67,72 These mice demonstrate that β-catenin regulates osteoclastogenesis through effects on expression of osteoprotegerin and RANKL.66
On the other hand, recent evidence has revealed that noncanonical Wnt signaling also plays a role in osteoblastogenesis. Takada et al. found that the noncanonical Wnt pathway through CaMKII-TAK1-TAB2-NLK promotes osteoblastogenesis by transcriptionally repressing PPARγ transactivation and inducing Runx2 expression.73 Tu et al. have shown that Wnt3a and Wnt7b each function in osteoblastogenesis through a protein kinase C delta (PKCδ) pathway.74 Wnt3a signals through G protein Gαq/11 subunits to activate PKCγ, and PKCγ-deficient mice exhibit a deficit in embryonic bone formation. Ablation of Wnt7b in skeletal progenitors also leads to reduced bone formation in mouse embryos.
2. TGF-β Superfamily Signaling
Once activated, TGF-β can interact with its receptor to induce signaling. All members of the TGF-β superfamily signal through a dual receptor system of type I and type II transmembrane serine/threonine kinases. The mothers against decapentaplegic homolog (Smad) signaling turned out to play a central role in the transmission of signals from all receptors activated by the TGF-β superfamily members to target genes in the nucleus. Several members of the TGF-β superfamily, such as BMPs, have potent osteogenic effects. BMPs are a group of phylogenetically conserved signaling molecules, and were initially identified by their capacity to induce endochondral bone formation.75–77 BMP-1 through BMP-7 are expressed in skeletal tissue, and BMP-2, −4, and −6 are the most readily detectable BMPs in osteoblast cultures.75,78 BMPs are unique because they are implicated in the specification of both chondrocytes and osteoblasts,79 as well as in the subsequent modification of the osteogenic program, where some BMPs promote bone formation, such as BMP-2, BMP-7, BMP-6, and BMP-9,80 although BMP-3 acts as a negative regulator of bone formation.81
On receptor activation, BMPs transmit signals through Smad-dependent and Smad-independent pathways, including Jun N-terminal kinase (JNK), and p38 mitogen activated protein (MAP) kinase (MAPK) pathways.82 Smads are the major signal transducers for the serine/threonine kinase receptors.83 There are three classes of Smads: (1) receptor-regulated Smads (R-Smads) that can be BMP activated, such as Smad 1, 5, and 8 (referred to as BR-Smads in this article), or TGF-β activated, such as Smad 2 and 3 (TR-Smads); (2) common partner BMP and TGF-β mediator Smads (Co-Smads), such as Smad 4; and (3) inhibitory Smads (I-Smads), such as Smad 6 and 7. On ligand stimulation and activation by type II receptors, type I receptors phosphorylate R-Smads, which in turn form complexes with Co-Smads (Fig. 3).84 The R-Smad/Co-Smad complexes then translocate into the nucleus and regulate transcription of target genes by interacting with various transcription factors and transcriptional coactivators or corepressors. The third class of Smads, I-Smads, negatively regulates signaling by the R-Smads and Co-Smads. Runx2 and BR-Smads physically interact with each other on activation of BMP signaling, and cooperatively regulate the transcription of target genes, leading to osteoblast differentiation of mesenchymal progenitor cells.85–87 BMP induces Runx2 expression in mesenchymal progenitor cells through the action of BR-Smads,88 and BR-Smads in turn interact with Runx2 and further induce osteoblastic differentiation. BMP does not directly induce the expression of Runx2 in mesenchymal cells,89 but it facilitates expression of distalless homeobox 5 (Dlx5) in osteoblasts,90,91 and Dlx5 then induces expression of Runx2 in osteoprogenitor cells. Osteoprogenitor cells, for example, C2C12 cells, have been widely used for the identification of BMP target genes during osteoblastic differentiation. Hairy/enhancer of split related with YRPW motif 1 (Hey1; also termed HesR1 and Herp2) and Tcf7 are transcription factors specifically expressed in osteoblast cells by BMP-2 treatment, and are involved in Notch and Wnt signaling, respectively.92 Using constitutively active BMP type I receptors, Korchynskyi et al. identified several genes as targets of BMP receptors in C2C12 cells, including transcription factors Hey1, ITF-2 (Tcf4), and interferon regulatory factor 8 (ICSBP).93
Although the Smads are critical mediators in the TGF-β signaling pathway, a substantial body of evidence illustrates the existence of additional, Smad-independent pathways. BMP-2 can activate ERK, JNK, and p38 in osteoblastic cells and provide evidence that these MAP kinases have distinct roles in regulating alkaline phosphatase and osteocalcin expression (Fig. 3).94,95 Recent reports suggest that during osteoblast differentiation, BMP-2 activates JNK and p38 via protein kinase D (PKD), independent of PKC activity.96 It has been demonstrated that following TGF-β and BMP induction, both the Smad and p38 MAPK pathways converge at the Runx2 gene to control mesenchymal precursor cell differentiation.97 Runx2 plays a central role in the BMP-2–induced transdifferentiation of C2C12 cells at an early restriction point by diverting them from the myogenic pathway to the osteogenic pathway.89,98 It has been found that the homeobox gene Dlx5 is an upstream target of BMP-2 signaling and that it plays a pivotal role in stimulating the downstream osteogenic master transcription factor Runx2.90 In turn, Runx2 acts simultaneously or sequentially to induce the expression of bone-specific genes that represent BMP-2–induced osteogenic transdifferentiation. However, inhibition of BMP signaling was shown to disrupt the ability of RUNX2 to stimulate osteoblast differentiation and transactivate an osteocalcin gene promoter–luciferase reporter in C3H10T1/2 cells.99 In conclusion, we can state that the JNK, ERK, and p38 MAPK pathways contribute considerably to all TGF-β–induced responses, but further characterization is needed to assess their importance in relation to the Smad-dependent and other TGF-β–induced signaling pathways.
BMP-2 has been reported to induce Osterix (Osx) expression in mouse progenitor cells and chondrocytes.100,101 Moreover, BMP-2–induced Osx expression is mediated by Dlx5 but is independent of Runx2.102 In the bone microenvironment, BMPs act in conjunction with other growth factors. Celil et al. identified the involvement of BMP-2 and IGF-I in mediating Osx expression in human mesenchymal stem cells (hMSCs).103 The BMP-2–induced effect on Osx expression was mediated via p38 but not via Erk. Under osteogenic culture conditions, both Erk and p38 were involved in mediating Osx expression.103
In the past several years, ubiquitin-mediated proteasomal degradation has been implicated in the regulation of BMP-2 and TGF-β signaling pathways in various cell types.104,105 Recently, several studies highlighted the importance of this mechanism in regulating the in vivo effects of TGF-β (Fig. 3).106,107 Dupont et al. redefined the role of a previously identified Smad 1 ubiquitin ligase, Smurf-1. The absence of Smurf-1 causes the accumulation of MEK kinase 2 (MEKK2), resulting in activation of JNK, an event that is both necessary and sufficient for BMP sensitization in osteoblasts.106 Sapkota et al. found that MAPK-dependent linker phosphorylation of Smad 1 allows recognition by Smurf-1 and restricts Smad 1 activity, controlling BMP signaling during mouse osteoblast differentiation.108 BMP also triggers linker phosphorylation, creating feedback control. Yamashita et al. identified and characterized a novel Smad 4 ubiquitin ligase, Ectodermin (Ecto), and provided convincing evidence that Ecto represents the elusive determinant of ectoderm formation, acting as a critical inhibitor of all Smad-dependent TGF-β signaling during vertebrate development.107
Recently, Tsuji et al. found that BMP-2 is a necessary component of the signaling cascade that governs fracture repair.109 Mice lacking the ability to produce BMP-2 in their limb bones have spontaneous fractures that do not resolve with time and the earliest steps of fracture healing seem to be blocked. The presence of other osteogenic stimuli cannot compensate for the absence of BMP-2, identfying BMP-2 as an endogenous mediator necessary for fracture repair.
3. Hedgehog Signaling
Osteoblast progenitors can first be identified within the inner perichondrium adjacent to, and coincident with, the first appearance of hypertrophic chondrocytes. This tight linkage reflects a crucial role for Ihh signaling (Fig. 1).110 Ihh is produced by prehypertrophic chondrocytes and appears to act directly on perichondrially located osteoblast progenitors to specify the osteoblast precursors.111,112 The failure of activation of Runx2, a crucial early determinant of the osteoblast lineage, indicates that hedgehog (Hh) signaling acts to initiate an osteogenic program.113 Furthermore, Hh activates osteoblast development in a variety of mesenchymal and skeletogenic cell types in vitro.111,114,115 Genetic manipulation of smoothened (Smo), which encodes an obligatory component of the Hh signaling pathway, has revealed that cells devoid of Smo, and hence Hh signaling, fail to undergo osteoblast differentiation.111 Although Ihh signaling plays the crucial role in regulating the temporal and spatial program of early osteoblast commitment, Ihh does not play an ongoing role beyond this stage.6 When Smo activity is removed in Osx1+ osteoblast precursors, normal bone-secreting OcHigh osteoblasts are generated, and the endochondral skeleton at birth is indistinguishable from wild type.6 Whether this is also true in the adult is currently under investigation.
The interaction between Hh and Wnt signaling is probably complex. It has been demonstrated that nuclear localization of β-catenin as well as expression of target genes for the Wnt canonical pathway were abolished in the perichondrium in Ihh−/− embryos.68 This could, among other possibilities, be due to the downregulation of Wnt expression in the absence of Hh signaling. Indeed, expression of Wnt9a and Wnt7b was either reduced or abolished in the perichondrium in Ihh−/− embryos. In addition, both genes were induced by Hh signaling in C3H10T1/2 cells (Fig. 3). Alternatively, the Hh and Wnt signaling pathways could intersect intracellularly via common regulators such as Suppressor of fused [Su(fu)]116 and GSK3.117,118 Other pathways in addition to canonical Wnt signaling also contribute to Hh-induced osteogenesis. Of note, some groups reported that Hh-induced osteogenesis in C3H10T1/2 cells required BMP signaling.119,120
4. FGF Signaling
The fibroblast growth factors (FGFs) are a family of secreted polypeptides that act through four related tyrosine kinase receptors (Fgfr1–Fgfr4) to regulate a plethora of developmental processes, and they are critical for the control of endochondral and intramembranous ossification.121 Human diseases that manifest the precocious osseous obliteration of sutures, known as craniosynostosis, often result from gain-of-function mutations in FGF receptors 1–3 (Fig 3).122,123 Fgfrs 1–3 are expressed in the developing and mature skeleton in patterns suggestive of both unique and redundant function.121 In the developing growth plate, both Fgfr1 and Fgfr2 are expressed in condensing mesenchyme that will give rise to cartilage. Fgfr2 remains expressed in reserve chondrocytes and appears to be downregulated in proliferating chondrocytes, whereas Fgfr1 is expressed in hyper-trophic chondrocytes. Later in development, Fgfr1 and Fgfr2 are both expressed in the perichondrium and periosteum, tissues that give rise to osteoblasts and cortical bone. In contrast to Fgfr1 and Fgfr2, Fgfr3 is prominently expressed in proliferating chondrocytes where it regulates cell growth and differentiation124 and in differentiated osteoblasts where it regulates bone density and cortical thickness.125,126 Mutations in Fgfrs account for many of the craniosynostosis and chondrodysplasia syndromes in humans.121,127,128
Embryos lacking Fgfr1 (Fgfr1−/−) die shortly after gastrulation,129 necessitating a conditional knockout approach to address function later in development. Hypomorphic alleles of Fgfr1 or conditional inactivation of Fgfr1 prior to limb bud initiation affects digital patterning and the formation of some skeletal elements.130–132 Fgfr1 signaling in the osteoprogenitor cell normally acts to stimulate differentiation, whereas it functions to suppress differentiation in differentiated osteoblasts. Thus, Fgfr1 signaling has stage-specific effects on osteoblast maturation.133
The FGF ligands that signal to Fgfr1 in osteoblasts are not known; however, three FGFs (FGFs 2, 9, and 18) are likely candidates for this role. FGF9 and FGF18 are expressed in the perichondrium/periosteum, and mice lacking these FGFs show delayed ossification during midgestation skeletogenesis.134–136 FGF2 is expressed in periosteal cells and osteoblasts,137,138 and adult FGF2−/− mice showed a loss of trabecular bone volume; however, no skeletal dysmorphology was reported in neonatal FGF2−/− mice.139 These observations suggest that FGFs 2, 9, and 18 may act alone or redundantly to regulate osteoblast activity and physiology, and that FGFs 9 and 18 may constitute the predominant signals during embryonic development, whereas FGF2 may be more important during postnatal stages. Consistent with a role for FGF2 in more differentiated osteoblasts, bone marrow stromal cultures from FGF2−/− mice showed a significantly decreased ability to mineralize in vitro.139 It has been demonstrated that Runx2 is phosphorylated and activated by FGF2 via the MAPK pathway and suggests that FGF2 plays an important role in regulation of Runx2 function and bone formation.140
Fgfr2−/− mice die at embryonic day 10.5 (E10.5), prior to skeletal development.141 The contribution of Fgfr2 signaling to skeletal development has been clarified to some extent by using splice form–specific knockouts and conditional gene deletion approaches in mice. These studies demonstrated that Fgfr2 positively regulates bone growth and the anabolic function of osteoblasts. The resulting phenotype of mice lacking mesenchymal Fgfr2 included skeletal dwarfism, decreased bone density, incomplete formation of the dorsal vertebrae, and tarsal joint fusion.142,143 Alternative splicing of Fgfr2 is tissue specific, resulting in epithelial variants (b splice forms) and mesenchymal variants (c splice forms).144–146 Ligand-binding studies demonstrate that mesenchymally expressed ligands such as FGF7 and 10 activate Fgfr2b, whereas FGF2, 4, 6, 8, and 9 activate Fgfr2c.147,148
As adults, Fgfr3−/− mice were osteopenic, suggesting a role for Fgfr3 signaling in differentiated, Fgfr3-expressing osteoblasts.125 Mice lacking either FGF18 or Fgfr3 exhibited expanded zones of proliferating and hypertrophic chondrocytes and increased chondrocyte proliferation, differentiation, and Indian hedgehog signaling. These data suggest that FGF18 acts as a physiological ligand for Fgfr3.135 In addition, FGF18−/− mice had decreased endochondral and intramembranous bone formation, suggesting that FGF18 positively regulates osteogenesis and/or osteoblast function independent of Fgfr3.135
Two lines of evidence indicate that engrailed 1 (En1) regulates signaling mediated by Fgfrs. First, the activation ERK, normally restricted to the mature endosteal osteoblasts of wild-type calvarial bone, is severely impeded in En1 mutants. Second, En1 ablation results in loss of the FGF target gene sprouty homolog 2 (Drosophila) (Spry2) in ectoperiosteal osteoblasts. Furthermore, En1 may regulate alternative FGF-signaling effectors known to affect osteoblast differentiation, such as p38 MAPK or PKC.149–151 A precise temporal and spatial delineation of these intra-cellular pathways will enable a better understanding of how osteoblastic differentiation is coordinated by En1 and FGFs.
To study the effects of growth factors on hMSC, Kratchmarova et al. tested the effects of epidermal growth factor (EGF), platelet-derived growth factor (PDGF), FGF, and nerve growth factor (NGF) on cellular responses and observed that EGF and PDGF elicited the strongest responses.152 They also found that the differentiation of human mesenchymal stem cells into bone-forming cells is stimulated by EGF but not PDGF. Mass spectrometry–based proteomics analysis demonstrated that more than 90% of these signaling proteins were used by both ligands, whereas the phosphatidylinositol 3-kinase (PI3K) pathway was exclusively activated by PDGF, implicating it as a possible control point. Indeed, chemical inhibition of PI3K in PDGF-stimulated cells removed the differential effect of the two growth factors, bestowing full differentiation effects onto PDGF.
5. Ephrin Signaling
Ephrins have the capacity for bidirectional signaling. That is, when a cell expressing an ephrin receptor contacts a cell expressing an ephrin ligand, signals are transduced into both the ephrin receptor–expressing cell (forward signaling) and the ephrin ligand–expressing cell (reverse signaling). There are two classes of ephrins, the B class (ephrin B1 to B3) are ligands for EphB tyrosine kinase receptors (B1 to B6), whereas class A ephrins (A1 to A5) are ligands for glycosylphospha-tidylinositol (GPI)–anchored EphA receptors (A1 to A10).153 In bone biology, ephrinB and EphB receptors control patterning of the developing skeleton,154 and disruption of ephrin signaling is implicated in a syndrome called craniofrontonasal syndrome [CFNS (MIM 304110)].155 Zhao et al. now suggest that ephrin signaling is critical to the two-way communication between osteoclasts and osteoblasts.156 This bidirectional signaling is mediated by the transmembrane ephrinB2 ligand in osteoclasts and EphB4, a tyrosine kinase receptor, in osteoblasts (Fig. 3). Using osteoblast-osteoclast coculture assays, as well as loss- and gain-of-function studies in vitro, the authors demonstrated that reverse signaling from EphB4 in osteoblasts to ephrinB2 in osteoclast progenitors leads to the inhibition of osteoclast differentiation. On the other hand, EphB4 expression is constitutive and the forward signaling through EphB4 induces osteogenic regulatory factors, such as Dlx5, Osx, and Runx2, in calvarial osteoblasts, suggesting that EphB4 is at the top of the regulatory cascade during osteoblast differentiation. Zhao et al. demonstrated that forward signaling between the extracellular domains of ephrinB2 and EphB4 in osteoblasts stimulates their differentiation, a process that may be dependent on ras homolog gene family, member A (RhoA), in-activation in osteoblasts. Expressing active RhoA in osteoblasts may block the ability of ephrinB to promote osteoblast differentiation. By contrast, McBeath et al., using human mesenchymal stem cells, suggest that active RhoA enhances osteoblast differentiation (Fig. 3).157 Therefore, the involvement of RhoA in EphB4 forward signaling will need to be confirmed by future pharmacological or genetic studies. This study establishes the concept that ephrin-Eph signaling contributes to bone homeostasis.
6. PTH Signaling
PTH can be used pharmacologically to build bone. PTH has been used as an effective treatment for osteoporosis due to the fact that PTH exerts either a catabolic or anabolic effect, depending on the method of administration. New insight into the structure of PTH, PTHrP, and the PTH/PTHrP receptor has stimulated the field of calcium and bone biology and posed new questions about the role of PTH and PTHrP.158 Currently, PTH(1–34) is the only FDA-approved anabolic agent for the treatment of osteoporosis. Pettway et al. aimed to gain an understaning of the mechanism allowing PTH(1–34) to work as a treatment.159 They found that three weeks of PTH treatment on bone marrow stromal cells implanted in immunocompromised mice resulted in an anabolic response. PTH-treated ossicles showed increased donor cell proliferation. When zoledronic acid was combined with PTH, PTH-induced proliferation was reduced without reducing bone volume, indicating that combining PTH and bisphosphonate therapy warrants further investigation.
The PTH signaling pathway involves Gα proteins that bind to the PTH receptor.160 Gα proteins activate cAMP, which binds the regulatory subunits of protein kinase A (PKA) to release the catalytic subunits of the enzyme. PKA then phosphorylates proteins to cause changes in the structure and function of Runx2. Other Gα proteins on the PTH receptor interact with phospholipase C (PLC) to activate PKC.
