Abstract
To facilitate clinical research, clinical data needs to be stored in a machine processable and understandable way. Manual annotating clinical data is time consuming. Automatic approaches (e.g., Natural Language Processing systems) have been adopted to convert such data into structured formats; however, the quality of such automatically extracted data may not always be satisfying. In this paper, we propose Semantator, a semi-automatic tool for document annotation with Semantic Web ontologies. With a loaded free text document and an ontology, Semantator supports the creation/deletion of ontology instances for any document fragment, linking/disconnecting instances with the properties in the ontology, and also enables automatic annotation by connecting to the NCBO annotator and cTAKES. By representing annotations in Semantic Web standards, Semantator supports reasoning based upon the underlying semantics of the owl:disjointWith and owl:equivalentClass predicates. We present discussions based on user experiences of using Semantator.
1. Introduction
In recent years, computerized approaches have been widely adopted to conduct clinical research, particularly by using data that is organized in a machine processable and understandable way. Louit et al. [1] reviewed the current methods and problems for genomic medicine data integration. Bodenreide [2] described how Semantic Web ontologies [3] are being used for knowledge management, data integration, etc. in the biomedical field. Tenenbaum et. al. [4] proposed the Biomedical Resource Ontology (BRO) for semantic annotation and discovery of biomedical resources.
One prerequisite here is to have decent structured clinical data, i.e., converting originally free text based data into certain structured formats. Automatic approaches that are based upon Natural Language Processing (NLP) techniques have been well studied [5]. According to the reported performance of the clinical Text Analysis and Knowledge Extraction Systems (cTAKES) [6], one of the state-of-the-art NLP systems in the medical domain, it has been able to achieve a F-score of 82.4% for Named Entity Recognition (NER) [7] which is a fundamental task for transforming free text to structured data. However, clinical research usually requires more precise results. Fully automatic approaches for data extraction are preferred but they do not always give satisfying results; while it may not be realistic to only rely on manual annotation to construct structured data due to the large volume of clinical notes that are needed to process. Therefore, semi-automatic data extraction could be one choice where we automatically extract information from clinical narratives and then manual efforts are used to refine such automatic annotations. The results from this semi-automatic process could potentially serve as the training sets to help automatic systems to further improve their performance.
Another important problem is that what structured format we should use to store the data. In clinical research, a variety of data formats have been adopted, such as Comma-separated Values (CSV) [8], relational databases, eXtensible Markup Language (XML), etc. Jeronimo et al. [9] constructed a multimedia database tool containing cervical cancer related patient records. The data is stored in a relational database and can be exported as CSV files. The HL7 Clinical Document Architecture (CDA) [10] is an XML based markup standard intended to specify the encoding, structure and semantics of clinical documents for exchange. The advantage of these data formats is that they provide a structured way for data storage and thus provide the possibility for more convenient data management compared to free text based data representation. Compared to the existing formats, the Semantic Web [3] and its corresponding knowledge representation standards (e.g., the Resource Description Framework (RDF)2 and the Web Ontology Language (OWL)3) adds an expressive framework for semantics-enabled data representation and knowledge inference.
In the Semantic Web, an ontology is an explicit and formal specification of a conceptualization, formally describing a domain of discourse. It consists of a set of terms (classes) and their relationships (class hierarchies and predicates). RDF is a graph based data model for describing resources and their relationships. Two resources are connected via one or more predicates in the form of triple. A triple, < s, p, o >, consists of three parts: subject, predicate and object. The subject is an identifier (e.g., a URI) and the object can either be an identifier or a literal value, such as strings, numbers, dates, etc. One advantage of the Semantic Web is that it supports knowledge inferencing. For instance, two classes A and B are defined to be disjoint in an ontology and if an automatic system or a human annotator annotates a piece of free text with both classes, the system should be able to report a potential annotation error. For automatic systems, such formal representation of semantics could be helpful to improve their performance under specific scenarios [11].
In this paper, we propose Semantator4, a semi-automatic annotation system for annotating clinical narratives using semantic web ontologies. Here, semantic annotation is to annotate entities with ontology classes, such as instances and their relationships creation/deletion. Although Semantator is designed for annotating clinical documents, it can also be applied to documents in other domains. Semantator is developed as a plugin of Protégé5, a well known tool for building and interacting with Semantic Web ontologies. The current version of Semantator provides: 1) the basic manual annotation functionalities, including ontology instance creation/deletion, relationship creation/deletion, linking equivalent instances and exporting/reloading annotation; 2) automatic annotation by connecting to the NCBO annotator [12] and cTAKES [6]; 3) Semantic Web based reasoning by utilizing the underlying semantics of the owl:disjointWith and owl:equivalentClass predicates.
