Abstract
The ability to conduct genome-wide association studies (GWAS) has enabled new exploration of how genetic variations contribute to health and disease etiology. One of the key requirements to perform GWAS is the identification of subject cohorts with accurate classification of disease phenotypes. In this work, we study how emerging Semantic Web technologies can be applied in conjunction with clinical data stored in electronic health records (EHRs) to accurately identify subjects with specific diseases for inclusion in cohort studies. In particular, we demonstrate the role of using Resource Description Framework (RDF) for representing EHR data and enabling federated querying and inferencing via standardized Web protocols for identifying subjects with Diabetes Mellitus. Our study highlights the potential of using Web-scale data federation approaches to execute complex queries.
Introduction
In the past decade, there has been a huge splurge of discoveries in genomic sciences involving complex, non-Mendelian diseases that relate single-nucleotide polymorphisms (SNPs) to clinical conditions and measurable traits1. This has become feasible due to the advances in high-throughput genotyping technologies and genome-wide association studies (GWAS) that allow studying the entire human genome in thousands of unrelated individuals regarding genetic associations with different diseases. However, due to stringent requirements for achieving acceptable levels of statistical significance and replication of findings in GWAS, one of the key aspects in conducting such studies is the “subject sample size” (for cases and controls)—the larger the size of the cohort, the higher the probability for attaining genome-wide significant results and discovering genetic variants that influence diseases.
To address this need, the U.S. National Institutes of Health initiated the Electronic Medical Records and Genomics (eMERGE2,3) consortium that aims to determine whether patient data stored in electronic health records (EHRs) can identify disease phenotypes for application in GWAS. Especially in the era of Meaningful Use4 that promotes wide scale adoption of EHRs, such an approach for identification of disease phenotype cohorts using EHR data, if successful, has the potential to enable and rapidly scale genetic discoveries and research. Early results from the eMERGE network, of which Mayo Clinic is a member, has demonstrated the applicability of EHR-derived phenotyping algorithms for cohort identification to conduct genomic studies for several diseases, including peripheral arterial disease5, red blood cells6, and atrioventricular conduction7. A common thread across the library of algorithms8 is access to different types and modalities of data for algorithm execution, which includes billing and diagnoses information, laboratory measurements, patient procedure encounters, medication and prescription management data, and co-morbidities (e.g., smoking history, socio-economic status). This naturally presents us with the problem of representing and integration of data from the EHR and public knowledge bases (e.g., a knowledgebase for drug side effects) in a form that would allow federated querying, reasoning and efficient information retrieval across multiple sources of information.
Semantic Web9 technologies provide such a rigorous mechanism for defining and linking heterogeneous data using Web protocols and a simple data model called Resource Description Framework (RDF10). By representing data as labeled graphs, RDF provides a powerful framework for expressing and integrating any type of data. As of March 2011, under the auspices of an initiative called the Linked Open Data (LOD11,12), more than 215 public datasets from multiple domains (e.g., gene and disease relationships, drugs and side effects) are available in RDF, and have been integrated by specifying approximately 350 million links between the RDF graphs. Not only such efforts provides tremendous opportunities to devise novel approaches for combining private, and institution-specific EHR data with public knowledgebases for phenotyping, but also presents several challenges in representing EHR data using RDF, creating linkages between multiple disparate RDF graphs, and developing mechanisms for executing federated queries analyzing information spanning genes, proteins, pathways, diseases, drugs and adverse events.
To this end, in this paper, we describe our efforts in representing real patient data from EHR systems at Mayo Clinic as RDF graphs. In particular, we leverage open-source tooling and infrastructure developed by the Linked Data community for demonstrating Web-scale federated querying and answering for information about Diabetes Mellitus using public knowledgebases. Our tool highlights the potential of combining and integrating private-public information to answer complex queries in a robust, uniformed, and scalable way.