It was recently found that there is a link between PTH and noncanonical Wnt4 signaling.161 Wnt4 signaling in response to PTH implicates cross talk of multiple signaling pathways, showing PTH’s anabolic effect in bone. PTH stimulation of Wnt4 is primarily through the PKA pathway. Swarthout et al. found that PTH regulates genes and proteins through the PKA pathway,162 which was supported in vivo by microarray data from peptides PTH(1–34), PTH(1–31), and PTH(3–34).163 Guo et al. generated mice with mutant PTH/PTHrP receptor that does not activate PLC.164 The mice exhibited abnormalities in embryonic endochondral bone development including delayed ossification and increased chondrocyte proliferation, indicating that PLC signaling through the PTH/PTHrP receptor slows proliferation and promotes differentiation of chondrocytes.
7. Sympathetic Signaling
Neural control of bone metabolism, both trophic and atrophic, has been suggested by numerous experimental and clinical observations. Osteoblasts have been reported to express receptors for several neuropeptides, suggesting that they could indeed integrate multiple neuronal signals.165 Immunolabeling studies have revealed a close association between glutamate-, catecholamine-, or peptide-containing nerve fibers and osteoblasts or osteoclasts in the endosteum.166 Blockade of glutamate receptors was reported to reduce the DNA-binding activity and expression of Runx2 in cultured osteoblasts.167 The effect of the sympathetic nervous system (SNS) on bone formation has only recently been elucidated using genetic models.168 These studies revealed that leptin induced bone loss through SNS-derived signals originating in the ventromedial hypothalamic nuclei.166 Sato et al. found that leptin- or SNS-mediated inhibition of bone formation was abolished in neuromedin U null mice, which show high bone mass due to an increase in bone formation.169
Fu et al. indicated that an important, new regulator of bone remodeling is the circadian cycle.170 The model that emerges from the results of Fu et al. suggests that signaling by β2-adrenergic receptors first activates the transcription factor cAMP responsive element binding protein (CREB) (Fig. 3). CREB in turn stimulates expression of clock genes, which mediate the antiproliferative function by inhibiting G1 cyclin expression, and activator protein 1 (AP1) genes, which stimulate proliferation of osteoblasts. Fu et al.’s work provided evidence that the inhibition of osteoblast proliferation by clock proteins is the dominant effect. Pharmacological or genetic ablation of adrenergic neurotransmission indicates that norepinephrine (NE) signaling controls granulocytecolony stimulating factor (G-CSF)–induced osteoblast suppression.171 Based on studies describing leptin-mediated neuronal control of osteoblast function166,172 and the fact that leptin and G-CSF receptors display a high degree of homology,173 it has been proposed that G-CSF signals directly in the hypothalamus through the leptin receptor.171
Sequence-specific DNA-binding proteins are frequently encoded by gene families. Such proteins display highly conserved DNA-binding properties, yet are assumed to retain promoter selectivity. Yamamoto et al. addressed the factor-specificity issue with his update on the varied roles of the glucocorticoid receptor (GR).174 They presented evidence that two ligands, the small nuclear hormone and the larger DNA molecule, act to direct the activities of GR. Most fascinating were the data showing that different hormone ligands could affect promoter selectivity.174
8. Osteocyte Signaling
A recent study has shed light on the role of osteocytes in bone remodeling: osteocytes deep inside the bone serve an important function in regulating the activities of osteoclasts and osteoblasts at the bone surface. Tatsumi et al. ablated osteocytes in vivo using diphtheria toxin and found that mice exhibited osteoporosis characterized by fragile bone with intracortical porosity and microfractures, osteoblastic dysfunction, and trabecular bone loss.175 They also found that osteocytes play a role in bone homeostasis by sending signals to osteoclasts to maintain bone mass or initiate bone loss. Osteocytes may therefore comprise an important target for the development of diagnostics and therapeutics for bone disease, as exemplified by osteoporosis.
B. Osteoclast Signaling
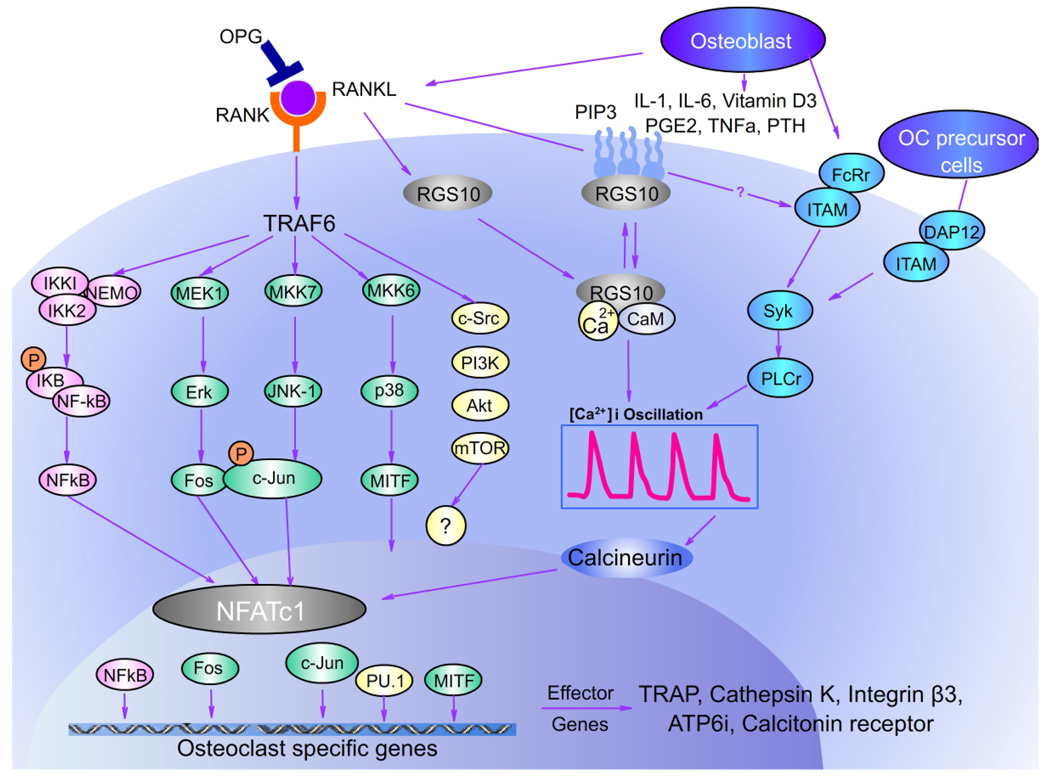
Osteoblasts, chondrocytes, and their mineralized matrices, together with stromal and endothelial cells, provide the microenvironment for homing of osteoclast precursor cells. Osteoblasts/stromal cells produce cytokines including M-CSF and RANKL that induce and modulate growth and differentiation of the precursors to mature osteoclasts. M-CSF binds to its receptor, colony stimulating factor 1 receptor (c-Fms), present on osteoclast precursors, providing the signals for macrophage survival and proliferation. Intracellular RANK signaling by its interaction with RANKL induces recruitment and activation of cytoplasmic tumor necrosis factor receptor–associated factors (TRAFs), leading to the activation of multiple signaling cascades such as MAP, NF-κB, Rous sarcoma oncogene (Src), and Akt (Fig. 4).176 Of the several TRAF proteins that have been described in conjunction with RANKL, including TRAF-1, −2, −5, and −6, TRAF-6 is indeed an essential adaptor required for RANK-associated signaling. It is necessary for RANK-induced NF-κB activation and in vitro osteoclastogenesis and the deletion of TRAF-6 leads to osteopetrosis. By contrast, the contributions of TRAF-2 and TRAF-5 to osteoclastogenesis are minor.177,178 Osteoclast signaling molecules also play a role in bone disease. Macrophage inflammatory protein-1α (MIP-1α) has been found to be an important osteoclast stimulatory factor in multiple myeloma.179,180 MIP-1α enhances osteoclast formation induced by IL-6, PTHrP, and RANKL, all of which are implicated in myeloma bone disease.
FIGURE 4.
RANK signaling network of osteoclast differentiation. Binding of RANKL to its receptor RANK induces various intracellular signaling cascades through TRAF-6, such as MAP (ERK, p38, JNK), NF-κB, Src, and NFATc1. Several transcription factors have been found crucial for osteoclast differentiation downstream of RANKL/RANK signaling such as NF-κB, NFATc1, c-Fos, c-Jun, and Mitf. RGS10 functions downstream of RANKL to regulate calcium oscillations and NFATc1 expression. These transcription factors are essential for differentiation of mononuclear precursors into multinucleated osteoclasts and are essential for the transcription of osteoclast-specific genes.
1. M-CSF
After commitment to the OC lineage, the entrance of mononuclear cells into the early preOC pathway is a response to M-CSF (Fig. 2). M-CSF is required for the proliferation, differentiation, and survival of hematopoietic cells in the mono-cytic lineage.20,181,182 Treatment with M-CSF alone results in the formation of macrophagelike cells. It also contributes to their differentiation and regulates the cytoskeletal changes that accompany bone resorption. The absence of a functional M-CSF in the op/op mouse causes not only a macrophage deficiency, but also a lack of OCs, resulting in an osteopetrotic phenotype.183,184 These deficiencies can be restored with injections of M-CSF.183,185 Op/op marrow stromal cells can support the differentiation and proliferation of OC progenitors from inoculated stem cells, as shown by experimental evidence that M-CSF is not essential for the early stages of OC development.186 Furthermore, M-CSF, although necessary for entry of precursors into the early preOC pathway, was found to inhibit osteoclastogenesis at high doses.
Binding of M-CSF to its receptor c-Fms recruits adapter proteins and cytosolic kinases, thereby activating a variety of intracellular signals. Tyrosines 559 and 807 in the cytoplasmic tail of c-Fms play distinct roles in OC differentiation and function. Changes in M-CSF–receptor expression appear to modulate the final lineage selection of the pluripotent monoblastic progenitor.187 M-CSF–induced genes are necessary for a direct response to RANKL and interleukins. Cappellen et al. reported that M-CSF induced the receptor for RANKL (RANK) and other RANK/NF-κB pathway components [TRAF2A, PI3-kinase, MEKK3, and receptor (TNFRSF)–interacting serine-threonine kinase 1 (RIPK1)], providing a molecular explanation for the synergy of M-CSF and RANKL. Interleukins, interferons, and their receptors [IL-1α, IL-18, interferon (IFN)-β, IL-11Rα2, IL-6/11R gp130, IFNγR] were also induced by M-CSF.188 Furthermore, M-CSF acts as a survival factor for the OC precursors through Bcl-X(L)–induced inhibition of caspase-9 activation, which inhibits apoptosis of OC precursors.189 Akiyama et al. found that ubiquitylation of the B-cell leukemia/lymphoma 2 (Bcl-2) family member BCL2-like 11 (apoptosis facilitator) (Bim) regulates apoptosis in osteoclasts and bone marrow cells from Bim-null mice show prolonged survival in the absence of M-CSF, but the bone-resorbing activity of osteoclasts was reduced.190
2. RANKL
RANKL, also called TNF-related activation-induced cytokine (TRANCE), osteoprotegrin ligand (OPGL), or OC differentiation factor (ODF), is a member of the TNF family RANK−/− mice demonstrated profound osteopetrosis resulting from an apparent block in OC differentiation, revealing that RANK provides critical signals necessary for OC differentiation.191 RANK connects to its extracellular signal factor RANKL and induces recruitment and activation of its adaptor TRAF6, leading to multiple downstream cascades. There are six known main signal pathways: MAP (ERK, p38, and JNK), NF-κB, Src, and Akt, shown by recent research results. OPG, encoded by Tnfrsf11b, as a decoy receptor of RANKL, is also expressed by OCs and preOCs. It is secreted and competes with RANK by binding to RANKL. Overexpression of OPG causes osteoclast-deficient osteopetrosis, while deletion of OPG leads to osteoporosis due to increased OC number and activity. Autosomal recessive osteopetrosis in humans involves mutations in the RANKL gene, which result in a lack of osteoclasts.192
The role of RANKL in rheumatoid arthritis has recently been elucidated at sites of pannus invasion into bone. Pettit et al. found that RANKL protein expression and RANK-expressing osteoclast precursor cells were confined to sites of osteoclast-mediated erosion at the pannus-bone interface and subchondral bone erosion.193 OPG protein expression, on the other hand, was limited at sites of bone erosion. These results implicate RANKL in the pathogenesis of rheumatoid arthritis and show that it contributes to the generation of a local environment that favors osteoclast differentiation and activity.
3. Src
Src is a member of a family of nine nonreceptor tyrosine kinases (NRTKs) that associate with the cytoplasmic surface of cellular membranes.194 Deletion of the gene encoding c-Src produces an osteopetrotic skeletal phenotype that is the consequence of the inability of the mature OC to efficiently resorb bone. Src−/− OCs exhibit reduced motility and abnormal organization of the apical secretory domain (the ruffled border) and attachment-related cytoskeletal elements that are necessary for bone resorption. A key function of Src in OCs is to promote the rapid assembly and disassembly of the podosomes, the specialized integrin-based attachment structures of OCs and other highly motile cells.195 Dynamin, a GTPase, colocalizes with Casitas B–lineage lymphoma (Cbl), an adaptor protein, in these actin-rich podosomes and forms a complex in osteoclasts, which is decreased by Src tyrosine kinase activity.196 Phosphorylation of Cbl and the subsequent recruitment and activation of PI3K may be critical signaling events downstream of Src in osteoclasts.197,198 In addition to Cbl, Src forms a complex with protein tyrosine kinase 2 (Pyk2). Src osteoclastic bone resorption requires both c-Src kinase activity and the targeting of Src kinase by Pyk2.197 Src−/− osteoclasts were examined to determine the specific functions of Src in the organization and dynamics of podosomes.199 Src was found to regulate the formation, structure, life span, and rate of actin polymerization in podosomes and in the actin cloud. Kinase activity and the SH2 or SH3 binding domain are required for Src to restore normal podosome organization and dynamics. The absence of Src affects the bone-resorbing activity of mature OCs, but does not affect OC formation.200,201 In fact, the number of OCs in bones of Src−/− mice is more than twice that in normal mice, suggesting that in vivo OC differentiation and/or survival is enhanced in the absence of Src. Recently, it was reported that c-Src kinase activity, not only on the plasma membrane but also within mitochondria where it phosphorylates cytochrome c oxidase (Cox), is essential for the regulation of osteoclastic bone resorption.202 Of the several signaling mechanisms that are activated downstream of RANK, only the sequential activation of PI3K and Akt is known to involve Src and the Cbl proteins.
Besides these pathways, c-Src interaction with TRAF-6 was found to enhance the kinase activity of c-Src, activating the Akt/PI3K pathway, skeletal rearrangement, and cell motility. The molecular adapter growth factor receptor-bound protein 1 (Grb-1)–associated binder-2 (Gab2) was also found recently to associate with RANK and mediate RANK-induced activation of NF-κB, Akt, and Jnk (Fig. 4).203 Src homology 2–containing inositol-5-phosphatase (SHIP) was shown in negatively regulated osteoclast formation and function. SHIP blunts PI3K signaling by dephosphorylating its major substrate, phosphatidylinositol-3,4,5-trisphosphate (PIP3). SHIP−/− mice show a twofold increase in osteoclast number, due to the prolonged life span of these cells and to hypersensitivity of precursors to M-CSF and RANKL.204 Similar to pagetic osteoclasts, SHIP−/− osteoclasts are enlarged and exhibit enhanced resorptive activity, and serum levels of interleukin-6 are markedly increased.
4. NF-κB
NF- dimers are normally rendered inactive in the cytosol. Activation of this transcription factor is a consequence of phosphorylation of its inhibitory protein, IκB proteins (IκBa, IκBb, IκBe, IκBg, Bcl-3), by an upstream kinase complex termed IκB kinase (IKK), resulting in the release and translocation of NF-κB to the nuclear compartment. The NF-κB pathway is relevant for RANKL-RANK–regulated osteoclast development and osteoclast function not only in mice, but also in humans as can be seen from patients with X-linked osteopetrosis, lymphedema, anhidrotic ectodermal dysplasia, and immunodeficiency (OLEDA-ID syndrome), which carry a X420W point mutation in IKKg and have osteopetrosis.205 Besides, NF-κB, TRAF proteins also activate the upstream MAP kinase (MAPK) MEKK1, which is followed by activation of a large number of MAP kinase mediators, eventually leading to induction of three distinct MAP kinase families, namely, JNK/c-Jun, ERK1/2, and p38 kinases. Dominant-negative forms of various MAP kinases and selective inhibitors of the MAP kinase pathways inhibited osteoclastogenesis or reduced osteoclast survival.206–208 Activated p38-MAPKs downstream of RANK can directly phosphorylate signal transducer and activator of transcription 1 (STAT1) and hence control expression of a variety of target genes.209 JNKs and their direct upstream kinase MKK7 have been shown to be involved in osteoclastogenesis through in vitro cell culture.210,211 JNK activation facilitates the phosphorylation of c-Jun and increases its transcriptional activity, leading to AP-1 activation. ERK, which activates the other component of AP-1, c-Fos, is also activated on RANK stimulation. Inhibition of MEKs (ERK kinases) by PD98059 or U0126 does not, however, attenuate osteoclast differentiation, but rather increases osteoclastogenesis.
Expression of IL-1 is regulated by NF-κB and IL-1 mediates bone resorption in a variety of diseases affecting bone. TNF induces IL-1 expression and activates c-Fos in osteoclast formation. Yao et al. found that IL-1 is capable of inducing osteoclast formation directly from osteoclast precursors overexpressing c-Fos without addition of cytokines.212 IL-1 was secreted from osteoclast precursors on bone, stimulated by bone matrix proteins dentin sialoprotein and osteopontin, to induce c-Fos–mediated differentiation, which was more prevelant in eroding inflamed joints. These results indicate that osteoclast progenitors expressing c-Fos may interact with bone matrix to produce IL-1 by an autocrine mechanism and induce their differentiation into osteoclasts. McHugh et al. have proponed the idea that certain osteoclast genes and transcriptional pathways are induced by interaction of osteoclast precursors with specific components of the bone matrix.213 Xing et al. found that NF-κB p50 or p52 is required in osteoclasts and their precursors for IL-1-mediated bone resorption.214
5. ITAM-Dependent Costimulatory Signals
Most recently, Koga et al. reported that mice lacking immunoreceptor tyrosine-based activation motif (ITAM)–harboring adaptors, Fc receptor common γ subunit (FcRγ) and DNAX-activating protein (DAP) 12, exhibit severe osteopetrosis owing to impaired osteoclast differentiation. In osteoclast precursor cells, FcRγ and DAP12 associate with multiple immunoreceptors and activate calcium signaling through PLCγ. Thus, ITAM-dependent costimulatory signals activated by multiple immunoreceptors are essential for the maintenance of bone homeostasis (Fig. 4).215 However, in ITAM-mediated costimulatory signaling, the ligands for the Ig-like receptors are not identified. The molecules that connect RANK with ITAM signaling are also unknown. DAP12−/− FcRγ−/− bone marrow cells show impaired phosphorylation of spleen tyrosine kinase (Syk), in addition to failure to differentiate into multi-nucleated osteoclasts or resorb bone.216 Syk−/− progenitors are also defective in osteoclast development and bone resorption, indicating that recruitment of Syk to phosphorylated ITAMs is critical for osteoclastogenesis. SH3-domain binding protein 2 (Sh3bp2) mutation plays a role in “cherubism” mice through the Syk pathway.217 Sh3bp2 mutant myeloid cells show increased responses to M-CSF and RANKL stimulation through increased Syk phosphorylation/activation, forming unusually large, hyperactive osteoclasts, reflecting Sh3bp2 gain of function and leading to trabecular bone loss, TNF-α-dependent inflammation, and cortical bone erosion.