The rest of the paper is organized as following. Section 2 discusses related work. Section 3 and Section 4 introduce the manual and automatic annotation functionalities respectively. We present some discussions on Semantator based on our user studies in Section 5 and conclude in Section 6.
2. Related Work
Existing annotation systems can be generally categorized into pattern based and machine learning based systems. Pattern based systems, such as PANKOW [13] and Armadillo [14], find entities by discovering patterns that are either manually identified or semi-automatically induced with a set of initial manually tagged seed patterns. SemTag [15] and KIM [16] adopt a set of pre-defined rules to locate the information of interest. Differently, S-CREAM [17], MnM [18], GATE [19] and cTAKES [6] explore machine learning algorithms and natural language processing techniques to identify entities. BioNLP UIMA6 provides a framework for users to plugin tools for different components for document annotation. Although machine learning based systems do not need to have human identified rules, they require certain amount of training data that may not always be available for each domain such systems would be applied to.
As the emergence of Semantic Web, annotation systems based on Semantic Web techniques have been proposed. Semantic-document [20] and GoNTogle [21] support semantic annotation on documents with ontology classes. In addition to annotation, GoNTogle also supports searching within document annotation results. Compared to these two systems, our proposed Semantator further supports instance relationship creation and has basic reasoning capabilities. Knowtator [22] is a plugin for Protégé. It facilitates manual creation of annotation to user’s text. However, the biggest drawback of Knowtator is that it only works with Protégé-Frames7 and can only export its annotations in XML; while our system supports annotation with OWL ontologies, stores annotations in RDF, and therefore can leverage the semantic web reasoners directly. Textpresso [23] is a text mining tool to extract terms from academic articles of a given domain for ontology population and this populated ontology can then be used to enable searching.
3. Manual Annotation with Semantator
In this section, we introduce the manual annotation functionalities of Semantator, including instance creation/deletion, relationship creation/deletion, linking equivalent instances and exporting/reloading existing annotations. Accompanying with our descriptions, we will also illustrate these functionalities with figures to make them more clear. A user first can load a plain text document that contains the clinical notes to annotate by simply clicking File and then Open in the menu to choose the file. The contents of the chosen document will be displayed in Semantator.
Instance Creation
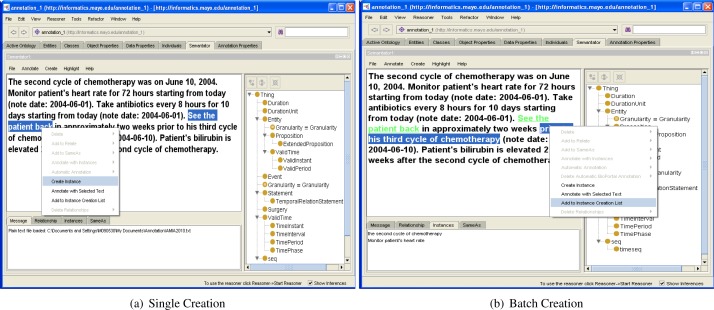
In Semantator, we provide two ways for a user to create instances: One-at-a-Time and Batch Creation. Figures 1(a) and 1(b) demonstrate the two alternatives respectively.
Figure 1:
Instance Creation
One-at-a-Time
For the first option, a user can create one instance each time. To create an instance, a user will need to highlight a piece of text in the loaded document and select a class from the loaded ontology in Protégé. The system allows the user to pick a color to be used for highlighting all instances of the selected class when this class is first used for annotation. In Figure 1(a), we are creating an instance of the Event class with the document fragment See the patient back; and Figure 1(b) shows that it is highlighted in green. This instance is added to the ontology with the triple: < event1 rdf : type Event > and is also associated with an rdfs:label8 by adding the triple: < event1 rdfs : label See the patient back >. Note that by default, Semantator stores the highlighted text using rdfs:label; but the user can choose to use any properties as needed.