Background
Semantic Web and related technologies
A key benefit of using Semantic Web technologies is a rigorous mechanism of defining and linking data using Web protocols in a way, such that, the data can be used by machines not just for display, but for automation, integration and reuse of across various applications. Examples of adoption of Semantic Web technologies include both the US13 and UK14 governments for making multiple types of governmental data publicly available. Specifically, an “attractive” element of the Semantic Web is its simple data model, called Resource Description Framework (RDF10), that represents data as a labeled graph connecting resources and their property values with labeled edges representing properties. The graph can be structurally parsed into a set of triples (subject, predicate, object), making it very general and easy to express any type of data. Such a model coupled with (i) dereferenceable Uniform Resource Identifiers (URI’s) for creating globally unique names, and (ii) standard languages such as RDFS15, OWL16, and SPARQL17 for creating ontologies as well as modeling and querying data, provides a very powerful framework for heterogeneous data integration. Of particular relevance to this study is the Linked Open Data (LOD11) initiative from the World Wide Web Consortium (W3C) that aims to bootstrap the Web of data by publishing existing data sets in RDF on the Web and creating numerous links between them. As of March 2011, the LOD project has more than 215 public datasets from multiple domains (e.g., genes, drugs and side effects, diseases, anatomy) with approximately 25 billion triples connected via more than 350 million links, and comprises resources such as DBpedia18) that provide an RDF representation of Wikipedia.
It should be noted that while most of the clinical and research data is typically stored using relational databases (e.g., Oracle, MySQL) and queried using Structured Query Language (SQL), such technologies have several inherent limitations compared to RDF: (i) First, when database schemas are changed in a relational model, the whole repository, table structure, index keys etc. have to be reorganized—a task that can be quite complex and time-consuming. RDF, on the other hand, does not distinguish between schema (i.e., ontology classes and properties) and data (i.e., instances of the ontology classes) changes—both are merely addition or deletion of RDF triples, making such a model very nimble and flexible for updates. (ii) Second, RDF resources are identified by (globally) unique URI’s, thereby allowing anyone to add additional information about the resource. For example, via RDF links, it is possible to create references between two different RDF graphs, even in completely different namespaces, enabling much easier data linkage and integration. This is rather difficult to achieve in the classical relational database paradigm. (iii) Third, a relational data model lacks any inherent notion of a hierarchy. For instance, simply because a particular drug is an Angiotensin Receptor Blocker (ARB), a typical SQL query engine (without any ad-hoc workarounds) cannot reason that it belongs to a class of Anti-hypertensive drugs. Such queries are natively supported in RDFS and OWL. (iv) Finally, due to the lack of a formal temporal model for representing relational data, SQL provides minimal support for temporal queries natively19. Such extensions are already in place for SPARQL20.
In summary, linked data, and its enabling technologies such as RDF, provide a more robust, flexible, yet scalable model for integrating and querying data, thereby warranting investigation on how such technologies can applied in a clinical and translational research environment. However, while on one hand, such a huge integrated-network dataset provides exciting opportunities to execute expressive federated queries and integrating and analyzing information spanning genes, proteins, pathways, diseases, drugs and adverse events, several questions remain unanswered about its applicability to its integration with patient data in EHR systems to enable EHR-driven high-throughput phenotyping. In the remainder of this paper, we provide a brief overview of RDF and SPARQL—the building blocks for linked data, and then proceed to propose our methods and present preliminary result in using Semantic Web technologies for EHR-derived high-throughput phenotyping.
RDF and RDF-S
Resource Description Framework (RDF10) is a World Wide Web Consortium (W3C) standardized data model for representing semantic Web resources. It uses graphs to represent information using a triple-based notation comprising a subject, predicate and an object. All these entities can be uniquely identified by Internationalized Resource Identifiers (IRIs). As an example Figure 1 shows an instance of RDF graph for “United States of America” from DBPedia.org. The subject at (line 5) (“http://dbpedia.org/page/United_States”) is stated (at line 6) to belong to the category “Country” using the predicate “rdf:type”. The same subject is given a label (via the predicate “rdfs:label”; line 7) of “USA”. The graph further states that “Washington D.C.” is its capital (via the predicate “dbpedia-owl:capital”; line 8) and the total area square miles is “3794101” (via the predicate “dbpediaowl:areaSqMi; line 9).
Figure 1.