6. RGS10 and RGS12
Despite the importance of the calcium-nuclear factor of activated T-cells (NFAT) pathway, it remained unclear how RANKL activates calcium signals leading to the induction of NFAT, cytoplasmic, calcineurin-dependent 1 (NFATc1). Recent work in our lab reveals more details of the RANKL-induced signaling pathway controlling calcium oscillations and NFATc1 activation. The regulator of G-protein signaling (RGS) proteins are a family of 21 proteins, all containing the RGS domain. We found that RGS10A is important in osteoclast signaling.218 This isoform of RGS10 is specifically expressed in human osteoclasts and RNAi silencing of RGS10A blocked calcium oscillations, NFATc1 expression, and osteoclast terminal differentiation. Target components of RGS10A are different from those of RGS12; RGS10A interacts with calmodulin while RGS12 interacts with the calcium-sensing receptor. We also found that RGS10A acts upstream of calcineurin in the RANKL-calcium oscillation-calcineruin-NFATc1 pathway. The specificity RGS10A as a key component in the osteoclast differentiation signaling pathway makes it an excellent target for potential therapies.
To further define the role of RGS10 in osteoclast signaling, we went on to generate RGS10-deficient mice, which exhibited severe osteopetrosis.219 We found that RGS10 competitively interacts with the calcium/calmodulin complex and PIP3 in a calcium-dependent manner to mediate PLCγ activation and calcium oscillations. Our results provide in vivo evidence that RGS10 specifically regulates the RANKL-evoked RGS10/calmodulin-calcium oscillation-calineurin-NFATc1 signaling pathway in osteoclast differentiation. Based on our data from this study, we proposed an RGS10 working model (Fig. 4): RANKL mediates DAP12 and FcRγ, the membrane adaptor molecules that contain an ITAM motif and that activate PLCγ. PLCγ hydrolyzes PIP2 to generate inositol 3-phosphate (IP3). IP3 then triggers a transient initial release of calcium from intracellular stores. Intracellular calcium release allows an increase in intracellular calcium to reach peak concentration and leads to formation of the calcium/calmodulin complex. The calcium/calmodulin complex competes for the PIP3-binding site on RGS10 and frees the bound PIP3. Once the calcium concentration reaches its peak formation, intracellular calcium begins to reload into the endoplasmic reticulum (ER) in the absence of further PLCγ activation, and the combination of calcium reloading in the ER and binding to calmodulin causes the calcium concentration to decrease. The calcium/calmodulin complex dissociates from RGS10 at the low calcium concentration. Free PIP3 activates PLCγ and then binds RGS10 again without calcium/calmodulin complex competition. PLCγ activation triggers a release of calcium from intracellular stores by generating IP3 to cause a second peak. This process continues to cycle, causing calcium oscillations. In this way, RGS10 mediates PLCγ activation and calcium oscillations through its calcium-dependent dual interaction with calcium/calmodulin and PIP3. The RGS10-mediated intracellular calcium oscillations activate calcineurin and NFATc1 expression for osteoclast terminal differentiation (Fig. 4).
In addition to our work on RGS10, we identified RGS12 as a new signaling protein in osteoclasts.220 RNA interference (RNAi) silencing of RGS12 expression impaired phosphorylation of PLCγ and blocked calcium oscillations, NFATc1 expression, and osteoclast differentiation. RGS12 was further found to directly interact with N-type calcium channels. Our results revealed that RGS12 is essential for RANKL-induced terminal differentiation of osteoclasts.
7. Cathepsin K Regulates Osteoclast Apoptosis and Senescence
Cathepsin K was cloned from human osteoclastomas in our lab32 and from rabbit and human osteoclasts in other labs.221–224 The important role of cathepsin K in osteoclast function was first suggested by the finding of clinical research that mutations in this gene caused pycnodysostosis, a rare, autosomal, recessive, skeletal disorder caused by mutations in the cathepsin K gene at 1q21, which codes for cathepsin K protein, a lysosomal cysteine protease. Mutation in this gene affects the metabolism of the skeletal system, causing defects in bone resorption and bone remodeling.225,226 Recently, we discovered a novel pycnodysostosis mouse model in the 129/Sv background that exhibited many characteristics of the human pycnodysostosis phenotype.227 This model revealed that cathepsin K−/− osteoclasts lacked normal apoptosis and senescence and have reduced expression levels of p19, p53, and p21, leading to unusually high osteoclast numbers. Cathepsin K−/− osteoclasts did not begin to shrink or exhibit condensed nuclei, had little to no chromosome degradation, and survived through 36 hours, while no wild-type cells survived that period. Cathepsin K was associated with senescence in wild-type cells, which indicates that impaired senescence might be the major cause of the extraordinarily high number of osteoclasts in cathepsin K−/− mice. This is the first evidence of the role of cathepsin K in osteoclast apoptosis and senescence. The mechanism by which cathepsin K regulates osteoclast apoptosis and senescence is under characterization.
8. Vav3
The regulation of osteoclast activation has remained unclear; however, significant progess has recently been made by Faccio et al.228 They reported that Rho family guanine nucleotide exchange factor Vav3 is essential for stimulated osteoclast activation and bone density. Osteoclasts from Vav3-deficient mice show defective actin cytoskeleton organization, polarization, spreading, and resorptive activity resulting from impaired signaling downstream of the M-CSF receptor and αvβ3 integrin. These mice also have increased bone mass and are protected from bone loss induced by systemic bone resorption stimuli such as parathyroid hormone or RANKL. In addition, Faccio et al. discovered a role for Syk tyrosine kinase as a crucial upstream regulator of Vav3 in osteoclasts.228
IV. OSTEOBLAST GENE TRANSCRIPTION: TRANSCRIPTION FACTORS THAT REGULATE OSTEOBLAST DIFFERENTIATION
Transcription factors that regulate osteoblasts include a range of homeodomain proteins: the AP family members Jun, Fos, and Fra, Smads, C/EBPβ and C/EBPd, lymphoid-enhancing factor (a Wnt effector), twist, activating transcription factor 4, Runx2, and osterix, the last three of which are considered master genes for osteoblast differentiation.
Commitment of mesenchymal stem cells (MSCs) to tissue-specific cell types is orchestrated by transcriptional regulators that serve as “master switches.” A central regulator of bone formation is the Runx2 transcription factor, which fulfills its role as a master regulatory switch through unique properties for mediating the temporal activation and/or repression of cell growth and phenotypic genes as osteoblasts progress through stages of differentiation.229 Although Runx2 is essential for osteoblast differentiation, this differentiation program also requires other genes, such as osterix, which encode a transcription factor genetically “downstream” of Runx2.230 Thus, multiple genes regulate Runx2 activity and the effectiveness of Runx2 in stimulating osteoblast formation. It is perhaps not surprising then that Runx2 expression in osteoblast precursors predates by several days the first evidence for osteoblast activity.
A. Runx2
Runx2 is a member of the Runx family of transcription factors [previously known as acute myeloid leukemia (AML) factor, polyomavirus enhancer binding protein 2 (PEBP2), and core binding factor (CBF)]. The family members, Runx1 (also called PEBP2aB, CBFA2, and AML1), Runx2 (also called PEBP2aA, CBFA1, and AML3), and Runx3 (also called PEBP2aC, CBFA3, and AML2), are encoded by distinct unlinked genes but share a common DNA recognition motif (TGTGGT) and heterodimerize with the ubiquitous subunit CBFβ for stable DNA binding.231 Their highly conserved DNA binding domain is homologous to that from the Drosophila segmentation gene runt.232 In addition to the Runx DNA binding domain, Runx2 contains an active transactivation domain, rich in glutamine and alanine residues, and activates the Osteocalcin and Col1a1 genes.233–236 Thus, Runx2 is an initial marker of the osteogenic cell lineage.
Runx2 is expressed in the thymus and testes (in T-lymphocytes tendon), is abundantly expressed in calcified cartilage and bone tissues, and is absent from the brain, heart, lung, gut, and liver.237–239 The function of Runx2 in bone formation (Fig. 1) has been demonstrated by analyzing its role in regulating the expression of the principal osteoblast-specific genes and by studying Runx2 null mice.233,238,240 Targeted disruption of Runx2 results in the complete lack of bone formation by osteoblasts, revealing that Runx2 is essential for both endochondral and membranous bone formation.238 The haploinsufficiency of the Runx2 gene, which leads to cleidocranial dysplasia, a genetic disease in humans that is characterized by hypoplastic clavicles, large open spaces between the frontal and parietal bones of the skull, and other skeletal dysplasias, is caused by heterozygous mutations in the Runx2 gene.241 Moreover, Runx2 is sufficient to induce osteoblast differentiation. This is true in cell culture, where forced expression of Runx2 in skin fibroblasts leads to osteoblast-specific gene expression,233 and in vivo, since ectopic expression of Runx2 leads to endochondral ossification in parts of the skeleton that would normally never ossify.242,243 Importantly, Runx2 may function as an inhibitor of proliferation of progenitors, thus providing a mechanism for regulating the transition from growth to a postproliferative stage as a component of cellular commitment to the osteogenic lineage.244 Thus, Runx2 may be expressed in early osteoprogenitors to induce a program of gene expression required for lineage determination and differentiation of mesenchymal cells. Finally, Runx2 is also required for osteoblast function beyond differentiation.245,246 Overexpression of Runx2 inhibits osteocyte formation from osteoblasts, showing that Runx2 maintains the supply of osteoblasts (Fig. 1).247 These functions, along with its role during hypertrophic chondrocyte differentiation and vascular invasion, identify Runx2 as the most pleiotropic regulator of skeletogenesis.230 The literature now embraces the concept that Runx2 functions as a scaffold for the interaction with coregulatory proteins at subnuclear foci to provide an architectural basis for accommodating the requirements of biological control.
B. Runx2 Upstream Proteins
1. Msx2 and Bapx1
Msh homeobox 2 (Msx2), which encodes a homeobox-containing transcription factor, is expressed in osteoblasts during development. The role of Msx2 during skull formation was first uncovered by human genetic studies. Indeed, one syndrome characterized by increased bone formation around the cranial suture, Boston-type craniosynostosis, is caused by an activating mutation in MSX2.248 Msx2 inactivation in mice causes a marked delay of ossification in the bones of the skull and an overall decrease in bone volume. This phenotype is accompanied by a downregulation of Runx2 expression, indicating that Msx2 directly or indirectly regulates Runx2 expression (Fig. 1).249 Recently, Cheng et al. and Ichida et al. reported that a homeobox gene, the Msx2 gene, stimulates the commitment of mesenchymal cells into an osteoblast lineage in association with inhibition of adipogenesis.250,251 NK3 homeobox 2 (Bapx1), another homeobox protein encoding gene, is required for axial skeleton formation. In Bapx1-deficient mice Runx2 expression is downregulated in the axial skeleton,252 suggesting that Bapx1 is another activator of Runx2 expression.
2. Dlx5
Dlx5 expression in osteoblasts is facilitated by BMP, and Dlx5 goes on to induce expression of Runx2 in osteoprogenitor cells.90,91 Dlx5 has been found to enhance Runx2 P1 promoter activity.90,253 Furthermore, there is increasing evidence that Dlx5 promotes activation of Osteocalcin by forming heterodimers with Msx2, which antagonizes the Msx2-mediated repression of Osteocalcin.254,255
3. Twist
Twist-1 (previously called Twist) and Twist-2 (previously called Dermo-1) encode vertebrate basic helix-loop-helix transcription factors homologous to Drosophila Twist, a mediator of dorsal-ventral patterning and mesoderm formation. Knockout of Twist-1 in mice leads to lethality at E10.5 due to failure of neural tube closure.256 Twist-1 heterozygotes (both in mice and in humans) exhibit craniosynostosis, a disease caused by premature osteoblast differentiation in the skull. Bialek et al. show that Runx2-induced osteoblast gene expression only occurs when expression of Twist genes disappears in osteoblast precursors.257 Twist-1 heterozygosity reverses skull abnormalities in Runx2+/− mice, and Twist-2−/− reverses clavicular abnormalities in Runx2+/− mice and accelerates osteoblast differentiation in bones formed through endochondral bone formation. Twist proteins’ antiosteogenic function is mediated by a novel domain, the Twist box, which interacts with the Runx2 DNA binding domain to inhibit its function. They conclude that Twist-1 and Twist-2 regulate the developmental action of Runx2 in bone formation through the direct interaction of these proteins. Bialek et al., by bringing together the actions of Twist and Runx proteins, have clarified an important stage in bone formation and set a research agenda for the future.258
4. p53
It has long been recognized that the p53 tumor suppressor plays a pivotal role in preventing cancer. Two independent studies have addressed the role of p53 in bone differentiation in mouse models.259,260 In one case, Wang et al. examined skeletal structure and bone metabolism in p53 knockout mice.260 Conversely, Lengner et al. analyzed the effects of hyperactive p53 on bone formation caused by the conditional deletion of transformed mouse 3T3 cell double minute 2 (Mdm2) in osteoblasts.259 Surprisingly, and in contrast to the in vitro studies,261 both groups came to the same conclusion that p53 suppresses differentiation. Specifically, p53−/− osteoblasts displayed a marked propensity to differentiate, which was manifested by a modest but significant increase in bone formation and bone density in adult p53 knockout mice. Consistent with these results, the conditional deletion of Mdm2 in osteoblasts interfered with terminal differentiation, leading to late-stage embryonic lethality, where the embryos displayed more porous and shorter bones. These findings suggest that the interplay between p53 and Mdm2 could either positively or negatively impact bone development. The studies of both Lengner et al. and Wang et al. provide compelling evidence that p53 suppresses osteoblast differentiation by repressing the expression of either Runx2 or Osterix.259,260 The subtle discrepancy that exists between the two studies (whether Runx2 or Osterix is the target of p53 action) may be related to how p53 activity is targeted and whether this mechanism alters the stage of cell differentiation. In either case, the concept that the absence of a tumor suppressor gene can enhance cell proliferation while favoring the differentiation of mesenchymal stem cells is intriguing but counterintuitive. It is likely that p53-deficient osteoprogenitors can still exit the cell cycle on terminal differentiation, which may be enhanced as a result of the elevated expression of Runx2 and Osterix. These findings clearly establish p53 as a negative regulator of osteoblast differentiation both in vitro and in vivo.
5. Schnurri-3
Schnurri-3 (Shn3), a large zinc finger protein, was originally identified as a DNA binding protein of the heptameric recombination signal sequence required for V(D)J recombination of immunoglobulin genes262; however, it also functions as an adapter protein in the immune system.263 Jones et al. found that Shn3 is an essential regulator of adult bone formation.264 Mice lacking Shn3 display adult-onset osteosclerosis with increased bone mass due to augmented osteoblast activity. Shn3 was found to control protein levels of Runx2 by promoting its degradation through recruitment of the E3 ubiquitin ligase WWP1 to Runx2. By this means, Runx2-mediated extracellular matrix mineralization was antagonized, revealing an essential role for Shn3 as a central regulator of postnatal bone mass.264
6. Cyclin-D1-Cdk4
Cyclin D1-cyclin–dependent kinase 4 (Cdk4) also induces Runx2 degradation by ubiquitination. Shen et al. studied the functional significance of post-translational modification of Runx2 to find that mutation of Runx2 in a consensus Cdk site increases the half-life of Runx2 and causes loss of sensitivity to cyclin D1–induced Runx2 degradation.265 This presents a novel mechanism through which Runx2 activity is regulated together with the cell cycle in bone cells.
7. Hoxa10 and Hoxa2
Homeobox A10 (Hoxa10) has recently been shown to activate Runx2 and directly regulate osteoblast phenotype genes.266 Hoxa10 is induced by BMP-2 and, in addition to Runx2, activates alkaline phosphatase, osteocalcin, and bone sialoprotein. Hoxa10 associates with the promoters of these genes through chromatin remodeling prior to Runx2 recruitment. Hassan et al. propose that Hoxa10 activates Runx2 in mesenchymal cells and contributes to the onset of osteogenesis.266 On the other hand, besides defects in branchial arch patterning, Hoxa2−/− mice show an upregulation of the cartilage- and bone-specifying genes SRY (sex determining region Y)–box containing gene 9 (Sox9) and Runx2.267 This area of osteoblast biology is still in its infancy, and the transcription factors that act upstream of Runx2 to control its expression remain to be identified.
C. Runx2 Coactivators and Corepressors
1. Cbfβ
Runx2 protein is shown to interact with a number of transcriptional coactivators. The most important coregulatory protein, essential for enhancement of Runx DNA binding, is Cbfβ (also known as PEBP2β), the non–DNA-binding partner of all three Runx proteins. Inactivation of Cbfβ causes embryo lethality in mice between E11.5 and E13.5. This results from hemorrhaging in several tissues and the absence of liver hematopoiesis because Cbfβ is a heterodimerizing partner of Runx1 and Runx3, which are essential for haematopoiesis. The timing of embryonic lethality precludes examining the role of the Cbfβ subunit in osteoblast differentiation. Transgenic rescue of embryonic lethal Cbfβ-null mice and “knock-in” of Cbfβ fused in-frame to a cDNA encoding green fluorescent protein resulted in mice that exhibited delayed ossification, indicating a role for Cbfβ in bone.268 However, unlike Runx2-null mice that completely lack bone and osteoblasts, ossification is initiated in these mice, suggesting that Runx2 can act in the absence of Cbfβ. Similar observations were made simultaneously by two other groups.269,270
2. p300, CBP, MOZ, MORF
Runx2 also interacts physically and/or functionally with other well-characterized coactivators, including p300, CREB binding protein (CBP), MYST histone acetyltransferase monocytic leukemia 3 (MOZ), and MYST histone acetyl-transferase monocytic leukemia 4 (MORF). p300 and CBP physically interact with various R-Smads on ligand stimulation and enhance Smad-dependent transcription of target genes.271,272 Neither p300 nor CBP has been coprecipitated with Runx2. Two members of the MYST family of HATs, MOZ and MORF, however, do interact with the activation domain of Runx2 and enhance activation of the osteocalcin promoter.273 This interaction may be functionally relevant to intramembranous bone formation, since mice with a mutation in the MYST gene have craniofacial defects.274 Amino-terminal enhancer of split (Grg5), retinoblastoma 1 (pRb), and tafazzin (TAZ) are other proteins that enhance Runx2-mediated transactivation.275–278 Although most Grg/transducin-like enhancer (TLE) proteins are corepressors, Grg5 appears to be a dominant negative form of longer Grg/TLE proteins, and thereby enhances Runx2 activity in vivo.278 The 14-3-3– binding protein, TAZ (transcriptional coactivator with PDZ-binding motif), coactivates Runx2-dependent gene transcription while repressing PPARγ-dependent gene transcription, indicating that TAZ functions as a molecular rheostat to fine-tune the balance between osteoblast and adipocyte development.276
3. Histone Deacetylases
Histone deacetylases (HDACs) remove acetyl groups from lysine residues on many proteins, including histones. The elimination of the acetyl group alters chromatin structure by removing a mark needed to recruit coactivating proteins and by facilitating chromatin condensation to promote transcriptional repression.279 General HDAC inhibitors, such as trichostatin A, increase Runx2-mediated activation, and Runx2 associates with several HDACs, including HDAC3, HDAC4, and HDAC6.280–282 HDAC3 interacts with the N terminus of Runx2. Suppression of HDAC3 expression in differentiating MC3T3-E1 cells accelerates matrix mineralization and the expression of bone marker genes such as osteopontin, bone sialoprotein, and osteocalcin.280 Vega et al. claimed that HDAC4 inhibits Runx2 activity by blocking Runx2 DNA binding.281 HDAC4-null mice display premature ossification due to early onset chondrocyte hypertrophy, and overexpression of HDAC4 inhibits chondrocyte hypertrophy, suggesting that Runx2 activity is controlled by HDAC4 in prehypertrophic chondrocytes.281 On the other hand, muscle and cardiovascular transcription factor myocyte enhancer factor 2C (Mef2c) has been found to control bone development by impairing hypertrophy, cartilage angiogenesis, ossification, and longitudinal bone growth in mice.283 Bone deficiency of Mef2c mutant mice can be rescued by an Hdac4 mutation, showing that endochondral bone formation is sensitive to the balance between Mef2c and HDAC4. Runx2 expression was also dramatically diminished in the endochondral cartilage of Mef2c mutant mice, suggesting that Mef2c is required, either directly or indirectly, for Runx2 expression. HDAC6 was identified as a Runx2 binding protein in co-immunoprecipitation experiments designed to identify corepressors that bind to the potent C terminus repression domain of Runx2.282 Moreover, several other groups have reported that HDAC inhibitors increased Runx2-dependent activation of the osteocalcin promoter280 and osteoblast maturation and differentiation.284,285 However, results from a new study support a requirement for HDAC4 and −5 deacetylase activity to regulate acetylation, abundance, and activity of Runx2.286 As the phenotypes of other HDAC mutations are probed in detail, it will be exciting to learn whether these proteins have a general role controlling osteoblast differentiation. Recently, Runx2 was found to form complexes with RNA polymerase I transcription factors upstream binding transcription factor, RNA polymerase I (UBF1), and TATA box binding protein (TBP)–associated factor, RNA polymerase I (SL1) in osteoblasts, co-occupy the rRNA gene promoter, and affect chromatin histone modifications at rDNA regulatory regions, whereby Runx2 directly represses rDNA promoter activity and so rRNA synthesis.287 This shows another mechanism of lineage commitment and cell proliferation control by Runx2.
