Abstract
Extracting and encoding clinical information captured in free text with standard medical terminologies is vital to enable secondary use of electronic medical records (EMRs) for clinical decision support, improved patient safety, and clinical/translational research. A critical portion of free text is comprised of ‘summary level’ information in the form of problem lists, diagnoses and reasons of visit. We conducted a systematic analysis of SNOMED-CT in representing the summary level information utilizing a large collection of summary level data in the form of itemized entries. Results indicate that about 80% of the entries can be encoded with SNOMED-CT normalized phrases. When tolerating one unmapped token, 96% of the itemized entries can be encoded with SNOMED-CT concepts. The study provides a solid foundation for developing an automated system to encode summary level data using SNOMED-CT.
Introduction
Much of the data in electronic medical records (EMRs) is in free text format because compared to structured data, free text is a more efficient way to express concepts and events as a result of dictation transcription, direct entry, or deployment of speech recognition applications.1 Extracting and encoding clinical information captured in free text with standard medical terminologies is critical to enable secondary use of EMRs for clinical and translational research. Medical documentation tends to be organized around problems.2 The summary level information related to problems has been used by health care personnel to concisely convey a patient’s problems, and they are important for clarifying and reasoning at the point of care. Encoding summary level information with standard medical terminology is an important step towards secondary uses of EMRs.
One of the popular medical terminologies for coding clinical information is SNOMED-CT.3,4 It provides more granular coding of clinical information found in EMRs than terminologies such as the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM). SNOMED-CT allows compositional encoding of clinical concepts and multiple concepts can be combined to form a more detailed representation of the clinical problem. For example, the medical condition described as “Hypertrophic actinic keratosis with focus of squamous cell carcinoma in-situ, right dorsal hand” can be represented by an expression containing four SNOMED-CT concepts (underlined). Compositional expressions allow more complex descriptions and therefore provide more complete representation of medical concepts.
We are currently in the process of improving Mayo production automated encoding system, Clinical Notes Indexing (CNI). Since it is critical to encode summary level information correctly, we conducted a systematic analysis on a large collection of summary level data in the form of itemized entries extracted from Mayo Clinic’s Enterprise Data Trust (EDT).5 Specifically, we would like to find out how summary level information is distributed. Additionally, one fundamental problem faced by medical terminologies when used for encoding text is their coverage. SNOMEDCT is empowered by adopting compositional schemes in encoding. We also would like to know how comprehensive SNOMED-CT is in representing summary level information found in clinical notes. Furthermore, as a large and heterogeneous medical terminology, it is impossible to maintain, audit, and assure the quality of SNOMED-CT in a completely manual way. Observing physicians tend to organize closely related concepts as one itemized entry, we wanted to see if it is feasible to uncover some missing relationships using the acquired summary level data. The findings of our systematic analysis are reported in this paper.
Background and Related Work
Compositional Scheme in SNOMED-CT
There are two types of concepts in SNOMED-CT, primitive or non-primitive, where primitive concepts form the building block to compose complex concepts. Encoding using compositional scheme terminologies may introduce nonsense combinations and multiple combinations of the same concept, creating difficulties in finding problems when compositional scheme is not carefully designed. In the other words, if we simply combine multiple concepts without specific attributes, it is still very difficult for automated systems to interpret the concepts. For example, when representing “Hypertrophic actinic keratosis with focus of squamous cell carcinoma in-situ, right dorsal hand” as a list of “Hypertrophic actinic keratosis”, “squamous cell carcinoma in-situ”, “right”, and “dorsal hand”, we lose the information that right and dorsal hand are connected. It would be interesting to see the co-occurrence statistics between concepts and identify significant co-occurring pairs.
Related work
As a reference terminology system, there are multiple efforts in evaluating or encoding summary level concepts using SNOMED-CT. One such effort is the UMLS Clinical Observations Recording and Encoding (CORE) project which defines a subset of SNOMED-CT concepts occurred frequently on summary level datasets collected from several large-scale institutions (including Mayo Clinic). There are several related studies that map summary level terms to SNOMED-CT. For example, Wade and Rosenbloom mapped 1,510 terms representing legacy interfacing concepts used at the Vanderbilt EMR systems to SNOMED-CT and reported that it is critical for terminologists to have considerable clinical background when mapping interfacing concepts to SNOMED-CT.6,7 They also found some quality issues related to SNOMED-CT such as redundancies or deficiencies. Nadkarni and Darer also conducted a study which maps legacy ICD-9CM problem lists used in their organization to SNOMEDCT.8 Among 2,194 ICD-9CM codes, 784 (35.7%) required manual mapping and searching SNOMED for the correct concepts often required extensive application of knowledge of both English and medical synonymy. Peter Elkin et al. evaluated the content coverage of SNOMED-CT with 4,996 most common summary level unduplicated text strings associated with inpatient and outpatient episodes of care.9 Their study indicates SNOMED-CT had a sensitivity of 92.3%, a specificity of 80.0%, and a positive predictive value of 99.8% when automatically encoding the summary level at Mayo Clinic.