Batch Creation
There may exist many instances that could actually be categorized of the same class in a document and creating each instance individually could be time-consuming. Therefore, we provide the second option to create instances, Batch Creation. A user can add different pieces of document fragments that describe instances for the same class into a list. When finishing selecting all document fragments to annotate, the user can then choose an ontology class and annotate all document fragments in the list to be instances of this chosen class. Note that here we assume all selected document fragments represent instances of the same class. Document fragments in the candidate list can be easily removed by right clicking on it and choosing to remove.
Instance Deletion
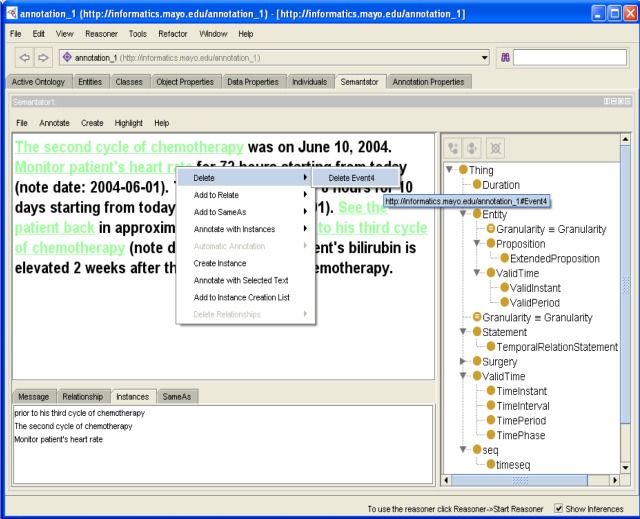
Semantator allows users to delete any previously created instances if desired. The same document fragment could have been annotated with ontology instances of different classes. For instance, the text See the patient back could be annotated to be an Event and an owl:Thing (the most general class in any ontology). When a user chooses to delete the instance(s) of a document fragment, the system detects all instances for which this fragment has been created. Then, the user can choose to delete any of these instances individually. Figure 2 demonstrates this deletion operation.
Figure 2:
Instance Deletion
Generating Instance Equivalences
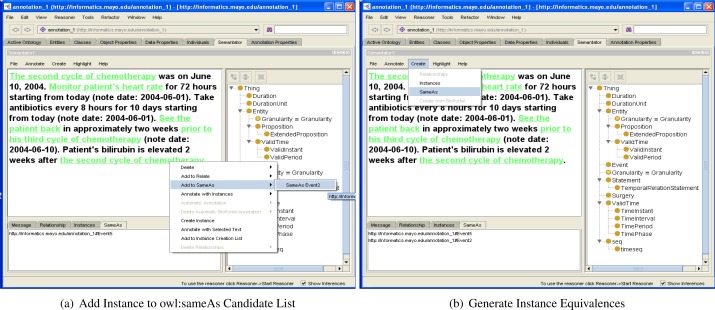
In a clinical document, it is possible that instances that occur at different places in the document could actually refer to the same real world entity9. For instance, in the following document, two instances (in bold) of the Event class have been created, and they actually represent the same event in real world.
The second cycle of chemotherapy was on June 10, 2004. Patient’s bilirubin is elevated 2 weeks after the second cycle of chemotherapy.
Such annotations on equivalences could be important to infer new knowledge from existing data and could be useful for medical care related applications [25]. Therefore, in Semantator, we provide the functionality to generate equivalences between two or more instances. In a similar fashion to instance batch creation, users will select an arbitrary number of annotated instances and add them to a sameAs candidate list to make them pairwisely equivalent. Figures 3(a) and 3(b) demonstrate this process. Although they only show generating equivalences between two instances, an arbitrary number of instances can be added to the sameAs candidate list to generate the linkages. Users can remove any instance from the candidate list before making them all equivalent.
