Abstract
Researchers estimate that electronic health record systems record roughly 2-million ambulatory adverse drug events and that patients suffer from adverse drug events in roughly 30% of hospital stays. Some have used structured databases of patient medical records and health insurance claims recently—going beyond the current paradigm of using spontaneous reporting systems like AERS—to detect drug-safety signals. However, most efforts do not use the free-text from clinical notes in monitoring for drug-safety signals. We hypothesize that drug–disease co-occurrences, extracted from ontology-based annotations of the clinical notes, can be examined for statistical enrichment and used for drug safety surveillance. When analyzing such co-occurrences of drugs and diseases, one major challenge is to differentiate whether the disease in a drug–disease pair represents an indication or an adverse event. We demonstrate that it is possible to make this distinction by combining the frequency distribution of the drug, the disease, and the drug-disease pair as well as the temporal ordering of the drugs and diseases in each pair across more than one million patients.
Introduction
Detecting adverse drug events in clinical records remains a challenging problem. The U.S. Food and Drug Administration (FDA) Amendments Act of 2007 requires the FDA to develop a system for using health care data to identify risks of marketed drugs and other medical products. In 2008 the FDA launched the Sentinel Initiative, which would enable the FDA to query diverse healthcare data actively—like EHRs, insurance claims databases, and registries—to evaluate possible medical product safety issues quickly and securely [1]. The Observational Medical Outcomes Partnership (OMOP) was recently designed to establish requirements for a viable national program of active drug safety surveillance by using observational data [2].
The current paradigm of drug safety surveillance is based on spontaneous reporting systems, which are databases containing voluntarily submitted reports of suspected adverse drug events encountered during clinical practice. In the USA, the primary database for such reports is the public Adverse Event Reporting System (AERS) [3] database hosted by the FDA. Researchers typically mine the reports in these structured databases for drug-adverse event associations (called safety signals) via statistical methods based on disproportionality measures, which quantify the magnitude of difference between observed and expected rates of particular drug-adverse event pairs [4]. The reporting odds ratio (ROR) is a commonly used measure of the strength of the association between a drug and an adverse event [4], and it is usually qualified with a 95% confidence interval (CI) to convey its accuracy and a p-value to convey the significance.
Researchers have estimated that roughly 30% of hospital stays experience an adverse drug event [5]. On average, preventable adverse drug events amongst inpatients cost $4685 per occurrence [6]—extrapolated for inflation and 34.4 million inpatient discharges in the USA, their monetary cost was estimated to be 4.4 billion dollars in 2007 [7, 8]. With advances in natural language processing (NLP) [9–14], testing as well as de novo detection of drug safety signals using textual clinical notes as well as literature is becoming possible [12, 15–18]. For example, recently, researchers showed that roughly half of the drug-safety signals can be detected in the literature before an official alert is issued [18]. The advantage of focusing on textual clinical notes is that they do not suffer from the reporting biases seen in spontaneous reporting systems—thus providing reliable denominator data for risk estimation.
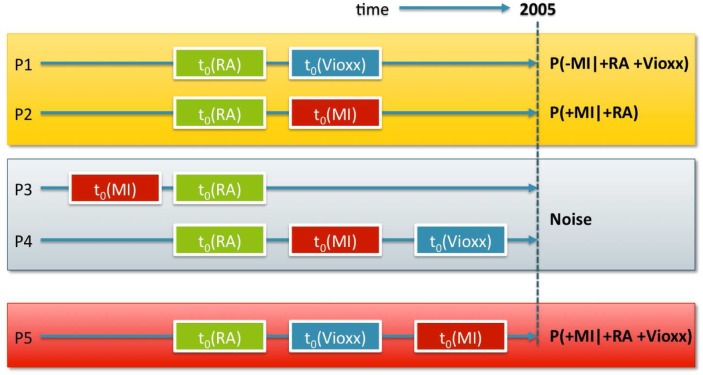
In our previous work, we tested the association between Vioxx and myocardial infarction (MI) using textual clinical notes and taking the temporal constraints depicted in Figure 1 into consideration [19]. From the patient counts, corresponding to the different patterns in Figure 1, we constructed a contingency table and calculated the ROR as described in [4]. Using first mentions of Vioxx and MI in the clinical notes before Vioxx was taken off the market in 2005, we obtained an ROR of 2.06 with a CI of [1.80, 2.35] and an uncorrected X2 p-value lower than 10−7. This result demonstrates that it is possible to analyze annotations of clinical notes for testing drug safety signals [19].
Figure 1:
The Vioxx risk pattern (P5) occurs within a background of Rheumatoid Arthritis (RA) patients (P1 & P2) that either don’t have an myocardial infarction (MI) incident (t0 denotes first occurrence) after Vioxx or never take Vioxx; and among records in which MI occurs prior to the diagnosis of RA (P3), or prior to Vioxx use (P4).
Despite successes in testing a known drug safety signal, when examining drug–disease co-occurrences in clinical notes to discover new adverse events, discerning indications from adverse events (AEs) for a given drug–disease pair remains a challenge.