Snapshot of RDF graph representing United States of American (DBPedia.org)
A key aspect in order to define such graphs is to establish a vocabulary that provides an ontological foundation and semantic definition for the properties (i.e., p redicates) and concepts (i.e., subjects and objects) used in the graph. Resource Description Framework Schema (RDFS15) provides a lightweight language for describing such a vocabulary. Examples of properties from above Figure 1 include “rdfs:label” which is used to attach a textual label to a resource. Furthermore, W3C has proposed standards for more expressive languages, such as the Web Ontology Language (OWL16) to model vocabularies with higher-order logics and complexity.
SPARQL: The query language for RDF
Simple Protocol and RDF Query Language (SPARQL17) is a W3C recommend standard for querying RDF data. Similar in spirit to SQL, a SPARQL query is composed of five parts (Figure 2): zero or more prefix declarations for abbreviating IRIs, zero or more FROM or FROM NAMED clauses stating what RDF graph(s) are being queried, a query result clause specifying what information to return from the query, a WHERE clause specifying what to query for in the underlying dataset, and zero or more query modifiers to slice, order, and otherwise rearrange the query results.
Figure 2.
SPARQL query components
SPARQL specifies one of the four forms of query result clauses: SELECT, CONSTRUCT, ASK and DESCRIBE, such that SELECT result clause returns a table of result values, CONSTRUCT returns an RDF graph, ASK returns a boolean true or false depending on whether or not the query pattern has any matches in the dataset, and DESCRIBE allows the server to return whatever RDF it wants that describes the given resource(s). The optional set of FROM or FROM NAMED clauses define the dataset against which the query is executed. The WHERE clause is core for any SPARQL query, and is specified in terms of triple patterns. Finally, the optional set of modifiers operate over the result set of the WHERE clause before generating the final query results.
Methods
System Architecture: Representing Patient Records as RDF graphs and Linked Data
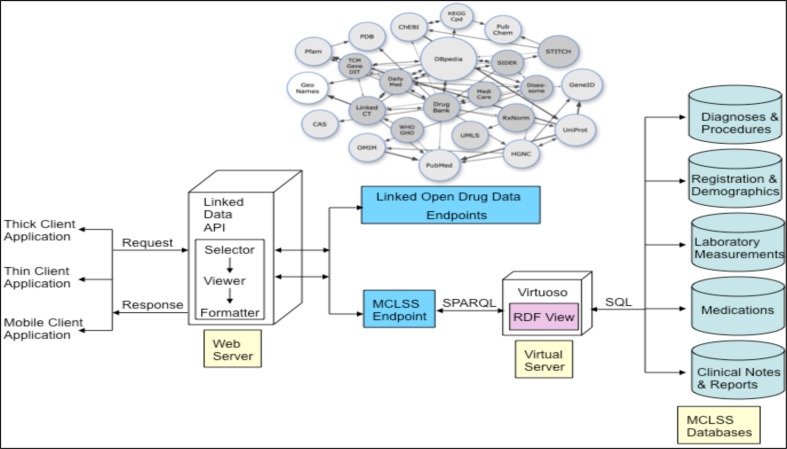
Figure 3 shows our proposed architecture for representing patient health records at Mayo Clinic using RDF, linked data and related technologies. It comprises of three main components: (1) data access and storage, (2) RDF virtualization and ontology mapping, and (3) SPARQL-based querying interface. Here we provide a brief overview of these components, and more details were described in our prior work21.
Figure 3.
System architecture for representing patient records using RDF
Data Access and Storage.
This component comprises the patient demographics, diagnoses, procedures, laboratory results, and free-text clinical and pathology notes generated during a clinician encounter. For our purposes in this study, we leverage the Mayo Clinic Life Sciences System (MCLSS22) which is a rich clinical data repository maintained by the Enterprise Data Warehousing Section of the Department of Information Technology. MCLSS contains patient demographics, diagnoses, hospital, laboratory, flowsheet, clinical notes and pathology data obtained from multiple clinical and hospital source systems within Mayo Clinic at Rochester, Minnesota. Data in MCLSS is accessed via the Data Discovery and Query Builder (DDQB) toolset, consisting of a web-based GUI application and programmatic API. Investigators, study staff and data retrieval specialists can utilize DDQB and MCLSS to rapidly and efficiently search millions of patient records. Data found by DDQB can be exported into CSV, TAB or Microsoft® Excel files for portability. It implements full data authorization and audit logging to ensure data security standards are met.
