Abstract
Cancers are genetic diseases, driven by somatic mutations that perturb cellular signaling systems. In this study, we aim to reveal the signal transduction pathways that are perturbed by mutations in ovarian cancer. Our approach searches for genetic mutations that lead to a common cellular response, e.g., differential expression of a set of functional related genes. To this end, we first developed a knowledge mining approach to identify functional expression modules; we then developed a graph-based data mining approach to identify mutations that are highly related to the functional modules, as a means to re-constitute signal pathways. Our results indicate that unification of knowledge mining with data mining significantly enhance identification of potential signaling pathways in ovarian cancers.
1. Introduction
Cancers are caused by somatic mutations that lead to hallmark changes in cellular signaling systems1, e.g., sustaining proliferative signaling, evading growth suppressors, activating invasion and metastasis, enabling replicative immortality, inducing angiogenesis, and resisting cell death. Large-scale efforts have been devoted to identify somatic and germ-line mutations from a large number of tumor samples, including the Cancer Genome Atlas (TCGA) project and the international network of cancer genomic projects 2–4. A major thrust in cancer genomic research is to identify driving mutations and reconstruct perturbed signaling pathways that underlie the hallmark behaviors. This body of information will shed light on the disease mechanisms of cancers, reveal novel drug targets, and more importantly guide patient treatment based on personal genetic information.
It is not uncommon that cancer cells accumulate a large number of mutations during development; some are cancer-causing (driving mutations) while others have no relation to cancers (passenger mutations)5,6. It is challenging to delineate a pathway based on the mutations because mutations of genes on the same pathway usually are mutually exclusive in a tumor6,7 and thus one has to reconstruct such a pathway by compiling the mutations across multiple tumor samples. This in turn requires one to identify the tumor samples in which a common pathway is perturbed. Furthermore, the nature, that multiple aberrant signaling pathways underlie each tumor, makes more challenge to the task of de-convoluting large-scale cancer genomic data solely based on genetic mutation7.
In this study, we address the task of revealing perturbed signaling pathways by integrating genomic mutation data with functional genomic data, i.e., gene expression data. The main idea underlying our approach is to use differential expression gene modules as the readouts of signaling pathway perturbations, which enable us to reconstruct a signaling pathway by finding the mutations that are strongly associated with a gene expression module. We developed a framework that unifies ontology-guided knowledge mining and graph-based data mining to achieve the goal. Our methods were able to discover perturbation of many well-known cancer signaling pathways, and we conjecture that some of our results may help to discover novel pathways in cancers. In the paper, we applied our methods in the ovarian cancer data from TCGA as a test, and we believe this general technology can also be used to study other types of cancer data.
2. Methods
2.1. Data sets and preprocessing
TCGA data
The gene expression data and somatic mutation data of ovarian serous (OV) were collected from the TCGA project website. These data were used to identify functionally coherent gene expression modules and the somatic mutations that are strongly associated with them. In the TCGA data, gene expressions of tumor and normal cells have been measured using two different platforms – Affymetrix and Agilent.
In this study, we identified differentially expressed genes in each tumor sample so that we could study the association between somatic mutations and gene expression modules at an individual sample level. To this end, we first pooled the expression data from 8 normal control samples collected by the TCGA and determined the average expression value of each gene. Then for each tumor sample, we identified differential expressed genes by calculating the fold change between the sample and the average expression value of the gene in the control group. If a gene demonstrated an over 3-fold change (up or down) in both Affymetrix and Agilent platforms, it was deemed as differentially expressed. We divided genes into up-regulated and down-regulated groups for further analysis.
We labeled a gene as being perturbed if somatic mutation analysis by the TCGA reported a missense, deletion, insertion or frame-shift mutation of the gene. Due to the large volume of data, we did not perform further function analysis to determine if mutations resulted in a loss or gain of function.
The Gene Ontology
We collected the Gene Ontology (GO) annotation of genes/proteins from the GO Consortium8. The relationships between the concepts (GO terms) were programmatically represented as a directed acyclic graph using a software package referred to as GOGrapher9.