4. Grg/TLE and YAP
Grg/TLE proteins are coexpressed with Runx2 in skeletal cells.288 The induction of the OCN gene correlates with downregulation of the level of Grg/TLE in mice skeletal tissues between E14 and birth, and Grg/TLEs were shown to inhibit Runx2-dependent activation of OCN gene transcription.289 However, Grg5, a dominant negative form of long Grg/TLE proteins, can enhance Runx2 transcriptional activity in vitro.278 Depletion of Grg5 alone, with normal activity of Runx2, causes postnatal growth retardation in about 50% of the mice, and in Grg5 null Runx2+/− mice, the lack of Grg5 function combined with the heterozygous loss of Runx2 activity resulted in a growth deficiency that was more pronounced than would have been expected. This finding suggests that Grg5 and Runx2 interact with each other in vivo and that their combined activity is necessary for the activation of another factor important for bone and cartilage development. It is highly probable that the factor regulated by Grg5-Runx2 interaction is Ihh.278 Runx2 also interacts with Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 (yes)–associated protein (YAP), a mediator of Src/Yes signaling, in the cytoplasm and translocates it to the nuclear matrix where YAP represses Runx2-mediated activation of the osteocalcin promoter.290 It is not yet known whether Runx2 associates with multiple corepressor complexes, or whether all of the corepressors mentioned above are components of the same complex.
5. Smurf1
One of the mechanisms by which transcription factors are regulated is by modulation of degradation. The proteosome degradation pathway decreases Runx2 protein levels to slow osteoblast differentiation.291 Smad-specific E3 ubiquitin protein ligase 1 (Smurf-1) induces Runx2 degradation292 and Smad 6 enhances Smurf-1–induced Runx2 degradation.293 For example, TNF-α attenuates osteoblast differentiation by promoting Runx2 proteasomal degradation through upregulation of Smurf-1 and Smurf-2 expression.294 Transgenic overexpression of Smurf-1 in murine osteoblasts suppresses their differentiation and bone formation, while Smurf-1–deficient mice develop age-dependent increases in bone mass.107,295 HDAC4 and HDAC5 deacetylate Runx2, allowing the protein to undergo Smurf-mediated degradation.286
6. Other Runx2 Interaction Partners
Many transcription factors involved in regulation of the osteoblast differentiation process exert their action by interacting with Runx2. Some provide costimulatory signals, while others directly repress Runx2 function by affecting its DNA binding activity and/or transactivation potential. DNA binding proteins that interact and cooperate with Runx2 to activate gene expression include AP1 [FBJ osteosarcoma oncogene (c-Fos) and c-Jun],296 BMP-responsive Smads (Smad 1 and Smad 5),89,297,298 E26 avian leukemia oncogene 1, 5′ domain (Ets1),299 C/EBPβ and -δ,300,301 hairy and enhancer of split 1 (Drosophila) (Hes1),302 and Menin.303 Most of these proteins interact with either the DNA binding domain or the activation domain of Runx2, although the binding sites for some have not been defined. It is generally believed that these transcription factors cooperate with Runx2 to facilitate the recruitment of coactivators and the assembly of higher-order transactivation complexes. Some proteins, including Hes1, may perturb TLE-Runx interaction both by competing with TLE corepressors for the binding site on Runx2 and by titrating TLE away from Runx2.302 Other cooperating transcription factors, such as AP1, Smad 1, and Smad5, integrate Runx2 with cell signaling pathways and to the extracellular environment.304,305 δFosB is a naturally occurring truncated form of AP1 family member FosB that is expressed in osteoblasts.306 Overexpression of δFosB causes increased bone formation, leading to osteosclerosis while down-regulating early markers of adipocyte differentiation, indicating that δFosB transcriptionally regulates osteoblastogenesis. Mutations that affect Runx2-Smad interactions are found in cleidocranial dysplasia (CCD) patients and inhibit the ability of Runx2 to induce osteoblast differentiation after BMP stimulation.87 Runx2 null cells do not react to BMP-2 signaling and a triple mutation in the C-terminal domain of Runx2, HTY (426– 428), disrupts Runx2-Smad interaction.307 The HTY mutation also causes cells to be unable to integrate the BMP-2/TGF-β signal on promoter reporter assays, is almost nonfunctional in promoting early stages of osteoblast differentiation, and exhibits reduced subnuclear targeting. Therefore, subnuclear targeting is inseparable from the Runx2/Smad osteogenic complex.
Transcription factors that inhibit Runx2-dependent activation in mesenchymal cells or osteoblasts include Dlx3,308 Lef1,309 Msx2,255 PPARγ,310 Smad 3,311 Hey1,312 and Stat1.313 These proteins repress Runx2 via several mechanisms, including binding the Runt domain and preventing DNA binding (e.g., Lef1, PPARγ),309,310 sequestering Runx2 in the cytoplasm (e.g., C/EBPδ, Stat1),313,314 or unknown mechanisms that involve binding in or around the nuclear matrix targeting domain of Runx2 (e.g., Dlx3).308
D. Osterix
The discovery of a BMP-2–inducible gene, Osx, a Kruppel-like specificity protein 1 transcription factor (Sp1) binding factor, identified a second transcriptional regulator for the final stages of bone tissue formation.100 Osx contains a DNA-binding domain consisting of three C2H2-type zinc fingers at its C terminus that share a high degree of sequence identity with similar motifs in Spl, Sp3, and Sp4. In addition, Osx also contains a proline- and serine-rich transactivation domain and activates the OCN and Col1a1 genes. In Osx-null mutant mice, no endochondral or intramembranous bone formation occurs.100 The mesenchymal cells in Osx-null mutant mice do not deposit bone matrix, and cells in the periosteum and the condensed mesenchyme of membranous skeletal elements cannot differentiate into osteoblasts, although these cells express normal levels of Runx2. Interestingly, Osx-null osteoblast precursors in the periosteum of membranous bones express chondrocyte markers, such as Sox9 and Col2a1, suggesting that Runx2-expressing preosteoblasts are still bipotential cells and Osx acts downstream of Runx2 to induce osteoblastic differentiation in bipotential osteochondroprogenitor cells (Fig. 1).100 Currently, there is no evidence to indicate whether Runx2 and Osx functionally or physically interact. Milona et al. have reported that there is an OSE2 element in the specificity protein-7 (Sp7; the human homologue of the mouse Osterix gene) regulatory region, so the Osx promoter may be a direct target of Runx2.315 Further analysis on the relationship between Runx2 and Osx will be the focus of future studies.
Several studies implicate the presence of Runx2-independent mechanisms for ossification.60,250 These studies implicate that additional signaling pathways may act in parallel to, or independent of, Runx2 during osteoblast differentiation. It has been shown that MAPK and PKD signaling pathways serve as points of convergence for mediating the BMP-2–and IGF-I–induced effects on Osx expression in mesenchymal stem cells.316 Additionally, Runx2 was required but not sufficient for the BMP-2–mediated Osx induction. This result indicated that additional factors (e.g., Dlx5) acting downstream of BMP-2 could induce Osx independent of the levels of Runx2 activity. Mechanistic analyses of the effect of FK506 on bone mass showed that NFAT cooperates with Osterix and accelerates osteoblast differentiation and bone formation.317 Overexpression of NFATc1 stimulates Osterix-dependent activation of the Col1a1 promoter, but not Runx2-dependent activation of the bone gamma carboxyglutamate protein 1 (Bglap1; encoding osteocalcin) promoter.
En1 expression temporally precedes that of the osteogenic determinant Osx, and, in the absence of En1, the onset of Osx expression is delayed. Since Osx is necessary for potentiating the osteogenic fate of the skeletogenic mesenchyme,100 its perturbed expression provides a mechanistic basis for the delayed calvarial ossification in En1−/− mice.318 Furthermore, that Osx expression remains impaired in En1-null osteoblasts suggests that En1 also lies upstream of Osx during later phases of calvarial osteogenesis, and thus mediates distinct functions in osteoblast differentiation. En1−/− osteoblasts were deficient in mediating osteoid mineralization and exhibited reduced ALP activity, an enzyme that is essential for this process.319,320 Moreover, ablation of En1 results in impaired OCN and BSP expression, genes that are normally associated with advanced osteoblast differentiation. OCN expression has also been shown to be dependent on Osx.100
E. ATF4
Using a combination of human genetic information, analysis of mutant mouse strains, and molecular studies, Yang et al. identified an activating transcription factor (ATF4) as a critical substrate of ribosomal protein S6 kinase polypeptide 3 (RSK2) that is required for the timely onset of osteoblast differentiation, for terminal differentiation of osteoblasts, for Bsp, and for Osteocalcin expression.321 Additionally, RSK2 and ATF4 posttranscriptionally regulate the synthesis of type I collagen, the main constituent of the bone matrix. These findings identify ATF4 as a critical regulator of osteoblast differentiation and function, and indicate that lack of ATF4 phosphorylation by RSK2 may contribute to the skeletal phenotype of Coffin-Lowry syndrome (CLS).321 Elefteriou et al. found that mice lacking neurofibromin in their osteoblasts show increased ATF4-dependent collagen synthesis, RSK2 activity, and bone formation.322 On the other hand, ATF4 phosphorylation by PKA is also increased in neurofibromin null mice, causing enhanced RANKL expression, osteoclast differentiation, and bone resorption. These changes could be corrected by a low-protein diet, which normalized bone formation and bone mass, showing that ATF4-dependent skeletal dysplasiae are treatable by dietary manipulations.
In addition, treatment of nonosteoblastic cells with MG115, a proteasome inhibitor, induced ATF4 accumulation and resulted in activation of an Osteocalcin promoter luciferase construct as well as expression of endogenous Osteocalcin, a molecular marker of differentiated osteoblasts.323 This study establishes that ATF4, like other osteoblast differentiation factors, such as Runx2 and Osterix, has the ability to induce osteoblast-specific gene expression in nonosteoblastic cells. Cooperative interactions between ATF4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression.324
F. SATB2
The patterning of skeletal elements and bone formation are generally thought to represent distinct pathways; however, evidence is emerging for cross talk between these processes. This is illustrated in studies that establish the functional role of Hoxa2 in skeletal development, an inhibitor of bone formation and regulator of branchial arch patterning.267,325 Understanding the mechanisms that mediate these dual roles of Hoxa2 will provide valuable insight into coordination of pathways governing bone patterning and differentiation.326 Grosschedl and colleagues make an important stride toward this goal by demonstrating that the nuclear matrix protein special AT-rich sequence binding protein 2 (Satb2) represses Hoxa2 expression and is an activator of multiple steps of Runx2-dependent osteoblast differentiation (Fig. 1).327
Satb2 is a recently cloned member of the family of special AT-rich binding proteins that binds to nuclear matrix-attachment regions (MARs) and activates transcription in a MAR-dependent manner.328,329 In humans, translocations that involve the chromosomal region 2q32-q33 and are associated with a cleft palate under conditions of haploinsufficiency have been found to interrupt the Satb2 gene.330 Using targeted mutagenesis of Satb2 in mice, Dobreva et al. provide insight into how the nuclear matrix, chromatin remodeling, and gene activation come together to regulate osteoblast differentiation in development.327 The most striking phenotypes detected in mice lacking Satb2 are craniofacial defects in skeletal elements and the inhibition of normal osteoblast differentiation. By combining an impressive series of molecular and genetic approaches, the authors reveal that Satb2 represses Hoxa2 expression in osteoblasts through direct recognition of an MAR-like sequence. Chromatin immunoprecipitation (ChIP) and transactivation experiments reveal that Satb2 can bind to and regulate BSP and OCN genes, themselves critical components in osteoblast formation. This strongly suggests that Satb2 has multiple inputs into transcriptional control of osteoblast differentiation. On the basis of genetic synergy between mouse mutants, protein interaction, and transactivation analyses, Dobreva et al. discovered that Satb2 directly interacts with and enhances the activity of both Runx2 and ATF4.327 The interaction of Satb2 with ATF4 and Runx2 augment their binding to the cognate DNA-recognition elements, although Satb2 does not bind itself to the OSE1 and OSE2 sequences. Satb2 has been shown to recognize specific sequences, termed MAR sequences,329 and indeed, Satb2 was found to regulate the Bsp promoter by binding to a site that resembles a bona fide MAR element. Therefore, Satb2 can act not only as an activating or repressing DNA bound protein, but also as a protein scaffold that enhances the activity of other DNA binding proteins. By its ability to regulate the expression or activity of multiple key determinants of skeletal development, Satb2 appears to represent a molecular node of a transcriptional network underlying this process.327 Distinguishing between the different modes of Satb2 activity will be important for a detailed understanding of how the nuclear matrix, chromatin structure, and transcriptional activity coordinate the regulation of multiple steps during osteoblast differentiation.326
V. OSTEOCLAST GENE TRANSCRIPTION: TRANSCRIPTION FACTORS AND CYTOKINES THAT REGULATE OSTEOCLAST DIFFERENTIATION
The transcription of eukaryotic genes is regulated by specific DNA binding proteins (transcription factors) that assemble on cis-acting DNA sequences in promoters and enhancers. Our lab has cloned osteoclast-specific genes cathepsin K and OC116/ATP6i/a3,32,331 analyzed thier promoters332 that provided gene models for osteolcast gene regulation, and characterized their function using mouse knockout.227,333 Many of these DNA binding proteins are ubiquitous in their expression and serve a general role in gene transcription. Others are restricted in expression to one or a few cell types. Cell-specific transcription factors have been identified that activate lineage-specific gene expression in skeletal muscle, neuronal, erythroid, myeloid, and lymphoid lineages.233,334–337
Gene-targeted disruption studies have revealed that various transcription factors are essential at different stages of osteoclast differentiation, activation, and survival. PU.1 is thought to be responsible for the earliest established event in osteoclastogenesis. PU.1 disruption in embryonic stem (ES) cells can make ES cells fail to differentiate into macrophages in vitro. Several other transcription factors have been found crucial for osteoclast differentiation downstream of M-CSF and RANKL/RANK signaling. Development of macrophages is preserved in NF-κB null mice, suggesting that NF-κB functions later than PU.1 during osteoclast differentiation. A deficiency in c-fos causes severe osteopetrosis with lack of osteoclasts.338,339 NFATc1 is a transcription factor that plays an essential and sufficient role in osteoclastogenesis. It is a master switch for regulating terminal differentiation of OCs, functioning downstream of RANKL.340 Overexpression of a constitutively active form of NFATc1 in c-fos null cells restores expression of osteoclast-specific genes, demonstrating that NFATc1 is a critical transcriptional regulator downstream of c-fos during osteoclast differentiation.341 Microphthalmia-associated transcription factors (MITFs) and transcription factor E (TFE) 3, TFEB, and TFEC are essential for differentiation of mononuclear precursors into multinucleated osteoclasts.342 Most of these transcription factors are involved in both the RANK-associated signaling and the expression of typical osteoclastic genes, including calcitonin receptor, TRAP, cathepsin K, osteoclast-associated receptor (OSCAR), and αvβ3 class of integrins.
How a multipotential cell chooses a single pathway of OC differentiation is a central problem in OC biology. Spontaneous and genetically engineered osteopetrotic mutant mice have yielded important insights into the regulation of OC differentiation.
A. PU.1
PU.1 is an EST-domain transcription factor essential for the development of myeloid and lymphoid lineage cells. Mice homozygous for a null mutation in the PU.1 gene die during fetal development by 18.5 days postcoitum (d.p.c.) and lack lymphoid and myeloid lineages.343 PU.1−/− ES cells fail to differentiate into macrophages in vitro.344 This failure is complemented by a PU.1 transgene, providing evidence that PU.1 promotes macrophage differentiation from pluripotent ES cells.345 The role of PU.1 in macrophage differentiation was further determined by the discovery that commitment to the monocytic lineage occurs in the absence of PU.1 and a low percentage of monocytic precursors are produced in the PU.1 null mice.346 The major role of PU.1 in lymphoid and myeloid development is thought to be the regulation of lineage-specific cytokine receptor genes, such as M-CSF receptor (c-fms), granulocyte-macrophage colony stimulating factor receptor low-affinity subunit (GM-CSFRα), G-CSF receptor (G-CSFR), and IL-7Rα.347 The combined data show that PU.1 is absolutely required for macrophage development during the differentiation from promonocyte to the monocyte stage.
OCs are bone-resorbing cells of hematopoietic origin. Hematopoietic transcription factor PU.1 is also critical for osteoclastogenesis; PU.1 expression is detected at all stages of OC differentiation and PU.1 mRNA increases threefold as cells differentiate into OCs.348 PU.1−/− knockout mice are osteopetrotic and are devoid of both OCs and macrophages, indicating a differentiation block at a common MO-CSF precursor stage.348 PU.1 also binds to the corresponding sequences in the promoters and enhancers of many OC-specific genes. RANK gene, which has been shown to be crucial for osteoclastogenesis, is a transcriptional target of PU.1. The PU.1−/− progenitor cells failed to express the RANK gene and reconstitution of PU.1 in these cells induced RANK expression.349 RANKL-induced cathepsin K gene expression is cooperatively regulated by the combination of PU.1 and NFATc1.350 The transcription factors Mitf and PU.1 interact with the TRAP gene promoter and activate TRAP gene expression in OCs.351 Thus, PU.1 appears to be a master regulator, critical for the development of a common progenitor for the lymphoid myeloid cell lineages in the hematopoietic system.