Figure 3:
Generating Instance Equivalences
Managing Instance Relationships
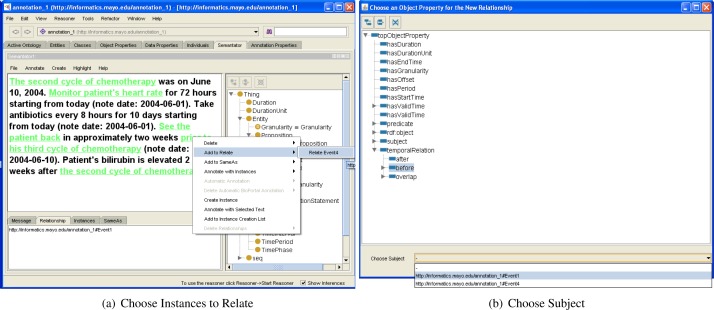
Another equally important type of annotation is instance relationships annotation. As described in Section 1, each ontology instance is a resource in the Semantic Web and different instances can be connected with one or more properties from ontologies. For example, we have two events Event1 and Event2; and they can be connected to form the following triple: < Event1 before Event2 >. Semantator allows users to create a single relationship between two instances at a time. Users can select two instances to be related and add them to the relationship candidate list that holds a maximum of two instances (in the Semantic Web, all relationships are binary). Then, they can choose any object property from the loaded ontology to connect the selected candidates. Finally, users need to decide the subject of this new relationship. Choosing an incorrect subject will sometimes totally change the underlying semantics of a relationship. In the triple example given above, Event1 is the subject and Event2 is the object. If we reverse their ordering, the triple will mean that Event2 happened before Event1, which then deviates from the fact. Figures 4(a) and 4(b) give an overview of this relationship creation process. A relationship between two instances can be easily deleted following a similar procedure as deleting an instance. Note that the instances need to be created first before they can be related.
Figure 4:
Connecting Instances with Ontological Properties
Exporting and Reloading Document Annotations
Till now, we have presented how to perform manual annotations. In Semantator, users can export their annotations to an RDF file together with a XML file that contains annotation related metadata, such as the position of each annotated instance, the color used to highlight instances of each class, etc. Next time when a user opens the same document, Semantator will ask the users whether or not they would like to load their previous annotations on this document. Once the correct RDF and XML files are selected, the created instances, relationships and their relevant metadata will be reloaded to Semantator for further manipulations.
4. Semi-automatic Annotation
As discussed earlier, a clinical document may be long and there exists a large volume of such documents. Therefore, manual annotating such documents could be time consuming. To facilitate the entire annotation process, in this section, we introduce the semi-automatic annotation capability of Semantator by connecting to back end services, including the NCBO annotator [12] and cTAKES [6].
NCBO Annotator and cTAKES Supported Automatic Annotation
BioPortal [12] is a Semantic Web based platform designed for the biomedical domain. It allows users to search for specific ontologies that match certain user provided keywords. It provides an online annotation tool, the NCBO annotator, that takes user inputs (free text), chooses relevant ontologies for annotation, recognizes relevant biomedical ontology terms in the text, and finally returns such annotations to the users. The NCBO annotator can also be used as a web service, which enables us to utilize it from within Semantator. The NCBO annotator can be called from Semantator with simply one click. Before actually starting doing any automatic annotation, we provide a list of ontologies (by querying BioPortal) that are currently supported by BioPortal and a user can choose an arbitrary number of ontologies from this list against which the annotator will match the words and phrases in a loaded document. All automatically annotated entities are treated as potential ontology instances and are highlighted in Semantator. Users can then examine such results and only retain those correctly identified instances from their perspective.
cTAKES is another tool used in Semantator that supports automatic annotation in a similar way to that supported by NCBO annotator. Different from the NCBO annotator, cTAKES 1) is designed for clinical domain; 2) adopts NLP techniques and supports negation and time constraints. Currently, cTAKES performs annotation with the SNOMED CT and RxNorm [26] dictionaries but users can add their own dictionaries as needed. Although we currently use cTAKES as a packaged Java library, a web service for cTAKES is under development.
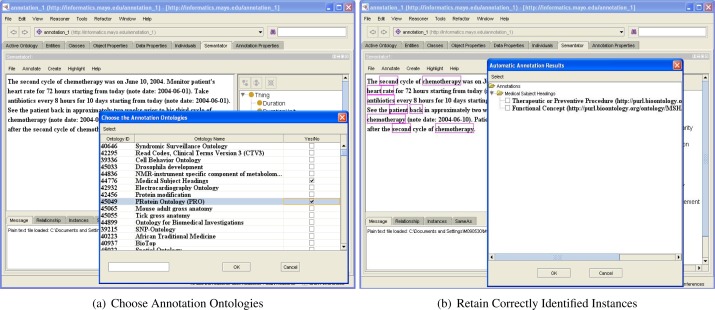
With such automatic processes, a document can be firstly annotated with the available domain knowledge provided by the chosen ontologies in BioPortal and dictionaries of cTAKES respectively to recognize candidate instances. Next, such automatic results can be corrected and augmented by human annotators. Users could potentially benefit from employing such automatic annotation services assuming decent performances can be achieved. We hope that using such automatic tools could facilitate the entire process by reducing the time needed for document annotation. In the current version of Semantator, we support automatic annotation by using BioPortal and cTAKES because they are well-acknowledged tools in the biomedical domain; however, the key idea here is that users can plug in any annotation tool that is suitable for a particular domain. In future work, we would like to provide APIs for our end users to support this. As shown in Figure 5(a), a user has selected the NCBO annotator and is choosing the ontologies to use; in Figure 5(b), we see that the automatically annotated instances are highlighted when human annotators can then decide which ones to retain.