In this study, we conducted a systematic corpus analysis on the ability of SNOMED-CT to representing summary level data at Mayo Clinic. Different from Elkin’s study which focused on most common itemized entries, we evaluate concept coverage and degree of compositions required for all itemized entries appearing in clinical notes. We also assess the relationship coverage in SNOMED-CT based on significant co-occurring pairs.
Materials
SNOMED-CT
We used the descriptions, concepts, and relationships tables available in the latest release of SNOMED-CT (International release 08/2011). The descriptions table provides several synonymous terms for each concept, and among them, a unique fully specified name (FSN) is provided which includes the corresponding semantic types (e.g., disorder, event, or attribute etc).
The Unified Medical Language Systems (UMLS)
To enable aggressive mapping, we used the Unified Medical Language System (UMLS), developed and maintained by the National Library of Medicine (NLM).10 The goal of the UMLS is to overcome retrieval problems caused by differences in terminologies and the scattering of relevant information across many databases, by integrating different electronic biomedical terminologies into one concept-oriented knowledge base. It contains three knowledge sources: the Metathesaurus (META), the Specialist Lexicon, and the Semantic Network. The META provides a uniform, integrated distribution format for about 160 biomedical vocabularies and classifications, and links many different terms for the same concepts. Each distinct concept has been assigned a unique concept identifier (CUI). Concept names corresponding to the same concept are assigned the same CUI. The Specialist Lexicon contains syntactic information for many terms, component words, and English words, including verbs, which do not appear in the META. The Semantic Network contains information about the types or categories (e.g., Disease or Syndrome, Virus) to which all META concepts have been assigned and the permissible relationships among these types (e.g., Virus causes Disease or Syndrome).
Experimental Methods
Extraction of a corpus consisting of summary level entries
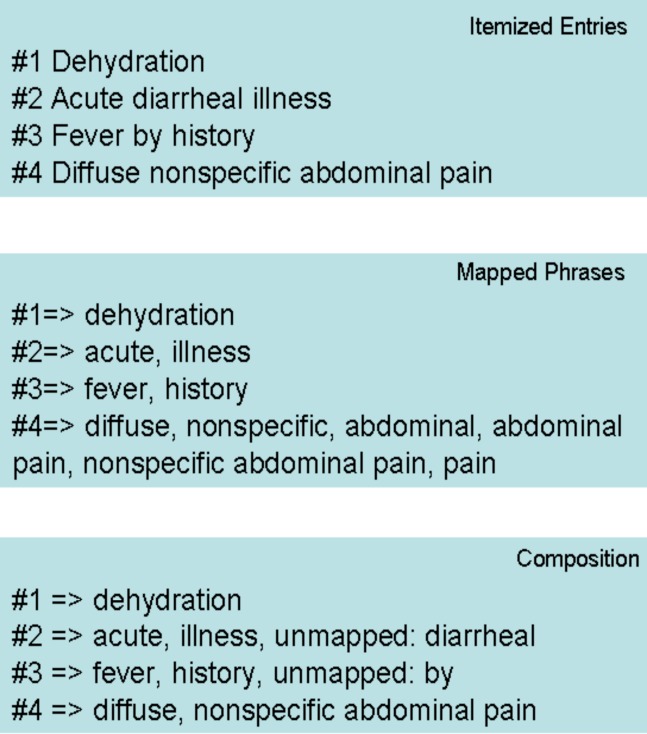
Summary level information in documents generally appears as itemized entries in our EMRs. The top panel in Figure 1 shows an example of a piece of summary level information in a report which contains four itemized entries. We extracted a corpus consisting of itemized summary level entries from Mayo Enterprise Data Trust (EDT) which contains over 15 years’ EMRs at Mayo Clinic10.
Figure 1.
An example of summary level data in our study.
