Abstract
RNA splicing is one of the fundamental processes in gene expression in eukaryotes. Splicing of pre-mRNA is catalysed by a large ribonucleoprotein complex called the spliceosome, which consists of five small nuclear RNAs and numerous protein factors. The spliceosome is a highly dynamic structure, assembled by sequential binding and release of the small nuclear RNAs and protein factors. DExD/H-box RNA helicases are required to mediate structural changes in the spliceosome at various steps in the assembly pathway and have also been implicated in the fidelity control of the splicing reaction. Other proteins also play key roles in mediating the progression of the spliceosome pathway. In this review, we discuss the functional roles of the protein factors involved in the spliceosome pathway primarily from studies in the yeast system.
Keywords: protein splicing factor, RNA splicing, small nuclear ribonucleoprotein (snRNP), spliceosome, yeast
Abbreviations: BBP, branchpoint-binding protein; CWC, complexed with Cef1; eIF4G, eukaryotic initiation factor 4G; NTC, NineTeen Complex; RRM, RNA-recognition motif; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein particle
INTRODUCTION
Most eukaryotic genes are disrupted by intervening sequences (introns or IVS) that are removed after transcription by RNA polymerases in a process called RNA splicing. Different classes of RNAs use different chemical strategies and different machineries to splice out their introns. Among them, the process of pre-mRNA splicing is the most complicated. Splicing of pre-mRNA takes place on a large ribonucleoprotein particle called the spliceosome, within which two transesterification reactions take place (Figure 1) [1–3]. The spliceosome comprises five snRNAs (small nuclear RNAs), U1, U2, U4, U5 and U6, and many other protein factors. It is assembled through ordered binding of these factors to the pre-mRNA for each round of splicing, and then disassembled after completion of the reaction to recycle the splicing factors (Figure 2) (reviewed in [4,5]). Since the splicing reactions are mechanistically identical with those of Group II self-splicing introns [6,7], it is widely believed that pre-mRNA splicing is also an RNA-catalysed reaction [8]. The spliceosome presumably plays a role in folding the pre-mRNA for proper alignment of the two splice sites so that the splicing reaction can take place. While the RNA–RNA interactions involved in spliceosome formation have been extensively studied [9,10], the functional roles of protein factors have only been better studied more recently.
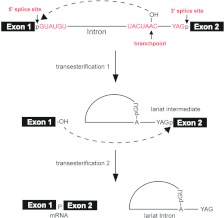
Figure 1. Two-step transesterification reaction of pre-mRNA splicing.
Yeast splice site conserved sequences are shown in red. The splicing reaction takes place in two steps. The first step is cleavage of the 5′ splice site and formation of lariat intron–exon 2 via a 2′–5′ phosphodiester linkage. The second step is cleavage of the 3′ splice site and ligation of the two exons.
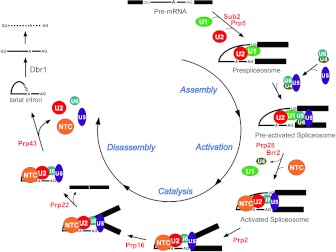
Figure 2. Schematic representation of the spliceosome pathway in yeast.
The pathway can be divided into four stages: spliceosome assembly, spliceosome activation, catalysis and spliceosome disassembly. Spliceosome assembly involves ordered interactions of snRNPs with pre-mRNA. Spliceosome activation starts with the release of U1 and U4, followed by binding of the NTC. After the two catalytic reactions, the spliceosome is disassembled first by releasing the mature mRNA, and the spliceosome is then dismantled.
OVERVIEW OF THE SPLICEOSOME
Most introns have common consensus sequences near their 5′ and 3′ ends that are recognized by the spliceosomal components (Figure 1). Metazoan genes, however, have been found to contain a different class of rare introns that have non-canonical consensus sequences and use a distinct set of snRNAs for their recognition [11]. These snRNAs, U11, U12, U4atac and U6atac, are functionally analogous to U1, U2, U4 and U6 used in the splicing of the major class of introns [12]. This review focuses on the U2-dependent spliceosome with emphasis on findings from yeast. The known spliceosomal protein components are listed in Table 1.
Table 1. Protein splicing factors.
SF3a and SF3b are subcomplexes of U2 snRNP. Asterisks mark proteins not found on the spliceosome. Non-snRNP proteins are sorted by their functions.
| (a) snRNP proteins | ||||||
|---|---|---|---|---|---|---|
| U1 | U2 | U4 | U5 | U6 | U4/U6 | U4/U6.U5 |
| SmB, D1, D2, D3, E, F and G | SmB, D1, D2, D3, E, F and G | SmB, D1, D2, D3, E, F and G | SmB, D1, D2, D3, E, F and G | Lsm2–8 | SmB, D1, D2, D3, E, F and G | SmB, D1, D2, D3, E, F and G |
| Snp1 | Lea1 | Prp3 | Dib1 | *Prp24 | Lsm2–8 | Lsm2–8 |
| Mud1 | Msl1 | Prp4 | Prp8 | Prp3 | Prp3 | |
| Yhc1 | *Cus2 | Snu13 | Prp28 | Prp4 | Prp4 | |
| Luc7 | SF3a | Brr2 | Snu13 | Snu13 | ||
| Nam8 | Prp9 | *Lin1 | Prp31 | Prp31 | ||
| Prp39 | Prp11 | Snu114 | Dib1 | |||
| Prp40 | Prp21 | Prp6 | Prp8 | |||
| Prp42 | SF3b | *Aar2 | Prp28 | |||
| Snu56 | Cus1 | Brr2 | ||||
| Snu71 | Rse1 | Snu114 | ||||
| Hsh49 | Prp6 | |||||
| Hsh155 | Prp38 | |||||
| Rds3 | Snu23 | |||||
| Ysf3 | Snu66 | |||||
| RES | Spp381 | |||||
| Ist3 | *Sad1 | |||||
| Bud13 | ||||||
| Pml1 | ||||||
| (b) Non-snRNP proteins | ||||||
| Assembly | Activation | First reaction | Second reaction | Disassembly | Unknown | |
| Msl5 | Prp19 | Cwc22 | Prp17 | Spp382 | Bud31 | |
| Mud2 | Snt309 | Spp2 | Prp16 | Ntr2 | Cwc15 | |
| Sub2 | Cef1 | Prp2 | Slu7 | Prp43 | Cwc24 | |
| Prp5 | Syf1 | Cwc25 | Prp18 | Cwc27 | ||
| Clf1 | Yju2 | Prp22 | Urn1 | |||
| Syf2 | ||||||
| Isy1 | ||||||
| Ntc20 | ||||||
| Cwc2 | ||||||
| Prp45 | ||||||
| Prp46 | ||||||
| Ecm2 | ||||||
| Cwc21 |
snRNPs (small nuclear ribonucleoprotein particles) and their protein components
Each of the snRNAs, except for U6, is in complex with Sm core proteins and several other proteins unique to the snRNA to form an snRNP [13,14]. U4 forms base-pairs with U6 to form a single RNP, the U4/U6 di-snRNP, which can further interact with U5 snRNP to form the U4/U6.U5 tri-snRNP [15–18]. The protein components of the snRNPs are evolutionarily conserved, but some mammalian proteins contain additional domains, presumably to increase their capacity for protein–protein interactions and/or for regulation. The protein components of snRNPs not only function in maintaining the structure of the snRNPs but also in regulating the splicing activity by interacting with other spliceosomal components.