Figure 5:
Semi-automatic Annotation with BioPortal Web Services
Semantic Web based Reasoning
One advantage of putting annotation results in Semantic Web formats is that users can benefit from the reasoning capabilities provided by Semantic Web techniques. In this section, we discuss the two types of reasoning that are currently supported in Semantator based upon class disjointness and class equivalence.
In the Semantic Web, classes can be defined to be disjoint with each other10, i.e., Ci and Cj are two classes and Ci ⊑ ¬Cj. Disjoint classes have no instances in common. For example, two classes Male and Female are disjoint and one person instance can only be declared to belong to either of these two classes [27]. Classes can also be defined to be equivalent11, i.e., Ci ≡ Cj. For example, two classes Man (Ci) and the intersection of Human and Male (Cj) are equivalent and thus any instance that is declared to be a Man should also be an instance of the other class.
In Semantator, we support reasoning based upon the two types of class relationship discussed above. Using the automatic annotation services, the same document fragment might be annotated to be candidate instances of disjoint classes. Take the following sentence as an example:
I was pleased to inform Mr. Smith that his PSA today is undetectable.
In this example, by calling the NCBO annotator with the SNOMED CT ontology, the word today is annotated to be an Organic Chemical; however, a human annotator may simply annotate it to be an instance of the TimeInstant class from the CNTRO ontology [28]. Assuming we have the knowledge about the disjointness between the two classes: Organic Chemical and TimeInstant, Semantator will report an inconsistency. Similarly, if we annotate this sentence with both NCI Thesaurus and the International Classification Nursing Practice (ICNP) ontologies, BioPortal will annotate the word today to be an instance of the Antibiotic class in both ontologies. If we assume that the two Antibiotic classes from the two ontologies are equivalent and a user only annotated today to be an instance of one of them, Semantator will then suggest the user to also annotate it to be an instance of the other. For future work, we will explore how to provide a more general reasoning capable framework within Semantator.
5. Discussion
In this section, we present discussions based upon some user experience on Semantator. Two of our annotators are experts on ontologies. Semantator was also evaluated by one student intern at Mayo Clinic without any background in the areas of medical informatics and the Semantic Web, and adopted by an Engineer at Boston Scientific who had some experience with ontologies [29] and Knowtator [22], a manual annotation tool for Protégé-Frames.
Compared to Knowtator, the biggest advantage of Semantator is its semi-automatic annotation capability. Semantator and Knowtator share the same objective to annotate clinical narratives to create ontology instances and their relationships with properties in an ontology. A human annotator can generally finish annotating a document faster using Semantator because the automatic annotation capability at least helps the users to focus their attention on specific parts rather than the entire document. Second, it was mentioned by one of our annotators that the Semantator relationships are easier to follow as the system simply asks the user to identify the two instances, choose an object property, and specify the subject. Another notable aspect of Semantator is that it works with OWL and thus can leverage the state-of-the-art Semantic Web based reasoning techniques for inconsistency checking and automatic classification.
One drawback of Semantator is the number of files needed. Semantator requires 3 files: the text file, the annotated OWL file, and a metadata XML file storing the information (e.g., positions and colors) of the annotations for users to reload and visualize their previous annotations. There is therefore more file organization required, and it required more practice to understand how to save and re-open files. Another problem mentioned by our annotators was that if an ontology class is first used for annotating a document, Semantator will ask the user to choose a color for highlighting its instances throughout this document. However, it would be good if the system could reuse the chosen colors for the same classes across different documents. This might be feasible by establishing a user repository. Whenever a user wants to use Semantator, the user could choose to log in so that all history information can be loaded; thus all the choices about colors made by this user before can automatically apply.