2.2. Identifying functionally coherent gene expression modules
Given a set of genes and their functional annotations in the form of GO terms, we grouped the genes into non-disjoint, functionally coherent modules so that the function of each module was represented by a GO term that summarized the function represented by the original GO annotations. Our framework is to utilize the directed acyclic graph (DAG) structure of the GO and the IS_A semantic relationship among GO terms to find an informative GO term that captures/retains as much as possible the semantic information of the original annotations of a module of genes. For each tumor sample, we represented the differentially expressed genes and their annotations using a graph referred to as GO-Gene graph9, in which GO terms were organized based on the definition of the ontology and genes were attached to their annotation terms. We assessed if the genes annotated by a given term were significantly enriched among the differential expressed genes, based on a p-value calculated according to a hypergeometric distribution. If it is not significant, we trimmed the GO term from the graph and merged its associated genes to the ancestor term that would result in the least semantic information loss10. It should be noted that the edges in the original GO structure have no weights. Using the information-theory-based approach10, to assign weights for edges in the GO structure, we were able to selectively merge the collapsed term to its closest parent noted. We stopped merging until a GO term was deemed as significant and designated the genes annotated by such a GO term as a coherent expression module. Given a differentially expressed gene list, our algorithm would return a collection of disjoint, coherent modules or a empty collection if no significantly enriched GO terms were identified. We further produced a large collection of gene modules by unioning the modules annotated by a common GO term across multiple tumor samples.
2.3. Finding tumors with a common perturbed signaling pathway
In this study, we aimed to identify the tumors that shared a common differentially expressed module, which served as the manifestation that these tumors share a common perturbed signaling pathway. We organized the data into a bipartite graph in which nodes on one side were the genes of a functional module, while the nodes on the other side were tumors; an edge between a tumor and a gene indicated that the gene was differentially expressed in the tumor. This approach not only ensured that the gene modules were functionally coherent, but also significantly reduced the complexity of the search of highly connected subgraph in the bipartite graph. We formulated the computational task as follows: find a sub-graph of k genes and l tumors such that a) the size of the subgraph (kl) was maximized, b) each gene was connected to at least rl tumors and each tumor was connected to at least rk genes in the sub-graph. The task of finding such a subgraph is NP-hard and we designed a greedy algorithm to solve it. In the application, we let r to be 0.75, k>10 and l>4, where variable r (0 < r ≤ 1) can be thought of as a noise-tolerating parameter, which allows the algorithm to include tumors and genes into a module even when they are not fully connected to their counterparts in the bipartite graph. Due to the space limit, the algorithm is not presented.
2.4. Identifying mutation-prone networks
Given a cluster of tumor samples associated with a functional module, we collected all the mutated genes in these samples. In order to reveal the connections among proteins encoded by the mutated genes, we constructed a graph based on the global PPI network obtained from the BioGrid11 database, in which nodes were proteins and an edge indicated physical interaction between a pair of proteins. We further added weights to the proteins, which equaled the number of tumors in which the gene was muted and referred to such a network as PPI-mutation network. A subnetwork consisting heavily mutated genes was extracted as follows. We first applied the k-path algorithm developed by Kelley et al12, to search for the heaviest paths of length k within a graph. We further constrained that a path should start with a mutated gene and end with a TF protein, so that the subgraph would contain TFs that were strongly connected to the mutated genes, thus likely to be part of the signaling pathway regulating gene expression. We then reconstructed a mutation-prone subnetwork by joining the top 200 paths.
3. Results and Discussion
The overarching goal of this study was to delineate signaling pathways that underlie the disease mechanism and different subtypes of ovarian cancers. Our strategy was to use functional genomic data as readouts of signaling pathway perturbations, identify tumors sharing a common perturbation, and then reconstruct protein networks and constitute the perturbed signaling pathway.
3.1. Identifying tumors sharing a common signaling perturbation
The fundamental assumption underlying our approach is that genetic perturbations along a signaling pathway often leads to differential expression of a set module of genes that perform coherently related functions—differential expression of function modules. This allows us to transform the task of searching for tumors that share a common perturbation in signaling systems into searching for shared expression component.