B. AP-1
AP-1, which is composed mainly of Fos (c-Fos, FosB, Fra-1, and Fra-2) and Jun (c-Jun, JunB, and JunD) proteins, is important in the osteoclastogenic process.352 Differentiation of the common precursors into either bone or immune lineages is determined by ligands binding to cell-surface receptors, RANK for OCs or Toll-like receptors (TLRs) for mononuclear phagocytes. Both RANK and TLRs activate the dimeric transcription factors NF-κB and AP-1. Yet, c-Fos/AP-1 plays a positive role in OCs, but a negative role in macrophages and dendritic cells.353
Disruption of the c-fos proto-oncogene also leads to an osteopetrotic phenotype.183,338,354,355 However, the number of EGF-like module containing, mucin-like, hormone receptor-like sequence 1 (F4/80) and lectin, galactose binding, soluble 3 (Mac-2) positive cells, presumably macrophages, is increased, and cells positive for the 92-kD type IV collagenase, proposed as a relatively early marker of the OC lineage, are evident at the capillary invasion front of the metaphysis. Thus, the absence of c-fos blocks OC development at the point of divergence from the common MO-CSF precursor, prior to the expression of TRAP-positivity. RANKL signaling to the cell interior also leads to fos-dependent transcription of target genes such as fos-like antigen 1 (Fra-1) and NFATc1. NFATc1 seems to be the critical downstream target gene of fos in osteoclastogenesis, because differentiation of fos-deficient bone marrow monocytes (BMMs) into OCs is rescued by ectopic expression of NFATc1.341 Recently, it was found that the calcium/calmodulin–dependent protein kinase (CaMK)-CREB pathway functions to regulate the transcriptional program of osteoclastic bone resorption by enhancing induction of NFATc1 and facilitating NFATc1-dependent gene regulation once its expression is induced.356 Genetic ablation of Camk4 reduced CREB phosphorylation and downregulated the expression of c-fos, which is required for the induction of NFATc1. Additionally, PPARγ conditional knockout mice were found to exhibit osteopetrosis and PPARγ was revealed to be a regulator of osteoclastogenesis by directly regulating c-fos expression.357
Transgenic mice expressing dominant-negative c-Jun specifically in the OC lineage manifested severe osteopetrosis due to impaired osteoclastogenesis.358 c-Jun is clearly RANKL activated via TRAF6 by a process involving JNK1, but not JNK2. JNK1 appears to modulate osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms.359,360 Recently, c-Jun signaling was found to be in cooperation with NFAT in RANKL-regulated OC differentiation.358 When transcriptionally active, c-Jun associates with members of the fos family. Several cytokine genes are cooperatively regulated by NFATs and AP-1, which bind to a composite recognition element in their promoter regions.
C. NF-κB
NF-κB is a family of five transcription factors that are expressed in most cell types and play an essential role in immune and inflammatory responses by regulating the expression of a variety of proinflammatory cytokines and other inflammatory mediators.361 The five transcription factors p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), c-Rel, and RelB each contain an Rel homology domain that allows these factors to dimerize and bind DNA. Unlike most transcriptional activators, this family of proteins resides in the cytoplasm and must therefore translocate into the nucleus. Once in the nucleus, they attach to κB binding sites in the promoter region of a large number of genes including IL-1, IL-6, TNF-α, and GM-CSF and, after stimuli-induced IκB protein degradation in response to extracellular signals, induce their transcription.362
Mice lacking NF-κB1 and NF-κB2 (double-knockout mice) developed osteopetrosis because of a defect in OC differentiation, suggesting redundant functions of NF-κB1 and NF-κB2 proteins in the development of this cell lineage.363 The complete absence of OC precursors in these mice indicates that OC differentiation was arrested at a relatively early stage. Further studies have demonstrated that NF-κB1 and NF-κB2 expression is not required for formation of RANK-expressing OC progenitors but is essential for RANK-expressing OC precursors to differentiate into TRAP+ OCs in response to RANKL and other osteoclastogenic cytokines.364
D. NFATc1
Recently, Takayanagi et al. reported that NFATc1 plays an essential and sufficient role in osteoclastogenesis. They demonstrated that RANKL induces and activates NFATc1 through calcium signaling. Both the transient initial release of Ca2+ from intracellular stores and the influx through specialized Ca2+ channels control the dephosphorylation of the cytoplasmic components (NFATc1 proteins) and lead to their nuclear localization, which is followed by the activation of osteoclast-specific genes.365 They reported that inhibiting NFATc1 activity using dominant negative alleles blocks osteoclastogenesis, whereas overexpression of the wild-type protein stimulates osteoclast development from embryonic stem cells in a RANKL-independent manner.
The nuclear factor of activated T-cells cytoplasmic (NFATc) is a family of transcription factors originally identified in T cells. The gene family is currently known to have four members (NFATc1 through NFATc4), which have roles both within and outside the immune system. NFATc1 is the major induced NFATc in human OCs, with expression greatly exceeding that of NFATc2 through NFATc4.366 NFATc1-deficient embryonic stem cells fail to differentiate into OCs in response to RANKL stimulation, and ectopic expression of NFATc1 causes precursor cells to undergo efficient differentiation without RANKL signaling, indicating that NFATc1 is a master switch for regulating terminal differentiation of OCs, functioning downstream of RANKL.340 RANKL induces and activates NFATc1 through calcium signaling, and calcineurin inhibitors such as FK506 and cyclosporin A strongly inhibit osteoclastogenesis.340 FK506-mediated inhibition of NFATc1 activity results in a defective induction of the mRNA of NFATc1, indicating that NFATc1 induction is dependent on its own activity: NFATc1 autoamplifies its own gene, possibly by binding to its own promoter.367,368 RANKL/TRAF-6 signaling, rather than CD40/TRAF-6, leads to NFATc1 activation and osteoclastogenesis.369 Transcriptional activation of NFATc1 in OCs is mediated by a RANKL/TRAF-6/Fos signaling pathway because NFATc1 rescues osteoclastogenesis in precursors lacking c-Fos.340,341 c-Jun signaling in cooperation with NFAT is crucial for RANKL-regulated OC differentiation. Osteoclastogenic activities of NFAT were abrogated by overexpression of dominant-negative c-Jun and overexpression of NFAT in transgenic mice expressing dominant-negative c-Jun rescues differentiation of OC precursor cells into TRAP-positive multinucleated OC-like cells even in the absence of RANKL.358 At the final stage of OC differentiation, NFATc1 cooperates with Fos and Jun proteins to induce OC-specific genes such as TRAP, calcitonin receptor, cathepsin K and αvβ3 integrin gene.350,358,370–373 αvβ3 integrin is crucial to bone resorption and αvβ3 integrin null mice exhibit osteosclerosis and an increased number of dysfunctional osteoclasts, which have an abnormal cytoskeleton and fail to spread in vitro, to form actin rings ex vivo, or to form normal ruffled membranes in vivo.373
E. Mitf
Mitf is a member of the basic/helix-loop-helix/leucine zipper (b-HLH-ZIP) transcription factor subfamily, which also includes Tfe3, Tfeb, and Tfec. Mitf is required for the proper development of several cell lineages including OCs, melanocytes, retinal pigment epithelial cells, mast cells, and natural killer cells. Mutations in Mitf result in osteopetrosis in several organisms due to defective OC development.342 Mononuclear OCs can be detected in mi/mi mice but these cells are incapable of fusing to form multinucleate cells, lack a distinct ruffled border, and are defective in bone resorption.374–377 Compared with many other osteopetrotic mouse models, such as c-fos, PU.1, and NF-κB mouse knockout models, in which earlier steps of OC differentiation are affected resulting in lower numbers of OCs in vivo,348,355,378 defects in the OCs of mi/mi mutant mouse models appear to occur late in differentiation.
The interaction of Mitf with either PU.1 or PU.1-interacting protein allows efficient induction of an OC-specific marker of the TRAP gene in a synergistic manner.351 Through three consensus elements in the cathepsin K promoter, Mitf and Tfe3 could regulate the expression of cathepsin K on the transcriptional level. PU.1 and Mitf transcription factors also synergistically activate the expression of the OSCAR gene, which has two Ig-like domains and functions as a bone-specific regulator of OC differentiation.379 Furthermore, Mitf is a target for the RANKL signaling pathway in OCs, and phosphorylation of Mitf leads to an increase in OC-specific gene expression.380 M-CSF could induce phosphorylation of Mitf and Tfe3 via a conserved MAPK consensus site, thereby triggering their recruitment of the coactivator p300.381
F. Myc
Genes of the myelocytomatosis oncogene (Myc) family contribute to the genesis of many human tumors. In mammals, there are four related genes in the Myc family, namely, c-Myc, N-Myc, L-Myc, and S-Myc. They are key regulators of cell proliferation, and their deregulation contributes to the genesis of most human tumors.382 The number of target genes that are regulated by Myc is surprisingly large. C-Myc was also found to be strongly upregulated in RANKL-induced osteoclast-like cells (OCLs) but was absent in undifferentiated cells. Dominant negative Myc in RAW264.7 cells was able to block RANKL-induced OCL formation. Therefore, C-Myc is a downstream target of RANKL and its expression is required for RANKL-induced osteoclastogenesis.383 It was also found that transcription from the TRAP promoter could be negatively regulated by Myc.384 Compared to the understanding of osteoblast transcriptional regulation, osteoclast transcriptional regulation remains relatively unclear.
VI. SUMMARY
Bone is constantly being remodeled in a dynamic process where osteoblasts are responsible for bone formation and osteoclasts for its resorption. Formation of skeletal elements during embryogenesis and the dynamic remodeling of bone in the adult involve an exquisite interplay of developmental cues, signaling proteins, transcription factors, and their coregulatory proteins that support differentiation of osteogenic lineage cells from the initial mesenchymal progenitor cell to the mature osteocyte in mineralized connective tissue.229 Wnt/β-catenin signaling is an enticing target for developing drugs to combat skeletal diseases since the canonical pathway is composed of a series of molecular interactions that offer potential places for pharmacological intervention.69 Several unresolved issues are offered here in the hopes of stimulating their resolution. Although we have learned much in recent years regarding the role of canonical Wnt signaling in bone formation, some important questions remain to be addressed. For example, we know that most Wnts and FZDs are expressed in bone, but is there a role for Wnt and FZD specificity in the control of osteoblast physiology? Recent evidence has shown that non-canonical pathways also play a role in bone metabolism but there is more research to be done in this area. Finally, canonical Wnt signaling appears to control osteoclastogenesis through actions on osteoblasts, but do Wnts also have direct effects on osteoclasts? This and other questions are likely to be answered in the coming years.
Like all developmentally important proteins, multiple mechanisms actively control Runx2 activity. Further studies are required to clarify the detailed mechanism of the temporal and spatial regulation of Runx2 for the normal development and homeostatic regulation of the skeletal system. Additional proteins will be added to the list of Runx2-interacting proteins in the next decade. Mechanisms regulating expression and subsequent activity of Osx, ATF, and SATB2 at different stages of osteoblast maturation are only beginning to be understood. The role of Msx2 remains to be defined since some studies have reported that Msx2 enhances osteoblast differentiation while other studies report that Msx2 inhibits it (Fig. 1).385 Finally, insights into the mechanisms by which proteins and developmental signals affect these transcription factors’ subcellular localization to the nuclear matrix will be needed to understand how they regulate gene expression.
Recent studies provide evidence that the skeleton plays a role in energy metabolism by exerting an endocrine regulation of sugar homeostasis.386,387 Lee et al. found that mice lacking OST-PTP, a gene expressed in osteoblasts encoding a receptorlike protein tyrosine phosphatase, are hypoglycemic and show an increase in β-cell proliferation, insulin secretion, and insulin sensitivity.386 On the other hand, mice lacking osteocalcin showed diminished β-cell proliferation, glucose intolerance, and insulin resistance. Osteocalcin was found to improve glucose tolerance in vivo and stimulate CylcinD1 and insulin expression in β cells and an insulin-sensitizing adipokine in adipocytes ex vivo.386 Several questions remain to fully demonstrate that the skeleton is a ductless gland secreting osteocalcin from osteoblasts as a hormone.387 How is the skeletal regulation of glucose and fat connected to the control of bone by leptin through either the hypothalamus or the sympathetic nervous system? Where does osteocalcin sit in the hierarchy of hormones that influence glucose and fat metabolism? How the osteoblast senses the need for metabolic control must be understood, as well as how the cell translates this information into osteocalcin release. The mechanism of osteocalcin signaling will need to be investigated. If osteocalcin deficiency plays a role in human disease such as type 2 diabetes mellitus, there will be several clinical and therapeutic implications.
Osteoclasts are the principal bone-resorbing cells, and their activity has a profound impact on skeletal health. A more complete understanding of the mechanisms by which OCs differentiate from their precursors is therefore critical to developing therapies for often debilitating diseases. In this review, we have summarized the transcription factors and cytokines that have been shown through genetic studies to be important in bone development. The PU.1−/− mouse lacks both OCs and macrophages; it represents the earliest developmental form of osteopetrosis yet described. PU.1 is known to be required for macrophage development during differentiation from promonocyte to monocyte. Recently, it was also reported that Tal-1 may lie upstream of PU.1 in a regulatory hierarchy during osteoclastogenesis.388 Differentiation of mature OCs from committed OC progenitors was blocked by disruption of the Mitf, NF-κB, c-Fos, and NFATc1 genes. Although these transcription factors play an important role in OC differentiation, the expression of these transcription factors in various cell types suggests that they are unlikely to be the switch that dominates the OC differentiation process. Further study is needed to determine which transcription factors function together with ubiquitous transcription factors (i.e., c-Fos, PU.1, NF-κB, and NFATc1) to control OC differentiation. Cytokines including M-CSF and RANKL also induce and modulate growth and differentiation of the precursors to mature osteoclasts. M-CSF binds to its receptor to provide the signals for macrophage survival and proliferation. RANKL interaction with RANK induces recruitment and activation of TRAFs, leading to the activation of multiple signaling cascades. Although much is understood about RANK signaling, several questions remain. The in vivo significance of MAPKs, along with other genes activated by RANKL, has not yet been established. In ITAM-mediated costimulatory signaling, the ligands for the Ig-like receptors are not identified. The molecules that connect RANK with ITAM signaling are also unknown. Among the NFATc1 target genes, the genes that directly promote the differentiation process remain to be elucidated. Additional RANKL/RANK signaling questions include: How does RANKL/RANK signaling regulate osteoblast function and the anabolic effects of PTH or other potential anabolic agents? Does RANKL/RANK signaling play an important role in cancer cell growth and interaction with other cells in bone? How exactly do immune cells influence osteoclast and osteoblast/stromal cell function in normal and disease states? How important are osteoclasts and their precursors in regulating their own formation and function relative to macrophages and other immune cells? Taken together, osteoclastogenesis is likely to be a complicated process, controlled by several regulatory mechanisms at several differentiation transition points. Thus, further detailed studies are necessary for the complete understanding of the osteoclast signaling pathways.
Molecular understanding of these signaling pathways in osteoblasts and osteoclasts will provide unprecedented therapeutic strategies for bone disease. One promising therapeutic target is Atp6v0d2 in osteoclasts. This gene encodes a subunit of the vacuolar ATPase necessary for osteoclastic bone resorption and the d2 isoform is predominantly expressed in osteoclasts.389 Atp6v0d2-deficient mice exhibit markedly increased bone mass due to defective osteoclasts and enhanced bone formation, revealing that Atp6v0d2 is required for efficient preosteoclast fusion. In addition, Atp6v0d2 was shown to inhibit osteoblasts, although the mechanism for this finding has not been revealed. These results raise the possibility of therapeutic intervention for osteoporosis by targeting a single gene product in osteoclasts to inhibit bone destruction and stimulate bone formation.389,390 The new findings raise hope that the limitations of current drugs for bone disease might be overcome.
ACKNOWLEDGMENTS
We would like to thank Ning Li for his assistance with the manuscript. We apologize to the many researchers whose work could not be cited due to space limitations. Work in our laboratory is supported by grants by NIH grant AR44741 (Y.-P. Li) and AR-48133 (Y.-P. Li).