Prp8, Brr2 and Snu114 are integral components of the U5 snRNP. Prp8 is a large protein of more than 250 kDa in size and is one of the most highly conserved nuclear proteins [19,20]. Prp8 interacts with many spliceosomal proteins, with snRNAs and with both 5′ and 3′ splice sites [21–26]. Prp8 contains an RRM (RNA-recognition motif) domain in the middle of the protein (amino acid residues 1059–1151 in yeast), and a Jab1/MPN domain at its C-terminus (amino acid residues 2173–2310 in yeast) [25,27]. Several mutations linked to human RP (retinitis pigmentosa) are located at the extreme C-terminus of Prp8. They were found to affect splicing and U5 snRNP maturation [28]. Prp8 lacks the catalytic residues in its Jab1/MPN domain required for the activity of deubiquitination, but has been shown to bind ubiquitin in vitro [29]. Although ubiquitin has been implicated in the spliceosome pathway [30], the functional significance of ubiquitin binding or whether this function is related to Prp8 is not known. Upstream from the Jab1/MPN domain, a segment of ~250 amino acids has been demonstrated to form an RNase H-like structure (1833–1950 in yeast) [31–34]. Since the putative catalytic residues of the RNase H enzyme are displaced in the Prp8 domain, whether Prp8 retains its nuclealytic function is questionable. A C-terminal fragment of Prp8 comprising the RNase H, Jab1/MPN and RP domains has been shown to stimulate the unwinding activity of Brr2 [35–37].
Brr2 is a DExD/H-box RNA helicase that mediates the unwinding of U4/U6 helices to release U4 from the spliceosome [38,39]. It has also been implicated in the mediation of spliceosome disassembly [40]. Unlike the other spliceosomal DExD/H-box proteins, Brr2 has two helicase domains and remains associated with the spliceosome throughout the reaction after its association as a component of tri-snRNP [41]. The N-terminal helicase domain is responsible for the unwinding activity [42]. The C-terminal helicase domain is not catalytically active, but serves as a platform for interaction with many spliceosomal components [26,37].
Snu114 is a GTPase that is highly homologous with translation elongation factor G [43] and has been suggested to be involved in spliceosome activation [44,45]. Despite never having been shown to hydrolyse GTP in vitro, Snu114 has been demonstrated to bind GTP by UV-cross-linking [43,46], and its binding of GTP/GDP has been proposed to regulate spliceosome dynamics mediated by Brr2 during spliceosome activation and disassembly [40]. Furthermore, a functional GTPase domain is required for assembly of Snu114 into U5 snRNP [47].
Prp24 is an RNA-binding protein containing four RRMs [48]. Prp24 has been shown to interact with U6 and U4/U6, and stimulate annealing of U4 and U6 for formation of the di-snRNP [49–52]. In extracts, while U4 always complexes with U6 as di-snRNP or tri-snRNP, U6 also exists as a distinct RNP particle, which contains Prp24. A role for Prp24 in recycling the spliceosome has been demonstrated [49].
DExD/H-box RNA helicases
DExD/H-box proteins are a ubiquitous class of enzymes that mediate rearrangement of RNA–RNA or RNA–protein interactions in an ATP-dependent manner [53,54]. Eight DExD/H-box proteins are required for the splicing process [55,56]. They function in mediating structural rearrangements of the spliceosome at different steps in the spliceosome pathway. Several of them have been shown to unwind RNA duplexes in vitro [38,39,57–60], but no direct evidence shows that they catalyse RNA unwinding in the spliceosome. Two of these proteins, Sub2 and Prp5, are involved in the early steps of spliceosome assembly. Sub2 has been proposed to displace Mud2 and BBP (branchpoint-binding protein), which bind to the 3′ splice site and branch site respectively (see the section ‘Spliceosome assembly’ for details), to facilitate the association of U2 snRNP with the spliceosome [61]. Prp5 also functions in facilitating U2 binding to the spliceosome, in part by ATP-dependent displacement of Cus2 from U2 snRNP to convert U2 into a functional form [62,63]. Prp28 and Brr2 are required for spliceosome activation in mediating the release of U1 and U4 respectively [38,39,64]. The requirement of Prp28 can be bypassed if the interaction of U1C protein (Yhc1 in yeast) or U1 snRNA with the 5′ splice site is destabilized by mutations [65]. Brr2 is required for unwinding of U4/U6 during spliceosome activation [38,39] and also for separation of U2/U6 interaction in disassembly of the spliceosome [40]. Prp2 and Prp16 are involved in catalytic steps [66,67]. The action of Prp2 is associated with the release of SF3a/b after the activation of the spliceosome [68,69], and that of Prp16 is associated with the release of Yju2 and Cwc25 after lariat formation [70]. After completion of splicing, two DExD/H-box proteins are required to disassemble the spliceosome. Prp22 first mediates the release of mature mRNA from the spliceosome, and Prp43 then catalyses disassembly of the intron-containing spliceosome [71,72].
In addition to these ATP-dependent functions, several DExD/H-box splicing factors have been shown to have an ATP-independent role in splicing. Although Prp5 is required for displacement of Cus2 from U2 snRNP before the binding of U2 to the spliceosome, it is still needed for pre-spliceosome formation in the absence of Cus2 independent of its ATPase function [63]. Prp2 has been implicated in a novel ATP-independent conformational change [68]. Prp16 has been shown to stabilize the association of Cwc25 with the spliceosome formed on pre-mRNA carrying mutations at the branchpoint [70]. Prp22 also participates in the second transesterification reaction independent of ATP in addition to its role in mRNA release [59]. It will be interesting to see whether it is a general property for splicing DExD/H-box proteins to have both ATP-dependent and ATP-independent functions.
The Prp19 complex
The Prp19 complex [or NTC (NineTeen Complex)] was identified as a large RNA-free protein complex associated with essential splicing factor Prp19 [73,74]. Eight proteins, Prp19, Syf1/Ntc90, Cef1/Ntc85, Clf1/Ntc77, Syf2/Ntc31, Isy1/Ntc30, Snt309/Ntc25 and Ntc20, have been shown to be components of the complex, and several others have also been suggested to be putative NTC components [74–80]. The NTC is recruited to the spliceosome after the release of U1 and U4 and remains associated with the spliceosome until completion of the catalytic reactions. The NTC is required for stabilizing the association of U5 and U6 with the spliceosome [81]. It promotes specific interactions of U5 and U6 with pre-mRNA within several bases, and also triggers the release of Lsm from binding to the 3′-end of U6 snRNA to allow further interaction of U6 with the intron sequence near the 5′ splice site [81,82]. Proteomic analysis of proteins associated with Cef1/Ntc85 has also identified a complex, called CWC (complexed with Cef1), containing U2, U5 and U6 snRNAs and more than 25 proteins, including all the NTC components [83]. Cwc2 directly interacts with U6 snRNA, and has been suggested a role in linking the NTC to the spliceosome [84]. In contrast, Cwc22 and Cwc25 are required for the catalytic steps, and only bind to the spliceosome after its activation [85,86].