It is to be noted that while DDQB provides graphical user and application programming interfaces for accessing the warehouse database, our goal is to represent the data stored in the MCLSS database as RDF. In particular, our goal is to create “virtual RDF graphs” which essentially wraps one or more relational databases into a virtual, read-only RDF graph. This will allow us to access the content of large, live, non-RDF databases without having to replicate all the information into RDF. Consequently, for this study, we obtained appropriate approvals from Mayo’s Institutional Review Board (IRB) for accessing patient information in the MCLSS database using programmatic API and JDBC calls (see more details below).
RDF Virtualization and Ontology Mapping.
The RDF virtualization and ontology mapping component is based on the open-source Virtuoso Universal Server23 which acts a mediator in the creation of virtual RDF graphs as well as provides a SPARQL endpoint for querying the graphs. In particular, a declarative language is used to describe the mappings between the relational schema and RDFS/OWL ontologies to create the virtual RDF graphs. This language generates a mapping file from table structures of the databases in MCLSS, which can then be customized by replacing the auto-generated terms with concepts from standardized ontologies. In our case, we use the Translational Medicine Ontology (TMO24) for creating these mappings. In particular, we created extensions to TMO via mappings to NCI Thesaurus and SNOMED to provide a larger coverage for clinical concepts. For example, concepts relevant to a subject’s vital measurements (e.g., body mass index), interventions and procedures, laboratory measurements etc. were not specified as part of the current release of TMO (version 1.0). Consequently, leveraging existing ontologies, namely the Ontology for Biomedical Investigations25 and Prostate Cancer Ontology26, we created several new concepts and properties that were mapped to the NCI Thesaurus and extended the current release of TMO.
SPARQL endpoint.
The virtual RDF graphs created from MCLSS using the above approach were exposed via a SPARQL endpoint in the Virtuoso server. This allows humans or software application clients to query the MCLSS RDF graphs using the SPARQL query language. Given that our overarching goal is to integrate the MCLSS RDF graphs with the RDF data that is part of the Linked Open Data cloud, our objective is to execute federated queries across multiple SPARQL endpoints. We discuss the details of SPARQL-based federated querying in the next section.
SPARQL-based federated querying for Diabetes Mellitus cohort identification
Diabetes Mellitus (DM) is an increasing public health problem, and several environmental factors including diet and physical activity as well as genetic makeup contribute to the disease etiology. It is a major cause of heart disease and stroke, as well as the most common cause of blindness, kidney failure and amputations in U.S. adults. Given the high prevalence of DM patients in the U.S. adult population, our demonstration use case for this study is to enable a clinician or a researcher to ask questions about DM, ranging from its diagnosis, to side effects and adverse reactions, to clinical trials, that span across multiple RDF data sources. In particular, we want to investigate federated querying capabilities across twelve interlinked RDF datasets as part of the Linked Open Drug Data (LODD27) cloud. Table 1 shows the list of the LODD datasets and provides a brief description. These datasets are periodically refreshed, and the HTTP-based unique identifiers (i.e., uniform resource locators) for representing entities in the linked datasets are stable and are chosen by the LODD participants. The LODD datasets are linked with each other, as well as with datasets provided by other Linked Data projects, such as Bio2RDF28 and Chem2Bio2RDF29, as well as other data providers that expose the information in RDF, such as UniProt30.
Table 1.