We hypothesize that statistically enriched cooccurrences of drug–disease mentions in the clinical notes can be used not only to test, but also to detect new adverse drug event signals. The ability to distinguish indications from AEs directly in a given drug-disease co-occurrence pair is a first-step towards direct data driven detection of safety signals from unstructured EMR data. Using the methods we describe below, we show that by using co-occurrence frequencies and by keeping track of the time at which a drug or disease is mentioned we can discriminate between drug-adverse events pairs from drug-indication pairs.
In this study, we build our co-occurrence frequency models by analyzing over 9-million clinical notes for more than one million patients from the Stanford Clinical Data Warehouse (STRIDE). The patient records include both inpatient and outpatient notes, the records are from 620,946 female patients, 424,060 male patients, and 2330 cases where the sex information is missing; we include all note types. In terms of the age distribution, for each 10-year age range from 0 to 70, there are between 90,000 and 170,000 patients in each age range—in terms of age at first visit.
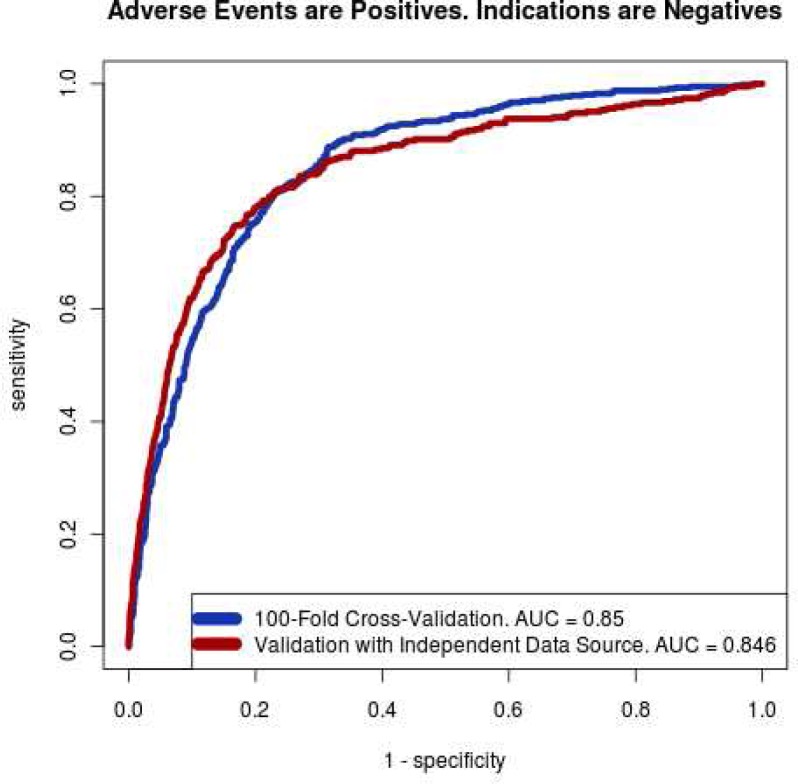
We use a sample of 1,550 drug-disease pairs from Medi-Span® Adverse Drug Effects Database™ (from Wolters Kluwer Health, Indianapolis, IN), AERS, and the National Drug File ontology (NDFRT) as gold standard. We train a support-vector machine (SVM) [20] classifier using the empirical data from STRIDE. Finally, we validate the results against an independent set of drug-indication and drug-AE pairs from the external sources. The classifier performs well in cross-validation (AUC=0.85) and independent validation (AUC=0.846).
Methods
Overview:
Our study consists of two broad components: an NLP workflow that annotates textual medical records with relevant drug and disease terms, and a statistical framework under which we organize and classify drug-disease pairs. For the NLP workflow, existing work from the 2009 i2b2 medication extraction challenge appears particularly promising. However, because medical records are known to vary from institution to institution in style and structure, it is uncertain whether the medical records from STRIDE would be directly usable with the methods described in [12]. Moreover, our dataset is about 10,000 times larger than those used in the competition [12]. Thus, for performance reasons, we needed a more scalable method. In contrast, our annotator workflow, which we describe below, performs a heavily optimized exact string matching which is computationally efficient. Finally, the primary purpose of this study is to demonstrate that it is possible to distinguish drug-indication pairs from drug-AE pairs. Once feasibility is established, we can focus on the question of identifying the “best” NLP system to use—which is a complicated task.
For our statistical framework, we apply a novel combination of regression and classification techniques to address a handful of basic but salient sources of confounding so as to achieve improved accuracy in discerning drug-AE pairs from drug-indication pairs. Methods based purely on association strength are unable to make that distinction among drug-disease pairs created based on co-occurrence.