The functions of NTC and several of its putative components in splicing have been characterized. Prp19 contains an U-box motif for ubiquitin ligase at its N-terminus, a coiled-coil domain for tetramerization at central region and a WD40 repeat domain as a splicing scaffold at its C-terminus [87–90]. Prp19 has been demonstrated to promote ubiquitination of Prp3 during the splicing reaction [91]. Snt309/Ntc25 is required for the integrity of the NTC. It interacts with Prp19 to regulate the interaction of Prp19 with Cef1/Ntc85, which further interacts with Syf1/Ntc90 and Clf1/Ntc77 in the formation of the NTC [75,92]. Although Clf1/Ntc77 has been shown to bind to the spliceosome as an integral component of the NTC, it has also been reported to be required for the addition of tri-snRNP to the spliceosome [75,93]. Syf1/Ntc90, Syf2/Ntc31, Isy1/Ntc30 and Ntc20 form a stable subcomplex, which is not required for spliceosome activation, but is required for the recruitment of step-one factor Yju2 [94]. Deletion of ISY1/NTC30 gene was found to partially rescue the cold-sensitive growth phenotype of prp16-302 mutant and restore fidelity of branchpoint usage in prp16-302 cells to wild-type level, leading to the hypothesis that Isy1/Ntc30 can regulate the function of Prp16 in splicing fidelity control [95]. Ecm2 was shown to be a component of the CWC complex, and was also identified as Slt11 on the basis of synthetic lethality with a mutation in the 5′-end of U2 snRNA that disrupts U2/U6 helix II [83,96]. Ecm2 contains two conserved zinc-finger motifs and two regions of the RRM, and has been shown to bind specific RNA structures reminiscent of U2/U6 helix II, suggesting a role for Ecm2 in the formation of helix II [97]. Cwc2 has been shown to interact with Prp19 directly by two-hybrid assays [79] and is likely to be the Ntc40 detected to interact with Prp19 by far Western blotting [74]. Cwc2 also contains an RRM and a zinc-finger motif, and can directly bind U6 snRNA [84], but how binding of Cwc2 to U6 mediates spliceosome activation remains to be investigated.
SPLICEOSOME ASSEMBLY
The spliceosome is assembled via sequential binding of snRNPs to the pre-mRNA in the order of U1, U2 and then U4/U6.U5 (as the preformed tri-snRNP) [17,98]. U1 snRNP binds to the 5′ splice site through base-pairing of U1 with the 5′ splice site to form the commitment complex [99–101]. Binding of U1 to the pre-mRNA is ATP-independent and can occur without incubation [98,99]. U2 binds to the branch site also through base-pairing to form the pre-spliceosome, which requires ATP [98,102–105]. The U4/U6.U5 tri-snRNP is recruited to the spliceosome following binding of U2 [17,98] and interacts with the 5′ splice site, but such interaction can occur independent of prior binding of U2 to the branch site in HeLa or nematode cell extracts [106,107]. The BBP [SF1 (splicing factor 1) in humans] is thought to bind the branchpoint sequence early and can bridge the 5′ splice site and the 3′ splice site by interacting with U1 components Prp40 and Mud2 (U2AF65 in humans) [108–110]. BBP is displaced by U2 snRNP in an ATP-dependent step upon formation of the pre-spliceosome [111]. A large 45S particle containing all five snRNAs, called penta-snRNP, has been detected in yeast cell extracts and demonstrated to be functional for splicing upon addition of pre-mRNA substrate [112]. It has also long been known that large 200S complexes, containing all five snRNAs and protein splicing factors exist in HeLa nuclear extracts [113,114]. These observations raise the possibility that splicing occurs on preassembled spliceosomes in vivo. However, in vivo studies of spliceosome formation in yeast by chromatin immunoprecipitation analysis suggest spliceosome assembly to be co-transcriptional and occur in an ordered manner [115,116].
SPLICEOSOME ACTIVATION
After the binding of tri-snRNP, the spliceosome undergoes a major conformational rearrangement in which U1 and U4 are dissociated from the spliceosome [17,117]. The release of U1 and U4 requires the unwinding of U4/U6 duplexes and destabilization of U1–5′ splice site base-pairing, both of which require ATP. DExD/H-box RNA helicases Prp28 and Brr2 have been implicated in the release of U1 and U4 respectively [38,39,64]. Brr2 is a component of the U5 snRNP [41] and has been demonstrated to catalyse the unwinding of the U4/U6 RNA duplexes in vitro [38,39]. Human, but not yeast, Prp28 is associated with the tri-snRNP [118], which requires phosphorylation of Prp28 by SRPK2 (serine/arginine protein-specific kinase2) [119]. Genetic studies have revealed that Prp28 is required to destabilize U1–5′ splice site base-pairing [64], but the mechanism underlying the action of Prp28 is not clear. Prp28 may act by directly unwinding U1–5′ splice site duplex, although RNA unwinding activity has not been demonstrated in vitro. Alternatively, Prp28 may displace stabilizing proteins that bind to the 5′ splice site. Supporting this notion, U1 component U1C has been shown to directly bind to the 5′ splice site [120], and mutation in U1C was able to bypass the requirement of Prp28 [65].
The release of U1 and U4 allows new base-pair formation between U6 and the 5′ splice site and between U6 and U2, as demonstrated by extensive genetic and UV-cross-linking analyses [9,10] (Figure 3). Interactions between U5 and the exon sequence at the splice junction have also been demonstrated [121–123]. Although such RNA base-pairings constitute the framework of the catalytic core of the spliceosome, they are usually in short stretches and require protein factors to stabilize their structure. Prp8 has been shown to cross-link to both 5′ and 3′ splice site regions, suggesting its binding to the pre-mRNA may stabilize the base-pairing of U5 with the exon sequences for splice site alignment [22,121].
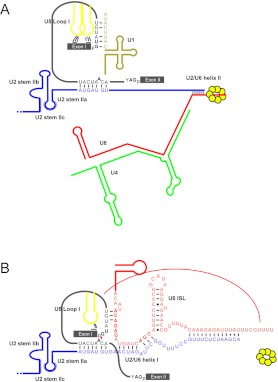
Figure 3. RNA–RNA interactions in pre-activated and activated spliceosomes.
(A) The pre-activated spliceosome contains five snRNAs. U1 snRNA base-pairs with the 5′ splice site and U2 snRNA base-pairs with the branch site with a stem IIa structure. U2/U6 helix II has been shown to be required for tri-snRNP recruitment in mammalian cells. Loop I of U5 snRNA interacts with two broad regions of exon 1. (B) On the activated spliceosome, base-pairings of the 5′ splice site with U1 are displaced by U6. In addition, the Lsm complex is dissociated from U6 to allow interactions of its 3′ end with the intron sequence in a region downstream from the 5′ splice site.
The NTC was shown to be required for spliceosome activation after the release of U1 and U4 [81]. The binding of NTC does not require much of the sequence downstream of the branchpoint [124], but how it is recruited to the spliceosome remains unknown. The NTC is required for stabilizing the association of U5 and U6 with the spliceosome in formation of the active spliceosome [81]. In the absence of the NTC, both U5 and U6 interact with the pre-mRNA in a dynamic manner after U1 and U4 are released, as revealed by UV-cross-linking analysis. The presence of the NTC renders base-pairings of U5 and U6 with defined residues of the pre-mRNA [81,82]. The binding of the NTC also promotes the release of Lsm proteins from U6 to allow for the interaction of the Lsm-binding site near the 3′-end of U6 snRNA with the intron in a region approximately 30 bp downstream from the 5′ splice site [81].
