Abstract
Alzheimer’s and Parkinson’s diseases (AD and PD) are two common neurodegenerative diseases primarily affecting memory and motor functions, respectively. In this study, we integrated data from various sources, and took a systems-biology approach to compare and contrast the molecular and network based dysregulation associated with AD and PD and we integrated these data with known pathways of drug treatment. First, we identified genes that exhibit consistent prior evidence of association with each disease. Then, we extracted disease-specific sub-networks from a human interactome database using associated genes as seeds. To rank the sub-networks we used existing gene expression data from cases and controls. Comparison of resulting disease-associated genes and networks revealed significant overlap between AD and PD. In addition, the identified sub-networks correlated with known drug interdiction pathways, and suggested new potential targets for intervention.
Introduction
Alzheimer’s disease (AD) and Parkinson’s disease (PD) are caused by progressive degeneration and/or death of nerve cells. In AD, the patient’s memory and ability to think and carry out tasks is slowly destroyed; while PD mainly affects the patient's physical abilities: patients lose body control and have difficulty with movement and coordination1,2. Both diseases are strongly linked with the process of aging3,4. For most patients with AD and PD, symptoms first appear after age 60. Since the average age of the population is increasing, the numbers of AD and PD patients are expected to grow rapidly: e.g., it is estimated that 5.4 million Americans have AD in 2010 and this number will increase to 11–16 million in 20505.
To date, FDA has approved several drugs to slow the progression of AD and PD. Most drugs attempt to prevent the breakdown of critical chemicals whose levels are decreased in patients, e.g., cholinesterase inhibitors for AD patients slow the metabolic breakdown of acetylcholine that is involved in nerve cell communication; carbidopa for PD delays the conversion of levodopa. However, almost all of the currently available drugs are effective for short periods (a few months to a few years). For both diseases, in order to develop more efficient treatments and drugs, it is important to investigate and understand the molecular mechanisms and molecular networks that are altered in the development and progression of diseases.
A large number of studies, including recent genome-wide associate studies (GWAS)6–11, traditional association, linkage, and gene expression studies, have been conducted to identify genes associated with AD and PD incidence and progression. Consequently, hundreds of potentially associated genes have been identified. A few of these genes have very strong connections with disease. In particular, APOE is linked to approximately 50% of AD patients12, and alpha-synuclein is associated with PD in members of a large Italian family13. However, for both diseases, the majority of the hundreds of identified genes are likely to individually have both small and potentially complex effects on the development and progression of disease14,15. To uncover the underlying molecular mechanisms in these two diseases, comprehensive analysis of identified genes and their interactions within a network framework might provide many important insights beyond the traditional single-gene or single-marker analyses16,17.
Network and pathway analysis are relatively new approaches to the study and identification of dysregulated components in diseases18. The underlying principle is that human diseases are caused by perturbations of the complex networks/pathways that link molecular components (such as genes and proteins) in a human cell. Pathways emphasize what is known and relatively well understood, thus the results can be easily integrated into familiar biological frameworks. On the other hand, current knowledge of pathways is incomplete, and network analysis explores new connections, and connects what are often perceived as distinct pathways. Network analysis has been employed to detect the networks associated with many complex diseases, such as cancer19,20, schizophrenia16 and addiction21. Recently, we have used integrated genome wide mRNA expression with protein-protein interaction (PPI) networks to detect sub-networks that are dysregulated in colon cancer and sleep disorders22,23. The aim of such studies is to identify the functionally related genes that exhibit coordinate differential expression between healthy and diseased patients. Due to the computational complexity of examining the actions of multiple genes simultaneously, such studies are currently focused on small subnetworks as markers of disease. However, multiple studies have demonstrated that the results are biologically meaningful and can provide testable hypothesis for further validation19,22,23. The framework of PPI permits scoring of multiple gene combinations within a highly functional context, while the reduced search space limits multiple testing corrections. These PPI sub-networks can reveal critical nodes and edges reflecting both biomarkers of disease, e.g. molecular beacons of the condition, as well as pinpoint critical nodes for potential functional intervention, e.g. important drug targets. We wished to test these properties of molecular network assessment in the context of known information on AD and PD, including the known biomarkers and known targets of drug treatment.