REFERENCES
- 1.Rubin J, Rubin C, Jacobs CR. Molecular pathways mediating mechanical signaling in bone. Gene. 2006;367:1–16. doi: 10.1016/j.gene.2005.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, Harada Y, Azuma Y, Krust A, Yamamoto Y, Nishina H, Takeda S, Takayanagi H, Metzger D, Kanno J, Takaoka K, Martin TJ, Chambon P, Kato S. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007;130(5):811–823. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 3.Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest. 2005;115(12):3318–3325. doi: 10.1172/JCI27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokstad E. Bone quality fills holes in fracture risk. Science. 2005;308(5728):1580–1581. doi: 10.1126/science.308.5728.1580. [DOI] [PubMed] [Google Scholar]
- 5.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 6.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 7.Watt FM. Stem cell fate and patterning in mammalian epidermis. Curr Opin Genet Dev. 2001;11:410–417. doi: 10.1016/s0959-437x(00)00211-2. [DOI] [PubMed] [Google Scholar]
- 8.Gronthos S, Zannettino AC, Graves SE, Ohta S, Hay SJ, Simmons PJ. Differential cell surface expression of the STRO-1 and alkaline phosphatase antigens on discrete developmental stages in primary cultures of human bone cells. J Bone Miner Res. 1999;14:47–56. doi: 10.1359/jbmr.1999.14.1.47. [DOI] [PubMed] [Google Scholar]
- 9.Stein GS, Lian JB, Stein JL, Van Wijnen AJ, Montecino M. Transcriptional control of osteoblast growth and differentiation. Physiol Rev. 1996;76:593–629. doi: 10.1152/physrev.1996.76.2.593. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Blair HC, Peng Y, Zaidi N, Adebanjo OA, Wu XB, Wu XY, Iqbal J, Epstein S, Abe E, Moonga BS, Zaidi M. Calcineurin regulates bone formation by the osteoblast. Proc Natl Acad Sci U S A. 2005;102(47):17130–17135. doi: 10.1073/pnas.0508480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundberg P, Allison SJ, Lee NJ, Baldock PA, Brouard N, Rost S, Enriquez RF, Sainsbury A, Lamghari M, Simmons P, Eisman JA, Gardiner EM, Herzog H. Greater bone formation of Y2 knockout mice is associated with increased osteoprogenitor numbers and altered Y1 receptor expression. J Biol Chem. 2007;282(26):19082–19091. doi: 10.1074/jbc.M609629200. [DOI] [PubMed] [Google Scholar]
- 12.Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, Enriquez RF, McDonald MM, Zhang L, During MJ, Little DG, Eisman JA, Gardiner EM, Yulyaningsih E, Lin S, Sainsbury A, Herzog H. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem. 2007;282(26):19092–19102. doi: 10.1074/jbc.M700644200. [DOI] [PubMed] [Google Scholar]
- 13.Coccia PF, Krivit W, Cervenka J, Clawson C, Kersey JH, Kim TH, Nesbit ME, Ramsay NK, Warkentin PI, Teitelbaum SL, Kahn AJ, Brown DM. Successful bone-marrow transplantation for infantile malignant osteopetrosis. N Engl J Med. 1980;302(13):701–708. doi: 10.1056/NEJM198003273021301. [DOI] [PubMed] [Google Scholar]
- 14.Baron R, Neff L, Tran Van P, Nefussi JR, Vignery A. Kinetic and cytochemical identification of osteoclast precursors and their differentiation into multinucleated osteoclasts. Am J Pathol. 1986;122:363–378. [PMC free article] [PubMed] [Google Scholar]
- 15.Kurihara N. The origin of osteoclasts. Bull Kanagawa Dent Coll. 1990;18:161–164. [PubMed] [Google Scholar]
- 16.Schneider GB, Relfson M. A bone marrow fraction enriched for granulocyte-macrophage progenitors gives rise to osteoclasts in vitro. Bone. 1988;9:303–308. doi: 10.1016/8756-3282(88)90014-2. [DOI] [PubMed] [Google Scholar]
- 17.Udagawa N, Takahashi N, Akatsu T, Tanaka H, Sasaki T, Nishihara T, Koga T, Martin TJ, Suda T. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc Natl Acad Sci U S A. 1990;87(18):7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagenaars CE, van der Kraan AA, Kawilarang-De Haas EW, Visser JW, Nijweide PJ. Osteoclast formation from cloned pluripotent hemopoietic stem cells. Bone Miner. 1989;6:179–189. doi: 10.1016/0169-6009(89)90049-4. [DOI] [PubMed] [Google Scholar]
- 19.Burger EH, Van der Meer JW, Nijweide PJ. Osteoclast formation from mononuclear phagocytes: role of bone-forming cells. J Cell Biol. 1984;99:1901–1906. doi: 10.1083/jcb.99.6.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suda T, Takahashi N, Martin TJ. Modulation of osteoclast differentiation. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [published erratum appears in Endocr Rev. 1992;13:191] [DOI] [PubMed] [Google Scholar]
- 21.Menaa C, Devlin RD, Reddy SV, Gazitt Y, Choi SJ, Roodman GD. Annexin II increases osteoclast formation by stimulating the proliferation of osteoclast precursors in human marrow cultures. J Clin Invest. 1999;103:1605–1613. doi: 10.1172/JCI6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jilka RL, Hangoc G, Girasole G, Passeri G, Williams DC, Abrams JS, Boyce B, Broxmeyer H, Manolagas SC. Increased osteoclast development after estrogen loss: mediation by interleukin-6. Science. 1992;257:88–91. doi: 10.1126/science.1621100. [DOI] [PubMed] [Google Scholar]
- 23.Suda T, Takahashi N, Abe E. Role of vitamin D in bone resorption. J Cell Biochem. 1992;49:53–58. doi: 10.1002/jcb.240490110. [DOI] [PubMed] [Google Scholar]
- 24.Usui M, Xing L, Drissi H, Zuscik M, O’Keefe R, Chen D, Boyce BF. Murine and chicken chondrocytes regulate osteoclastogenesis by producing RANKL in response to BMP2. J Bone Miner Res. 2008;23(3):314–325. doi: 10.1359/JBMR.071025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Peng Y, Sharrow AC, Iqbal J, Zhang Z, Papachristou DJ, Zaidi S, Zhu LL, Yaroslavskiy BB, Zhou H, Zallone A, Sairam MR, Kumar TR, Bo W, Braun J, Cardoso-Landa L, Schaffler MB, Moonga BS, Blair HC, Zaidi M. FSH directly regulates bone mass. Cell. 2006;125:247–260. doi: 10.1016/j.cell.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 26.Suda T, Udagawa N, Nakamura I, Miyaura C, Takahashi N. Modulation of osteoclast differentiation by local factors. Bone. 1995;17(2 Suppl):87S–91S. doi: 10.1016/8756-3282(95)00185-g. [DOI] [PubMed] [Google Scholar]
- 27.Ejiri S. The preosteoclast and its cytodifferentiation into the osteoclast: ultrastructural and histochemical studies of rat fetal parietal bone. Arch Histol Jpn. 1983;46:533–557. [PubMed] [Google Scholar]
- 28.Rifkin BR, Brand JS, Cushing JE, Coleman SJ, Sanavi F. Fine structure of fetal rat calvarium; provisional identification of preosteoclasts. Calcif Tissue Int. 1980;31:21–28. doi: 10.1007/BF02407163. [DOI] [PubMed] [Google Scholar]
- 29.Scheven BA, Kawilarang-De Haas EW, Wassenaar AM, Nijweide PJ. Differentiation kinetics of osteoclasts in the periosteum of embryonic bones in vivo and in vitro. Anatom Record. 1986;214:418–423. doi: 10.1002/ar.1092140413. [DOI] [PubMed] [Google Scholar]
- 30.Scott BL. The occurrence of specific cytoplasmic granules in the osteoclast. J Ultrastruct Res. 1967;19(5):417–431. doi: 10.1016/s0022-5320(67)80071-6. [DOI] [PubMed] [Google Scholar]
- 31.Wucherpfennig AL, Li YP, Stetler-Stevenson WG, Rosenberg AE, Stashenko P. Expression of 92 kD type IV collagenase/gelatinase B in human osteoclasts. J Bone Miner Res. 1994;9:549–556. doi: 10.1002/jbmr.5650090415. [DOI] [PubMed] [Google Scholar]
- 32.Li YP, Alexander M, Wucherpfennig AL, Yelick P, Chen W, Stashenko P. Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J Bone Miner Res. 1995;10:1197–1202. doi: 10.1002/jbmr.5650100809. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, Moseley JM, Martin TJ, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123:2600–2662. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Li YP. Generation of mouse osteoclastogenic cell lines immortalized with SV40 large T antigen. J Bone Miner Res. 1998;13:1112–1123. doi: 10.1359/jbmr.1998.13.7.1112. [DOI] [PubMed] [Google Scholar]
- 35.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 36.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 37.Rawadi G, Roman-Roman S. Wnt signalling pathway: a new target for the treatment of osteoporosis. Expert Opin Ther Targets. 2005;9:1063–1077. doi: 10.1517/14728222.9.5.1063. [DOI] [PubMed] [Google Scholar]
- 38.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 39.Hay E, Faucheu C, Suc-Royer I, Touitou R, Stiot V, Vayssiere B, Baron R, Roman-Roman S, Rawadi G. Interaction between LRP5 and Frat1 mediates the activation of the Wnt canonical pathway. J Biol Chem. 2005;280(14):13616–13623. doi: 10.1074/jbc.M411999200. [DOI] [PubMed] [Google Scholar]
- 40.Clevers H. Wnt signaling: Ig-norrin the dogma. Curr Biol. 2004;14:R436–R437. doi: 10.1016/j.cub.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaur T, Rich L, Lengner CJ, Hussain S, Trevant B, Ayers D, Stein JL, Bodine PV, Komm BS, Stein GS, Lian JB. Secreted frizzled related protein 1 regulates Wnt signaling for BMP2 induced chondrocyte differentiation. J Cell Physiol. 2006;208:87–96. doi: 10.1002/jcp.20637. [DOI] [PubMed] [Google Scholar]
- 44.Bodine PV, Zhao W, Kharode YP, Bex FJ, Lambert AJ, Goad MB, Gaur T, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol Endocrinol. 2004;18:1222–1237. doi: 10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- 45.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280(29) doi: 10.1074/jbc.M504308200. 26770--5. [DOI] [PubMed] [Google Scholar]
- 46.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417(6889):664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 47.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411(6835):321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 48.Diarra D, Stolina M, Polzer K, Zwerina J, Ominsky MS, Dwyer D, Korb A, Smolen J, Hoffmann M, Scheinecker C, van der HD, Landewe R, Lacey D, Richards WG, Schett G. Dickkopf-1 is a master regulator of joint remodeling. Nat Med. 2007;13:156–163. doi: 10.1038/nm1538. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Liu P, Liu W, Maye P, Zhang J, Zhang Y, Hurley M, Guo C, Boskey A, Sun L, Harris SE, Rowe DW, Ke HZ, Wu D. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat Genet. 2005;37:945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- 50.Abe E, Marians RC, Yu W, Wu XB, Ando T, Li Y, Iqbal J, Eldeiry L, Rajendren G, Blair HC, Davies TF, Zaidi M. TSH is a negative regulator of skeletal remodeling. Cell. 2003;115(2):151–162. doi: 10.1016/s0092-8674(03)00771-2. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130:1003–1015. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- 53.Ai M, Holmen SL, Van HW, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346(20):1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 55.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, rslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De PA, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van HW, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 56.Van WL, Cleiren E, Gram J, Beals RK, Benichou O, Scopelliti D, Key L, Renton T, Bartels C, Gong Y, Warman ML, De Vernejoul MC, Bollerslev J, Van HW. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartikka H, Makitie O, Mannikko M, Doria AS, Daneman A, Cole WG, la-Kokko L, Sochett EB. Heterozygous mutations in the LDL receptor-related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J Bone Miner Res. 2005;20:783–789. doi: 10.1359/JBMR.050101. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris S, Wu D. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balemans W, Devogelaer JP, Cleiren E, Piters E, Caussin E, Van HW. Novel LRP5 missense mutation in a patient with a high bone mass phenotype results in decreased DKK1-mediated inhibition of Wnt signaling. J Bone Miner Res. 2007;22:708–716. doi: 10.1359/jbmr.070211. [DOI] [PubMed] [Google Scholar]
- 60.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, Macdougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson A, Vayssiere B, Garcia T, Newell W, Baron R, Roman-Roman S, Rawadi G. Gene array analysis of Wnt-regulated genes in C3H10T1/2 cells. Bone. 2005;36:585–598. doi: 10.1016/j.bone.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 63.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 64.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 65.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 66.Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005;280(22):211624–211628. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 67.Glass DA, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 68.Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 69.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280(39):33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 71.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282(19):14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 72.Glass DA, Karsenty G. Canonical Wnt signaling in osteoblasts is required for osteoclast differentiation. Ann N Y Acad Sci. 2006;1068:117–130. doi: 10.1196/annals.1346.015. [DOI] [PubMed] [Google Scholar]
- 73.Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, Takezawa S, Yogiashi Y, Kitagawa H, Yamada G, Takada S, Minami Y, Shibuya H, Matsumoto K, Kato S. A histone lysine methyl-transferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 74.Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, McMahon AP, Long F. Noncanonical Wnt signaling through G protein-linked PKCdelta activation promotes bone formation. Dev Cell. 2007;12:113–127. doi: 10.1016/j.devcel.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 76.Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357:1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 78.Anderson HC, Hodges PT, Aguilera XM, Missana L, Moylan PE. Bone morphogenetic protein (BMP) localization in developing human and rat growth plate, metaphysis, epiphysis, and articular cartilage. J Histochem Cytochem. 2000;48:1493–1502. doi: 10.1177/002215540004801106. [DOI] [PubMed] [Google Scholar]
- 79.Hoffmann A, Gross G. BMP signaling pathways in cartilage and bone formation. Crit Rev Eukaryot Gene Expr. 2001;11:23–45. [PubMed] [Google Scholar]
- 80.Peng Y, Kang Q, Cheng H, Li X, Sun MH, Jiang W, Luu HH, Park JY, Haydon RC, He TC. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J Cell Biochem. 2003;90:1149–1165. doi: 10.1002/jcb.10744. [DOI] [PubMed] [Google Scholar]
- 81.Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 82.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 83.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 84.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 85.Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr Opin Genet Dev. 2003;13:43–47. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 86.Miyazono K, Maeda S, Imamura T. Coordinate regulation of cell growth and differentiation by TGF-beta superfamily and Runx proteins. Oncogene. 2004;23(24):4232–4237. doi: 10.1038/sj.onc.1207131. [DOI] [PubMed] [Google Scholar]
- 87.Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97(19):10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;23:552–563. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces os-teoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee MH, Kim YJ, Kim HJ, Park HD, Kang AR, Kyung HM, Sung JH, Wozney JM, Kim HJ, Ryoo HM. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem. 2003;278(36):34387–34394. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 91.Miyama K, Yamada G, Yamamoto TS, Takagi C, Miyado K, Sakai M, Ueno N, Shibuya H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev Biol. 1999;208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 92.de Jong DS, Vaes BL, Dechering KJ, Feijen A, Hendriks JM, Wehrens R, Mummery CL, van Zoelen EJ, Olijve W, Steegenga WT. Identification of novel regulators associated with early-phase osteoblast differentiation. J Bone Miner Res. 2004;19:947–958. doi: 10.1359/JBMR.040216. [DOI] [PubMed] [Google Scholar]
- 93.Korchynskyi O, Dechering KJ, Sijbers AM, Olijve W, ten DP. Gene array analysis of bone morphogenetic protein type I receptor-induced osteoblast differentiation. J Bone Miner Res. 2003;18:1177–1185. doi: 10.1359/jbmr.2003.18.7.1177. [DOI] [PubMed] [Google Scholar]
- 94.Guicheux J, Lemonnier J, Ghayor C, Suzuki A, Palmer G, Caverzasio J. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J Bone Miner Res. 2003;18:2060–2068. doi: 10.1359/jbmr.2003.18.11.2060. [DOI] [PubMed] [Google Scholar]
- 95.Lai CF, Cheng SL. Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-beta in normal human osteoblastic cells. J Biol Chem. 2002;277(18):15514–15522. doi: 10.1074/jbc.M200794200. [DOI] [PubMed] [Google Scholar]
- 96.Lemonnier J, Ghayor C, Guicheux J, Caverzasio J. Protein kinase C-independent activation of protein kinase D is involved in BMP-2-induced activation of stress mitogen-activated protein kinases JNK and p38 and osteoblastic cell differentiation. J Biol Chem. 2004;279:259–264. doi: 10.1074/jbc.M308665200. [DOI] [PubMed] [Google Scholar]
- 97.Lee KS, Hong SH, Bae SC. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene. 2002;21(47):7156–7163. doi: 10.1038/sj.onc.1205937. [DOI] [PubMed] [Google Scholar]
- 98.Lee MH, Javed A, Kim HJ, Shin HI, Gutierrez S, Choi JY, Rosen V, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Ryoo HM. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem. 1999;73:114–125. [PubMed] [Google Scholar]
- 99.Phimphilai M, Zhao Z, Boules H, Roca H, Franceschi RT. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J Bone Miner Res. 2006;21:637–646. doi: 10.1359/JBMR.060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 101.Yagi K, Tsuji K, Nifuji A, Shinomiya K, Nakashima K, DeCrombrugghe B, Noda M. Bone morphogenetic protein-2 enhances osterix gene expression in chondrocytes. J Cell Biochem. 2003;88:1077–1083. doi: 10.1002/jcb.10467. [DOI] [PubMed] [Google Scholar]
- 102.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 103.Celil AB, Hollinger JO, Campbell PG. Osx transcriptional regulation is mediated by additional pathways to BMP2/Smad signaling. J Cell Biochem. 2005;95:518–528. doi: 10.1002/jcb.20429. [DOI] [PubMed] [Google Scholar]
- 104.Datto M, Wang XF. Ubiquitin-mediated degradation a mechanism for fine-tuning TGF-beta signaling. Cell. 2005;121:2–4. doi: 10.1016/j.cell.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 105.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 106.Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 107.Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 109.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38:1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- 110.Chung UI, Schipani E, McMahon AP, Kronenberg HM. Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J Clin Invest. 2001;107:295–304. doi: 10.1172/JCI11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- 112.St Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Razzaque MS, Soegiarto DW, Chang D, Long F, Lanske B. Conditional deletion of Indian hedgehog from collagen type 2alpha1-expressing cells results in abnormal endochondral bone formation. J Pathol. 2005;207:453–461. doi: 10.1002/path.1870. [DOI] [PubMed] [Google Scholar]
- 114.Nakamura T, Aikawa T, Iwamoto-Enomoto M, Iwamoto M, Higuchi Y, Pacifici M, Kinto N, Yamaguchi A, Noji S, Kurisu K, Matsuya T. Induction of osteogenic differentiation by hedgehog proteins. Biochem Biophys Res Commun. 1997;237:465–469. doi: 10.1006/bbrc.1997.7156. [DOI] [PubMed] [Google Scholar]
- 115.van der Horst G, Farih-Sips H, Löwik CW, Karperien M. Hedgehog stimulates only osteoblastic differentiation of undifferentiated KS483 cells. Bone. 2003;33:899–910. doi: 10.1016/j.bone.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 116.Meng X, Poon R, Zhang X, Cheah A, Ding Q, Hui CC, Alman B. Suppressor of fused negatively regulates beta-catenin signaling. J Biol Chem. 2001;276(43):40113–40119. doi: 10.1074/jbc.M105317200. [DOI] [PubMed] [Google Scholar]
- 117.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416(6880):548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- 118.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 119.Spinella-Jaegle S, Rawadi G, Kawai S, Gallea S, Faucheu C, Mollat P, Courtois B, Bergaud B, Ramez V, Blanchet AM, Adelmant G, Baron R, Roman-Roman S. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J Cell Sci. 2001;114(Pt 11):2085–2094. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- 120.Yuasa T, Kataoka H, Kinto N, Iwamoto M, Enomoto-Iwamoto M, Iemura S, Ueno N, Shibata Y, Kurosawa H, Yamaguchi A. Sonic hedgehog is involved in osteoblast differentiation by cooperating with BMP-2. J Cell Physiol. 2002;193:225–232. doi: 10.1002/jcp.10166. [DOI] [PubMed] [Google Scholar]
- 121.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 122.Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- 123.Wilkie AO. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- 124.Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125(24):4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 125.Valverde-Franco G, Liu H, Davidson D, Chai S, Valderrama-Carvajal H, Goltzman D, Ornitz DM, Henderson JE. Defective bone mineralization and osteopenia in young adult FGFR3−/− mice. Hum Mol Genet. 2004;13:271–284. doi: 10.1093/hmg/ddh034. [DOI] [PubMed] [Google Scholar]
- 126.Xiao L, Naganawa T, Obugunde E, Gronowicz G, Ornitz DM, Coffin JD, Hurley MM. Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J Biol Chem. 2004;279(26):27743–27752. doi: 10.1074/jbc.M314323200. [DOI] [PubMed] [Google Scholar]
- 127.Chen L, Deng CX. Roles of FGF signaling in skeletal development and human genetic diseases. Front Biosci. 2005;10:1961–1976. doi: 10.2741/1671. [DOI] [PubMed] [Google Scholar]
- 128.Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16:205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8(24):3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 130.Li C, Xu X, Nelson DK, Williams T, Kuehn MR, Deng CX. FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development. 2005;132(21):4755–4764. doi: 10.1242/dev.02065. [DOI] [PubMed] [Google Scholar]