CATALYTIC STEPS
After pairing of U6 to the 5′ splice site and to U2 snRNA, the catalytic core of the spliceosome is established and stabilized by the binding of the NTC and protein components of snRNPs. The splicing reaction is completed via two consecutive transesterification reactions on the activated spliceosome. The first step is cleavage at the 5′ splice site and formation of the lariat intron–exon 2 via a 2′–5′ phosphodiester linkage. The second step is cleavage at the 3′ splice site and ligation of the two exons (Figure 1) [125–127]. Although transesterification reactions do not require energy, both catalytic steps require DExD/H-box ATPases, Prp2 for the first step and Prp16 for the second step, and several other proteins [56] (Figure 4). Much of the work concerning the mechanism of the catalytic steps has been conducted using the yeast Saccharomyces cerevisiae.
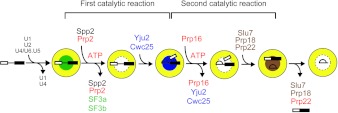
Figure 4. Spliceosome dynamics during the catalytic steps.
Both catalytic steps can be divided into ATP-dependent and ATP-independent steps. Prp2 and Prp16 are required in ATP-dependent steps. Hydrolysis of ATP results in the release of SF3a/b and Yju2/Cwc25 in the first and second step respectively. This allows Yju2/Cwc25 and Slu7/Prp18/Prp22 to bind to the spliceosome and promote the first and second transesterification reactions respectively in an ATP-independent manner. Yellow circles with different colours in the centre represent spliceosomes with different sets of splicing factors binding at the catalytic centre.
First catalytic step
It has been shown that the 3′ splice site is not required for the first reaction [128], but the lack of most nucleotides between the branchpoint and the 3′ splice site blocks the first reaction without affecting spliceosome activation, indicating that this region is important for the first reaction [81,124,129]. The first catalytic step can be divided into two stages based on the ATP requirement, the Prp2-mediated ATP-dependent step with no chemical change to the RNA substrate, followed by the ATP-independent step, which requires additional protein factors to promote the chemical reaction [130].
Prp2 can associate with the spliceosome and directly interact with pre-mRNA in the absence of ATP prior to the first reaction [66,131,132]. Upon ATP hydrolysis, Prp2 is dissociated from the spliceosome [132]. The recruitment of Prp2 to the spliceosome requires a co-factor Spp2, originally identified as a high-copy suppressor of temperature-sensitive prp2-1 mutation [133,134]. The C-terminal region of Prp2 is important for its interaction with Spp2 and for spliceosome binding [135,136]. Spp2 interacts with Prp2 through its G-patch domain, which has been proposed to mediate RNA binding [136,137]. Spp2 can associate with the spliceosome prior to the first reaction and is dissociated from the spliceosome with Prp2 upon ATP hydrolysis [134].
The function of Prp2 in the first catalytic step was obscure until recently, when it was demonstrated that the ATPase function of Prp2 is associated with the release of SF3a and SF3b from the spliceosome [68,69]. SF3a and SF3b are subunits of U2 snRNP, comprising three and six or seven proteins respectively. SF3b subunit SAP155 (Hsh155 in yeast) has been shown to cross-link to intron sequences flanking the branchpoint, suggesting a role for SF3b in stabilizing U2-branch site base-pairing during spliceosome assembly [138–140]. Conceivably, the removal of SF3b from binding to the branch site is necessary to expose the branchpoint to initiate the chemical reaction. Prp2 and Spp2 are dissociated from the spliceosome together with SF3a/b. More recently, another splicing factor, Cwc22, was shown to be required for the Prp2-dependent step [86]. Cwc22 is an eIF4G (eukaryotic initiation factor 4G)-like protein and contains both MIF4G (middle of eIF4G) and MA3 domains found in eIF4G. It binds to the spliceosome after the binding of NTC and is not required for spliceosome activation. Although Cwc22 is not required for the binding of Prp2 to the spliceosome, in the absence of Cwc22, Prp2 is released from the spliceosome upon ATP hydrolysis without promoting the release of SF3a/b [86].
The release of SF3a/b, however, is not sufficient for the first reaction to take place. A protein factor of unknown identity, named HP (heat-resistant protein) for its heat-resistant property, was previously shown to be required for the first catalytic reaction independent of ATP [130]. Two proteins, Yju2 and Cwc25, were later found to be required for the first reaction after the action of Prp2. Yju2 has been shown to interact with the NTC via its interaction with Ntc90 and Ntc77, and can bind to the spliceosome before or after the action of Prp2 [141]. In contrast, Cwc25 is recruited to the spliceosome exclusively after the action of Prp2 but only in the presence of Yju2 [85], and is the last protein factor recruited to the spliceosome before the first catalytic reaction [69,85]. Both Yju2 and Cwc25 become stably associated with the spliceosome after the first reaction, but are released prior to the second reaction [70] (see below).
Second catalytic step
The second catalytic step concerns the identification of the 3′ splice site to align with the 5′ splice site for exon ligation. U5 snRNP plays an important role in the alignment of exons. Loop 1 of U5 snRNA contains nine evolutionarily invariant nucleo-tides that can interact with both exons at the splice junction [122,142–144]. In yeast, loop 1 of U5 is required only for the second reaction in vitro [145,146], but can play a role in the first step [144]. Surprisingly, loop 1 is totally dispensable for the human in vitro splicing reaction [147]. The U5 component Prp8 has been shown to cross-link to both exons at sites near the splice junction, and proposed to stabilize the interaction of U5 with the exon sequences during the second catalytic reaction [21,22,148]. Genetic interactions between Prp8 and the 5′ splice site, 3′ splice site, and almost all the other second-step factors (see below) have also been demonstrated [148–151]. In addition, the non-Watson–Crick interaction between the first and last intron nucleotides has been shown to be required for the second reaction [152,153], and the interactions between both splice site consensus sequences and U6 snRNA also play an important role to juxtapose the two exons in the second step [154]. Mutations that affect these interactions can be suppressed by allele-specific prp8 mutants [149,150,155], suggesting that Prp8 may modulate the conformational transition between the two transesterification reactions [156,157].
Like the first step, the second catalytic step also involves an ATP-dependent step, which requires DExD/H-box protein Prp16, followed by an ATP-independent step, in which transesterification occurs [158]. Prp16 was originally identified as a suppressor of branchpoint mutant A259C (or brC) of the actin intron [159,160], but was found to be only required for the second reaction in vitro [67]. It has been shown that Prp16 can associate with the spliceosome in the absence of ATP, and upon ATP hydrolysis, Prp16 is released from the spliceosome [67]. The N-terminus of Prp16 is required and sufficient for its association with the spliceosome [161]. UV-cross-linking analysis has revealed that Prp16 interacts with pre-mRNA in the intron sequence near the 3′ splice site [23,138]. Prp17 has also been demonstrated to function in the ATP-dependent step, but the binding of Prp16 to the spliceosome is not affected in the absence of Prp17 [23,162]. Prp16 has been proposed to mediate a conformational change in the spliceosome prior to the second reaction, since the 3′ splice site becomes protected from RNase H cleavage after Prp16 action [163]. This protected region was later shown to cross-link with Prp22, suggesting that the binding of Prp22 may confer the nuclease protection phenotype [59,164].