We represented genetic mutation and expression data as a bipartite graph, see Methods. Then the task was to find a cluster of tumors that are densely connected to a cluster of differentially expressed genes, a task also referred to as biclustering problem13,14, which was NP-hard and one of state-of-the-art algorithm, referred to as SAMBA, was developed by Tanay et al14. However the challenge confronting us was that none of the contemporary bioclustering algorithms explicitly pursue functional coherence of genes during module searching, hence modules identified by those approaches usually contained diverse, functionally unrelated genes. To address this issue, we developed an ontology-based knowledge mining approach to first identify functional modules among the differentially expressed genes and then searched for the tumors that were highly connected to a given module, see Methods for details. After applying our methods to the TCGA ovarian cancer data, we identified 178 function modules (70 up-regulated and 108 down-regulated) and their corresponding tumor clusters. We also applied the SAMBA algorithm to the same data set, which identified a total of 443 gene modules (196 up-regulated and 247 down-regulated) together with their corresponding tumor clusters. We evaluated and compared the functional coherence of gene modules returned by the two clustering methods in the following aspects.
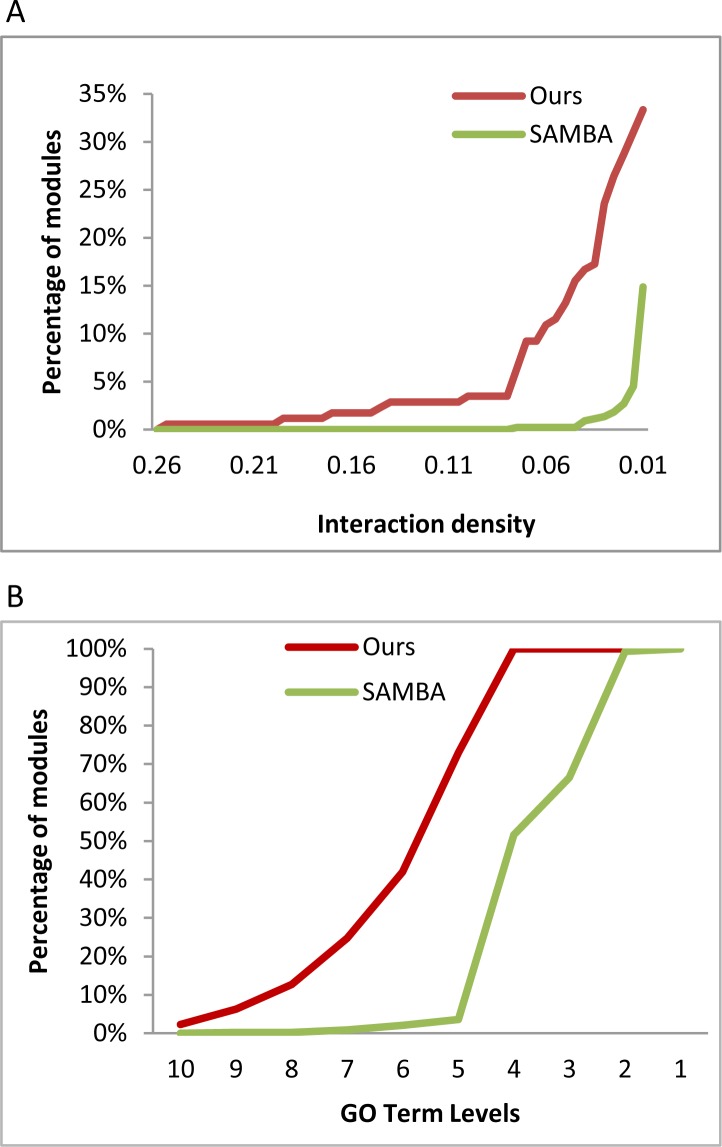
First, we identified all intro-module PPIs and assessed the interaction density by calculating the ratio of the number of actual interactions within module PPIs over the number of all possible pairwise interactions within a module. The results are shown in Figure 1A, which shows the cumulative percent of modules that have a given interaction density or better. The figure shows that the modules by our method have higher intro-module physical interactions. Hence the results indicate that the member proteins of our modules are more likely to perform coherently related functions.
Figure 1.
Comparison of functional coherence. A. The cumulative function of intro-module PPI. B. The cumulative function of specificity of functions of gene modules.
Second, we inspected the specificity of the functional annotations of the gene modules returned by both methods. By nature of our algorithm, each module returned by our method is annotated by a GO term that summarizes the function of its member genes. For each module derived by the SAMBA, we tried to find a common ancestor GO term that covers at least over 50% genes in a module as the representative GO term. We noted that it often took the root of the Biological Process (BP) branch of the GO ontology to fully cover all the genes in the module by the SAMBA. Figure 1B shows the results of the specificities of the summary GO terms, measured as the number of steps away from the root of BP. The figure shows that the GO terms annotating our modules are significantly more specific than those from the SAMBA; in fact, almost all GO terms from our methods are 5 steps away from the root of the BP, while the majority of the GO terms annotating SAMBA modules are within 5 steps.