List of Linked Open Drug Data datasets
| Dataset | Topic | Dataset description |
|---|---|---|
| DrugBank | Drugs | Provides drug (e.g., pharmacological) with drug target (e.g., pathway) data |
| LinkedCT | Clinical Trials | Clinical trials registry |
| DailyMed | Drugs | FDA approved drugs |
| DBPedia | Drugs, diseases, proteins | RDF data extracted from the Wikipedia |
| Diseasome | Diseases, genes | Links diseases and genes by known associations |
| RDF-TCM | Drugs, genes, diseases | Traditional Chinese medicine gene and disease associations dataset |
| RxNorm | Drugs | NLM’s RxNorm vocabulary |
| SIDER | Drug side effects | Marketed drugs and their adverse effects |
| STITCH | Chemicals, Proteins | Chemical, proteins, and their interactions |
| ChEMBL | Chemicals, Assays, Literature | Trial drugs, literature, drug targets |
| WHO Global Health Observatory | Infectious diseases, environmental factors, socioeconomic conditions | Data and statistics for infectious diseases at country, regional and global levels |
| Medicare | Drug formulary | Medicare D approved drugs |
Our objective in this study is to demonstrate the potential and utility of seamless integration and federated querying of distributed and heterogeneous publicly available data sources as part of the LODD with patient-specific data stored within the MCLSS environment for DM cohort identification. To this end, we have created a list of sample queries (Table 2) demonstrating the SPARQL-based federated querying infrastructure. These queries were informed by the criteria (inclusion and exclusion criteria) defined within the eMERGE consortium for identifying DM subjects3.
Table 2.
Sample federated queries for Diabetes Mellitus (*Bio2RDF is not part of LODD)
| Query description | LODD datasets to be queried |
|---|---|
| Q1. Find all patients having a side effect of Prandin after being administered. | RxNorm, SIDER |
| Q2. Find all FDA approved drugs for DM, and identify the patients that were being administered those drugs by drug class. | RxNorm, DailyMed |
| Q3. What genes or biomarkers are associated with DM as published in the literature and find their interaction pathway information? | Diseasome, Bio2RDF* |
| Q4. A variant of HNF4α is known to have a strong correlation with DM predisposition. Find all patients administered with drugs that target HNF4α. | RxNorm, DrugBank, Diseasome, ChemBL |
| Q5. Find all patients that are on Sulfonylureas, Metformin, Metglitinides, and Thiazolinediones, or combinations of them? | RxNorm, DrugBank, DailyMed |
| Q6. Find all patients taking Alpha-glucosidase inhibitors. | RxNorm, DrugBank |
Results
Figure 4 shows the SPARQL query for execution of query #1 (from Table 2) to determine the side-effects of Prandin using the SIDER (Drug Side Effect Resource31) SPARQL endpoint in the LODD cloud, and then find subjects with those side-effects who have been prescribed Prandin using the MCLSS and RxNorm SPARQL endpoints. The query is based on using an extension defined as part of SPARQL 1.1 recommendation that allows federated querying between multiple SPARQL endpoints using the “SERVICE” keyword. In essence, the query is divided into three main segments: the first segment is querying the SIDER SPARQL endpoint to find out all the known side-effects of Prandin. The list of side-effects is used as an input for the second segment of the query which is divided into two parts: the first part simply queries the RxNorm SPARQL endpoint to determine the RxNorm code for Prandin. This code is then used to find patients in MCLSS who have been prescribed Prandin, and the results are further filtered to only those patients who have been assigned an ICD-9 code for one or more side-effects of Prandin. In our execution of query at the time of this writing, 1456 unique patients were returned.
Figure 4.
SPARQL federated query for drug side-effects of Prandin
Similarly, Figure 5 shows the SPARQL query for execution of query #3 (from Table 2) to determine the published associations between genes or biomarkers with DM. This query relies on 2 different public sources: Diseasome and Bio2RDF. Diseasome publishes a network of approximately 5,000 disorders and disease genes linked by known disorder-gene associations, and Bio2RDF provides a federated repository of large databases in RDF, including KEGG32 which contains gene pathway information. This query is divided into two main segments: the first segment is querying the Diseasome SPARQL endpoint to find out all the genes associated with “Diabetes Mellitus”. The list of genes is then used for finding the KEGG pathway information in Bio2RDF. In our execution of this query at the time of this writing, 27 unique genes were returned. One of the genes is AKT2 that encodes the enzyme RAC-beta serine/threonine-protein kinase. This gene is associated to 35 unique KEGG pathways.
Figure 5.