Dictionary Lookup
Due to the high diversity of natural language expressions in terms of inflection, derivation, and synonymy, we take advantage of a comprehensive list of synonyms provided by the UMLS to find SNOMED-CT codes. Both the input text and dictionary entries are normalized to facilitate practical flexible matching. The lookup normalization step includes a) changing upper case letters to lower case, b) converting tokens (words) to their base forms according to the UMLS SPECIALIST lexicon, and c) ignoring punctuation marks. After normalization, dictionary entries with at most 10 words, and at most 100 and at least 3 characters are kept. All matching occurrences, including overlapping matches, are recorded during dictionary lookup. The second panel in Figure 1 demonstrates the dictionary lookup results. For example, the fourth item in the itemized entries “Diffuse nonspecific abdominal pain” is mapped to six normalized phrases.
Assessment
We processed the acquired corpus using the dictionary lookup procedure. Statistics of the occurrences of the itemized entries (i.e., the raw summary level data) and the mapped phrases (i.e., UMLS normalized phrases with at least one occurrence in the corpus) were obtained. Note some of the terms in SNOMED-CT are composition terms. We assume less granular SNOMED-CT terms are more accurate representations of the itemized entries because they capture not only the information in more granular terms but also the relationships among them. We represented each itemized entry with the minimum number of SNOMED-CT normalized phrases to measure the degree of composition required We also checked the number of words that failed to be mapped to SNOMED-CT. The bottom panel in Figure 1 indicates the composition level for the fourth itemized entry is 2 since two normalized phrases, “diffuse” and “nonspecific abdominal pain” can represent its meaning.
The assessment of the co-occurrence information of SNOMED-CT concepts is based on concepts that co-occurred together in an itemized entry. We hypothesized that concepts semantically related have higher probability to appear in a single itemized entry than those semantically unrelated. Let C1 and C2 be two concepts. We used χ2 test to rank the dependency of C1 and C2 in itemized entries:
where O(C1, C2) is the observed frequency and E(C1, C2) is the expected frequency which can be estimated as:
where O(·) is the observed frequency and TOTAL is the total number of entries. We use χ2 scores to rank concept pairs where higher scores indicate more dependency. Since SNOMED-CT provides a relationship table, we use it to see how those existing relationships are ranked.
Results and Discussion
Distribution of itemized entries and mapped phrases based on the UMLS
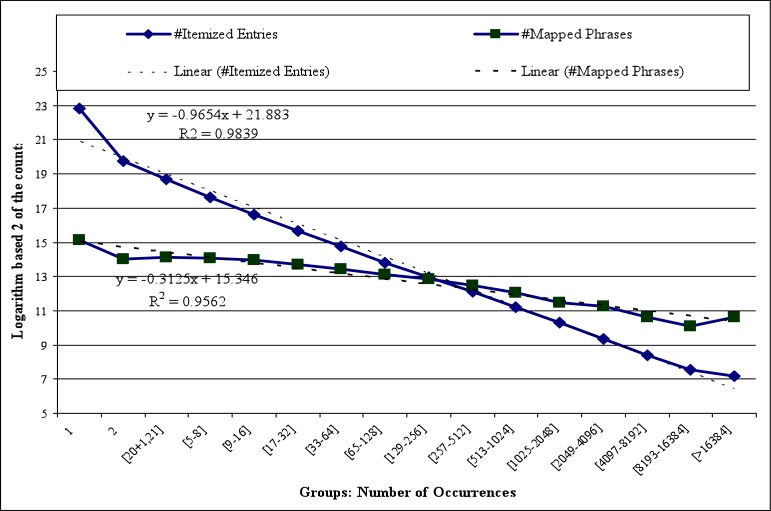
There are 36m itemized entries extracted from 14.7m documents that contain summary level data, with an average of 2.43 entries per document. The number of unique itemized entries is 9.16m with an average of 3.93 occurrences per entry. About 7.4m entries occur only once in the corpus. Entries with over 100K occurrences in our corpus are shown in the second column of Table 1 (round to the closest thousands). There are 170m occurrences of mapped phrases corresponding to 164k unique normalized phrases. The last column of Table 1 shows the total number of occurrences of the corresponding normalized phrases in the corpus (i.e., including ones that occurred alone as itemized entries and those that co-occurred with other phrases). Figure 2 shows the statistics of entries and the statistics of mapped phrases. The x-axis represents 16 occurrence groups, where group 1 and group 2 include those occurring once or twice, respectively, followed by groups [2i+1, 2i+1], for i from 1 to 13, and the last group includes those occurring more than 214=16284 times. The y-axis is the logarithm base 2 of the number of itemized entries or mapped phrases. From Figure 2, we can clearly see the distribution of itemized entries follows Zipf’s law almost perfectly (R2 is close to 1). The distribution of mapped phrases also follows Zipf’s law.