The NCBO Annotator Workflow:
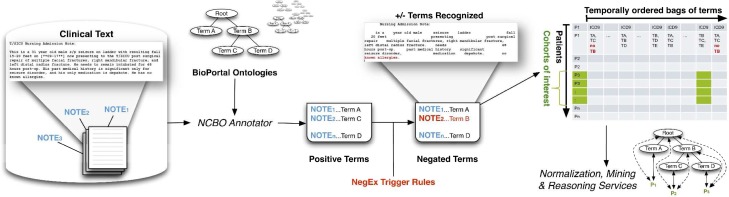
Figure 2 illustrates the workflow to annotate the clinical text from electronic health record systems and to extract disease and drug mentions from the EHR. We created a standalone Annotator Workflow based upon the existing National Center for Biomedical Ontology (NCBO) Annotator Web Service [21]. The Annotator uses biomedical terms from the NCBO BioPortal library and matches them against input text. The annotation process utilizes the NCBO BioPortal ontology library of over 250 ontologies to identify biomedical concepts from text using a dictionary of terms generated from those ontologies. For this study, we specifically configured the workflow to use SNOMED-CT and RxNORM. The resulting lexicon contains 1.6 million terms. Negation detection is based on trigger terms used in the NegEx algorithm [22].
Figure 2.
The NCBO Annotator Workflow tags clinical records with terms from ontologies: (1) We obtain a lexicon of 1.6 million terms from the NCBO BioPortal library. (2) We use the NCBO Annotator to rapidly find those terms in clinical notes—which we call annotations (3) We apply NegEx trigger rules to determine negated concepts. (4) We compile terms (both positive and negative) into a temporally ordered series of sets for each patient and combine them with coded and structured data when possible. (5) We reason over the structure of the ontologies to normalize and to aggregate terms for further analysis.
The output of the annotation workflow is a set of negated and non-negated terms from each note. As a result, for each patient we end up with a temporal series of “symbols” or tags comprising of the terms derived from the notes (red denotes negated terms in Figure 2) and the coded data collected at each patient encounter. Because each encounter’s date is recorded, we can order each set of terms for a patient to create a timeline view of their record. Using the tags as features, we can define patterns of interest such patients with rheumatoid arthritis, who took rofecoxib, and then suffered from myocardial infarction, which we studied previously [19]. Our goal in this study is to focus on discriminating the drug-adverse event pairs from the drug-indication pairs.
Creating Drug–disease Associations:
For every patient, we scan through their notes chronologically and record the first mention of every drug and disease. Drugs and diseases will re-appear throughout a patient’s timeline, yet we only note the very first occurrence (denoted T0 for initial time). All subsequent mentions of the noted term are ignored. This simplifies computation and captures the temporal ordering between the first mentions of drugs and diseases.
For the brevity of subsequent explanations, we introduce two definitions: co-mentions, and drug-first fractions. For any drug-disease pair, the co-mention count is the number of distinct patients for whom both the drug and disease are mentioned in their record—in any chronological order. For such co-mentions, there is one first-mention for the drug and one first-mention for the disease in a patient’s record. There are three possible cases for each drug–disease pair when examining the first mentions in a single patient’s record: either the drug is mentioned before the disease, or the disease is mentioned before the drug; or the drug and the disease are mentioned at the same time (Figure 3). A fraction of the patients will support the first case: where the first mention of the drug precedes the first mention of the disease. The numerical fraction of patients with this specific temporal ordering is defined as the drug-first fraction for a particular drug-disease pair. The drug-first fraction characterizes the temporal ordering between the first mentions of the drugs versus the first mentions of the diseases. This study shows that disproportionalities in the counts of co-mentions and the drug-first fractions will sufficiently characterize drug-disease pairs to classify them into drugs-AEs and drug-indications.
Figure 3.
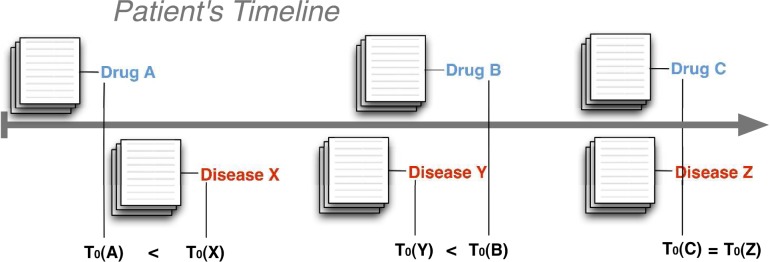
Associating diseases with drugs using first occurrences: for every patient, note the first occurrence (denoted T0) of a drug or disease mention by scanning forward in time through all of their sets of notes. The three cases for a drug–disease pair are: drug mentioned before disease, drug mentioned after disease, and drug mentioned at the same time as disease.