- 131.Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12(15):2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Verheyden JM, Lewandoski M, Deng C, Harfe BD, Sun X. Conditional inactivation of Fgfr1 in mouse defines its role in limb bud establishment, outgrowth and digit patterning. Development. 2005;132(19):4235–4245. doi: 10.1242/dev.02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast growth factor receptor 1 signaling in the osteochondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 135.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hurley MM, Tetradis S, Huang YF, Hock J, Kream BE, Raisz LG, Sabbieti MG. Parathyroid hormone regulates the expression of fibroblast growth factor-2 mRNA and fibroblast growth factor receptor mRNA in osteoblastic cells. J Bone Miner Res. 1999;14:776–783. doi: 10.1359/jbmr.1999.14.5.776. [DOI] [PubMed] [Google Scholar]
- 138.Sabbieti MG, Marchetti L, Abreu C, Montero A, Hand AR, Raisz LG, Hurley MM. Prostaglandins regulate the expression of fibroblast growth factor-2 in bone. Endocrinology. 1999;140:434–444. doi: 10.1210/endo.140.1.6442. [DOI] [PubMed] [Google Scholar]
- 139.Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277(39):36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- 141.Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 142.Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129(16):3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 143.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130(13):3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 144.Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM, Aaronson SA. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci U S A. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Naski MC, Ornitz DM. FGF signaling in skeletal development. Front Biosci. 1998;3:d781–d794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 146.Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 147.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271(25) doi: 10.1074/jbc.271.25.15292. 15292--7. [DOI] [PubMed] [Google Scholar]
- 148.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3) doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kozawa O, Tokuda H, Matsuno H, Uematsu T. Involvement of p38 mitogen-activated protein kinase in basic fibroblast growth factor-induced interleukin-6 synthesis in osteoblasts. J Cell Biochem. 1999;74:479–485. [PubMed] [Google Scholar]
- 150.Lemonnier J, Delannoy P, Hott M, Lomri A, Modrowski D, Marie PJ. The Ser252Trp fibroblast growth factor receptor-2 (FGFR-2) mutation induces PKC-independent downregulation of FGFR-2 associated with premature calvaria osteoblast differentiation. Exp Cell Res. 2000;256:158–167. doi: 10.1006/excr.2000.4820. [DOI] [PubMed] [Google Scholar]
- 151.Lomri A, Lemonnier J, Delannoy P, Marie PJ. Increased expression of protein kinase Calpha, interleukin-1alpha, and RhoA guanosine 5'-triphosphatase in osteoblasts expressing the Ser252Trp fibroblast growth factor 2 receptor Apert mutation: identification by analysis of complementary DNA microarray. J Bone Miner Res. 2001;16(4):705–712. doi: 10.1359/jbmr.2001.16.4.705. [DOI] [PubMed] [Google Scholar]
- 152.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308(5727):1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 153.Mundy GR, Elefteriou F. Boning up on ephrin signaling. Cell. 2006;126:441–443. doi: 10.1016/j.cell.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 154.Compagni A, Logan M, Klein R, Adams RH. Control of skeletal patterning by ephrinB1-EphB interactions. Dev Cell. 2003;5:217–230. doi: 10.1016/s1534-5807(03)00198-9. [DOI] [PubMed] [Google Scholar]
- 155.Wieland I, Jakubiczka S, Muschke P, Cohen M, Thiele H, Gerlach KL, Adams RH, Wieacker P. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 2004;74:1209–1215. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao C, Irie N, Takada Y, Shimoda K, Miyamoto T, Nishiwaki T, Suda T, Matsuo K. Bidirectional ephrinB2-EphB4 signaling controls bone homeostasis. Cell Metab. 2006;4:111–121. doi: 10.1016/j.cmet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 157.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 158.Potts JT, Gardella TJ. Progress, paradox, and potential: parathyroid hormone research over five decades. Ann N Y Acad Sci. 2007;1117:196–208. doi: 10.1196/annals.1402.088. [DOI] [PubMed] [Google Scholar]
- 159.Pettway GJ, Meganck JA, Koh AJ, Keller ET, Goldstein SA, McCauley LK. Parathyroid hormone mediates bone growth through the regulation of osteoblast proliferation and differentiation. Bone. 2008;42(4):806–818. doi: 10.1016/j.bone.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Boumah CE, Selvamurugan N, Partridge NC. Transcription in the osteoblast: regulatory mechanisms utilized by parathyroid hormone and transforming growth factor-beta. Prog Nucleic Acid Res Mol Biol. 2005;80:287–321. doi: 10.1016/S0079-6603(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 161.Bergenstock MK, Partridge NC. Parathyroid hormone stimulation of noncanonical Wnt signaling in bone. Ann N Y Acad Sci. 2007;1116:354–359. doi: 10.1196/annals.1402.047. [DOI] [PubMed] [Google Scholar]
- 162.Swarthout JT, D’Alonzo RC, Selvamurugan N, Partridge NC. Parathyroid hormone-dependent signaling pathways regulating genes in bone cells. Gene. 2002;282:1–17. doi: 10.1016/s0378-1119(01)00798-3. [DOI] [PubMed] [Google Scholar]
- 163.Partridge NC, Li X, Qin L. Understanding parathyroid hormone action. Ann N Y Acad Sci. 2006;1068:187–193. doi: 10.1196/annals.1346.024. [DOI] [PubMed] [Google Scholar]
- 164.Guo J, Chung UI, Kondo H, Bringhurst FR, Kronenberg HM. The PTH/PTHrP receptor can delay chondrocyte hypertrophy in vivo without activating phospholipase C. Dev Cell. 2002;3:183–194. doi: 10.1016/s1534-5807(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 165.Togari A. Adrenergic regulation of bone metabolism: possible involvement of sympathetic innervation of osteoblastic and osteoclastic cells. Microsc Res Tech. 2002;58:77–84. doi: 10.1002/jemt.10121. [DOI] [PubMed] [Google Scholar]
- 166.Takeda S, Elefteriou F, Levasseur R, Liu X, Zhao L, Parker KL, Armstrong D, Ducy P, Karsenty G. Leptin regulates bone formation via the sympathetic nervous system. Cell. 2002;111:305–317. doi: 10.1016/s0092-8674(02)01049-8. [DOI] [PubMed] [Google Scholar]
- 167.Hinoi E, Fujimori S, Yoneda Y. Modulation of cellular differentiation by N-methyl-D-aspartate receptors in osteoblasts. FASEB J. 2003;17:1532–1534. doi: 10.1096/fj.02-0820fje. [DOI] [PubMed] [Google Scholar]
- 168.Chien KR, Karsenty G. Longevity and lineages: toward the integrative biology of degenerative diseases in heart, muscle, and bone. Cell. 2005;120:533–544. doi: 10.1016/j.cell.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 169.Sato S, Hanada R, Kimura A, Abe T, Matsumoto T, Iwasaki M, Inose H, Ida T, Mieda M, Takeuchi Y, Fukumoto S, Fujita T, Kato S, Kangawa K, Kojima M, Shinomiya K, Takeda S. Central control of bone remodeling by neuromedin U. Nat Med. 2007;13:1234–1240. doi: 10.1038/nm1640. [DOI] [PubMed] [Google Scholar]
- 170.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 171.Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, Frenette PS. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 172.Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, Clement K, Vaisse C, Karsenty G. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434(7032):514–520. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- 173.Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, Muir C, Sanker S, Moriarty A, Moore KJ, Smutko JS, Mays GG, Wool EA, Monroe CA, Tepper RI. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 174.Wang JC, Shah N, Pantoja C, Meijsing SH, Ho JD, Scanlan TS, Yamamoto KR. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tatsumi S, Ishii K, Amizuka N, Li M, Kobayashi T, Kohno K, Ito M, Takeshita S, Ikeda K. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007;5:464–475. doi: 10.1016/j.cmet.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 176.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 177.Kanazawa K, Azuma Y, Nakano H, Kudo A. TRAF5 functions in both R. J Bone Miner Res. 2003;18:443–450. doi: 10.1359/jbmr.2003.18.3.443. [DOI] [PubMed] [Google Scholar]
- 178.Kanazawa K, Kudo A. TRAF2 is essential for TNF-alpha-induced osteoclastogenesis. J Bone Miner Res. 2005;20:840–847. doi: 10.1359/JBMR.041225. [DOI] [PubMed] [Google Scholar]
- 179.Choi SJ, Cruz JC, Craig F, Chung H, Devlin RD, Roodman GD, Alsina M. Macrophage inflammatory protein 1-alpha is a potential osteoclast stimulatory factor in multiple myeloma. Blood. 2000;96(2):671–675. [PubMed] [Google Scholar]
- 180.Han JH, Choi SJ, Kurihara N, Koide M, Oba Y, Roodman GD. Macrophage inflammatory protein-1alpha is an osteoclastogenic factor in myeloma that is independent of receptor activator of nuclear factor kappaB ligand. Blood. 2001;97:3349–3353. doi: 10.1182/blood.v97.11.3349. [DOI] [PubMed] [Google Scholar]
- 181.Sherr CJ, Rettenmier CW, Roussel MF. Macrophage colony-stimulating factor, CSF-1, and its protooncogene-encoded receptor. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):521–530. doi: 10.1101/sqb.1988.053.01.060. [DOI] [PubMed] [Google Scholar]
- 182.Tanaka S, Takahashi N, Udagawa N, Tamura T, Akatsu T, Stanley ER, Kurokawa T, Suda T. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J Clin Invest. 1993;91:257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 184.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Jr, hmed-Ansari A, Sell KW, Pollard JW, Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci U S A. 1990;87:4828–4832. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kodama H, Yamasaki A, Abe M, Niida S, Hakeda Y, Kawashima H. Transient recruitment of osteoclasts and expression of their function in osteopetrotic (op/op) mice by a single injection of macrophage colony-stimulating factor. J Bone Miner Res. 1993;8:45–50. doi: 10.1002/jbmr.5650080107. [DOI] [PubMed] [Google Scholar]
- 186.Lee TH, Fevold KL, Muguruma Y, Lottsfeldt JL, Lee MY. Relative roles of osteoclast colony-stimulating factor and macrophage colony-stimulating factor in the course of osteoclast development. Exp Hematol. 1994;22:66–73. [PubMed] [Google Scholar]
- 187.Fan X, Biskobing DM, Fan D, Hofstetter W, Rubin J. Macrophage colony stimulating factor down-regulates MCSF-receptor expression and entry of progenitors into the osteoclast lineage. J Bone Miner Res. 1997;12:1387–1395. doi: 10.1359/jbmr.1997.12.9.1387. [DOI] [PubMed] [Google Scholar]
- 188.Cappellen D, Luong-Nguyen NH, Bongiovanni S, Grenet O, Wanke C, Susa M. Transcriptional program of mouse osteoclast differentiation governed by the macrophage colony-stimulating factor and the ligand for the receptor activator of NFkappa B. J Biol Chem. 2002;277(24):21971–21982. doi: 10.1074/jbc.M200434200. [DOI] [PubMed] [Google Scholar]
- 189.Woo KM, Kim HM, Ko JS. Macrophage colony-stimulating factor promotes the survival of osteoclast precursors by up-regulating Bcl-X(L) Exp Mol Med. 2002;34:340–346. doi: 10.1038/emm.2002.48. [DOI] [PubMed] [Google Scholar]
- 190.Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, Fukuda A, Hikita A, Seto H, Okada T, Inaba T, Sanjay A, Baron R, Kawaguchi H, Oda H, Nakamura K, Strasser A, Tanaka S. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22(24):6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De ST, Daro E, Smith J, Tometsko ME, Maliszewski CR, Armstrong A, Shen V, Bain S, Cosman D, Anderson D, Morrissey PJ, Peschon JJ, Schuh J. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13(18):2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sobacchi C, Frattini A, Guerrini MM, Abinun M, Pangrazio A, Susani L, Bredius R, Mancini G, Cant A, Bishop N, Grabowski P, Del FA, Messina C, Errigo G, Coxon FP, Scott DI, Teti A, Rogers MJ, Vezzoni P, Villa A, Helfrich MH. Osteoclast-poor human osteopetrosis due to mutations in the gene encoding RANKL. Nat Genet. 2007;39:960–962. doi: 10.1038/ng2076. [DOI] [PubMed] [Google Scholar]
- 193.Pettit AR, Walsh NC, Manning C, Goldring SR, Gravallese EM. Rheumatology (Oxford) 9. Vol. 45. 2006. RANKL protein is expressed at the pannus-bone interface at sites of articular bone erosion in rheumatoid arthritis; pp. 1068–1076. [DOI] [PubMed] [Google Scholar]
- 194.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 195.Horne WC, Sanjay A, Bruzzaniti A, Baron R. The role(s) of Src kinase and Cbl proteins in the regulation of osteoclast differentiation and function. Immunol Rev. 2005;208:106–125. doi: 10.1111/j.0105-2896.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 196.Bruzzaniti A, Neff L, Sanjay A, Horne WC, De CP, Baron R. Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol Biol Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne WC, Baron R. Src kinase activity is essential for osteoclast function. J Biol Chem. 2004;279(17):17660–17666. doi: 10.1074/jbc.M311032200. [DOI] [PubMed] [Google Scholar]
- 198.Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy JB, Baron R. c-Cbl is downstream of c-Src in a signalling pathway necessary for bone resorption. Nature. 1996;383(6600):528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- 199.Destaing O, Sanjay A, Itzstein C, Horne WC, Toomre D, De CP, Baron R. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol Biol Cell. 2008;19:394–404. doi: 10.1091/mbc.E07-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Boyce BF, Yoneda T, Lowe C, Soriano P, Mundy GR. Requirement of pp60c–src expression for osteoclasts to form ruffled borders and resorb bone in mice. J Clin Invest. 1992;90:1622–1627. doi: 10.1172/JCI116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 202.Miyazaki T, Tanaka S, Sanjay A, Baron R. The role of c-Src kinase in the regulation of osteoclast function. Mod Rheumatol. 2006;16:68–74. doi: 10.1007/s10165-006-0460-z. [DOI] [PubMed] [Google Scholar]
- 203.Wada T, Nakashima T, Oliveira-dos-Santos AJ, Gasser J, Hara H, Schett G, Penninger JM. The molecular scaffold Gab2 is a crucial component of RANK signaling and osteoclastogenesis. Nat Med. 2005;11:394–399. doi: 10.1038/nm1203. [DOI] [PubMed] [Google Scholar]
- 204.Takeshita S, Namba N, Zhao JJ, Jiang Y, Genant HK, Silva MJ, Brodt MD, Helgason CD, Kalesnikoff J, Rauh MJ, Humphries RK, Krystal G, Teitelbaum SL, Ross FP. SHIP-deficient mice are severely osteoporotic due to increased numbers of hyper-resorptive osteoclasts. Nat Med. 2002;8:943–949. doi: 10.1038/nm752. [DOI] [PubMed] [Google Scholar]
- 205.Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, Wood P, Rabia SH, Headon DJ, Overbeek PA, Le DF, Holland SM, Belani K, Kumararatne DS, Fischer A, Shapiro R, Conley ME, Reimund E, Kalhoff H, Abinun M, Munnich A, Israel A, Courtois G, Casanova JL. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 206.Lee ZH, Lee SE, Kim CW, Lee SH, Kim SW, Kwack K, Walsh K, Kim HH. IL-1alpha stimulation of osteoclast survival through the PI 3- kinase/Akt and ERK pathways. J Biochem. 2002;131(1):161–166. doi: 10.1093/oxfordjournals.jbchem.a003071. [DOI] [PubMed] [Google Scholar]
- 207.Li X, Udagawa N, Itoh K, Suda K, Murase Y, Nishihara T, Suda T, Takahashi N. p38 MAPK-mediated signals are required for inducing osteoclast differentiation but not for osteoclast function. Endocrinology. 2002;143(8):3105–3113. doi: 10.1210/endo.143.8.8954. [DOI] [PubMed] [Google Scholar]
- 208.Matsumoto M, Sudo T, Saito T, Osada H, Tsujimoto M. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL) J Biol Chem. 2000;275(40):31155–31161. doi: 10.1074/jbc.M001229200. [DOI] [PubMed] [Google Scholar]
- 209.Kwak HB, Lee SW, Jin HM, Ha H, Lee SH, Takeshita S, Tanaka S, Kim HM, Kim HH, Lee ZH. Monokine induced by interferon-gamma is induced by receptor activator of nuclear factor kappa B ligand and is involved in osteoclast adhesion and migration. Blood. 2005;105:2963–2969. doi: 10.1182/blood-2004-07-2534. [DOI] [PubMed] [Google Scholar]
- 210.David JP, Sabapathy K, Hoffmann O, Idarraga MH, Wagner EF. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J Cell Sci. 2002;115(Pt 22):4317–4325. doi: 10.1242/jcs.00082. [DOI] [PubMed] [Google Scholar]
- 211.Yamamoto A, Miyazaki T, Kadono Y, Takayanagi H, Miura T, Nishina H, Katada T, Wakabayashi K, Oda H, Nakamura K, Tanaka S. Possible involvement of IkappaB kinase 2 and MKK7 in osteoclastogenesis induced by receptor activator of nuclear factor kappaB ligand. J Bone Miner Res. 2002;17:612–621. doi: 10.1359/jbmr.2002.17.4.612. [DOI] [PubMed] [Google Scholar]
- 212.Yao Z, Xing L, Qin C, Schwarz EM, Boyce BF. Osteoclast precursor interaction with bone matrix induces osteoclast formation directly by an interleukin-1-mediated autocrine mechanism. J Biol Chem. 2008;283(15):9917–9924. doi: 10.1074/jbc.M706415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McHugh KP, Shen Z, Crotti TN, Flannery MR, Fajardo R, Bierbaum BE, Goldring SR. Role of cell-matrix interactions in osteoclast differentiation. Adv Exp Med Biol. 2007;602:107–111. doi: 10.1007/978-0-387-72009-8_14. [DOI] [PubMed] [Google Scholar]
- 214.Xing L, Carlson L, Story B, Tai Z, Keng P, Siebenlist U, Boyce BF. Expression of either NF-kappaB p50 or p52 in osteoclast precursors is required for IL-1-induced bone resorption. J Bone Miner Res. 2003;18:260–269. doi: 10.1359/jbmr.2003.18.2.260. [DOI] [PubMed] [Google Scholar]
- 215.Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T, Taniguchi T, Takayanagi H, Takai T. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428(6984):758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 216.Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, Majumdar S, Lanier LL, Lowell CA, Nakamura MC. The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci U S A. 2004;101(16):6158–6163. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, Saheki Y, Kawai T, Mukherjee PM, Reichenberger E, Olsen BR. Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice. Cell. 2007;128:71–83. doi: 10.1016/j.cell.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 218.Yang S, Chen W, Stashenko P, Li YP. Specificity of RGS10A as a key component in the RANKL signaling mechanism for osteoclast differentiation. J Cell Sci. 2007;120(Pt 19):3362–3371. doi: 10.1242/jcs.008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang S, Li YP. RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 2007;21(14):1803–1816. doi: 10.1101/gad.1544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yang S, Li YP. RGS12 is essential for RANKL-evoked signaling for terminal differentiation of osteoclasts in vitro. J Bone Miner Res. 2007;22:45–54. doi: 10.1359/jbmr.061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bromme D, Okamoto K. Human cathepsin O2, a novel cysteine protease highly expressed in osteoclastomas and ovary molecular cloning, sequencing and tissue distribution. Biol Chem Hoppe Seyler. 1995;376:379–384. doi: 10.1515/bchm3.1995.376.6.379. [DOI] [PubMed] [Google Scholar]
- 222.Inaoka T, Bilbe G, Ishibashi O, Tezuka K, Kumegawa M, Kokubo T. Molecular cloning of human cDNA for cathepsin K: novel cysteine proteinase predominantly expressed in bone. Biochem Biophys Res Commun. 1995;206:89–96. doi: 10.1006/bbrc.1995.1013. [DOI] [PubMed] [Google Scholar]
- 223.Shi GP, Chapman HA, Bhairi SM, DeLeeuw C, Reddy VY, Weiss SJ. Molecular cloning of human cathepsin O, a novel endoproteinase and homologue of rabbit OC2. FEBS Lett. 1995;357:129–134. doi: 10.1016/0014-5793(94)01349-6. [DOI] [PubMed] [Google Scholar]
- 224.Tezuka K, Tezuka Y, Maejima A, Sato T, Nemoto K, Kamioka H, Hakeda Y, Kumegawa M. Molecular cloning of a possible cysteine proteinase predominantly expressed in osteoclasts. J Biol Chem. 1994;269:1106–1109. [PubMed] [Google Scholar]
- 225.Singh AR, Kaur A, Anand NK, Singh JR. Pyknodysostosis: visceral manifestations and simian crease. Indian J Pediatr. 2004;71:453–455. doi: 10.1007/BF02725641. [DOI] [PubMed] [Google Scholar]
- 226.Li CY, Jepsen KJ, Majeska RJ, Zhang J, Ni R, Gelb BD, Schaffler MB. Mice lacking cathepsin K maintain bone remodeling but develop bone fragility despite high bone mass. J Bone Miner Res. 2006;21:865–875. doi: 10.1359/jbmr.060313. [DOI] [PubMed] [Google Scholar]
- 227.Chen W, Yang S, Abe Y, Li M, Wang Y, Shao J, Li E, Li YP. Novel pycnodysostosis mouse model uncovers cathepsin K function as a potential regulator of osteoclast apoptosis and senescence. Hum Mol Genet. 2007;16:410–423. doi: 10.1093/hmg/ddl474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, Ross FP, Swat W. Vav3 regulates osteoclast function and bone mass. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- 229.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 230.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 231.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 232.Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 233.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 234.Kern B, Shen J, Starbuck M, Karsenty G. Cbfa1 contributes to the osteoblast-specific expression of type I collagen genes. J Biol Chem. 2001;276:7101–7107. doi: 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- 235.Geoffroy V, Ducy P, Karsenty G. A PEBP2 alpha/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J Biol Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 236.Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, Stein J, Stein GS. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 237.Banerjee C, McCabe LR, Choi JY, Hiebert SW, Stein JL, Stein GS, Lian JB. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 238.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts [see comments] Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 239.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [see comments] [DOI] [PubMed] [Google Scholar]
- 241.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [see comments] [DOI] [PubMed] [Google Scholar]
- 242.Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, Kurisu K, Komori T. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol. 2001;153:87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of preosteo-blasts. Cancer Res. 2003;63(17):5357–5362. [PubMed] [Google Scholar]
- 245.Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kanatani N, Fujita T, Fukuyama R, Liu W, Yoshida CA, Moriishi T, Yamana K, Miyazaki T, Toyosawa S, Komori T. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296:48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 248.Ma L, Golden S, Wu L, Maxson R. The molecular basis of Boston-type craniosynostosis: the Pro148— >His mutation in the N-terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum Mol Genet. 1996;5:1915–1920. doi: 10.1093/hmg/5.12.1915. [DOI] [PubMed] [Google Scholar]
