Abstract
Parkinson's disease (PD) is the second most prevalent progressive neurological disorder commonly associated with impaired mitochondrial function in dopaminergic neurons. Although familial PD is multifactorial in nature, a recent genetic screen involving PD patients identified two mitochondrial Hsp70 variants (P509S and R126W) that are suggested in PD pathogenesis. However, molecular mechanisms underlying how mtHsp70 PD variants are centrally involved in PD progression is totally elusive. In this article, we provide mechanistic insights into the mitochondrial dysfunction associated with human mtHsp70 PD variants. Biochemically, the R126W variant showed severely compromised protein stability and was found highly susceptible to aggregation at physiological conditions. Strikingly, on the other hand, the P509S variant exhibits significantly enhanced interaction with J-protein cochaperones involved in folding and import machinery, thus altering the overall regulation of chaperone-mediated folding cycle and protein homeostasis. To assess the impact of mtHsp70 PD mutations at the cellular level, we developed yeast as a model system by making analogous mutations in Ssc1 ortholog. Interestingly, PD mutations in yeast (R103W and P486S) exhibit multiple in vivo phenotypes, which are associated with ‘mitochondrial dysfunction’, including compromised growth, impairment in protein translocation, reduced functional mitochondrial mass, mitochondrial DNA loss, respiratory incompetency and increased susceptibility to oxidative stress. In addition to that, R103W protein is prone to aggregate in vivo due to reduced stability, whereas P486S showed enhanced interaction with J-proteins, thus remarkably recapitulating the cellular defects that are observed in human PD variants. Taken together, our findings provide evidence in favor of direct involvement of mtHsp70 as a susceptibility factor in PD.
INTRODUCTION
Parkinson's disease (PD) is one of the most prevalent neurodegenerative disorders characterized by the progressive loss of pigmented dopaminergic neurons of the substantia nigra (1). This is accompanied by a deficiency of the neurotransmitter dopamine and the presence of cytoplasmic inclusions of α-synuclein, known as Lewy bodies (2). Although 95% of PD cases are sporadic in nature, mutations at various loci such as SNCA (encoding α-synuclein), PINK 1, PARK2 (encoding parkin), PARK7 (encoding DJ-1) and LRRK2 have been implicated in the relatively rare familial forms of the disease (3,4). In case of all these proteins, the exact molecular mechanisms leading to disease development have not been fully elucidated. However, most of them lead to impaired mitochondrial dynamics and accumulation of dysfunctional mitochondria, which is a characteristic feature of PD pathogenesis (5).
A recent, clinical, gene-specific screen involving a cohort of late-onset PD patients revealed two rare missense mutations in the human mitochondrial Hsp70 protein (mtHsp70, also referred as mortalin, Grp75 and PBP74) (6). Interestingly, these heterozygous PD variants of mtHsp70 showed impairment in mitochondrial function in neuronal and non-neuronal human cell lines, thus highlighting a potential role of mtHsp70 in PD pathology (7). Furthermore, both the PD variants exhibited accumulation of excessive cellular reactive oxygen species (ROS), mitochondrial morphological changes and reduction in mitochondrial membrane potential in heterozygous conditions, suggesting an autosomal dominant effect (7). However, it is unclear whether the observed PD phenotypes are due to a consequence of mutations resulting from impaired chaperone-specific functions or altered interaction between mtHsp70 and DJ-1, thus leading to mitochondrial dysfunction (7). In addition to that, a reduction in the expression level of human mtHsp70 is also reported in the affected brain regions of late-onset PD patients as well as in a cellular model of the disease, thus signifying its possible central involvement in PD progression (8,9).