Normalizing, Filtering and Aggregation:
To reduce the computation load, we normalize the drug and disease terms early on in our analysis workflow, as shown in Figure 4. For drugs, we normalize drugs into ingredients using RxNORM relations like “has_ingredient.” In many cases, such as rofecoxib, drugs contain only one ingredient. Alternatively, multiple drugs may share a common ingredient, and multiple ingredients may be present in a single drug. For example, Excedrin has acetaminophen, aspirin, and caffeine, whereas Midol Complete has acetaminophen, caffeine, and pyrilamine maleate. Although drug normalization is a many-to-many mapping, ultimately the resulting number of unique ingredients in subsequent analysis is smaller. Thus, in subsequent analysis, we are ultimately comparing ingredient-disease pairs. In our analysis and interpretation of indications and adverse events, we treat drug ingredients as drugs.
Figure 4.
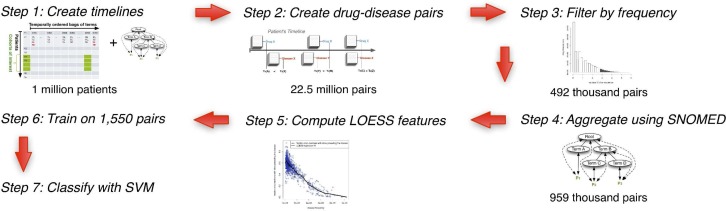
Analysis Workflow: (1, 2) We start by creating the patient timelines and generating drug-disease association frequencies. (3) Next, we filter drug-disease pairs to those having at least one thousand common patients. (4) This count of 492 thousand pairs roughly doubles when we aggregate up the SNOMED-CT hierarchy. (5) Then, we apply regression techniques to generate features. (6, 7) These features then feed into an SVM that classifies whether the disease in a given drug disease pair is an indication or adverse event.
In addition to normalizing drugs, we also normalize the diseases. Using UMLS provided “source-stated synonymy” relations, we normalize multiple disease terms into a single disease concept. Disease normalization constitutes a many-to-one mapping. Being part of UMLS, SNOMED-CT provides a subsumption hierarchy via “is-a” relations. Specialized child concepts (e.g., malignant melanoma) relate to their generalized parent concepts (e.g. malignant neoplasm) via this relation. When a specific child concept appears in text, we can count that mention as a mention of that concept’s parent terms. Using such hierarchical relations, we perform aggregation by accepting mentions of a child concept when searching for mentions of an ancestor concept—a process referred to as computing the transitive closure of the concept counts over the is-a hierarchy. Materializing this closure is computationally intensive, so we perform an optimization for speed: when a disease concept is never mentioned in STRIDE, we exclude that disease concept from being considered further. In separate work, our group has shown that the majority of UMLS concepts do not appear in clinical text and by removing them from the analysis, we gain computational efficiency [23].
From over 9 million notes, we detect 29,551 SNOMED-CT diseases and 2,926 drug ingredients, thus resulting in 86.5 million possible drug-disease pairs. Only 22.5 million actually occur in the data. We only consider pairs that occur in at least a thousand patients, which reduces our set to 492,115 pairs. After we perform aggregation based on SNOMED-CT, the count of pairs grows to 986,850 because of inclusion of general terms in the drug-disease pairs. These 986,850 pairs constitute the basis of all subsequent analysis.
Using LOESS Regression to Generate Z-scores for Co-mentions and Drug-first Fractions:
While the ROR is the traditional measure for disproportionlity [4], it does not adequately capture the temporal ordering, which is necessary in discerning an adverse event from an indication. Moreover, RORs assume independence, which is too restrictive; confounding factors can affect the frequencies of co-occurrences as well as temporal ordering. For example, neonatal diseases will appear disproportionately in the earlier parts of the medical record; thus, all temporal associations made subsequently as an adult will be skewed. To compensate, we fit local regression (LOESS) models [24] to define our baselines, substituting for the commonly used independence assumption (Step 5 in Figure 4; expanded in Figure 5).
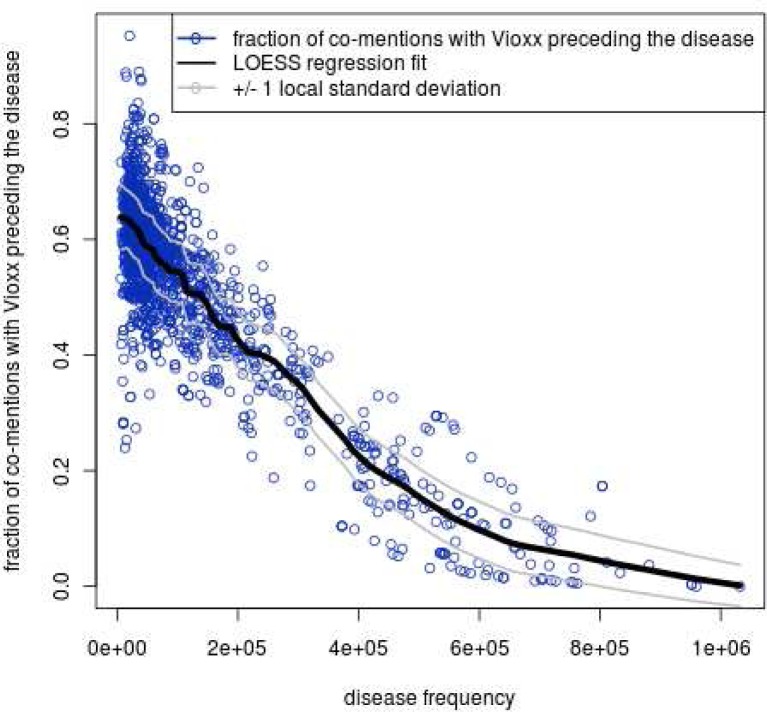
Figure 5.