Although AD and PD have their own unique neuropathological features; many patients with one disease later develop symptoms of the other24–28. This observation suggests the presence of common genetic variants that predispose individuals to both diseases and/or age related similarities in disease progression. Furthermore, beyond common genetic variants, perturbation of common pathways or network connections may also be shared in AD and PD. Studies show that the cerebral accumulation of beta-amyloid is associated with AD while alpha-synuclein is linked with PD29,30. Using a transgenic mouse model and NMR spectroscopy, a recent study has suggested an interaction between beta-amyloid and alpha-synuclein31–33 providing a potential molecular connection between AD and PD. Motivated by these observations, we conducted a systematic comparative analysis to investigate the molecular mechanisms and relationship of AD and PD, by taking advantage of the large amount of available molecular data for both diseases. We identified genes and networks strongly associated with AD and PD, and comparative analysis showed that AD and PD have strong connections and shared components at both molecular and network levels.
Methods and materials
To comprehensively investigate the molecular mechanisms of AD and PD, analyses at three levels were performed: (i) genes associated with AD and PD, (ii) networks connecting genes associated with AD and PD, and (iii) sub-networks dysregulated in AD and PD at the level of mRNA expression. This section describes the construction of genes and networks associated with each disease, and the detection of dysregulated sub-networks.
Construction of gene sets associated with diseases:
Genes associated with AD and PD (termed as AD genes and PD genes, respectively) were extracted from public databases constructed using literature searches10. For the purpose of comparison, gene sets related with three other common mental disorders (autism, multiple sclerosis (MS) and schizophrenia) were also generated using a similar approach. To extract associated genes with strong evidence, we only considered associations that are reported in at least four publications whenever possible. In the case of autism, only genes present in both databases were extracted. The data sources are summarized in Table 1, along with the number of extracted disease-associated genes for each disease.
Table 1.
Data source and number of genes used in this study
| Disease | Database | Website | # genes | Date of download |
|---|---|---|---|---|
| AD | ALZgene | www.alzgene.org | 193 | Sept. 20, 2010 |
| PD | PDgene | www.pdgene.org | 268 | Sept. 19, 2010 |
| MS | MSgene | www.msgene.org | 186 | Sept. 21, 2010 |
| SZ | SZGene | www.szgene.org | 155 | Sept. 21, 2010 |
| Autism | AutDB Autism Genetic Database |
www.mindspec.org wren.bcf.ku.edu |
131 | Sept. 21, 2010 |
AD: Alzheimer’s disease; PD: Parkinson’s diseases; MS: multiple sclerosis; SZ: schizophrenia.
The five resulting gene sets were then compared. In order to assess the significance of the overlap between the gene sets of each disease pair, we used a hypergeometric model. Namely, let N denote the total number of genes in the human genome (estimated to be around 21,000 at the time of this study). Let m and n respectively denote the number of genes associated with each disease (selected as described in the previous paragraph). If the number of genes that are common to both of these sets is k, then we computed the p-value of this overlap as follows:
To identify the pathways and enriched functional categories of genes shared between AD and PD, the commercial software Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/) was used.
Construction of gene networks associated with diseases:
Pathway analysis provides useful information on known biological processes common to diseases; however, it is limited in extracting novel information. PPI networks are useful in this respect, since they provide a comprehensive map of functional relationships among products of genes in the human genome. For this reason, we also mapped AD genes and PD genes to the human interactome, extracted the networks associated with AD and PD, and investigated the relationship of two diseases at the network level. We acknowledge that this approach may neglect important interactions; as such databases are currently incomplete. However, we had two guiding principles in the study. First, we wished to use only well curated interactions with few false positives. Second, we wanted the interaction set used to be “open source” so the results could be replicated by other groups. Thus, to fulfill both these criteria, we constructed networks using the human interactome downloaded from Human Protein Reference Database (HPRD)34. The human interactome downloaded from the HPRD server (www.hprd.org, version 9.0, downloaded in September 2010) had 9,453 protein-coding genes (genes and their encoded protein are used interchangeably in this paper) and 36,867 interactions among these gene products. We applied the Steiner tree algorithm35 to identify sub-networks that could connect AD genes (or PD genes) effectively while minimizing the number of non-AD (or non-PD) genes added to construct the sub-network. In particular, 151 out of 193 AD genes and 181 out of 268 PD genes were present in the HPRD network, thus with the choice of HPRD our gene set included over 70% of the originally identified genes. These genes were used as “seeds” to construct each disease specific network using Steiner tree algorithm35. Steiner tree algorithm generated a sub-network by two steps: a) all seed genes are connected by adding a minimal number of non-seed genes; b), the network is simplified with the shortest paths between seed genes. This algorithm has been widely applied for the generation of various networks16,36,37. Once the Steiner tree algorithm was applied to each gene set, the resulting networks were considered as AD network and PD network respectively35. Finally, networks were visualized and analyzed using the Cytoscape38,39 software.