Several cold-sensitive prp16 alleles have been shown to suppress mutations in U2 and U6 snRNAs in the regions that are involved in the formation of RNA duplexes, suggesting a role of Prp16 in disruption of these helices [165–167]. Although Prp16 has been shown to unwind synthetic RNA duplexes, there is no direct evidence for such function on the spliceosome [57]. The recent demonstration of Prp16- and ATP-dependent release of the first step factors Yju2 and Cwc25 suggests that Prp16 may play a role in remodelling the spliceosome to prepare the spliceosome for the second catalytic reaction [70]. Whether destabilization of Yju2 and Cwc25 by Prp16 leads to destabilization of RNA structures, or vice versa, is unclear.
Following Prp16 action, Slu7, Prp18 and Prp22 join the spliceosome to promote the second reaction in an ATP-independent manner [59,158,168]. The fact that Yju2 and Cwc25 are removed from the spliceosome prior to the association of Slu7, Prp18 and Prp22 suggests that displacement of proteins binding to the catalytic centre is necessary for positioning of the 5′ and 3′ splice sites for exon ligation. These three proteins interact with the 3′ splice site, and both Prp22 and Slu7 directly contact the intron, as revealed by cross-linking analysis [23,164]. Prp22 is a DExD/H-box protein, but its role in the second catalytic step is ATP-independent [59,72]. Prp22 is further required for the release of mature mRNA after completion of splicing, which depends on its ATPase activity, and together with Slu7 and Prp18, it is released from the spliceosome along with mRNA [58,59,72,169]. Prp18 has been shown to stabilize the interaction between loop 1 of U5 snRNA and both exons during the second reaction [142,170]. The requirement of Slu7, Prp18 and Prp22 for the second reaction depends on the distance between the branch site and the 3′ splice site of precursor mRNA [59,171,172]. None of the three proteins is needed if the distance is less than 7 nt [171]. The binding of these proteins follows an order of Slu7, Prp18 and then Prp22 [169]. Together, the second-step factors bind to the 3′ splice site, and coordinate with Prp8 to stabilize interactions of U5 loop 1 and the exons for the second transesterification.
Similarities between the two steps
The two catalytic steps are mechanistically similar. Both steps can be divided into two stages, an ATP-dependent step, which requires a DExD/H-box protein, followed by an ATP-independent catalytic reaction. Each step starts with displacement of protein binding to the catalytic centre to allow binding of another set of proteins to promote the catalytic reaction. Prp2 is required to mediate the release of SF3a/b, which bind to the branch site, to allow binding of Yju2 and Cwc25 to promote the first transesterification. Prp16 is required to mediate the release of Yju2 and Cwc25, to allow binding of Slu7, Prp18 and Prp22 to promote the second transesterification. Cwc25 binds to the branch site as it can cross-link to substrate RNA near the branch site (H.-C. Chen and S.-C. Cheng, unpublished work), and mutations at the branchpoint prevent its binding to the spliceosome [70]. Conceivably, the removal of factors binding to the catalytic centre and functioning in the previous step converts the catalytic centre into an open state, allowing splice sites to interact in a dynamic manner. Subsequent binding of specific protein factors, Yju2/Cwc25 for the first step and Slu7/Prp18/Prp22 for the second step, facilitates and confines the interaction at specific sites. It is interesting that Cwc25 was shown to be dispensable for the first reaction if the purified spliceosome was incubated in the presence of Mn2+, suggesting that increasing the dynamics of splice–site interaction can bypass the need for Cwc25 [85]. Similarly, Slu7/Prp18/Prp22 can be dispensable when the 3′ splice site is very close to the branch site so that the 3′ splice site is more susceptible to interaction with the 5′ splice site [59,171,172]. Moreover, both Prp16 and Prp22 have two roles in the splicing pathway. Prp22 has a primary role in promoting mRNA release, which requires the ATPase function of the protein, and is conditionally required for the second reaction in positioning the 3′ splice site in an ATP-independent manner. Prp16 also has an ATP-dependent role in promoting the release of Yju2/Cwc25, and has an ATP-independent role in the first catalytic step in stabilizing the binding of Cwc25 when pre-mRNA carries a mutation at the branchpoint.
DISASSEMBLY OF THE SPLICEOSOME
mRNA release
Following exon ligation, the mature mRNA is released before the spliceosome is disassembled. Prp22 was initially identified from a genetic screen for factors defective in pre-mRNA splicing [173]. The prp22-1 mutant accumulates mRNA and excised lariat-intron on the spliceosome at the non-permissive temperature [72]. Prp22 has been demonstrated to unwind RNA duplexes in vitro [58,59], and the helicase activity is required for mRNA release [174]. Mutations in motif III (SAT) of the helicase domain are defective in unwinding RNA duplex and releasing mRNA with no effect on the ATPase activity or the binding of Prp22 to the spliceosome [174]. It has been shown that cold-sensitive growth defect elicited by Prp22 helicase-defective mutants can be suppressed by several mutant alleles of Prp8 at Arg1753, suggesting that Prp22 may disrupt RNA–RNA or RNA–protein interaction stabilized by Prp8 during mRNA release [175]. In agreement with this notion, specific mutant alleles of U5 loop 1, which stabilizes the interaction of U5 loop 1 with splicing intermediate and suppresses prp8-R1753 mutant, can aggravate the defect of Prp22 helicase-defective mutants [151]. Prp22 was demonstrated to bind to the 3′ splice site during the second step [164] and then translocate to mRNA downstream of exon–exon junction after exon ligation [176]. It is proposed that Prp22 functions to liberate mRNA by moving along mRNA in a 3′ to 5′ directionality to disrupt U5–mRNA interaction [176]. More than 13 nt of 3′ exon in length is necessary for mRNA release, presumably to allow stable binding of Prp22 after translocation [176].
Spliceosome disassembly
After mRNA is released, the spliceosome is disassembled to free the lariat-intron and all associated spliceosomal components for recycling of splicing factors. The free lariat-introns are debranched, catalysed by debranchase (Dbr1), to yield linear intron molecules, which are generally degraded [177,178]. Prp43, a DExD/H-box RNA helicase, is the key player in mediating disassembly of the spliceosome [71,178] and is also involved in the biogenesis of both large- and small-subunits of the ribosome [179–181]. Prp43 has been demonstrated to unwind RNA duplexes in vitro [60]. The ATPase activity of Prp43 is required for spliceosome disassembly, but the level of the helicase activity does not have any apparent correlation with the disassembly efficiency [60,178].
The function of Prp43 in mediating spliceosome disassembly requires two co-factors, Ntr1 and Ntr2 [182,183]. Ntr1 interacts with Ntr2 to form a stable complex, which further interacts with Prp43 to form a functional NTR complex [182]. Spliceosome disassembly can be assayed in vitro using purified NTR. The post-splicing spliceosome isolated from NTR-depleted extracts, containing the lariat-intron, is dissociated into U2, U5, U6 and NTC upon incubation with purified NTR and ATP [182]. Ntr1 interacts with Ntr2 via the middle region of the protein, and with Prp43 via the N-terminal G-patch domain [182]. The G-patch domain of Ntr1 has been shown to stimulate the helicase activity, but not the ATPase activity, of Prp43 [184]. Ntr2 interacts with U5 snRNP via its interaction with U5 component Brr2, and such interaction is suggested to mediate the recruitment of NTR to the spliceosome [185].