Mitochondrial Hsp70 is a highly conserved member of the HSP70 multigene family and plays multiple essential functions in the biogenesis of mitochondria across kingdoms. Mammalian mtHsp70 is a nuclear encoded protein, predominantly localized in the mitochondrial matrix compartment (10). However, in many cell types, differential levels of the protein have an extra-mitochondrial localization that includes cytosolic and endoplasmic reticulum compartments (11). Yeast Ssc1 is an ortholog of mammalian mtHsp70 essential for viability in Saccharomyces cerevisiae. Human mtHsp70 and yeast Ssc1 share an overall 66% identity and 82% similarity between them. Both mammalian mtHsp70 and yeast Ssc1 form the core of the ‘protein import motor’ of the inner mitochondrial membrane, which translocates a majority of the precursor proteins destined for the matrix compartment (12–14). Besides, they constitute the core of the ‘folding machine’ and assist in the folding of newly-matrix-translocated precursor proteins, thereby essentially maintaining the protein quality control within mitochondria (13–15). Yeast mitochondrial matrix consists of multiple paralogs of Ssc1 specialized for additional functions such as formation of iron–sulfur cluster assembly and mitochondrial DNA (mtDNA) maintenance (16–19). However, the human mitochondrial matrix comprises a single mtHsp70 critically required for all the generic as well as for the specialized functions, thus underlining its central importance in the mitochondrial biogenesis (20).
Mitochondrial Hsp70 consists of two conserved functional domains, namely the N-terminal nucleotide-binding domain (NBD) and C-terminal substrate-binding domain (SBD) (21). The NBD of mtHsp70 possesses a weak intrinsic ATP hydrolyzing activity, whereas the SBD is involved in binding to mitochondrial client proteins (21). The SBD and NBD regions are connected via a flexible linker region, which modulates the allosteric communication between the domains. Like other generic Hsp70s, mtHsp70s have coevolved with J-protein cochaperones to function together as a ‘molecular machine’, with mtHsp70 being the core component (22). Yeast mitochondria consist of two J-proteins, namely Pam18 and Mdj1. Pam18 is tethered to the inner mitochondrial membrane and is required for the translocation function, whereas Mdj1 plays a critical role in the folding of precursor protein in the matrix (23–28). Similarly, in humans, DnaJC19 is required for translocation, whereas hTid-1L and hTid-1S splice variants assist in the folding of imported proteins in the matrix compartment (29,30). These J-proteins often sequester and deliver unfolded proteins to mtHsp70 in the ATP-bound state and catalytically stimulate the ATPase activity of Hsp70, thereby stabilizing the interactions with client proteins in the ADP-bound state (31). The nucleotide exchange factors (Mge1 in yeast and GrpEL1 in humans) regulate the exchange of nucleotides from mtHsp70s, thus operating the molecular chaperone cycle in connection with various essential physiological functions in the event of mitochondria biogenesis (29,32).
Owing to multiple and critical functions played by mtHsp70, the PD variants are expected to contribute significantly toward PD, where mitochondrial dysfunction is an established feature. However, contribution of disease-associated homozygous variants of human mtHsp70 toward PD pathogenesis and progression is still enigmatic. Because of conserved cellular biology, yeast has been used as a well-established model system to understand the PD pathogenesis (33,34). To delineate the impact of PD mutations in the haploid state toward mitochondrial function, we utilized yeast as a model system to generate analogous mutations in Ssc1 protein. Our findings highlight that the PD mutations in yeast Ssc1 lead to similar cellular phenotypes as observed in the affected dopaminergic neurons. Strikingly, in the haploid state, PD mutations in yeast Ssc1 exhibit severe growth defects, sensitivity to oxidative stress, susceptibility to mtDNA loss, compromised protein stability and enhanced J-protein interaction, thus overall contributing significantly toward ‘mitochondrial dysfunction’, which is the hallmark of PD pathogenesis.