Table 1.
Most frequent itemized entries in summary level data.
| Term | # As a single itemized entry (in thousands) | # Total occurrences (in thousands) |
|---|---|---|
| Hypertension | 777 | 1260 |
| Hyperlipidemia | 566 | 814 |
| Health-maintenance | 230 | 402 |
| Depression | 175 | 456 |
| Obesity | 151 | 288 |
| Coronary artery disease | 147 | 380 |
| Osteoporosis | 124 | 199 |
| Hypothyroidism | 122 | 251 |
| Diabetes mellitus | 103 | 386 |
Figure 2.
Statistics of itemized entries and the number of mapped phrases.
Distribution of SNOMED-CT phrases in the corpus
There are 199,720 normalized UMLS phrases that have corresponding SNOMED concepts. Among them, 99,261 (49.7%) occurred at least once in our corpus. The most frequent phrases are various qualifiers including “history of”, “right”, “after”, “status post”, “left” etc. The most frequent finding is “pain” and the most frequent disorder is “hypertension”. Table 2 lists the number of mapped phrases (column 2) and their average number of occurrences (column 3) for SNOMED-CT semantic tags with at least 500 normalized phrases. For example, the second row indicates there are 44,116 normalized disorder phrases with an average number of occurrences as 1,221. Disorders, findings, and procedures top the number of normalized phrases and the average occurrences of qualifier value and attribute phrases are highest in the corpus.
Table 2.
Distribution statistics of SNOMED Semantic Tags.
| SNOMED-CT Semantic Tag | # normalized phrases | Average occurrence of normalized phrases |
|---|---|---|
| disorder | 44,116 | 1,221 |
| Finding | 16,030 | 1,724 |
| procedure | 12,825 | 1,195 |
| body structure | 8,528 | 2,521 |
| substance | 6,761 | 758 |
| morphologic abnormality | 6,280 | 2,078 |
| qualifier value | 6,025 | 9,795 |
| Situation | 4,077 | 3,346 |
| Product | 3,503 | 808 |
| observable entity | 3,137 | 2,074 |
| Organism | 1,975 | 692 |
| Physical | 1,550 | 940 |
| regime/therapy | 931 | 1,476 |
| Attribute | 739 | 13,242 |
| Event | 528 | 1,044 |
SNOMED-CT coverage statistics
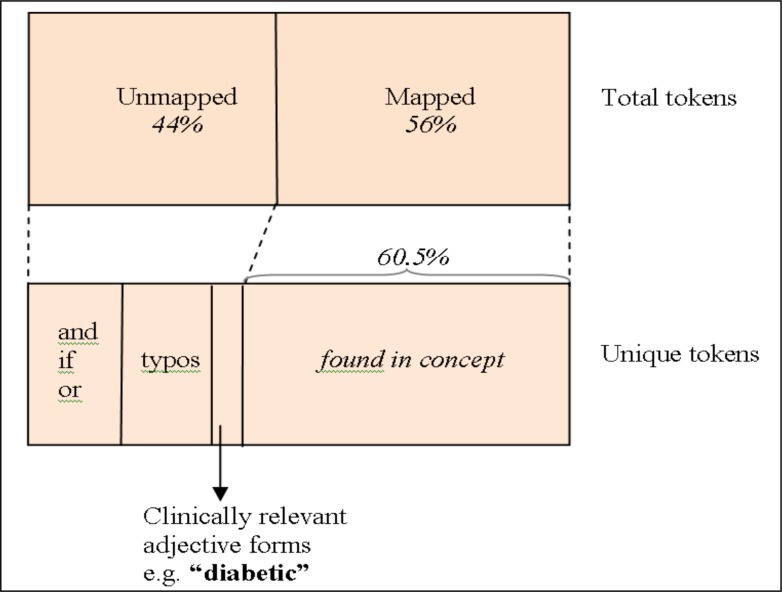
There are 68.3m tokens in our corpus and 56.8% of them are covered by mapped phrases. Most of the tokens that failed to be mapped (corresponding to 88k unique tokens) are prepositions or conjunctions or numbers such as “by”, “and”, “with”, “of”, “the” etc. Over half of the 88k unique tokens occurred less than three times, potentially typos (Figure 3). We notice a significant number of words are clinically relevant but appear in adjective form (e.g., “diarrheal”, “dystrophic”, “diabetic”, “mycotic”, “neuropathic”, “posttraumatic”, or “premenopausal”). When their associated concepts are not included in SNOMED-CT, they are considered as unmapped. The phrases “posttraumatic arthritis” and “diabetic ulcerations” are not included in SNOMED-CT, the best mappings found for them are “arthritis” and “ulcerations”. Therefore, they are considered as unmapped tokens for the corresponding entries. The creation of dictionary that maps the adjectival forms to the noun forms present in SNOMED-CT can resolve the mappings for adjectival forms. There are 19.5k unique unmapped tokens corresponding to a total of 2.45m tokens ending with three popular adjective suffixes (e.g., “ic”, “al”, or “ive”). Additionally, some of the undefined tokens are synonyms of known terms which will require manual curation for mapping them to correct codes.