Sub-network detection using Sub-network Analysis and Scoring System (SASSy):
The AD and PD networks were further used to identify dysregulated sub-networks in diseases by integrating network data with mRNA expression data. The aim of this procedure was to find sub-networks in which genes exhibit coordinate differential expression in the disease. Here, coordinate differential expression was assessed in terms of the ability of a group of genes in discriminating the samples from disease patients and healthy individuals when their expression profiles were considered together. For this purpose, we used two previous published mRNA expression data for AD and PD40,41. We scored sub-networks using a sub-network scoring method developed by Chuang et al19 and searched for high scoring sub-networks using a search algorithm (SASSy) developed by our group22,23. The principle and procedure implemented by SASSy is summarized as follows: Given a small sub-network, the “sub-network activity” for that sub-network in each sample is computed by aggregating the mRNA expression of the genes in the sub-network in that sample. Subsequently, the mutual information (MI) between phenotype (disease or control) and sub-network activity is computed as a measure of the capability of “sub-network activity” for that sub-network to discriminate the two groups. MI measures the reduction of uncertainty in phenotype upon observation of the sub-network activity (aggregate mRNA-level expression) of the genes in the sub-network. To this end, a high MI score for a sub-network is an indicator of the coordinate mRNA-level dysregulation of the genes in the sub-network. SASSy exhaustively searches for sub-networks (composed of up to 5 genes) of the AD (PD) network to identify sets of genes with high MI, which is guaranteed to find all sub-networks with a maximum MI, as opposed to a heuristic algorithm19. Since, in this study, the networks were restricted to the neighborhood of genes associated with each disease, such an exhaustive search was feasible. Limiting the size of the sub-network to five is in fact arbitrary, and is defined only to limit the computational search time.
Results and discussion
Comparison of AD genes and PD genes:
The 193 AD genes were compared with 268 PD genes to investigate the relationship between AD and PD. The results showed that 52 genes were shared in these two gene sets, and this overlap was highly significant by hypergeometric test (p-value < 10−10), indicating a strong connection between AD and PD at the molecular level. Functional analyses of the 52 overlapping genes using IPA indicated that the top functions associated with these genes are “death of normal cell and neurons” (p-value < 10−7) and “nervous system development and function” (p-value < 10−8).
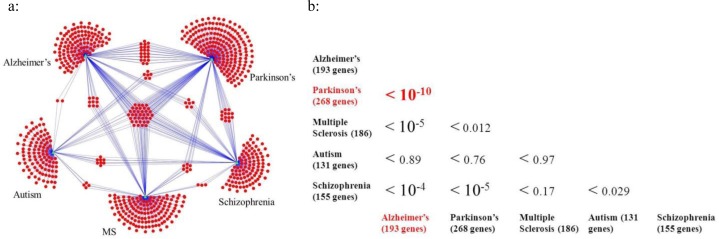
To elucidate the gene overlap with other mental disorders, in order to see if the AD/PD association was especially significant, additional gene sets for autism, MS and schizophrenia were collected, and compared in pair-wise fashion with AD and PD and each other. The results clearly showed that the genes that are confidently indentified with PD and AD have the most significant overlap (Figure 1a); the p-values for pairwise overlaps for all five diseases are summarized in Figure 1b. The genes confidently identified as being associated with autism had very modest overlap with other brain diseases (no p-value < 0.01) while MS and AD (p-value < 10−5), schizophrenia and PD (p-value < 10−5), schizophrenia and AD (p-value < 10−4) had significant overlap. The results also indicated that 38 genes were associated with at least three diseases (genes clustered in the center of Figure 1a) and 11 out of 38 genes are associated with four diseases (APOE, BDNF, GSTM1, IL1A, IL1B, IL10, MTHFR, MT-ND5, PON1, PTGS2, and SLC6A4), with the likely conclusion that these genes probably play a significant role for the proper functioning of brain. Among the 38 genes, seven are mitochondrial genes (Mt-ND1, Mt-ND2, Mt-ND3, Mt-ND5, Mt-COI, Mt-L2, and Mt-DLOOP), consistent with the important role of mitochondria in brain function and dysfunction42. Five of the 38 are interleukin or interleukin-related genes (IL1A, IL1B, IL6, IL10 and IL1rn) and it is well documented that IL1 is associated with behavior, neuroendocrine function, and sleep43; and dysregulated IL6 is linked with brain tumors44.