RESULTS
Human mtHsp70 PD variants display enhanced stimulation of ATPase activity by J-protein cochaperones
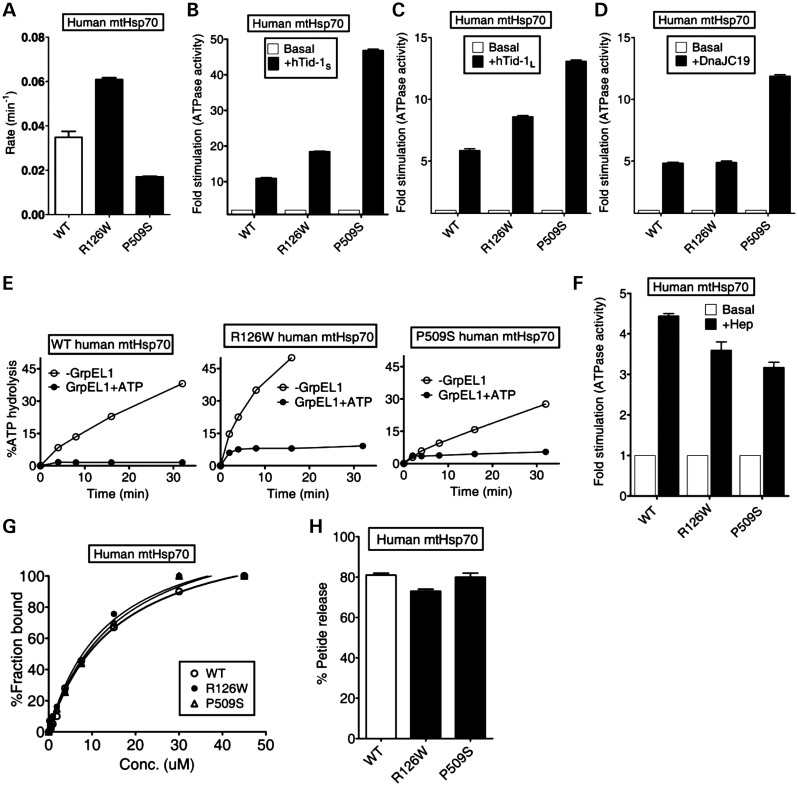
PD-associated human mtHsp70 variants R126W and P509S are located in the subdomain IB of the N-terminal ATPase domain and in the L5/6 loop of the SBD, respectively (35). To uncover the biochemical defects associated with PD variants in vitro, we purified the proteins from Escherichia coli by co-expressing with yeast Hep1. In order to determine whether these mutations lead to an alteration in the ATPase activity, we subjected the purified PD variant proteins to ATP hydrolysis using a well-established single-turnover ATPase assay. Interestingly, R126W showed an elevated catalytic rate constant (kcat, 0.060 min−1) for basal ATP hydrolysis when compared with wild-type protein (0.032 min−1). On the other hand, the P509S PD variant exhibited lower kcat of 0.017 min−1 for basal ATP hydrolysis (P < 0.05; Fig. 1A). To determine whether the mutations alter the regulation of the Hsp70 chaperone cycle, we studied the functional interaction of human mtHsp70 with its cochaperones. The human mitochondrial matrix contains two alternatively spliced isoforms of type-I J-proteins, known as hTid-1L (larger isoform) and hTid-1S (smaller isoform). Both the isoforms are involved in the folding of matrix proteins and thus play a critical role in maintaining the mitochondrial protein quality control (29). Strikingly, both the hTid-1 isoforms stimulated the ATPase activity of human mtHsp70 PD variants significantly higher than wild-type levels. At a 1:2 ratio of human mtHsp70 to hTid-1S, an 11-fold stimulation was observed for wild-type in comparison with 18-fold and 47-fold for R126W and P509S mutants, respectively (P < 0.01; Fig. 1B). Under similar conditions, the larger isoform hTid-1L stimulated wild-type to 6-fold when compared with the mutant levels of 8.5-fold (R126W) and 13-fold (P509S), respectively (P < 0.05, Fig. 1C). Besides, the PD variants were also tested for stimulation by mitochondrial inner-membrane-bound J-protein, DnaJC19, which is required for the protein translocation function (30). Interestingly, the P509S mutant was stimulated robustly by DnaJC19, which is consistent with hTid-1 isoforms (P < 0.01; Fig. 1D). In contrast, the nucleotide exchange ability with GrpEL1 as well as ‘Hep’-dependent stimulation remained relatively unaltered for the PD variants (P < 0.05; Fig. 1E and F).
Figure 1.