Figure 3.
Token distribution in the corpus. Mapping of distribution of total tokens to the distribution of unique tokens is shown using dashed lines.
After ignoring stop words or non-functional words, 5.55m (60.5%) unique entries corresponding to a total of 28.9m (80.3%) itemized entries can be mapped to a set of SNOMED-CT concepts (Figure 3). If we allow one unmapped token for an entry, 8.11m (88.5%) unique entries corresponding to a total of 34.5m (96%) itemized entries can be mapped. We notice that 32k unique entries corresponding to 306k itemized entries could not be mapped to any SNOMED-CT code. The most frequent one with no code is the string, “PAME”, which stands for “pre-anaesthetic medical evaluation”.
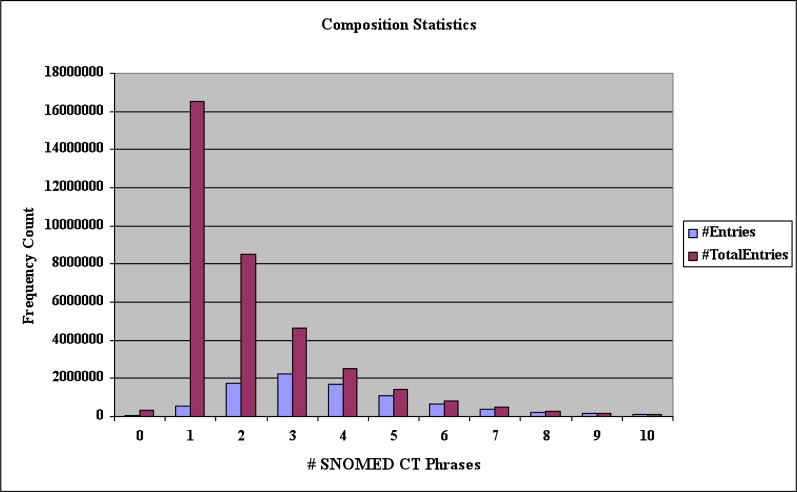
Compositional level statistics
Table 3 and Figure 4 show the statistics of the number of SNOMED-CT normalized phrases for representing each itemized entry. Most of the entries can be represented by one to three SNOMED-CT normalized phrases. There are 565k unique entries corresponding to a total 16,522k itemized entries with an average of 29.22 occurrences per unique entry that can be represented using one SNOMED code. 83% of the entries were mapped to 3 or fewer concepts. The proportion of phrases that can be encoded into three or less concepts would be much larger, given the fact that many itemized entries in the problem lists consists of several phrases. A limitation of the composition analysis we have performed is that we have not considered post-coordination rules described in SNOMEDCT, but have simply combined the concepts found by the dictionary lookup.
Table 3.
Statistics of the composition level for the entries.
| Composition | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #unique Entries (in thousands) | 32 | 565 | 1,772 | 2,218 | 1,715 | 1,103 | 671 | 403 | 244 | 152 | 95 |
| #Total Entries (in thousands) | 306 | 16,522 | 8,532 | 4,653 | 2,483 | 1,403 | 812 | 474 | 281 | 174 | 107 |
| Average occurrences | 9.70 | 29.22 | 4.82 | 2.10 | 1.45 | 1.27 | 1.21 | 1.18 | 1.15 | 1.14 | 1.13 |
| Cumulative % occurrence | 1 | 47 | 70 | 83 | 90 | 94 | 96 | 98 | 99 | 99 | 99 |
Figure 4.