LOESS provides a baseline (e.g., Vioxx versus all diseases): for each disease Z, we plot on the x-axis the total count of patients whose record ever mentions disease Z. The y-axis is the drug-first fraction for Vioxx-Z. Predictably, common disease concepts, have lower baseline expected value for drug-first fraction. The local one standard deviation lines are also shown.
As an example, suppose that we are trying to calculate the drug-first fraction of Vioxx versus myocardial infarction (MI). We have an observed drug-first fraction from the STRIDE data. Then, fixing the disease (MI) for every drug X in our vocabulary associated with MI, we count each drug-first fraction (X-MI) measured against the overall frequencies of each drug. We then fit a locally weighted smoothing regression (LOESS) [24] across all X-MI pairs (from the 986,850 pairs) to estimate the drug-first fraction for Vioxx-MI. This estimate serves as an expected value, which represents the null hypothesis that drugs with frequencies similar to Vioxx would have similar drug-first fractions against MI. We are interested in quantifying deviations from this expected value.
Given a LOESS estimate of drug-first fraction for MI across various drug frequencies, we define the observed error as difference between the LOESS estimate and the true observed value. The squares of these quantities are observed squared errors. We subsequently compute the observed local variance by running a separate LOESS fit on the observed squared errors with respect to the drug frequencies. The square roots of the local variances are the local standard deviations. Finally, we define the local z-scores as the quotients of the local errors divided by the local standard deviations.
In the previous step, we fix the disease – MI– and estimate the drug-first fraction with respect to drug frequencies. Next, we fix the drug – Vioxx– and analogously fit a LOESS regression of the drug-first fraction measured against disease frequencies across all diseases to generate a second estimate for the drug-first fraction for Vioxx-MI. This is illustrated in figure 5. We now have two distinct estimates, two distinct local variances, and two distinct local z-scores for the drug-first fraction of the pair Vioxx-MI. The two estimates, if compared to the actual observed drug-first fraction of Vioxx-MI, serve as baseline expected values. Moreover, the two local z-scores serve as measures by which the frequency of observed Vioxx-MI deviates from our expectations. The two LOESS local z-scores, alongside the observed drug-first fraction, capture the temporal ordering information implicit in the Vioxx-MI pairing.
In addition to the drug-first fraction, we also produce analogous estimates and analogous z-scores for the co-mention counts. Continuing the Vioxx-MI example, we would produce two estimates and two local z-scores of the Vioxx-MI co-occurrence frequency. One estimate is based on drug-MI co-occurrences across all drugs. The other estimate is based on Vioxx-disease co-occurrences across all diseases. In summary, we introduce six quantities: two LOESS estimates for drug-first fraction, the actual drug-first fraction, two estimates for the cooccurrence count, and the actual observed co-occurrence count.
Accounted Sources of Confounding:
The LOESS estimates are designed to alleviate drug-specific, disease-specific, and frequency-based sources of confounding. From purely a statistical point of view, for a fixed disease, and across many drugs, the more frequent the drug, the higher the drug-first fraction we should expect. Similarly, for a fixed drug, across many diseases, the more frequent the disease, the lower the drug-first fraction we should expect. For co-mentions, both increasing the drug frequency and increasing the disease frequency should lead to a higher co-mention estimate.
Our LOESS estimates account for these sources of confounding by anticipating their effects. Functions that only increase or only decrease are known as monotonic functions; in the aforementioned sources of confounding, the regression fits should be monotonic. When estimating monotonic functions, simply enforcing the monotone property by sorting the y-values improves the estimate [25] and does no harm. We apply this technique in our LOESS calculations.
Classification:
For each drug-disease pair, we have described six per-pair quantities as features. Three are based on the notion of the drug-first fraction—the fraction of pairs in which the mention of the drug precedes the mention of the disease; and the other three are based on the co-occurrence. We take the logarithm of the co-occurrences to place these quantities into logarithmic space. Table 1 lists all features we use for classification.
Table 1.