Biochemical properties of PD variants of human mtHsp70. (A–F) Effects of cochaperones on the ATPase activity. The preformed radiolabeled human mtHsp70-ATP complexes (1 µm) of wild-type (WT) and PD variants (R126W and P509S) were incubated with (A) buffer alone (P < 0.05), (B) 2 µm hTid-1S (P < 0.01), (C) 2 µm hTid-1L (P < 0.05), (D) 2 µm DnaJC19 (P < 0.01), (E) 1 µm GrpEL1, either alone (open circles) or in the presence of 250 µm ATP (closed circles) and (F) 4 µm Hep. ATP hydrolysis was monitored under single turn over conditions at different time intervals and the percentage of ATP-to-Pi conversion was determined. The rate of hydrolysis was calculated using GraphPad Prism 5. Fold stimulation was calculated by setting the basal rate to 1. All error bars are representative of standard deviation obtained from three independent experiments. (G) Substrate interaction studies. 25 nm fluorescein-labeled P5 peptide (F-P5) was incubated with increasing concentrations of the purified proteins and allowed to reach equilibrium. The observed anisotropy values were fitted to a one-site binding equation using the GraphPad Prism 5 software to obtain the Kd values. (H) ATP-induced peptide release. 1000-fold excess ATP was added to saturated protein-F-P5 complexes in the ADP-bound state. The resultant maximal decrease in fluorescence anisotropy of the bound F-P5 was noted to determine the percentage of bound F-P5 released due to ATP binding.
To determine client protein-binding affinities, the purified human PD variants were subjected to a fluorescence anisotropy-based binding assay, using the fluorescein-labeled P5 (F-P5) peptide. Importantly, the dissociation constants (Kd) obtained for R126W (13.61 ± 2.7 µm) and P509S (15.72 ± 1.9 µm) were comparable with the Kd measured for wild-type (15.63 ± 1.4 µm), suggesting that human PD variants retain the ability to bind client proteins to normal levels (Fig. 1G and Supplementary Material, Table S1). To determine any alterations in the inter-domain interface, we subjected the variants to the ATP-dependent peptide release assay as reported previously for bacterial DnaK (36). Upon addition of ATP to prebound F-P5-mtHsp70 complex, the wild-type exhibited 81% release of F-P5-peptide (P < 0.05; Fig. 1H). However, a 10% reproducible reduction in the release of prebound F-P5-peptide was observed for R126W (71% total release), indicating a minor defect associated with the allosteric communication between SBD and NBD domains. This observation is well correlated by the fact that the mutant also showed enhanced basal ATPase activity, which is an indication of inter-domain communication defects (29). Interestingly, P509S mutant did not show any alteration in the F-P5-peptide release upon ATP addition (P < 0.05; Fig. 1H). Overall, we conclude that the enhancement in J-protein stimulation is a unique gain of function associated with human mtHsp70 PD variants, thus altering the rate of the regulation of mitochondrial chaperone cycle and protein homeostasis.
Enhanced J-protein interaction of PD mutants of mtHsp70 leads to reduced chaperoning activity
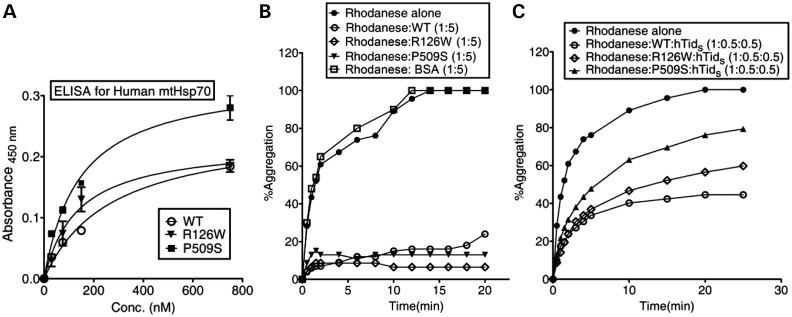
To test whether the increased stimulation of the ATPase activity is due to an enhanced physical interaction between the PD variants and J-proteins, we subjected them to an enzyme-linked immunosorbent assay (ELISA) using hTid-1S as a representative J-protein. As expected, both the human PD variants showed increased binding to hTid-1S. The P509S variant exhibited a much greater affinity for J-proteins when compared with wild-type, further validating the observed excessive stimulation of the ATPase activity (Figs 1B and 2A). The type 1 J-proteins (Hsp40s) deliver unfolded polypeptides to the SBD of Hsp70s through a transient interaction followed by coupling ATP hydrolysis with substrate capture in ADP conformation. To test whether the enhanced interaction of PD variants with J-proteins interferes with the normal chaperoning activity of the Hsp70/J-protein folding machine, we studied their ability to prevent the aggregation of a chemically denatured model substrate, rhodanese. As a control, in the absence of J-protein (hTid-1S), the PD variants alone showed a wild-type level of prevention of rhodanese aggregation, which is in agreement with their similar peptide-binding affinities (Fig. 2B). However, when an equimolar ratio of mtHsp70/J-protein (hTid-1S) was equilibrated to allow complex formation, followed by addition of rhodanese, wild-type mtHsp70 robustly prevented the aggregation of model substrate when compared with BSA control (Fig. 2C). On the other hand, the PD variants displayed significantly reduced ability to prevent premature aggregation of rhodanese. Strikingly, the P509S mutant exhibited a 40% reduction in the ability to prevent aggregation, whereas R126W showed a 20% lower ability in the prevention of rhodanese aggregation when compared with wild-type and BSA controls (Fig. 2B and C). Based on these observations, we conclude that abnormality in J-protein interaction associated with the PD variants reduces the chaperoning activity of the folding machine.