Taken together, we believe that our approach identifies functionally coherent gene expression modules, and as such the tumors that highly connected to these expression modules are more likely to share a common perturbed signaling pathway.
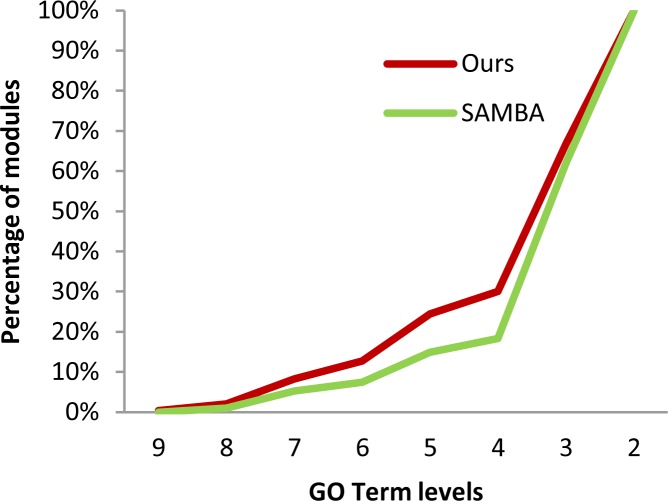
We then assessed if the mutated genes from the tumor clusters performed coherently related functions. We noted that the annotations for the mutated genes in tumor clusters tended to be diverse; therefore we compared the specificity of the top two most “enriched” GO terms between the two methods, and the results are shown in Figure 2. From the figure, we can see that functional annotations for the mutated genes in the tumor clusters by our methods are more specific than those derived by the SAMBA. A potential interpretation is that mutated genes retrieved by our method perform more coherently related functions.
Figure 2.
Function coherence of the mutated genes from tumor clusters
3.2. Identify perturbed signaling pathways
After retrieving the tumor samples that likely share a common signaling pathway perturbation—tumors share a common differentially expressed module, we further studied the mutated genes and their relationship to reveal a potential signaling pathway. We retrieved all the mutated genes from the tumor clusters, represented the union of the mutated genes on a PPI network11, and assigned a weight for each mutated gene, which was the number of tumors containing such a mutation. We refer to such a graph as PPI-mutation graph because it contains both the information regarding mutation and PPI. Then, we tried to identify a subgraph so that it contained the genes that were frequently mutated in the tumor cluster, plus the genes that provided connections among them. Here, we aimed to identify the subnetworks that contained as many highly mutated genes as possible while maintained relatively small size. This is a challenging computational task and the setup of commonly used prize-collecting Steiner tree15 does not fit our scenario because we do not know the Steiner nodes—which mutated genes should be included in the subnetwork. In this study, we developed a heuristic algorithm to identify a mutation-prone subgraph, see Methods section.
A mutation-prone subnetwork represents a potential signaling pathway, whose perturbation likely has contributed to the differential expression of a functional module. As a preliminary evaluation of the quality of the networks, we mapped the networks to well-known cancer signaling networks compiled by the NCI (http://pid.nci.nih.gov/index.shtml), by calculating the “enrichment” of the genes from an NCI network in our subnetworks. We found that all 178 mutation-prone subnetworks could be mapped to one or more NCI pathways, with p-values ranging from 1E-6 to 1E-16 and an average of 1E-12. The results indicate that indeed our networks contain significant amount of genes involved in cancers signaling. Our networks overlapped with 14 NCI pathways, a number significantly higher than the canonical pathways recently reported by the TCGA consortium on ovarian cancer3. The detailed evaluation of the mutation-prone networks is underway.