SPARQL federated query for genes and pathways associated with diabetes mellitus
There are several aspects in both these queries that are noteworthy: first, it demonstrates how data from public resources, such as SIDER and RxNorm, can be leveraged “on-the-fly” for a specific cohort identification search in Mayo’s EHR system (query #1) and how gene-disease-pathway information for diabetes can be retrieved from Diseasome and KEGG (query #3)–all within a single query. Secondly, it demonstrates powerful federated querying capabilities provided by SPARQL to query RDF data. Regardless of how the original data is stored and represented in the source (e.g., MySQL or DB2 database, XML files), once the data is represented as RDF and exposed via a SPARQL endpoint, the different storage modalities become irrelevant from a SPARQL query perspective. In a traditional RDBMS setting, one will have to address idiosyncratic issues about SQL implementations, JDBC drivers etc., and even then a purely federated query is non-trivial to formulate and execute. Finally, the query illustrates the potential to augment additional public/private RDF datasets (e.g., gene pathways, genotype-phenotype correlations) that provided unprecedented opportunities for translational science researchers to integrate and analyze multiple sources of data.
Note that due to space constraints, we exclude the discussion about remaining queries from Table 2 (query #3--#6) from this manuscript. The query details are available at: http://informatics.mayo.edu/LCD.
Discussion
Summary
Research in clinical and translational science demands effective and efficient methods for accessing, integrating, interpreting and analyzing data from multiple, distributed and often heterogeneous data sources in a unified way. Traditionally, such a process of data collection and analysis is done manually by investigators and researchers, which is not only time consuming and cumbersome, but in many cases, error prone. The emerging Semantic Web tools and technologies, and in particular W3C’s Linked Open Data project, is providing unprecedented opportunities by harnessing information from publicly available resources, such as Wikipedia and PubMed, and exposing the data as structured RDF that can be queried uniformly via SPARQL. Not only this provides the capabilities for interlinking and federated querying of diverse Web-based resources, but also enables fusion of private/local and public data in very powerful ways.
The overarching goal of this study is to investigate federated data integration and querying using public data sources from the Linked Open Data cloud, and private, identifiable patient data from Mayo Clinic’s EHR systems for cohort identification and phenotyping. Using open-source tooling and software, we developed a proof-of-concept system that allows representing patient data stored in Mayo’s enterprise warehouse system as RDF, and exposing it via a SPARQL endpoint for accessing and querying. We leveraged existing ontologies, such as the Translational Medicine Ontology and Ontology for Biomedical Investigations, for mapping the MCLSS database schema to standardized semantic concepts and relationships. Our use case for federated querying of Diabetes Mellitus information further demonstrated the applicability of such a system and the benefits of interlinking and querying multiple, heterogeneous Web data sources that are publicly available, with private (and institution-specific) patient information. We hypothesize that further development of such a system can immensely facilitate, and potentially accelerate scientific findings in clinical and translational research, including genomics and systems biology.
Limitations
There are several limitations in the proof-of-concept system developed as part of this study First, while we demonstrated the applicability of the system via sample use case queries, a more robust and rigorous evaluation along several dimensions (e.g., performance, query response, precision and recall of query results etc.) is required before it can be deployed within an enterprise environment. Note that since our use cases are based on federated querying of several public SPARQL endpoints, the system performance and query responses are dependent on the behavior of the endpoints (e.g., the endpoints may experience latency, denial of service). Nevertheless, we plan to perform a thorough system evaluation after the integration of additional MCLSS sources (e.g., laboratory, clinical and pathology reports) that contain large amounts of patient data. Second, we used the recently published Translational Medicine Ontology (TMO) in this study for mapping between MCLSS database schemas to standardized concepts and relationships. While TMO classes are mapped to more than 60 different standardized ontologies, including SNOMED CT and NCI Thesaurus, the scope and breadth of the current TMO release (Version 1.0) is significantly narrow for our purpose. Consequently, along with the creation new classes and relationships, we augmented TMO with Prostate Cancer Ontology and Relations Ontology. Since these extensions are not part of the official TMO release, our goal is to work closely with the TMO curators for content enhancement in future releases. Finally, formulating the complex SPARQL queries using existing SPARQL editors is cumbersome and error prone. Our current implementation lacks a more intuitive and user-friendly tool that can assist a “non-Semantic Web savvy” user in the query building process. We plan to address this issue within the timeframe of the project.