- 249.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 250.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278(46):45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 251.Ichida F, Nishimura R, Hata K, Matsubara T, Ikeda F, Hisada K, Yatani H, Cao X, Komori T, Yamaguchi A, Yoneda T. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J Biol Chem. 2004;279(32):34015–34022. doi: 10.1074/jbc.M403621200. [DOI] [PubMed] [Google Scholar]
- 252.Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126(24):5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 253.Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, Wozney JM, Chi XZ, Bae SC, Choi KY, Cho JY, Choi JY, Ryoo HM. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem. 2005;280(42):35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- 254.Newberry EP, Latifi T, Towler DA. Reciprocal regulation of osteocalcin transcription by the homeodomain proteins Msx2 and Dlx5. Biochemistry. 1998;37(46):16360–16368. doi: 10.1021/bi981878u. [DOI] [PubMed] [Google Scholar]
- 255.Shirakabe K, Terasawa K, Miyama K, Shibuya H, Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 2001;6:851–856. doi: 10.1046/j.1365-2443.2001.00466.x. [DOI] [PubMed] [Google Scholar]
- 256.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 257.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 258.Kronenberg HM. Twist genes regulate Runx2 and bone formation. Dev Cell. 2004;6:317–318. doi: 10.1016/s1534-5807(04)00069-3. [DOI] [PubMed] [Google Scholar]
- 259.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de CB, Yeh J, Li B. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 262.Wu LC, Mak CH, Dear N, Boehm T, Foroni L, Rabbitts TH. Molecular cloning of a zinc finger protein which binds to the heptamer of the signal sequence for V(D)J recombination. Nucleic Acids Res. 1993;21(22):5067–5073. doi: 10.1093/nar/21.22.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Oukka M, Kim ST, Lugo G, Sun J, Wu LC, Glimcher LH. A mammalian homolog of Drosophila schnurri, KRC, regulates TNF receptor-driven responses and interacts with TRAF2. Mol Cell. 2002;9:121–131. doi: 10.1016/s1097-2765(01)00434-8. [DOI] [PubMed] [Google Scholar]
- 264.Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312(5777):1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 265.Shen R, Wang X, Drissi H, Liu F, O’Keefe RJ, Chen D. Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J Biol Chem. 2006;281(24):16347–16353. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kanzler B, Kuschert SJ, Liu YH, Mallo M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development. 1998;125(14):2587–2597. doi: 10.1242/dev.125.14.2587. [DOI] [PubMed] [Google Scholar]
- 268.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 269.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32:639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 270.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32:645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 271.Miyazono K, ten DP, Heldin CH. TGF-beta signaling by Smad proteins. Adv Immunol. 2000;75:115–157. doi: 10.1016/s0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- 272.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Pelletier N, Champagne N, Stifani S, Yang XJ. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene. 2002;21(17):2729–2740. doi: 10.1038/sj.onc.1205367. [DOI] [PubMed] [Google Scholar]
- 274.Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 275.Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol Cell Biol. 2003;23:1004–1013. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 277.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 278.Wang W, Wang YG, Reginato AM, Glotzer DJ, Fukai N, Plotkina S, Karsenty G, Olsen BR. Groucho homologue Grg5 interacts with the transcription factor Runx2-Cbfa1 and modulates its activity during postnatal growth in mice. Dev Biol. 2004;270:364–381. doi: 10.1016/j.ydbio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 279.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14(14):R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 280.Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279(40):41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 281.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 282.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22(22):7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 284.Cho HH, Park HT, Kim YJ, Bae YC, Suh KT, Jung JS. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J Cell Biochem. 2005;96:533–542. doi: 10.1002/jcb.20544. [DOI] [PubMed] [Google Scholar]
- 285.Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. J Bone Miner Res. 2005;20:2254–2263. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- 286.Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, Ryoo HM, Choi JY, Yoshida M, Nishino N, Oh BC, Lee KS, Lee YH, Bae SC. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem. 2006;281(24):16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 287.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445(7126):442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 288.Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113(Pt 12):2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 290.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tintut Y, Parhami F, Le V, Karsenty G, Demer LL. Inhibition of osteoblast-specific transcription factor Cbfa1 by the cAMP pathway in osteoblastic cells. Ubiquitin/proteasome-dependent regulation. J Biol Chem. 1999;274(41):28875–28879. doi: 10.1074/jbc.274.41.28875. [DOI] [PubMed] [Google Scholar]
- 292.Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem. 2003;278(30):27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- 293.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O’Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem. 2006;281:3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–4333. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem. 2004;279(13):12854–12859. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- 297.Hanai J, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, Stavnezer J, Kawabata M, Miyazono K, Ito Y. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem. 1999;274(44):31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 298.Nishimura R, Hata K, Harris SE, Ikeda F, Yoneda T. Core-binding factor alpha 1 (Cbfa1) induces osteoblastic differentiation of C2C12 cells without interactions with Smad1 and Smad5. Bone. 2002;31:303–312. doi: 10.1016/s8756-3282(02)00826-8. [DOI] [PubMed] [Google Scholar]
- 299.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, Ochi T, Kitamura Y, Ito Y, Nomura S. Transcrip-tional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene. 1998;17:1517–1525. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 300.Gutierrez S, Javed A, Tennant DK, van RM, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 301.Hata K, Nishimura R, Ueda M, Ikeda F, Matsubara T, Ichida F, Hisada K, Nokubi T, Yamaguchi A, Yoneda T. A CCAAT/enhancer binding protein beta isoform, liver-enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Mol Cell Biol. 2005;25:1971–1979. doi: 10.1128/MCB.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 303.Sowa H, Kaji H, Hendy GN, Canaff L, Komori T, Sugimoto T, Chihara K. Menin is required for bone morphogenetic protein 2- and transforming growth factor beta-regulated osteoblastic differentiation through interaction with Smads and Runx2. J Biol Chem. 2004;279(39):40267–40275. doi: 10.1074/jbc.M401312200. [DOI] [PubMed] [Google Scholar]
- 304.Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 305.Selvamurugan N, Chou WY, Pearman AT, Pulumati MR, Partridge NC. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J Biol Chem. 1998;273(17):10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- 306.Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med. 2000;6:985–990. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 307.Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283(13):8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24(20):9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kahler RA, Westendorf JJ. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem. 2003;278(14):11937–11944. doi: 10.1074/jbc.M211443200. [DOI] [PubMed] [Google Scholar]
- 310.Jeon MJ, Kim JA, Kwon SH, Kim SW, Park KS, Park SW, Kim SY, Shin CS. Activation of peroxisome proliferator-activated receptor-gamma inhibits the Runx2-mediated transcription of osteocalcin in osteoblasts. J Biol Chem. 2003;278(26):23270–23277. doi: 10.1074/jbc.M211610200. [DOI] [PubMed] [Google Scholar]
- 311.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20(9):2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zamurovic N, Cappellen D, Rohner D, Susa M. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279(36):37704–37715. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- 313.Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17(16):1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.McCarthy TL, Ji C, Chen Y, Kim KK, Imagawa M, Ito Y, Centrella M. Runt domain factor (Runx)-dependent effects on CCAAT/enhancer-binding protein delta expression and activity in osteoblasts. J Biol Chem. 2000;275(28):21746–21753. doi: 10.1074/jbc.M002291200. [DOI] [PubMed] [Google Scholar]
- 315.Milona MA, Gough JE, Edgar AJ. Expression of alternatively spliced isoforms of human Sp7 in osteoblast-like cells. BMC Genomics. 2003;4(1):43–54. doi: 10.1186/1471-2164-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Celil AB, Campbell PG. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem. 2005;280(36):31353–31359. doi: 10.1074/jbc.M503845200. [DOI] [PubMed] [Google Scholar]
- 317.Koga T, Matsui Y, Asagiri M, Kodama T, de CB, Nakashima K, Takayanagi H. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- 318.Deckelbaum RA, Majithia A, Booker T, Henderson JE, Loomis CA. The homeoprotein engrailed 1 has pleiotropic functions in calvarial intramembranous bone formation and remodeling. Development. 2006;133:63–74. doi: 10.1242/dev.02171. [DOI] [PubMed] [Google Scholar]
- 319.Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millan JL, MacGregor GR, Whyte MP. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Murshed M, Harmey D, Millan JL, McKee MD, Karsenty G. Unique coexpression in osteoblasts of broadly expressed genes accounts for the spatial restriction of ECM mineralization to bone. Genes Dev. 2005;19:1093–1104. doi: 10.1101/gad.1276205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 322.Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–451. doi: 10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279(45):47109–47114. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- 324.Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblastspecific osteocalcin gene expression. J Biol Chem. 2005;280(35):30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 325.Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol. 2001;13:698–705. doi: 10.1016/s0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- 326.Ellies DL, Krumlauf R. Bone formation: The nuclear matrix reloaded. Cell. 2006;125:840–842. doi: 10.1016/j.cell.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 327.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofa-cial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 328.Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- 329.Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17(24):3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, Markham AF, Fantes JA, Bonthron DT. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12(19):2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- 331.Li YP, Chen W, Stashenko P. Molecular cloning and characterization of a putative novel human osteoclast-specific 116-kDa vacuolar proton pump subunit. Biochem Biophys Res Commun. 1996;218:813–821. doi: 10.1006/bbrc.1996.0145. [DOI] [PubMed] [Google Scholar]
- 332.Li YP, Chen W. Characterization of mouse cathepsin K gene, the gene promoter, and the gene expression. J Bone Miner Res. 1999;14:487–499. doi: 10.1359/jbmr.1999.14.4.487. [DOI] [PubMed] [Google Scholar]
- 333.Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i–deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- 334.Klemsz MJ, McKercher SR, Celada A, Van BC, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 335.Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- 336.Orkin SH. Transcription factors and hematopoietic development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 337.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 338.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos protooncogene. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 339.Wang ZQ, Liang J, Schellander K, Wagner EF, Grigoriadis AE. c-fos-induced osteosarcoma formation in transgenic mice: cooperativity with c-jun and the role of endogenous c-fos. Cancer Res. 1995;55(24):6244–6251. [PubMed] [Google Scholar]
- 340.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 341.Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279(25):26475–26480. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- 342.Hershey CL, Fisher DE. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34:689–696. doi: 10.1016/j.bone.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 343.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 344.Henkel GW, McKercher SR, Yamamoto H, Anderson KL, Oshima RG, Maki RA. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- 345.Olson MC, Scott EW, Hack AA, Su GH, Tenen DG, Singh H, Simon MCPU. 1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity. 1995;3:703–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 346.Henkel GW, McKercher SR, Leenen PJ, Maki RA. Commitment to the monocytic lineage occurs in the absence of the transcription factor PU.1. Blood. 1999;93:2849–2858. [PubMed] [Google Scholar]
- 347.Singh H, DeKoter RP, Walsh JC. PU.1, a shared transcriptional regulator of lymphoid and myeloid cell fates. Cold Spring Harb Symp Quant Biol. 1999;64:13–20. doi: 10.1101/sqb.1999.64.13. [DOI] [PubMed] [Google Scholar]
- 348.Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature. 1997;386:81–84. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- 349.Kwon OH, Lee CK, Lee YI, Paik SG, Lee HJ. The hematopoietic transcription factor PU.1 regulates RANK gene expression in myeloid progenitors. Biochem Biophys Res Commun. 2005;335:437–446. doi: 10.1016/j.bbrc.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 350.Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S, Hisatake K, Nogi Y. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem. 2004;279(44):45969–45979. doi: 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- 351.Partington GA, Fuller K, Chambers TJ, Pondel M. Mitf-PU.1 interactions with the tartrate-resistant acid phosphatase gene promoter during osteoclast differentiation. Bone. 2004;34:237–245. doi: 10.1016/j.bone.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 352.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 353.Matsuo K, Ray N. Osteoclasts, mononuclear phagocytes, and c-Fos: new insight into osteoimmunology. Keio J Med. 2004;53:78–84. doi: 10.2302/kjm.53.78. [DOI] [PubMed] [Google Scholar]
- 354.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 355.Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- 356.Sato K, Suematsu A, Nakashima T, Takemoto-Kimura S, Aoki K, Morishita Y, Asahara H, Ohya K, Yamaguchi A, Takai T, Kodama T, Chatila TA, Bito H, Takayanagi H. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med. 2006;12:1410–1416. doi: 10.1038/nm1515. [DOI] [PubMed] [Google Scholar]
- 357.Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- 358.Ikeda F, Nishimura R, Matsubara T, Tanaka S, Inoue J, Reddy SV, Hata K, Yamashita K, Hiraga T, Watanabe T, Kukita T, Yoshioka K, Rao A, Yoneda T. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J Clin Invest. 2004;114:475–484. doi: 10.1172/JCI19657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kim HH, Lee DE, Shin JN, Lee YS, Jeon YM, Chung CH, Ni J, Kwon BS, Lee ZH. Receptor activator of NF-kappaB recruits multiple TRAF family adaptors and activates c-Jun N-terminal kinase. FEBS Lett. 1999;443:297–302. doi: 10.1016/s0014-5793(98)01731-1. [DOI] [PubMed] [Google Scholar]
- 360.Teitelbaum SL. RANKing c-Jun in osteoclast development. J Clin Invest. 2004;114(4):463–465. doi: 10.1172/JCI22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Jimi E, Ghosh S. Role of nuclear factor-kappaB in the immune system and bone. Immunol Rev. 2005;208:80–87. doi: 10.1111/j.0105-2896.2005.00329.x. [DOI] [PubMed] [Google Scholar]
- 362.Thanos D, Maniatis T. NF-kappa B: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 363.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 364.Xing L, Bushnell TP, Carlson L, Tai Z, Tondravi M, Siebenlist U, Young F, Boyce BF. NF-kappaB p50 and p52 expression is not required for RANK-expressing osteoclast progenitor formation but is essential for. J Bone Miner Res. 2002;17:1200–1210. doi: 10.1359/jbmr.2002.17.7.1200. [DOI] [PubMed] [Google Scholar]
- 365.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109 Suppl:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 366.Day CJ, Kim MS, Lopez CM, Nicholson GC, Morrison NA. NFAT expression in human osteoclasts. J Cell Biochem. 2005;95:17–23. doi: 10.1002/jcb.20410. [DOI] [PubMed] [Google Scholar]
- 367.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, Takayanagi H. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202:1261–1269. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Takayanagi H. Mechanistic insight into osteoclast differentiation in osteoimmunology. J Mol Med. 2005;83:170–179. doi: 10.1007/s00109-004-0612-6. [DOI] [PubMed] [Google Scholar]
- 369.Gohda J, Akiyama T, Koga T, Takayanagi H, Tanaka S, Inoue J. RANK-mediated amplification of TRAF6 signaling leads to NFATc1 induction during osteoclastogenesis. EMBO J. 2005;24:790–799. doi: 10.1038/sj.emboj.7600564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Crotti TN, Flannery M, Walsh NC, Fleming JD, Goldring SR, McHugh KP. NFATc1 directly induces the human beta3 integrin gene in osteoclast differentiation. J Musculoskelet Neuronal Interact. 2005;5:335–337. [PMC free article] [PubMed] [Google Scholar]
- 371.Kim K, Kim JH, Lee J, Jin HM, Lee SH, Fisher DE, Kook H, Kim KK, Choi Y, Kim N. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem. 2005;280(42):35209–35216. doi: 10.1074/jbc.M505815200. [DOI] [PubMed] [Google Scholar]
- 372.Kim Y, Sato K, Asagiri M, Morita I, Soma K, Takayanagi H. Contribution of nuclear factor of activated T cells c1 to the transcriptional control of immunoreceptor osteoclast-associated receptor but not triggering receptor expressed by myeloid cells-2 during osteoclastogenesis. J Biol Chem. 2005;280(38):32905–32913. doi: 10.1074/jbc.M505820200. [DOI] [PubMed] [Google Scholar]
- 373.McHugh KP, Hodivala-Dilke K, Zheng MH, Namba N, Lam J, Novack D, Feng X, Ross FP, Hynes RO, Teitelbaum SL. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105:433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Glowacki J, Cox KA, Wilcon S. Impaired osteoclast differentiation in subcutaneous implants of bone particles in osteopetrotic mutants. Bone Miner. 1989;5:271–278. doi: 10.1016/0169-6009(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 375.Green PM, Marshall MJ, Nisbet NW. A study of osteoclasts on calvaria of normal and osteopetrotic (mi/mi) mice by vital staining with acridine orange. Br J Exp Pathol. 1986;67:85–93. [PMC free article] [PubMed] [Google Scholar]
- 376.Holtrop ME, Cox KA, Eilon G, Simmons HA, Raisz LG. The ultrastructure of osteoclasts in microphthalmic mice. Metab Bone Dis Relat Res. 1981;3:123–129. doi: 10.1016/0221-8747(81)90030-8. [DOI] [PubMed] [Google Scholar]
- 377.Thesingh CW, Scherft JP. Fusion disability of embryonic osteoclast precursor cells and macrophages in the microphthalmic osteopetrotic mouse. Bone. 1985;6:43–52. doi: 10.1016/8756-3282(85)90406-5. [DOI] [PubMed] [Google Scholar]
- 378.Franzoso G, Carlson L, Xing L, Poljak L, Shores EW, Brown KD, Leonardi A, Tran T, Boyce BF, Siebenlist U. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11(24):3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.So H, Rho J, Jeong D, Park R, Fisher DE, Ostrowski MC, Choi Y, Kim N. Microphthalmia transcription factor and PU.1 synergistically induce the leukocyte receptor osteoclast-associated receptor gene expression. J Biol Chem. 2003;278(26):24209–24216. doi: 10.1074/jbc.M302940200. [DOI] [PubMed] [Google Scholar]
- 380.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J Biol Chem. 2002;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- 381.Weilbaecher KN, Motyckova G, Huber WE, Takemoto CM, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol Cell. 2001;8:749–758. doi: 10.1016/s1097-2765(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 382.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 383.Battaglino R, Kim D, Fu J, Vaage B, Fu XY, Stashenko P. c-myc is required for osteoclast differentiation. J Bone Miner Res. 2002;17:763–773. doi: 10.1359/jbmr.2002.17.5.763. [DOI] [PubMed] [Google Scholar]
- 384.Daumer KM, Taparowsky EJ, Hall DJ, Steinbeck MJ. Transcription from the tartrate-resistant acid phosphatase promoter is negatively regulated by the Myc oncoprotein. J Bone Miner Res. 2002;17:1701–1709. doi: 10.1359/jbmr.2002.17.9.1701. [DOI] [PubMed] [Google Scholar]
- 385.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 386.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Martin TJ. A skeleton key to metabolism. Nat Med. 2007;13:1021–1023. doi: 10.1038/nm0907-1021. [DOI] [PubMed] [Google Scholar]
- 388.Tsuneto M, Tominaga A, Yamazaki H, Yoshino M, Orkin SH, Hayashi S. Enforced expression of PU.1 rescues osteoclastogenesis from embryonic stem cells lacking Tal-1. Stem Cells. 2005;23:134–143. doi: 10.1634/stemcells.2004-0154. [DOI] [PubMed] [Google Scholar]
- 389.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 390.Boyce BF, Xing L. Osteoclasts, no longer osteoblast slaves. Nat Med. 2006;12:1356–1358. doi: 10.1038/nm1206-1356. [DOI] [PubMed] [Google Scholar]