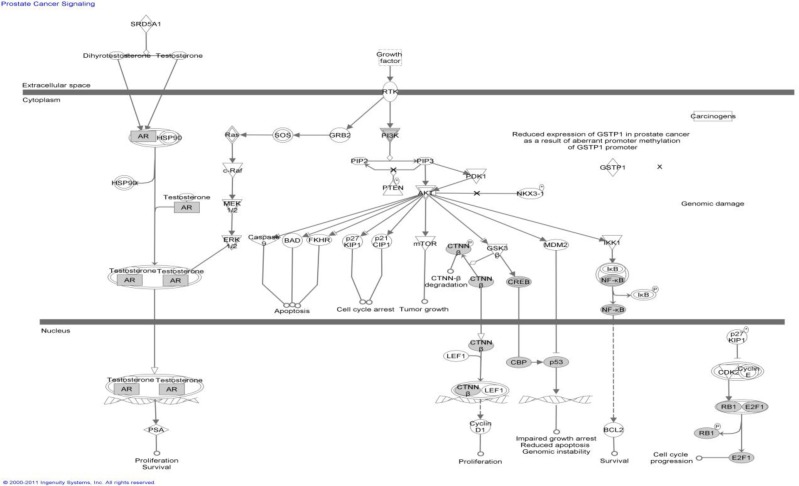
As an illustration, we searched the Ingenuity Pathway Database (http://www.ingenuity.com/) using the genes in the mutation-prone network corresponding to a module annotated with the term GO:0008285 (negative regulation of cell proliferation), which is down regulated in tumors. The result is shown in Figure 3. The results are biologically sensible in that, when a normal cell has undergone a DNA damage, the P53 pathway will be activated, which in turn induces genes that function to suppress cellular proliferation. Mutation in the pathway results in a loss of such suppression, often leading to cancers. The results here illustrate the main motivation of this study—revealing the signal pathway perturbations that lead to differentially expressed gene modules, thus providing the hypothesis regarding the disease mechanism.
Figure 3.
An example pathway. The shown ingenuity pathway (Prostate Cancer Signaling Pathway) overlaps significantly with the mutation-prone subnetwork corresponding to the expression module annotated by GO:0008285. The overlapping genes are shown as shaded nodes.
A list of known cancer pathways, which are overlapped with our subnetworks heavily, from NCI is given below:
Direct p53 effectors
CD40/CD40L signaling
Regulation of nuclear SMAD2/3 signaling
ATR signaling pathway
BCR signaling pathway
Signaling events mediated by HDAC Class I
E2F transcription factor network
Notch-mediated HES/HEY network
BARD1 signaling events
Regulation of Telomerase
IL2-mediated signaling events
Regulation of retinoblastoma protein
Regulation of nuclear beta catenin signaling and target gene transcription
Glucocorticoid receptor regulatory network
4. Conclusion and future work
In this study, we developed a novel approach to integrate genome data with functional genome data from the TCGA data to reveal perturbed signaling pathways in cancers. The main thrust of this study was to unify knowledge mining with data mining, so that we can use the functional genomic data as a readout to delineate specific signaling pathway perturbation underlying each tumor. The capability of revealing which pathways are perturbed in each tumor will enable researchers to investigate the disease mechanisms of different subtype of ovarian cancers.
In this paper, we only considered the perturbation of somatic mutations to the signaling pathways and did not take into account of the disturbance of other factors, such as gene copy number variance, methylation, and SNP. In next step, we will include such information into our research and study how SNP and methylation affect the cancer cells.
Acknowledgments
This research is partially supported by the following NLM grants: 5R01LM010144 and 5R01LM009153.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010 Apr 15;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 6.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Research. 2011:1–12. doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller B, Richards AJ, Jin B, Lu X. GOGrapher: A Python library for GO graph representation and analysis. BMC Res Notes. 2009;2:122. doi: 10.1186/1756-0500-2-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin B, Lu X. Identifying informative subsets of the Gene Ontology with information bottleneck methods. Bioinformatics. 2010 Oct 1;26(19):2445–2451. doi: 10.1093/bioinformatics/btq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011 Jan;39:D698–704. doi: 10.1093/nar/gkq1116. (Database issue): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley BP, Sharan R, Karp RM, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madeira SC, Oliveira AL. Biclustering algorithms for biological data analysis: a survey. IEEE/ACM Trans Comput Biol Bioinform. 2004 Jan-Mar;1(1):24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- 14.Tanay A, Sharan R, Shamir R. Discovering statistically significant biclusters in gene expression data. Bioinformatics. 2002;18(Suppl 1):S136–144. doi: 10.1093/bioinformatics/18.suppl_1.s136. [DOI] [PubMed] [Google Scholar]