Figure 1.
Disease gene network. a: Circles represent genes associated with five common mental disorders, triangles represent diseases, edges connecting circles and triangles represent the association between genes and diseases. Please note that genes located in the center of (a) are associated with at least three diseases, but are not necessarily associated with both AD and PD. b: Summary of p-values for overlapping of the pair-wise comparison
Comparison of networks associated with AD and PD:
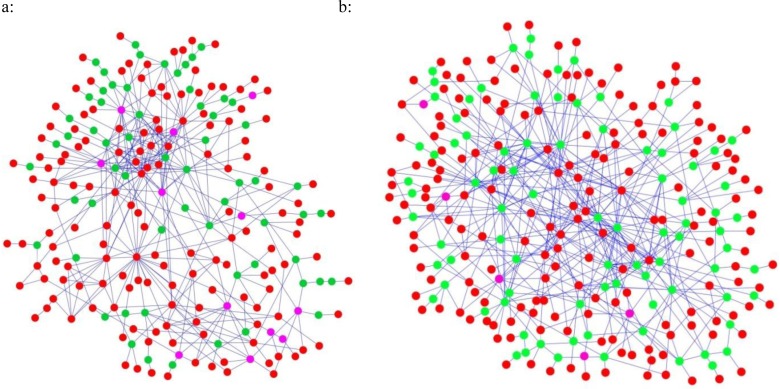
Based on AD genes and PD genes, two networks were extracted from the HPRD interactome using the Steiner tree algorithm. The resulting network for AD contained 225 genes and 387 interactions (AD network, Figure 2a). Among the initial 193 AD genes, 151 were retained in the network (the remaining 42 genes were absent in HPRD); to connect these 151 genes the algorithm added 74 additional genes. It is interesting to note that 13 out of these 74 new genes had prior evidence of association with AD in the database; they were not included in the AD genes as they did not satisfy the criterion of being present in at least four publications. The inclusion of these genes in the AD network provides further evidence for their functional association with other AD genes, and illustrates the power of a network-based approach. Similarly, the PD network had 273 genes and 502 interactions (Figure 2b), 185 PD genes were retained (the remaining 88 were absent in HPRD), and 5 of 88 added genes had evidence of PD association in the databases.
Figure 2.
Disease-specific networks extracted from HPRD using Steiner tree algorithm. a: AD network. b: PD network. Red nodes represent seed genes. Green nodes represent genes recruited by the algorithm. Pink nodes are recruited genes having evidence for the association with corresponding disease.