Features used in classification.
| Linear Space Features | Logarithmic Space Features |
|---|---|
|
| |
| Drug frequency | Disease frequency |
| Drug frequency | Disease frequency |
| Observed drug-first fraction | Observed co-mention count |
| Drug-first fraction z-score (fixed drug) | Co-mention count z-score (fixed drug) |
| Drug-first fraction z-score (fixed disease) | Co-mention count z-score (fixed disease) |
Our training set consists of 1,550 samples: 980 indications and 570 adverse events. Next, we normalize each feature to have mean zero and variance one for these 1,550 drug-disease pairs. Finally, we apply a support vector machine (SVM) on the ten features to produce a classifier that can classify any given drug-disease pair into drug-indication and drug-AE classes if given the ten feature quantities for that pair. We used SVMs for the primary reason that SVMs make fewer assumptions about the classification boundary than traditional methods like logistic regression. Another consideration was that our classifier should at least consider a strict superset of decision boundaries available to traditional ROR disproportionality studies. SVMs models can encompass linear relations with respect to its features. Intuitively, we see that the log reporting odds ratio is encoded by a linear combination of three of our log-space features: drug frequency, disease frequency, and observed co-mention count.
Validation:
To evaluate our results, we applied 100-fold cross-validation. We also applied independent validation using a set of known drug-indications and drug-adverse events—which were not used in training. The external source of indications was a list of indications from the Medi-Span Drug Indication Database™, which were not used in training. The external list of adverse events was taken from the public version of Adverse Event Reporting System (AERS). To filter out spurious relations, we restrict our attention to reports that contain either only one suspect drug, or only one adverse event. Furthermore, we restricted our attention to pairs that have a raw frequency of at least 500 to further filter spurious relations.
Results
The adverse events in our training set consist of 570 known adverse events taken from Medi-Span. We used only adverse events marked by Medi-Span to be in the most severe category and most frequent category. Our 980 indications consist of drug-disease pairs from the NDFRT ontology connected by “may_treat” relations. For both the adverse events and the indications, the only criterion of admittance into our training set was based on having at least 1,000 co-occurrences within STRIDE. This filter criterion applies to our independent validation set as well.
Figure 6 shows that we achieve good performance in distinguishing adverse events from indications. Our area under the receiver operating curve (AUC) was 0.85 in cross-validation and 0.846 in independent validation. To independently validate, we used a database of 79,966 pairs of known indications from Medi-Span (43,159 from FDA labels, 16,639 commonly accepted off-label uses, and 20,178 off-label uses having limited evidence). Subject to the 1,000 co-occurrences threshold in STRIDE, our analysis workflow retains 28,015 pairs. Analogously, from 851 AERS adverse event pairs that occurred at least 500 times in AERS, our analysis workflow retained 385 pairs. The classifier trained on the original training set achieved an AUC of 0.846 in this independent validation. The classifier uses only ten features and retains performance on independent validation; thus, our method does not suffer from significant over-fitting.
Figure 6.
Receiver operating curve (ROC) for our SVM Classifier shows that we can distinguish between drug– indication pairs from drug–AE pairs. The area under the curve (AUC) evaluates classification quality in a more comprehensive manner than accuracy.
Discussion
Given the amount of data available in AERS [26], researchers are developing methods for detecting new or latent multi-drug adverse events [27, 28], for detecting multi-item adverse events [28, 29], and for discovering drug groups that share a common set of adverse events [30]. Biclustering and association rule mining are able to capture many-to-many relations between drugs and adverse events [29, 30]. Increasingly there are efforts to use other data sources, such as EHRs, for the purpose of detecting potential new AEs [31] in order to counterbalance the biases inherent in AERS [32] and to discover multi-drug AEs [33]. Researchers have also attempted to use billing and claims data for active drug safety surveillance [14, 34], applied literature mining for drug safety [18], and tried reasoning over published literature to discover drug-drug interactions based on properties of drug metabolism [35].
We take a complementary approach that begins from the medical record. To our advantage, medical records provide backgrounds frequencies unaffected by some of the reporting biases that afflict AERS—thus providing reliable denominator data. We use the frequency distribution and the temporal ordering of drug-disease pairs in a large corpus to define ten features based on which we can identify known drug-indication and drug-AE pairs with high accuracy. Approaching the problem in this manner allows us to comprehensively track the drug and disease contexts in which the AE patterns occur; and use those patterns to evaluate putative new AEs. The ability to distinguish indications from adverse events directly opens up the possibility of detecting new drug-AE pairs. Finally, this capability is a first-step towards the data driven detection of multi-drug-multi-disease associations.
Our results hinge upon the efficacy of the annotation mechanism. We have previously conducted a comparative evaluation of the concept recognizer—Mgrep—which is used in the NCBO Annotator [21]. The precision of concept recognition varies depending on the text in each resource and type of entity being recognized: from 87% for recognizing disease terms in descriptions of clinical trials to 23% for PubMed abstracts, with an average of 68% across four different sources of text. We are currently conducting evaluations for text in clinical reports. Early results show, a 93% recall for detecting drug mentions in clinical text using RXNORM. In future work, we will perform manual chart review for random samples of reports to validate our ability to recognize drugs and diseases in medical records. As mentioned before, our dataset is about 10,000 times larger than those used in the i2b2 NLP challenges [12]. Thus, for performance reasons, we used our annotator workflow, which performs a heavily optimized exact string matching which is computationally efficient. Finally, the primary purpose of this study is to demonstrate that it is possible to distinguish drug-indication pairs from drug-AE pairs. Once feasibility is established, we can focus on the question of identifying the “best” NLP system to use. We expect better NLP methods to improve our results.