How NTR mediates spliceosome disassembly is not known. To disassemble the spliceosome, it is essential to disrupt base-pairings of U6/5′ splice site, U2/U6 and U2/branch site. Prp43 has been shown to cross-link to U6, but the precise cross-link sites have not been determined [186]. Prp43 may destabilize base-pairings of U6/5′ splice site or U2/U6 by unwinding RNA duplex or by displacing proteins binding to specific RNA structures, which then triggers disruption of other RNA duplexes. It has also been shown that mutations in Brr2 impede disassembly of the spliceosome isolated via its association with Prp43, and the ATPase activity of Brr2 is regulated by Snu114 through its binding of GTP versus GDP [40]. How Brr2 co-ordinates with Prp43 to mediate spliceosome disassembly remains unknown.
NTR1, previously identified as SPP382, was shown to function as a suppressor of prp38-1 mutation [187]. Prp38 is a component of the yeast tri-snRNP [188,189]. Several mutations in PRP43 affecting the ATPase activity have been shown to suppress the growth defect of prp38-1 with efficiencies inversely proportionate to the measured ATPase activities [187], suggesting that reducing the activity of Prp43 could partially compensate for impaired spliceosome assembly. The involvement of Prp43 in the discard of spliceosome intermediates is supported by the findings that the dissociation of pre-mRNA or the lariat intermediate from the impaired spliceosome depends on Prp43 [190,191].
SPLICING FIDELITY CONTROL
The mechanism of splicing fidelity control has been best studied in yeast. DExD/H-box protein Prp16 was first discovered to have a function in promoting the fidelity of branch site recognition [159,160]. Two other DExD/H-box proteins, Prp22 and Prp5, were later also implicated in the control of splicing fidelity [192,193]. Prp16 was identified in a genetic screen as a suppressor of branchpoint mutation brC (changing A to C). ATPase-defective mutants of Prp16 could suppress branchpoint mutation and accumulate lariat-intermediates with aberrant branch nucleotide, suggesting that the ATPase activity of Prp16 is required to discriminate mutations at the branchpoint [194]. Prp16 was further found to be essential in vitro for the second reaction but dispensable for the first reaction [67]. Based on these studies, a model for how splicing fidelity is controlled by a kinetic proofreading mechanism was formulated [194]. It was proposed that kinetic competition between a switch to the second-step conformation and the ATP-dependent rejection of substrate mediated by Prp16 serves as the basis for the control of fidelity [194]. The kinetic competition mechanism was further supported by a recent study using a mutant U6 snRNA with sulfur substitution for pro-Sp non-bridging oxygen at position U80 (U6–sU80) [190]. In the first reaction, splicing was impeded in the presence of Mg2+ using U6–sU80. Prp16 was able to bind to such spliceosomes and mediates rejection of the slow spliceosome in the way that requires ATP and the ATPase function of Prp16 [190].
An ATP-independent role for Prp16 in the first reaction has recently been demonstrated [70]. Cwc25 is known to bind to the spliceosome in the presence of Yju2 after the release of SF3a/b to promote the first reaction on wild-type pre-mRNA [69,85]. Mutations at the branchpoint prevent binding of Cwc25, and consequently inhibit the reaction [70]. It was found that the presence of Prp16, regardless of its ATPase activity, stabilizes binding of Cwc25 to the brC spliceosome and promotes the first reaction. This ATP-independent function of Prp16 counteracts its ATP-dependent function in mediating Cwc25 removal. The balance between the two activities determines the level of splicing of brC pre-mRNA. Reducing the ATPase activity of Prp16 promotes the reaction. In this view, although Prp16 is the key player in mediating fidelity control of the first reaction, Cwc25 plays an important role in enforcing specificity of branchpoint recognition [70,195].
Mutations at the 3′ splice site block exon ligation, which requires Prp22. Prp22 was shown to repress exon ligation at the mutated 3′ splice site in an ATP-dependent manner. Prp22 mutants that reduce ATPase activity compromise the fidelity of exon ligation both in vivo and in vitro [192]. These results suggest that Prp22 may play a role in proofreading exon ligation. It has also been shown that mutations within Prp5 in motif III of the helicase domain with reduced ATPase activity also increase the splicing activity of suboptimal substrate with less stable branch region-U2 snRNA pairing, suggesting a link between fidelity control in branch recognition and Prp5 ATPase activity [193].
CONCLUSIONS AND PERSPECTIVES
Pre-mRNA splicing is a complex process requiring assembly of a large ribonucleoprotein particle that involves a large number of factors. Over the years, genetic and biochemical studies have provided comprehensive insights into the mechanism of the splicing reaction. Compositional and structural analyses of the spliceosome and its components by MS, electron microscopy and X-ray crystallography have facilitated further understanding of the function of the spliceosome and its components at the structural level [196–201]. Approaches at the single-molecule level are also expected to result in new mechanistic insights [202,203], shedding further light on the molecular mechanism of nuclear pre-mRNA splicing.
ACKNOWLEDGEMENT
We thank M. Loney for editing the paper prior to submission.
FUNDING
Our own work was supported by Academia Sinica and the National Science Council (Taiwan) [grant number NSC99-2745-B-001-001-ASP].