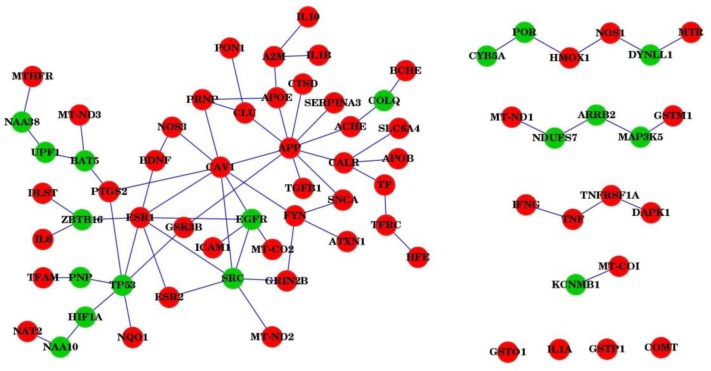
The comparison of AD network with PD network showed another level of relationship between the two diseases: 72 genes were shared in two networks; the network generation process added many genes that were common among the final disease networks. 51 out of 72 genes were clustered into a connected network, 4 genes were isolated from others, and the rest of genes formed four networks with 2, 4, 5, 6 genes respectively (Figure 3). Further analysis indicated that 54 out of these 72 genes were seed genes. Comparing these 54 seed genes with the AD and PD genes showed that 44 out of 54 genes were common in two disease gene sets. Furthermore, five of the AD genes (ACHE, APP, ATXN1, CLU and DAPK1) were included in the PD network by the Steiner tree algorithm, and five of PD genes (APOB, CALR, CAV1, NOS1, and TFRC) were included in AD network. In summary, more common genes were found when comparing at the network level due to the interactions between AD genes with PD genes.
Figure 3.
Common subnetworks between AD and PD networks. Red nodes represent seed genes in the AD and PD network construction. Green nodes represent genes recruited by the Steiner tree algorithm.
Several non-seed genes were added into the disease network due to their significant positions and connection to seed genes (Figure 3). For example, TP53, SRC, EGFR and ZBTB16 were not seed genes, they were included in the network because each of them had multiple interactions with seed genes. Three studies support the link between TP53 and AD45–47, while there have no evidence for the association of SRC, EGFR and ZBTB16 with AD or PD in the databases. Our findings suggest that they are likely associated with common neurodegenerative diseases.
Dysregulated sub-network detection using mRNA expression for AD and PD:
Integrating network information with mRNA expression data can help to identify sub-networks that are the most dysregulated in disease. We expect these highly dysregulated sub-networks (of five genes in this analysis) and their immediate neighbors, to include both important biomarkers of the disease process and to identify important points for drug interventions to reverse the dysregulations and thus reverse the disease process. To identify highly dysregulated sub-networks, the algorithm SASSy has been developed and tested to identify biomarkers and therapeutic targets related to cancer and sleep disorders22,23. These results have shown that SASSy is a powerful tool to assess the coordinate dysregulation of multiple functionally connected genes.
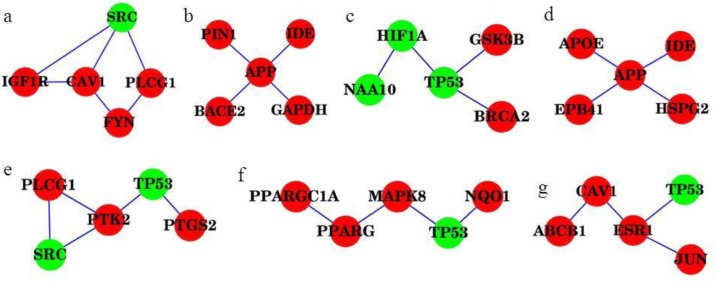
We applied SASSy to exhaustively search for small (up to five genes) dysregulated sub-networks in AD and PD. We report results here for the sub-networks that have the maximum MI in the analysis. The AD disease network was analyzed using mRNA expression data from hippocampal tissue (seven AD patients vs nine controls); this generated six sub-networks with equal MI (sub-networks a–f of Figure 4)40. Several genes well known to be associated with AD were found in these sub-networks, including APOE and APP (amyloid precursor protein). Most of the genes in the sub-networks were seeds from the original disease networks (colored red). This shows that the top sub-networks, as identified by SASSy, correctly identify known markers of disease. Several genes, like TP53 and APP, appeared multiple times, suggesting that they might mediate important crosstalk between different pathways. On the other hand, this is the first evidence for the association of AD with genes in the sub-networks such as TP53, SRC, PLCG1 and HIF1A, NAA10, etc. For example, NAA10 (N(alpha)-acetyltransferase 10) helps modulate HIF1A (hypoxia inducible factor 1) acetylation, thereby promoting its degradation48. These novel genes are potential targets for further experimental investigation in AD. Furthermore, six genes (APOE, CAV1, FYN, GSK3B, NQO1 and PTS2) from the AD sub-networks are also PD seed genes, which provides evidence that the most significantly dysregulated sub-networks in AD have strong connections to PD.