Future Work
In addition to addressing the limitations aforementioned, there are several activities that we plan to pursue in the future. Firstly, in this study, we performed simple mappings between the MCLSS database schema to classes and relationships in TMO (extended). A more rigorous approach will be to investigate reference information models, such as clinical archetypes33, that provide a mechanism to express data structures in a shared and interoperable way. Secondly, we will investigate existing Semantic Web querying visualization platforms such as SPARQLMotion34 and Triple Map35 which provide more intuitive and user-interactive interfaces for SPARQL query formulation and execution. Finally, we will investigate approaches for distributed and federated inferencing over RDF data. Recent studies36 have demonstrated that even simple subsumption inferences require significant computing power (such as Cray XMT37 supercomputer) when reasoning over massive RDF datasets. Since access to extremely high-performance computers is not readily available en masse, we will investigate distributed storage and indexing techniques using Apache Hadoop38 for addressing this problem.
Conclusion
This study demonstrates how Semantic Web technologies can be applied in conjunction with clinical data stored in EHRs and public knowledgebases to accurately identify subjects with specific diseases and phenotypes. Such an approach has the potential to immensely facilitate the tedious, cumbersome and error prone manual integration and analysis of data for clinical and translational research, including genomics studies and clinical trials.
Acknowledgments
This research is supported in part by the Mayo Clinic Early Career Development Award (FP00058504) and the eMERGE consortia (U01-HG-04599).
References
- 1.Pearson T, Manolio T. How to Interpret a Genome-wide Association Study. Journal of the American Medical Association. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 2.McCarty C, Chisholm R, Chute C, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Medical Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kho AN, Pacheco JA, Peissig PL, et al. Electronic Medical Records for Genetic Research: Results of the eMERGE Consortium. Science Translational Medicine. 2011 Apr 20;3(79) doi: 10.1126/scitranslmed.3001807. 2011. 79re71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal D, Tavenner M. The Meaningful Use Regulation for Electronic Health Records. New England Journal of Medicine. 2010;363(6):501–504. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 5.Kullo I, Fan J, Pathak J, Savova G, Ali Z, Chute C. Leveraging Informatics for Genetic Studies: Use of the Electronic Medical Record to Enable a Genome-Wide Association Study of Peripheral Arterial Disease. JAMIA. 2010;17(5):568–574. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A Genome-Wide Association Study of Red Blood Cell Traits Using the Electronic Medical Record. PLoS ONE. 2010;5(9):e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denny JC, Ritchie MD, Crawford DC, et al. Identification of Genomic Predictors of Atrioventricular Conduction / Clinical Perspective. Circulation. 2010 Nov 16;122(20):2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.eMERGE Library of Phenotyping Algorithms. https://www.gwas.net. Accessed August 8th, 2011.
- 9.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Scientific American. 2001 [Google Scholar]
- 10.Resource Description Framework (RDF) http://www.w3.org/RDF/. Accessed January 13, 2011, 2011.
- 11.Bizer C, Heath T, Berners-Lee T. Linked Data - The Story So Far. International Journal on Semantic Web and Information Systems. 2009;5(3):1–22. [Google Scholar]
- 12.Samwald M, Jentzsch A, Bouton C, et al. Linked open drug data for pharmaceutical research and development. Journal of Cheminformatics. 2011;3(19) doi: 10.1186/1758-2946-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.U.S. Data.gov Project. http://www.data.gov/. Accessed January 15, 2011, 2011.
- 14.U.K. Data.gov Project. http://data.gov.uk/. Accessed January 14, 2011, 2011.
- 15.Resource Description Framework (RDF) Schema Language 1.0. http://www.w3.org/TR/rdf-schema/. Accessed January 13, 2011, 2011.
- 16.Motik B, Patel-Schneider P, Horrocks I. OWL 2 Web Ontology Language. 2008. http://www.w3.org/2007/OWL/. Accessed January 13, 2011, 2011.