Temporal ordering of first mentions in medical records is subject to sources of confounding. Clinically, some diseases like dementia or cancer tend to afflict older populations, so their first mentions are more likely to temporally follow drugs in general. From purely a statistical perspective, common concepts are more likely to have an earlier first-mention than rare concepts. Our LOESS regression estimate explicitly accounts for the above sources of confounding. Confounding by co-morbidity is not addressed directly by our current method, and we plan to directly account for it in our future work.
Beyond indications and adverse events, we plan to generalize our method to recognize, for example, likely off-label drug usages. Concurrent with this work, we studied the use of a temporal sliding window (as opposed to first mentions) with promising results for detecting off-label drug usage [36]. Given that some adverse effects may surface only years after the treatment, while others are acute. Adjustable windowing may refine our ability to characterize and distinguish adverse events in the future. Clinical notes also contain rich contextual markers like section headings (e.g., family medical history) that could improve the precision of the analysis when taken into account. Thus, we plan to use this information in future iterations of our analysis workflows.
Several limitations apply to this work. We restricted our analysis to drug-disease pairs with at least 1000 co-mentions; while it ensures the statistical significance of our observations, this restriction also prevents us from detecting rare severe adverse events. This problem can be alleviated if we can apply our analysis to larger (e.g. regional and national level) databases, or if we apply computationally expensive algorithms that can statistically “salvage” some of the pairs that we exclude in this work. Another limitation is that our framework does not attempt to discern adverse drug events from drug side effects. Finally, we treated drugs as drug ingredients, which is at a very fine granularity; it may be valuable to perform aggregation and do analysis at the drug, drug class, and drug combination levels.
Our disease terms were restricted to SNOMED-CT because SNOMED-CT is the domain of disease concepts connected by “may_treat” relations as defined in NDFRT. Our workflow relied on the “may_treat” relations to train our SVM to recognize indications. In contrast to NDFRT, AERS specifies its diseases using the Medical Dictionary for Regulatory Activities (MedDRA) ontology. To map these AERS disease terms to SNOMED-CT, we applied our annotation workflow on the AERS text itself as well as used the synonymy relations between MedDRA and SNOMED-CT found in UMLS. In our annotation of the medical records, we used these synonymy relations so as to include additional synonyms and linguistically colloquial phrases offered by MedDRA.
In the current work, MedDRA terms unmapped to SNOMED-CT were excluded. We decided to choose a single ontology because this makes the hierarchical aggregation easier to interpret. Aggregation is one of the most computationally expensive tasks; having successfully applied our methods using SNOMED-CT, the largest of the ontologies, we are confident that we will be able to apply the same methods to reason simultaneously over many more ontologies in the future. Finally, compared to SNOMED-CT, MedDRA is not as exhaustive in enumerating plural forms and synonyms; using MedDRA would reduce the recall of our annotation workflow, which relies on exact matches. Thus, we ultimately chose SNOMED-CT as our primary ontology for disease terms and included MedDRA terms that could be mapped to it.
Conclusion
Statistically significant co-occurrences of drug–disease mentions in the clinical notes can potentially be used to detect drug safety signals. Currently, when examining pairs of drug–disease co-occurrences from textual clinical notes, a major challenge is to discern indications from adverse events (AEs) in a drug–disease pair. We demonstrate that it is possible to make this distinction by combining the frequency distribution of the drug, the disease, and the drug-disease pair as well as the temporal ordering of the drugs and diseases in each pair across more than one million patients.
By using LOESS regression models derived from one million patients’ records, which does not make independence assumptions built into traditional disproportionality based methods, we account for basic sources of confounding. Through a novel combination of using large datasets, annotation, and analytics, we discern drug indications from adverse events with good independent validation performance.
Acknowledgments
We acknowledge support from the NIH grant U54 HG004028 for the National Center for Biomedical Ontology. We acknowledge Nick Tatonetti for providing us with a copy of AERS data.
References
- 1.FDA; Jun, 2010. The Sentinel Initiative July 2010 Report. [Google Scholar]
- 2.Stang P, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Annals of internal medicine. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 3.Adverse Event Reporting System (AERS) 2011. [cited 2011 Oct 17]; Available from: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/default.htm.
- 4.Bate A, Evans SJW. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf. 2009;18(6):427–36. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- 5.Classen D, et al. ‘Global trigger tool’ shows that adverse events in hospitals may be ten times greater than previously measured. Health affairs (Project Hope) 2011;30(4):581–589. doi: 10.1377/hlthaff.2011.0190. [DOI] [PubMed] [Google Scholar]
- 6.Bates DW, et al. The costs of adverse drug events in hospitalized patients Adverse Drug Events Prevention Study Group. JAMA : the journal of the American Medical Association. 1997;277(4):307–311. [PubMed] [Google Scholar]
- 7.US Inflation Calculator. [cited 2011 Oct 30]; Available from: http://www.usinflationcalculator.com/.