Figure 4.
Highly significant dysregulated subnetworks detected by SASSy for (a–f) AD and (g) PD. The genes that were originally seeds, and thus have known significant association with disease are in red and the added nodes added by the Steiner Tree algorithm are in green.
In the case of PD, one significant sub-network (network g of Figure 4) was detected to be dysregulated using mRNA expression data from the substantia nigra region (16 PD patients and 9 healthy controls)41. Four out of five genes (ABCB1, CAV1, ESR1, and JUN) have strong evidence for the association with PD. This is the first evidence that TP53 has a significant association with PD. TP53 and CAV1 are present in dysregulated sub-networks of both AD and PD, which suggests another connection between AD and PD.
Comparison of disease networks with drug targets
We expect that our disease-based molecular networks are an informative framework for analyzing effects of AD and PD drug treatments. Also, we were interested to know if the dysregulated sub-networks identified by SASSy revealed any known drug targets, or were closely associated with known targets. AD and PD approved drugs and targets were identified by searching the IPA database (Table 2); we then compared the components of networks (Figure 2) with the targets of current drugs. The drugs and drug targets in the IPA database are shown in Table 2, the targets in bold were found in our disease-associated networks, and the disease networks included most of the targets. Several nodes of the disease associated networks link directly with more than one drug target, for example, CollagenQ (COLQ) directly links with ACHE and BCHE (targets for AD), these nodes are also in the common sub-networks for AD and PD (see Figure 3) while GIPC1 (PDZ domain-containing protein GIPC1) is linked with both DRD2 and DRD3 (dopamine receptor targets for PD).
Table 2.
Current drugs for AD and PD, and their targets.
| Drug | Disease | Targeta |
|---|---|---|
| Levodopa/carbidopa/entacapone | PD | COMT, DDC |
| Bromocriptine | PD | DRD1, DRD2 |
| Selegiline | PD | MAOB |
| Pramipexole | PD | DRD1,DRD2,DRD3,DRD4,DRD5 |
| Ropinirole | PD | DRD1,DRD2,DRD3,DRD4,DRD5 |
| Amantadine | PD | DRD2,DRD5 |
| Rasagiline | PD | MAOB |
| Donepezil | AD | ACHE |
| Rivastigmine | AD | ACHE,BCHE |
| Galantamine | AD | ACHE |
| Memantine | AD | CHRNA4,GRIN1,GRIN2A,GRIN2B, GRIN2C,GRIN2D, GRIN3A, GRIN3B, HTR3A |
full names of drug targets: COMT: catechol-O-methyltransferase; DDC: dopa decarboxylase; DRD: dopamine receptor D; MAOB: monoamine oxidase B; ACHE: acetylcholinesterase; BCHE: butyrylcholinesterase; CHRNA4: cholinergic receptor, nicotinic, alpha 4; GRIN: glutamate receptor, ionotropic, N-methyl D-aspartate; HTR3A: 5-hydroxytryptamine (serotonin) receptor 3A.
Although no drug target appeared in the SASSy scored sub-networks (Figure 4), as they are limited to five nodes, several direct interacting partners of drug targets are present in the sub-networks. For example, three nodes, SRC, FYN and PLCG1 of the subnetwork (a) in Figure 4 are direct partners of GRIN2B (glutamate receptor, ionotropic, N-methyl D-aspartate 2B), which is a target of Memantine, a treatment for AD. APP, found in a subnetwork of 4b and 4d, is an interacting partner of ACHE, the target of Donepezil and other agents. As SASSy was able to identify important nodes both dysregulated in disease and those directly connected to important drug targets, the sub-network nodes may include other potential important new targets that could become the focus of future studies. For example, FYN (a proto-oncogene tyrosine-protein kinase) and SRC related kinases, seen to be highly dysregulated in AD, are important candidates for new drug development in cancer and neuronal diseases49. IGF1R, (Insulin like growth factor receptor 1), a dysregulated target in AD (Figure 4a), is an important target for development of inhibitors in cancer50, interestingly it is up-regulated in AD and thus these agents could be explored for their effects in blocking or reversing the disease. Overall, agents mediating functions of SRC, FYN, and IGF1R represent logical drugs for validating the molecular networks seen in this study, as well for the proteins are potential new targets for AD therapies.