- 17.Prud'hommeaux E, Seaborne A. SPARQL Query Language for RDF. 2008. http://www.w3.org/TR/rdfsparql-query/. Accessed August 14th, 2010, 2010.
- 18.Bizer C, Lehmann J, Kobilarov G, et al. DBpedia - A crystallization point for the Web of Data. Web Semant. 2009;7(3):154–165. [Google Scholar]
- 19.Snodgrass R. TSQL2 and SQL3 Interactions. http://www.cs.arizona.edu/people/rts/sql3.html. Accessed January 12, 2011, 2011.
- 20.Tappolet J, Bernstein A. Applied Temporal RDF: Efficient Temporal Querying of RDF Data with SPARQL. 6th Annual European Semantic Web Conference. 2009. pp. 308–322. Vol 5554: Lecture Notes in Computer Science (LNCS)
- 21.Pathak J, Kiefer R, Chute C. Applying Linked Data Principles to Represent Patient’s Electronic Health Records at Mayo Clinic: A Case Report. 2nd ACM SIGHIT International Health Informatics Symposium. 2012.
- 22.Weiss T. IBM, Mayo Clinic to develop database for clinical trials, research. 2002. http://www.computerworld.com/s/article/69540/IBM_Mayo_Clinic_to_develop_database_for_clinical_trials_research. Accessed June 8, 2011.
- 23.Virtuoso Universal Server http://virtuoso.openlinksw.com/. Accessed January 16, 2011, 2011.
- 24.Dumoniter M, Andersson B, Batcherlor C, Domarew C, et al. The Translational Medicine Ontology: Driving Personalized Medicine by Bridging the Gap from Bedside to Bench. 13th ISMB SIG meeting on Bio-ontologies. 2010. pp. 120–123.
- 25.The Ontology for Biomedical Investigations. http://obi-ontology.org/. Accessed June 22, 2011.
- 26.Min H, Manion FJ, Goralczyk E, Wong Y-N, Ross E, Beck JR. Integration of prostate cancer clinical data using an ontology. J. of Biomedical Informatics. 2009;42(6):1035–1045. doi: 10.1016/j.jbi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linked Open Drug Data. http://www.w3.org/wiki/HCLSIG/LODD. Accessed February 15, 2011, 2011. [DOI] [PMC free article] [PubMed]
- 28.Belleau F, Nolin M-A, Tourigny N, Rigault P, Morissette J. Bio2RDF: Towards a mashup to build bioinformatics knowledge systems. Journal of Biomedical Informatics. 2008;41(5):706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Chen B, Dong X, Jiao D, et al. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinformatics. 2010;11(1):255. doi: 10.1186/1471-2105-11-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apweiler R, Bairoch A, Wu CH, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Research. 2004 Jan 1;32(suppl 1):D115–D119. doi: 10.1093/nar/gkh131. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Lee K, Song M, Lee D. Building the process-drug-side effect network to discover the relationship between biological Processes and side effects. BMC Bioinformatics. 2011;12(Suppl 2):S2. doi: 10.1186/1471-2105-12-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Hattori M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research. 34(suppl 1):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lezcano L, Sicilia M-A, Rodrìguez-Solano C. Integrating reasoning and clinical archetypes using OWL ontologies and SWRL rules. Journal of Biomedical Informatics. 2011;44(2):343–353. doi: 10.1016/j.jbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Waldman S. TopQuadrant: SPARQLMotion Visual Scripting Language. http://www.topquadrant.com/products/SPARQLMotion.html. Accessed June 28th, 2011.
- 35.Triple Map. http://www.triplemap.com/. Accessed August 19th, 2011, 2011.
- 36.Goodman E, Jimenez E, Mizell D, al-Saffar S, Adolf B, Haglin D. High-Performance Computing Applied to Semantic Databases. Extended Semantic Web Conference; Athens, Greece. 2011. pp. 31–45. [Google Scholar]
- 37.The Cray XMT Supercomputing System. http://www.cray.com/products/XMT.aspx. Accessed July 9, 2011.
- 38.Apache Hadoop Project. http://hadoop.apache.org/. Accessed July 15, 2011.