6. Conclusion and Future Work
In this paper, we introduce Semantator, a Semantic Web based semi-automatic annotation tool for annotating clinical documents. Although it is designed for the clinical domain, it can also be applied to annotate documents of other domains. Developed as a Protégé plugin, users can manually annotate documents with classes and properties from a loaded ontology in Protégé environment. To facilitate the annotation process, automatic annotation is supported by connecting to two back end services: NCBO annotator and cTAKES. Furthermore, the reasoning capability of Semantator could assist users in finding inconsistencies and incompleteness in their annotations. We present some discussion on Semantator based upon some user experiences.
For future work, it would be necessary to perform a comprehensive evaluation on the usability of Semantator and also provide some use cases. Next, it would be useful to provide a DIFF module to visualize the differences and calculate the inter-annotator agreement between annotations of different annotators. Furthermore, we would like to enhance Semantator with some query capability so that users can issue queries (e.g., SPARQL) to search within the annotation results. Also, we will explore how to provide a general framework to support reasoning within Semantator. In addition to automatic instance creation, automatic relation extraction could be one interesting research question to explore in the future.
Acknowledgments
This research is partially supported by the National Center for Biomedical Ontologies (NCBO) under the NIH Grant #N01-HG04028, and the NSF under Grant #0937060 to the CRA for the CIFellows Project. We would like to thank Kim Clark and Ian Chute for helping to test Semantator, and Vinod Kaggal and Dr. Hongfang Liu for helping setting up the cTAKES environment. We also thank Deepak Sharma and Donna Ihrke for their advice on improving Semantator.
Footnotes
This work was done while the first author was an intern at Mayo Clinic.
This is generally referred to as entity coreference [24], which is out of the scope of this paper.
References
- 1.Louie Brenton, Mork Peter, Martín-Sánchez Fernando, Halevy Alon Y, Tarczy-Hornoch Peter. Data integration and genomic medicine. Journal of Biomedical Informatics. 2007;40(1):5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008. pp. 67–79. [PMC free article] [PubMed]
- 3.Berners-Lee Tim, Hendler J, Lassila O. The semantic web. Scientific American. 2001;284(5):34–43. [Google Scholar]
- 4.Tenenbaum Jessica D, Whetzel Patricia L, Anderson Kent, Borromeo Charles D, Dinov Ivo D, Gabriel Davera, Kirschner Beth A, Mirel Barbara, Morris Timothy D, Noy Natasha Fridman, Nyulas Csongor, Rubenson David, Saxman Paul R, Singh Harpreet, Whelan Nancy, Wright Zach, Athey Brian D, Becich Michael J, Ginsburg Geoffrey S, Musen Mark A, et al. The biomedical resource ontology (bro) to enable resource discovery in clinical and translational research. Journal of Biomedical Informatics. 2011;44(1):137–145. doi: 10.1016/j.jbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demner-Fushman Dina, Chapman Wendy Webber, McDonald Clement J. What can natural language processing do for clinical decision support? Journal of Biomedical Informatics. 2009;42(5):760–772. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savova Guergana K, Masanz James J, Ogren Philip V, Zheng Jiaping, Sohn Sunghwan, Kipper-Schuler Karin C, Chute Christopher G. Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David Nadeau, Satoshi Sekine. A survey of named entity recognition and classification. Linguisticae Investigationes. 2007 Jan;30(1):3–26. [Google Scholar]
- 8.Shafranovich Y. RFC 4180: Common format and MIME type for Comma-Separated values (CSV) files, 2005.
- 9.Jeronimo Jose, Long L Rodney, Neve Leif, Michael Bopf, Antani Sameer, Schiffman Mark. Digital tools for collecting data from cervigrams for research and training in colposcopy. Journal of Lower Genital Tract Disease. 2006;10(1):16–25. doi: 10.1097/01.lgt.0000194057.20485.5a. [DOI] [PubMed] [Google Scholar]
- 10.Dolin RH, Alschuler L, Beebe C, Biron PV, Boyer SL, Essin D, Kimber E, Lincoln T, Mattison JE. The HL7 clinical document architecture. J Am Med Inform Assoc. 2001;8(6):552–569. doi: 10.1136/jamia.2001.0080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schäfer Ulrich. OntoNERdIE - Mapping and linking ontologies to named entity recognition and information extraction resources. Proceedings of the 5th International Conference on Language Resources and Evaluation (LREC) 2006. pp. 1756–1761.