Conclusion
In summary, we identified genes and sub-networks associated with AD and PD as well as sub-networks that connected the two diseases. These strong connections included three levels of evidence: significant overlap of AD and PD associated genes, large common regions among AD and PD networks, and shared nodes for highly dysregulated sub-networks. In addition, the dysregulated sub-networks identified by mutual information scoring of PPI sub-networks with gene expression data using SASSy provided powerful identification of both known and potential new biomarkers for AD and PD and potential novel drug targets for AD.
Acknowledgments
This work is supported in part by the Case Western Reserve University/Cleveland Clinic CTSA (Grant Number UL1 RR024989 including a supplement to support T1 research) from the National Center for Research Resources (NCRR) to MRC, a component of the National Institutes of Health and NIH roadmap for Medical Research. Support is also acknowledged from the National Science Foundation (CCF-053195 and IIS-0916102) to MK. Support for the development of SASSY is also provided by NCRR through a grant to Neo Proteomics, R43RR031932. The SASSy software tool is available exclusively through Neo Proteomics, Inc. (www.neoproteomics.net).
Reference
- 1.Jankovic J. Parkinson's disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. doi:79/4/368. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimers_Association Alzheimer's Association Report 2011 Alzheimer's disease facts and figures. Alzheimers Dement. 2011;7:208–244. doi: 10.1016/j.jalz.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Nagayama H, et al. Influence of ageing on the pharmacokinetics of levodopa in elderly patients with Parkinson's disease. Parkinsonism Relat Disord. 2011;17:150–152. doi: 10.1016/j.parkreldis.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Evans DA, et al. Prevalence of Alzheimer's disease in a community population of older persons. Higher than previously reported. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- 5.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 6.Simon-Sanchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 8.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 11.Bekris LM, Mata IF, Zabetian CP. The genetics of Parkinson disease. J Geriatr Psychiatry Neurol. 2010;23:228–242. doi: 10.1177/0891988710383572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders AM, et al. Apolipoprotein E epsilon 4 allele distributions in late-onset Alzheimer's disease and in other amyloid-forming diseases. Lancet. 1993;342:710–711. doi: 10.1016/0140-6736(93)91709-u. [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos MH, et al. Mapping of a gene for Parkinson's disease to chromosome 4q21–q23. Science. 1996;274:1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 14.Bertram L, Tanzi RE. Alzheimer's disease: one disorder, too many genes? Hum Mol Genet. 2004. pp. R135–141. 13 Spec No 1. [DOI] [PubMed]
- 15.Sutherland GT, et al. A cross-study transcriptional analysis of Parkinson's disease. PLoS One. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, et al. Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS One. 2010;5:e11351. doi: 10.1371/journal.pone.0011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Zhao Z. Functional features, biological pathways, and protein interaction networks of addiction-related genes. Chem Biodivers. 2010;7:1153–1162. doi: 10.1002/cbdv.200900319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Systems biology analyses of gene expression and genome wide association study data in obstructive sleep apnea. Pac Symp Biocomput. 2011:14–25. doi: 10.1142/9789814335058_0003. doi:9789814335058_0003. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marder K, et al. Risk of Alzheimer's disease in relatives of Parkinson's disease patients with and without dementia. Neurology. 1999;52:719–724. doi: 10.1212/wnl.52.4.719. [DOI] [PubMed] [Google Scholar]
- 25.Levy G, et al. Lack of familial aggregation of Parkinson disease and Alzheimer disease. Arch Neurol. 2004;61:1033–1039. doi: 10.1001/archneur.61.7.1033. [DOI] [PubMed] [Google Scholar]