Figure 2.
Study of J-protein interaction analysis and chaperoning activity of PD mutants. (A) Human mtHsp70 and J-protein interaction studies using ELISA. Three micrograms of hTid-1S was incubated with increasing concentrations of wild-type (WT), R126W and P509S mtHsp70 for 1 h at room temperature in a buffer containing Mg(OAc)2 (10 mm) and AMP-PNP (1 mm). Antiserum against mtHsp70, followed by horse-radish peroxidase-coupled secondary antibody was used to detect the bound-mtHsp70, utilizing a tetramethylbenzidine/H2O2 substrate-based reaction. Reactions were terminated with 2 N H2SO4 and absorbance measured at 450 nm. (B and C) Prevention of rhodanese aggregation. Chemically denatured rhodanese (0.46 µm) was added to (B) 2.3 µm mtHsp70 alone, and (C) a pre-incubated complex of mtHsp70 (0.23 µm) in combination with the J-protein hTid-1S (0.23 µm). The aggregation of rhodanese was measured at a wavelength of 320 nm. BSA was used as a negative control for chaperoning activity. The values were plotted by normalizing the values and setting the aggregation of rhodanese alone as 100%.
Analogous PD mutations in yeast Ssc1 leads to growth and protein translocation defects
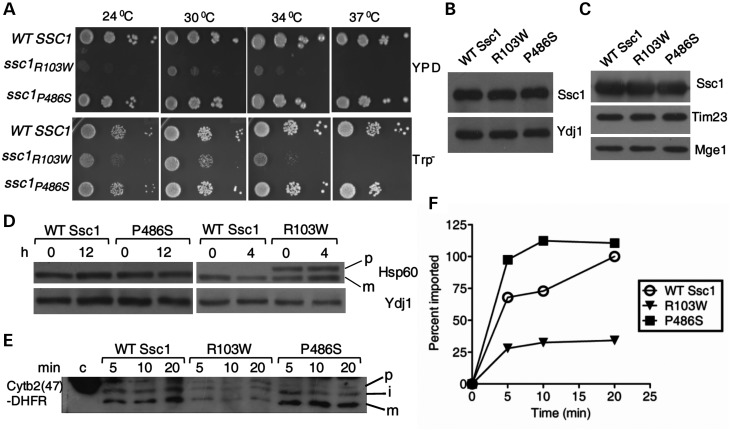
To delineate the molecular mechanism behind the mitochondrial dysfunction associated with the PD mutations in vivo, we carried out analogous amino acid substitutions in Saccharomyces cerevisiae, SSC1 gene. Yeast mitochondrial Ssc1 is an ortholog of human mtHsp70 essential for the import of precursor proteins and their subsequent folding in the matrix compartment. Human mtHsp70 reveals a 66% sequence identity at the amino acid level with yeast Ssc1. Importantly, PD-associated amino-acid residues of human mtHsp70 are conserved in yeast Ssc1 (Supplementary Material, Figs S1 and S2). We generated analogous mutations at the corresponding residues in Ssc1: arginine at 103rd position, substituted to tryptophan (R103W); and proline at 486th position, replaced by serine (P486S). To test whether PD mutants could rescue the inviability of the Δssc1 strain, centromeric plasmids carrying the ssc1 mutants were transformed into a Δssc1 strain carrying a wild-type copy of SSC1 on a plasmid having the URA3 gene. The transformed strains were selected on 5-fluoroorotic acid (5-FOA), which selects cells having lost the wild-type copy of the SSC1 gene. The resulting PD mutant strains were tested for growth phenotype, using drop test analysis on minimal (Trp−) and complete (YPD) media. Interestingly, the R103W ssc1 mutant showed a remarkable growth defect at the permissive temperature and was inviable at 37°C, thus highlighting the critical nature of the R103 residue for in vivo function in yeast (Fig. 3A). In contrast, P486S mutant displayed a comparable growth pattern with wild-type at all temperatures tested (Fig. 3A). In addition to that, both the PD mutants showed normal levels of protein expression in whole yeast cell lysates as well as in mitochondria upon immunoblot analysis using Ssc1-specific antibodies (Fig. 3B and C).