References
- 1.Brody E., Abelson J. The ‘spliceosome’: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- 2.Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985;42:355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985;42:345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- 4.Wahl M. C., Will C. L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Brow D. A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 6.Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 7.Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 8.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Madhani H. D., Guthrie C. Dynamic RNA–RNA interactions in the spliceosome. Annu. Rev. Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 10.Umen J. G., Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 11.Hall S. L., Padgett R. A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 12.Patel A. A., Steitz J. A. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 13.Lührmann R., Kastner B., Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 14.Raker V. A., Plessel G., Lührmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto C., Steitz J. A. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res. 1984;12:3283–3293. doi: 10.1093/nar/12.7.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bringmann P., Appel B., Rinke J., Reuter R., Theissen H., Lührmann R. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J. 1984;3:1357–1363. doi: 10.1002/j.1460-2075.1984.tb01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 18.Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 19.Jackson S. P., Lossky M., Beggs J. D. Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol. Cell. Biol. 1988;8:1067–1075. doi: 10.1128/mcb.8.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodges P. E., Jackson S. P., Brown J. D., Beggs J. D. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast. 1995;11:337–342. doi: 10.1002/yea.320110406. [DOI] [PubMed] [Google Scholar]
- 21.Dix I., Russell C. S., O'Keefe R. T., Newman A. J., Beggs J. D. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1675–1686. doi: 10.1017/s1355838298412998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teigelkamp S., Newman A. J., Beggs J. D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umen J. G., Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal V. P., Verdone L., Mayes A. E., Beggs J. D. Characterization of U6 snRNA–protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boon K. L., Norman C. M., Grainger R. J., Newman A. J., Beggs J. D. Prp8p dissection reveals domain structure and protein interaction sites. RNA. 2006;12:198–205. doi: 10.1261/rna.2281306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Nues R. W., Beggs J. D. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grainger R. J., Beggs J. D. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boon K. L., Grainger R. J., Ehsani P., Barrass J. D., Auchynnikava T., Inglehearn C. F., Beggs J. D. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat. Struct. Mol. Biol. 2007;14:1077–1083. doi: 10.1038/nsmb1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellare P., Kutach A. K., Rines A. K., Guthrie C., Sontheimer E. J. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellare P., Small E. C., Huang X., Wohlschlegel J. A., Staley J. P., Sontheimer E. J. A role for ubiquitin in the spliceosome assembly pathway. Nat. Struct. Mol. Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abelson J. Is the spliceosome a ribonucleoprotein enzyme? Nat. Struct. Mol. Biol. 2008;15:1235–1237. doi: 10.1038/nsmb1208-1235. [DOI] [PubMed] [Google Scholar]
- 32.Pena V., Rozov A., Fabrizio P., Lührmann R., Wahl M. C. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J. 2008;27:2929–2940. doi: 10.1038/emboj.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie D. B., Schellenberg M. J., Gesner E. M., Raithatha S. A., Stuart D. T., Macmillan A. M. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat. Struct. Mol. Biol. 2008;15:1199–1205. doi: 10.1038/nsmb.1505. [DOI] [PubMed] [Google Scholar]
- 34.Yang K., Zhang L., Xu T., Heroux A., Zhao R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeder C., Kutach A. K., Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat. Struct. Mol. Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pena V., Jovin S. M., Fabrizio P., Orlowski J., Bujnicki J. M., Lührmann R., Wahl M. C. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol. Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L., Xu T., Maeder C., Bud L. O., Shanks J., Nix J., Guthrie C., Pleiss J. A., Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat. Struct. Mol. Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghunathan P. L., Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 39.Laggerbauer B., Achsel T., Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small E. C., Leggett S. R., Winans A. A., Staley J. P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauber J., Fabrizio P., Teigelkamp S., Lane W. S., Hartmann E., Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D. H., Rossi J. J. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabrizio P., Laggerbauer B., Lauber J., Lane W. S., Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner T. J., Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartels C., Klatt C., Lührmann R., Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartels C., Urlaub H., Lührmann R., Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- 47.Brenner T. J., Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA. 2006;12:862–871. doi: 10.1261/rna.2319806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwan S. S., Brow D. A. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding. RNA. 2005;11:808–820. doi: 10.1261/rna.2010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raghunathan P. L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 50.Shannon K. W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 51.Ghetti A., Company M., Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- 52.Jandrositz A., Guthrie C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 1995;14:820–832. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pyle A. M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 55.Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- 56.Staley J. P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Wagner J. D., Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- 58.Wagner J. D., Jankowsky E., Company M., Pyle A. M., Abelson J. N. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwer B., Gross C. H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka N., Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kistler A. L., Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perriman R., Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- 63.Perriman R., Barta I., Voeltz G. K., Abelson J., Ares M., Jr ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staley J. P., Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 65.Chen J. Y., Stands L., Staley J. P., Jackups R. R., Jr, Latus L. J., Chang T. H. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 66.Kim S. H., Lin R. J. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1993;90:888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 68.Lardelli R. M., Thompson J. X., Yates J. R., III, Stevens S. W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warkocki Z., Odenwälder P., Schmitzová J., Platzmann F., Stark H., Urlaub H., Ficner R., Fabrizio P., Lührmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 70.Tseng C. K., Liu H. L., Cheng S. C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arenas J. E., Abelson J. N. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 73.Tarn W. Y., Lee K. R., Cheng S. C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tarn W. Y., Hsu C. H., Huang K. T., Chen H. R., Kao H. Y., Lee K. R., Cheng S. C. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C. H., Yu W. C., Tsao T. Y., Wang L. Y., Chen H. R., Lin J. Y., Tsai W. Y., Cheng S. C. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 2002;30:1029–1037. doi: 10.1093/nar/30.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsai W. Y., Chow Y. T., Chen H. R., Huang K. T., Hong R. I., Jan S. P., Kuo N. Y., Tsao T. Y., Chen C. H., Cheng S. C. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem. 1999;274:9455–9462. doi: 10.1074/jbc.274.14.9455. [DOI] [PubMed] [Google Scholar]
- 77.Chen C. H., Tsai W. Y., Chen H. R., Wang C. H., Cheng S. C. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 2001;276:488–494. doi: 10.1074/jbc.M006958200. [DOI] [PubMed] [Google Scholar]
- 78.Chen H. R., Jan S. P., Tsao T. Y., Sheu Y. J., Banroques J., Cheng S. C. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 1998;18:2196–2204. doi: 10.1128/mcb.18.4.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohi M. D., Gould K. L. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hogg R., McGrail J. C., O'Keefe R. T. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 2010;38:1110–1115. doi: 10.1042/BST0381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 82.Chan S. P., Cheng S. C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 83.Ohi M. D., Link A. J., Ren L., Jennings J. L., McDonald W. H., Gould K. L. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGrail J. C., Krause A., O'Keefe R. T. The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res. 2009;37:4205–4217. doi: 10.1093/nar/gkp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiu Y. F., Liu Y. C., Chiang T. W., Yeh T. C., Tseng C. K., Wu N. Y., Cheng S. C. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 2009;29:5671–5678. doi: 10.1128/MCB.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeh T. C., Liu H. L., Chung C. S., Wu N. Y., Liu Y. C., Cheng S. C. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol. Cell. Biol. 2011;31:43–53. doi: 10.1128/MCB.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Chazin W. J., Gould K. L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Ren L., Hirsch J. P., Chazin W. J., Walz T., Gould K. L. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 2005;25:451–460. doi: 10.1128/MCB.25.1.451-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vander Kooi C. W., Ren L., Xu P., Ohi M. D., Gould K. L., Chazin W. J. The Prp19 WD40 domain contains a conserved protein interaction region essential for its function. Structure. 2010;18:584–593. doi: 10.1016/j.str.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren L., McLean J. R., Hazbun T. R., Fields S., Vander Kooi C., Ohi M. D., Gould K. L. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS One. 2011;6:e16719. doi: 10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen H. R., Tsao T. Y., Chen C. H., Tsai W. Y., Her L. S., Hsu M. M., Cheng S. C. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5406–5411. doi: 10.1073/pnas.96.10.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chung S., McLean M. R., Rymond B. C. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA. 1999;5:1042–1054. doi: 10.1017/s1355838299990635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang K. J., Chen H. C., Cheng S. C. Ntc90 is required for recruiting first step factor Yju2 but not for spliceosome activation. RNA. 2009;15:1729–1739. doi: 10.1261/rna.1625309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villa T., Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu D., Field D. J., Tang S. J., Moris A., Bobechko B. P., Friesen J. D. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol. 1998;18:2055–2066. doi: 10.1128/mcb.18.