- 8.Hall M, et al. National Hospital Discharge Survey: 2007 summary. Hyattville, MD: 2010. [PubMed] [Google Scholar]
- 9.Ohno-Machado L. Realizing the full potential of electronic health records: the role of natural language processing. Journal of the American Medical Informatics Association: JAMIA. 2011;18(5):539. doi: 10.1136/amiajnl-2011-000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman WW, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. Journal of the American Medical Informatics Association: JAMIA. 2011;18(5):540–3. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association: JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. Journal of the American Medical Informatics Association. 2010;17(5):524–527. doi: 10.1136/jamia.2010.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patrick JD, et al. A knowledge discovery and reuse pipeline for information extraction in clinical notes. Journal of the American Medical Informatics Association : JAMIA. 2011;18(5):574–9. doi: 10.1136/amiajnl-2011-000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadkarni PM. Drug safety surveillance using de-identified EMR and claims data: issues and challenges. J Am Med Inform Assoc. 2010;17(6):671–4. doi: 10.1136/jamia.2010.008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harpaz R, et al. Mining electronic health records for adverse drug effects using regression based methods. Proceedings of the 1st ACM International Health Informatics Symposium. 2010. pp. 100–107.
- 16.Wang X, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. Journal of the American Medical Informatics Association : JAMIA. 2009;16(3):328–337. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. Journal of clinical epidemiology. 2005;58(4):323–337. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Shetty KD, Dalal S. Using information mining of the medical literature to improve drug safety. Journal of the American Medical Informatics Association : JAMIA. 2011 doi: 10.1136/amiajnl-2011-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LePendu P, et al. Bio-Ontologies SIG at ISMB 2011. Viena, Austria: 2011. Annotation Analysis for Testing Drug Safety Signals. [Google Scholar]
- 20.Boser BE, Guyon IM, Vapnik VN. Proceedings of the fifth annual workshop on Computational learning theory. ACM; Pittsburgh, Pennsylvania, United States: 1992. A training algorithm for optimal margin classifiers; pp. 144–152. [Google Scholar]
- 21.Shah NH, et al. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman WW, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of Biomedical Informatics. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, et al. AMIA Summit on Clinical Research Informatics. San Francisco, CA: 2012. UMLS Term Occurrences in Clinical Notes: A Large-scale Corpus Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleveland WS. Robust Locally Weighted Regression and Smoothing Scatterplots. Journal of the American Statistical Association. 1979;74(368):829–836. [Google Scholar]
- 25.Chernozhukov V, Fernández-Val I, Galichon A. Improving point and interval estimators of monotone functions by rearrangement. Biometrika. 2009;96(3):559–575. [Google Scholar]
- 26.Weiss-Smith S, et al. The FDA drug safety surveillance program: adverse event reporting trends. Arch Intern Med. 2011;171(6):591–3. doi: 10.1001/archinternmed.2011.89. [DOI] [PubMed] [Google Scholar]
- 27.Norén N, et al. A statistical methodology for drug-drug interaction surveillance. Statistics in medicine. 2008;27(16):3057–3070. doi: 10.1002/sim.3247. [DOI] [PubMed] [Google Scholar]
- 28.Tatonetti NP, et al. Detecting Drug Interactions From Adverse-Event Reports: Interaction Between Paroxetine and Pravastatin Increases Blood Glucose Levels. Clinical pharmacology and therapeutics. 2011. [DOI] [PMC free article] [PubMed]
- 29.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics. 2010;11(Suppl 9):S7. doi: 10.1186/1471-2105-11-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harpaz R, et al. Biclustering of adverse drug events in the FDA's spontaneous reporting system. Clin Pharmacol Ther. 2011;89(2):243–50. doi: 10.1038/clpt.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. AMIA. 2009;16(3):328–37. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneeweiss S, et al. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2009;20(4):512–22. doi: 10.1097/EDE.0b013e3181a663cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coloma PM, et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR Project. Pharmacoepidemiol Drug Saf. 2011;20(1):1–11. doi: 10.1002/pds.2053. [DOI] [PubMed] [Google Scholar]
- 34.Dore D, Seeger J, Arnold Chan K. Use of a claims-based active drug safety surveillance system to assess the risk of acute pancreatitis with exenatide or sitagliptin compared to metformin or glyburide. Current medical research and opinion. 2009;25(4):1019–1027. doi: 10.1185/03007990902820519. [DOI] [PubMed] [Google Scholar]
- 35.Tari L, et al. Discovering drug–drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010;26(18):i547. doi: 10.1093/bioinformatics/btq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LePendu P, et al. AMIA Summit on Clinical Research Informatics. San Francisco, CA: 2012. Analyzing Patterns of Drug Use in Clinical Notes for Patient Safety. [PMC free article] [PubMed] [Google Scholar]