- 26.Wilson RS, Schneider JA, Bienias JL, Evans DA, Bennett DA. Parkinsonianlike signs and risk of incident Alzheimer disease in older persons. Arch Neurol. 2003;60:539–544. doi: 10.1001/archneur.60.4.539. [DOI] [PubMed] [Google Scholar]
- 27.Ditter SM, Mirra SS. Neuropathologic and clinical features of Parkinson's disease in Alzheimer's disease patients. Neurology. 1987;37:754–760. doi: 10.1212/wnl.37.5.754. [DOI] [PubMed] [Google Scholar]
- 28.Galasko D, et al. Clinical-neuropathological correlations in Alzheimer's disease and related dementias. Arch Neurol. 1994;51:888–895. doi: 10.1001/archneur.1994.00540210060013. [DOI] [PubMed] [Google Scholar]
- 29.Masters CL, et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 31.Tsigelny IF, et al. Mechanisms of hybrid oligomer formation in the pathogenesis of combined Alzheimer's and Parkinson's diseases. PLoS One. 2008;3:e3135. doi: 10.1371/journal.pone.0003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandal PK, Pettegrew JW, Masliah E, Hamilton RL, Mandal R. Interaction between Abeta peptide and alpha synuclein: molecular mechanisms in overlapping pathology of Alzheimer's and Parkinson's in dementia with Lewy body disease. Neurochem Res. 2006;31:1153–1162. doi: 10.1007/s11064-006-9140-9. [DOI] [PubMed] [Google Scholar]
- 33.Masliah E, et al. beta-amyloid peptides enhance alpha-synuclein accumulation and neuronal deficits in a transgenic mouse model linking Alzheimer's disease and Parkinson's disease. Proc Natl Acad Sci U S A. 2001;98:12245–12250. doi: 10.1073/pnas.211412398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- 35.Klein P, Ravi R. A Nearly Best-Possible Approximation Algorithm for Node-Weighted Steiner Trees. J Algorithm. 1995;19:104–115. [Google Scholar]
- 36.Scott MS, et al. Identifying regulatory subnetworks for a set of genes. Mol Cell Proteomics. 2005;4:683–692. doi: 10.1074/mcp.M400110-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Bailly-Bechet M, et al. Finding undetected protein associations in cell signaling by belief propagation. Proc Natl Acad Sci U S A. 2011;108:882–887. doi: 10.1073/pnas.1004751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blalock EM, et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesnick TG, et al. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007;3:e98. doi: 10.1371/journal.pgen.0030098. doi:06-PLGE-RA-0572. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calabrese V, Scapagnini G, Giuffrida Stella AM, Bates TE, Clark JB. Mitochondrial involvement in brain function and dysfunction: relevance to aging, neurodegenerative disorders and longevity. Neurochem Res. 2001;26:739–764. doi: 10.1023/a:1010955807739. [DOI] [PubMed] [Google Scholar]
- 43.Rothwell NJ. Functions and mechanisms of interleukin 1 in the brain. Trends Pharmacol Sci. 1991;12:430–436. doi: 10.1016/0165-6147(91)90623-z. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells. 2009;27:2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emahazion T, et al. SNP association studies in Alzheimer's disease highlight problems for complex disease analysis. Trends Genet. 2001;17:407–413. doi: 10.1016/s0168-9525(01)02342-3. doi:S0168-9525(01)02342-3 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Rosenmann H, et al. An association study of the codon 72 polymorphism in the pro-apoptotic gene p53 and Alzheimer's disease. Neurosci Lett. 2003;340:29–32. doi: 10.1016/s0304-3940(03)00049-1. doi:S0304394003000491 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Scacchi R, Gambina G, Moretto G, Corbo RM. Association study between P53 and P73 gene polymorphisms and the sporadic late-onset form of Alzheimer's disease. J Neural Transm. 2009;116:1179–1184. doi: 10.1007/s00702-009-0276-z. [DOI] [PubMed] [Google Scholar]
- 48.Arnesen T, et al. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS Lett. 2005;579:6428–6432. doi: 10.1016/j.febslet.2005.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schenone S, et al. Fyn kinase in brain diseases and cancer: the search for inhibitors. Curr Med Chem. 2011;18:2921–2942. doi: 10.2174/092986711796150531. [DOI] [PubMed] [Google Scholar]
- 50.Lee AV, Yee D. Targeting IGF-1R: at a crossroad. Oncology (Williston Park) 2011;25:535–536. discussion 551. [PMC free article] [PubMed] [Google Scholar]