Compositional statistics. The x-axis shows the number of SNOMED CT phrases needed to encode an entry. The y-axis is the number of unique entries and the total number itemized entries.
Co-occurrence statistics
There are a total of 4.03m pairs of concepts with co-occurrences at least 100 times in the corpus. Only a very small portion (16,500 out of 1.44m or 1.14%) of pairs from the relationship table are found among those 4.03m pairs. Note that the actual coverage of SNOMED-CT relationships for co-occurred pairs can be much higher than 1.14% since certain relationships can be obtained through ontological propagation. On filtering out pairs with χ2 scores less than 10000, 0.86m pairs are kept including 14,499 (87.9%) out of the 16,500 pairs from the SNOMED-CT relationship table. We manually examined the top ranked pairs and found that they are semantically related. For example, the procedure “Tylectomy” and the disorder “Intraductal carcinoma in situ of breast” have a χ2 score of 217,318. When concepts co-occur significantly in itemized entries, it can indicate novel relationships among those concepts since physicians tend to group closely related problems as a single itemized entry.
One limitation of the study is that we excluded terms with more than 10 words or those with fewer than three letters or more than 100 letters during the dictionary lookup. Therefore, our study does not account for one or two-letter terms (mostly abbreviations) and very long phrases. We feel one or two-letter terms are highly ambiguous using our dictionary lookup procedure and it is not easy to disambiguate them. Another limitation of the study is that we use mapped phrases instead of mapped concepts for coverage statistics. Due to the fact that one string can be mapped to several concepts in SNOMED-CT (most of the time, those concepts are related concepts), it is sometimes infeasible or un-realistic to map one phrase to one SNOMED-CT code.
The list of adjectival forms that were mot mapped to SNOMEDCT can be utilized for improving the sensitivity of automated mapping tools. The concept co-occurrence method may be useful to discover concept relations for augmenting SNOMED-CT. Overall the statistics presented in this paper would benefit researchers to enhance use of SNOMEDCT for coding summary level clinical information.
Conclusion and Future Work
We have presented a study that analyzes the use of SNOMED-CT to encode summary level data. The results indicated that SNOMED-CT provides a good coverage for encoding summary level clinical information. The study provides a solid foundation for improving our automated system in using SNOMED-CT to encode summary level data. Future work includes adding adjective forms of the clinical terms to their noun phrases and discovering novel synonyms, and obtaining empirical compositional rules for a better encoding system.
Acknowledgments
This study was supported by the following grants: National Science Foundation ABI:0845523 and National Institute of Health R01LM009959A1.
References
- 1.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. Journal of the American Medical Informatics Association. 2011;18(2):181. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Vleck TT, Wilcox A, Stetson PD, Johnson SB, Elhadad N. Content and structure of clinical problem lists: A corpus analysis. AMIA Annu Symp Proc. 2008:753–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Spackman KA, Campbell KE. Compositional concept representation using SNOMED: towards further convergence of clinical terminologies. AMIA Annu Symp Proc. 1998:740–4. [PMC free article] [PubMed] [Google Scholar]
- 4.Spackman KA, Campbell KE, CÃ R. SNOMED RT: a reference terminology for health care. AMIA Annu Symp Proc. 1997:640–4. [PMC free article] [PubMed] [Google Scholar]
- 5.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. Journal of the American Medical Informatics Association : JAMIA. 2010 Mar-Apr;17(2):131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade G, Rosenbloom ST. Experiences mapping a legacy interface terminology to SNOMED CT. BMC Med Inform Decis Mak. 2008;8(Suppl 1):S3. doi: 10.1186/1472-6947-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade G, Rosenbloom ST. The impact of SNOMED CT revisions on a mapped interface terminology: terminology development and implementation issues. Journal of biomedical informatics. 2009 Jun;42(3):490–3. doi: 10.1016/j.jbi.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadkarni PM, Darer JA. Migrating existing clinical content from ICD-9 to SNOMED. Journal of the American Medical Informatics Association. 2010;17(5):602. doi: 10.1136/jamia.2009.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkin PL, Brown SH, Husser CS, et al. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists. Mayo Clin Proc. 2006 Jun;81(6):741–8. doi: 10.4065/81.6.741. [DOI] [PubMed] [Google Scholar]
- 10.Hubble J, Koller WC, Atchison P, et al. Linear pharmacokinetic behavior of ropinirole during multiple dosing in patients with Parkinson's disease. J Clin Pharmacol. 2000 Jun;40(6):641–6. [PubMed] [Google Scholar]