- 12.Noy Natalya Fridman, Shah Nigam H, Whetzel Patricia L, Dai Benjamin, Dorf Michael, Griffith Nicholas, Jonquet Clement, Rubin Daniel L, Storey Margaret-Anne D, Chute Christopher G, Musen Mark A. Bio-portal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research. 2009;37(Web-Server-Issue):170–173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimiano Philipp, Handschuh Siegfried, Staab Steffen. Towards the self-annotating web. Proceedings of the 13th international conference on World Wide Web (WWW) 2004. pp. 462–471.
- 14.Chapman Sam, Dingli Alexiei, Ciravegna Fabio. Armadillo: harvesting information for the semantic web. Proceedings of the 27th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval. 2004. p. 598.
- 15.Dill Stephen, Eiron Nadav, Gibson David, Gruhl Daniel, Guha Ramanathan V, Jhingran Anant, Kanungo Tapas, Rajagopalan Sridhar, Tomkins Andrew, Tomlin John A, Zien Jason Y. Semtag and seeker: bootstrapping the semantic web via automated semantic annotation. Proceedings of the 12th international conference on World Wide Web (WWW) 2003. pp. 178–186.
- 16.Popov Borislav, Kiryakov Atanas, Ognyanoff Damyan, Manov Dimitar, Kirilov Angel. Kim a semantic platform for information extraction and retrieval. Nat Lang Eng. 2004 Sep;10:375–392. [Google Scholar]
- 17.Handschuh Siegfried, Staab Steffen. Authoring and annotation of web pages in cream. Proceedings of the 11th international conference on World Wide Web (WWW) 2002. pp. 462–473.
- 18.Vargas-Vera Maria, Motta Enrico, Domingue John, Lanzoni Mattia, Stutt Arthur, Ciravegna Fabio. Mnm: Ontology driven semi-automatic and automatic support for semantic markup. 13th International Conference on Knowledge Engineering and Knowledge Management (EKAW) 2002. pp. 379–391.
- 19.Cunningham Hamish, Maynard Diana, Bontcheva Kalina, Tablan Valentin. Gate: A framework and graphical development environment for robust nlp tools and applications. Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL) 2002.
- 20.Eriksson Henrik. An annotation tool for semantic documents. The Semantic Web: Research and Applications, 4th European Semantic Web Conference (ESWC) 2007. pp. 759–768.
- 21.Giannopoulos Giorgos, Bikakis Nikos, Dalamagas Theodore, Sellis Timos K. Gontogle: A tool for semantic annotation and search. The Semantic Web: Research and Applications, 7th Extended Semantic Web Conference (ESWC) 2010. pp. 376–380.
- 22.Ogren Philip V. Knowtator: A protégé plug-in for annotated corpus construction. Human Language Technology Conference of the North American Chapter of the Association of Computational Linguistics (HLT-NAACL) 2006.
- 23.Müller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biology. 2004;2:e309+. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagga Amit, Baldwin Breck. Entity-based cross-document coreferencing using the vector space model. COLING-ACL. 1998. pp. 79–85.
- 25.Tao Cui, Solbrig Harold R, Sharma Deepak K, Wei Wei-Qi, Savova Guergana K, Chute Christopher G. Time-oriented question answering from clinical narratives using semantic-web techniques. 9th International Semantic Web Conference (ISWC) 2010. pp. 241–256.
- 26.Nelson Stuart J, Zeng Kelly, Kilbourne John, Powell Tammy, Moore Robin. Normalized names for clinical drugs: Rxnorm at 6 years. JAMIA. 2011;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosof Benjamin N, Horrocks Ian, Volz Raphael, Decker Stefan. Description logic programs: combining logic programs with description logic. Proceedings of the Twelfth International World Wide Web Conference (WWW) 2003. pp. 48–57.
- 28.Tao Cui, Wei Wei-Qi, Solbrig Harold R, Savova Guergana, Chute Christopher G. CNTRO: A semantic web ontology for temporal relation inferencing in clinical narratives. American Medical Informatics Assocication Annual Symposium (AMIA) 2010. pp. 787–91. [PMC free article] [PubMed]
- 29.Clark KK, Sharma DK, Chute CG, Tao C. Application of a temporal reasoning framework tool in analysis of medical device. American Medical Informatics Assocication Annual Symposium (AMIA), Accpeted. p. 2011. [PMC free article] [PubMed]