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu D., Friesen J. D. Splicing factor slt11p and its involvement in formation of U2/U6 helix II in activation of the yeast spliceosome. Mol. Cell. Biol. 2001;21:1011–1023. doi: 10.1128/MCB.21.4.1011-1023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987;6:2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA–protein complex selectively binds a 5′ splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- 100.Krämer A., Keller W., Appel B., Lührmann R. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- 101.Séraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 102.Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 103.Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 104.Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 105.Liao X. C., Colot H. V., Wang Y., Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992;20:4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maroney P. A., Romfo C. M., Nilsen T. W. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 107.Konforti B. B., Konarska M. M. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- 108.Abovich N., Liao X. C., Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 109.Abovich N., Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 110.Berglund J. A., Chua K., Abovich N., Reed R., Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 111.Rutz B., Séraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stevens S. W., Ryan D. E., Ge H. Y., Moore R. E., Young M. K., Lee T. D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 113.Ast G., Goldblatt D., Offen D., Sperling J., Sperling R. A novel splicing factor is an integral component of 200S large nuclear ribonucleoprotein (lnRNP) particles. EMBO J. 1991;10:425–432. doi: 10.1002/j.1460-2075.1991.tb07964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miriami E., Angenitzki M., Sperling R., Sperling J. Magnesium cations are required for the association of U small nuclear ribonucleoproteins and SR proteins with pre-mRNA in 200 S large nuclear ribonucleoprotein particles. J. Mol. Biol. 1995;246:254–263. doi: 10.1006/jmbi.1994.0081. [DOI] [PubMed] [Google Scholar]
- 115.Lacadie S. A., Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 116.Tardiff D. F., Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lamond A. I., Konarska M. M., Grabowski P. J., Sharp P. A. Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 1988;85:411–415. doi: 10.1073/pnas.85.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Teigelkamp S., Mundt C., Achsel T., Will C. L., Lührmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 119.Mathew R., Hartmuth K., Möhlmann S., Urlaub H., Ficner R., Lührmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 120.Du H., Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 121.Wyatt J. R., Sontheimer E. J., Steitz J. A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 122.Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 123.Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 124.Cheng S. C. Formation of the yeast splicing complex A1 and association of the splicing factor PRP19 with the pre-mRNA are independent of the 3′ region of the intron. Nucleic Acids Res. 1994;22:1548–1554. doi: 10.1093/nar/22.9.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984;39:611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- 126.Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 127.Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNAs as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 128.Rymond B. C., Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 129.Rymond B. C., Torrey D. D., Rosbash M. A novel role for the 3′ region of introns in pre-mRNA splicing of Saccharomyces cerevisiae. Genes Dev. 1987;1:238–246. doi: 10.1101/gad.1.3.238. [DOI] [PubMed] [Google Scholar]
- 130.Kim S. H., Lin R. J. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.King D. S., Beggs J. D. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:6559–6564. doi: 10.1093/nar/18.22.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Teigelkamp S., McGarvey M., Plumpton M., Beggs J. D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Last R. L., Maddock J. R., Woolford J. L., Jr Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics. 1987;117:619–631. doi: 10.1093/genetics/117.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roy J., Kim K., Maddock J. R., Anthony J. G., Woolford J. L., Jr The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- 135.Edwalds-Gilbert G., Kim D. H., Silverman E., Lin R. J. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA. 2004;10:210–220. doi: 10.1261/rna.5151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Silverman E. J., Maeda A., Wei J., Smith P., Beggs J. D., Lin R. J. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aravind L., Koonin E. V. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- 138.McPheeters D. S., Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Das B. K., Xia L., Palandjian L., Gozani O., Chyung Y., Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gozani O., Potashkin J., Reed R. A potential role for U2AF-SAP155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu Y. C., Chen H. C., Wu N. Y., Cheng S. C. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol. 2007;27:5403–5413. doi: 10.1128/MCB.00346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Crotti L. B., Bačíková D., Horowitz D. S. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frank D. N., Roiha H., Guthrie C. Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 145.O'Keefe R. T., Newman A. J. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.O'Keefe R. T., Norman C., Newman A. J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 147.Ségault V., Will C. L., Polycarpou-Schwarz M., Mattaj I. W., Branlant C., Lührmann R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Umen J. G., Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 149.Collins C. A., Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Siatecka M., Reyes J. L., Konarska M. M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aronova A., Bačíková D., Crotti L. B., Horowitz D. S., Schwer B. Functional interactions between Prp8, Prp18, Slu7, and U5 snRNA during the second step of pre-mRNA splicing. RNA. 2007;13:1437–1444. doi: 10.1261/rna.572807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Parker R., Siliciano P. G. Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 153.Chanfreau G., Legrain P., Dujon B., Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Collins C. A., Guthrie C. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA. 2001;7:1845–1854. [PMC free article] [PubMed] [Google Scholar]
- 155.Umen J. G., Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu L., Query C. C., Konarska M. M. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat. Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 157.Query C. C., Konarska M. M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 158.Horowitz D. S., Abelson J. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993;7:320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- 159.Burgess S., Couto J. R., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 160.Couto J. R., Tamm J., Parker R., Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- 161.Wang Y., Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 162.Jones M. H., Frank D. N., Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schwer B., Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McPheeters D. S., Schwer B., Muhlenkamp P. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 2000;28:1313–1321. doi: 10.1093/nar/28.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mefford M. A., Staley J. P. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–1397. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hilliker A. K., Mefford M. A., Staley J. P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Perriman R. J., Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ansari A., Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.James S. A., Turner W., Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bačíková D., Horowitz D. S. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol. Cell. Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brys A., Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang X., Schwer B. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 1997;25:2146–2152. doi: 10.1093/nar/25.11.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vijayraghavan U., Company M., Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 174.Schwer B., Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schneider S., Campodonico E., Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- 176.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chapman K. B., Boeke J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 178.Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 179.Combs D. J., Nagel R. J., Ares M., Jr, Stevens S. W. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lebaron S., Froment C., Fromont-Racine M., Rain J. C., Monsarrat B., Caizergues-Ferrer M., Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Leeds N. B., Small E. C., Hiley S. L., Hughes T. R., Staley J. P. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tsai R. T., Fu R. H., Yeh F. L., Tseng C. K., Lin Y. C., Huang Y. H., Cheng S. C. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Boon K. L., Auchynnikava T., Edwalds-Gilbert G., Barrass J. D., Droop A. P., Dez C., Beggs J. D. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell. Biol. 2006;26:6016–6023. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tanaka N., Aronova A., Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tsai R. T., Tseng C. K., Lee P. J., Chen H. C., Fu R. H., Chang K. J., Yeh F. L., Cheng S. C. Dynamic interactions of Ntr1–Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bohnsack M. T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pandit S., Lynn B., Rymond B. C. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xie J., Beickman K., Otte E., Rymond B. C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stevens S. W., Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Koodathingal P., Novak T., Piccirilli J. A., Staley J. P. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol. Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mayas R. M., Maita H., Semlow D. R., Staley J. P. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mayas R. M., Maita H., Staley J. P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu Y. Z., Query C. C. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Burgess S. M., Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 195.Horowitz D. S. The splice is right: guarantors of fidelity in pre-mRNA splicing. RNA. 2011;17:551–554. doi: 10.1261/rna.2577511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 197.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 198.Zhou Z., Licklider L. J., Gygi S. P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 199.Newman A. J., Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 200.Stark H., Lührmann R. Cryo-electron microscopy of spliceosomal components. Annu. Rev. Biophys. Biomol. Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 201.Will C. L., Lührmann R. Spliceosome structure and function. Cold Spring Harbor Perspect. Biol. 2011;3:pii: a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Crawford D. J., Hoskins A. A., Friedman L. J., Gelles J., Moore M. J. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14:170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Abelson J., Blanco M., Ditzler M. A., Fuller F., Aravamudhan P., Wood M., Villa T., Ryan D. E., Pleiss J. A., Maeder C., et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat. Struct. Mol. Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]