Figure 3.
Growth and protein import phenotypes of analogous PD mutations in yeast Ssc1. (A) Growth phenotype analysis. Equivalent number of cells from all the strains were subjected to 10-fold serial dilutions and spotted on rich and selective media. The plates were incubated at the indicated temperatures for 72 h. (B and C) Estimation of protein levels. Immunoblot analysis of (B) Ssc1 protein levels in the whole-cell lysates of the wild-type and mutant yeast strains. The cytosolic protein Ydj1 was used as a loading control; and (C) the amount of Ssc1 protein targeted to the mitochondria in the respective strains. The mitochondrial proteins Tim23 and Mge1 were used as loading controls. (D) In vivo precursor accumulation. The yeast cells were grown at 25°C in rich media till the early log phase and shifted to non-permissive temperatures for the indicated time periods. The whole-cell extracts were resolved by SDS–PAGE, followed by immunoblotting using anti-Hsp60 antibody; p, precursor form; m, mature form of Hsp60. The cytosolic protein Ydj1 was used as the loading control. (E and F) In vitro import of Cytb2(47)-DHFR. (E) 200 µg of mitochondria from wild-type (WT) and the PD mutant strains were pre-incubated in import buffer, and reaction was initiated by addition of saturating amount of purified Cytb2(47)-DHFR. The reaction mix from different time intervals was separated by SDS–PAGE and subjected to immunoblotting using a DHFR-specific antibody. One hundred percent of precursor protein offered to the mitochondria was loaded as control (c); precursor (p); mature (m); and intermediate (i) forms of Cytb2(47)-DHFR during import. (F) The amount of imported mature form was quantified by densitometry and plotted as a function of time, setting maximal import by WT as 100%.
To assess the efficiency of protein translocation in PD mutants, we analyzed the protein import into mitochondria by utilizing in vivo precursor accumulation analysis as demonstrated previously (12). Compromised Ssc1 function results in the cytosolic accumulation of non-processed precursor forms of nuclear-encoded mitochondrial proteins (12). By using this assay, we monitored the accumulation of non-processed form of an abundantly expressed matrix protein such as Hsp60 (12). To induce the import phenotype, the wild-type and mutant cells were grown at a permissive temperature, followed by heat shock at 37°C for 4–12 h. The cell lysates were analyzed for precursor accumulation by immunoblotting, using antibodies specific to Hsp60. Interestingly, R103W showed significant accumulation of non-processed form of Hsp60 at permissive temperature and 37°C, suggesting a strong in vivo import defect (Fig. 3D). On the other hand, P486S did not exhibit precursor accumulation even under prolonged heat shock conditions, thus correlating well with its growth phenotype (Fig. 3D). Additionally, for a detailed kinetic analysis of the protein translocation efficiencies, ex vivo import experiments were conducted using purified recombinant precursor proteins with isolated mitochondria (37). Mitochondria isolated from mutant yeast cells grown at permissive temperatures were pre-incubated for 15 min at 37°C to induce the mutant phenotype, and subjected to import reaction using saturating amounts of Cytb2(47)-DHFR precursor protein (Fig. 3E). When compared with wild-type, R103W mitochondria showed significant reduction in the kinetics of import of Cytb2(47)-DHFR precursor protein, whereas P486S showed an elevated rate of import (Fig. 3F). In summary, we find that the import kinetics associated with the yeast PD mutants is consistent with their growth phenotypes.
PD mutations in Ssc1 exhibit loss of mitochondrial mass, membrane potential and mtDNA
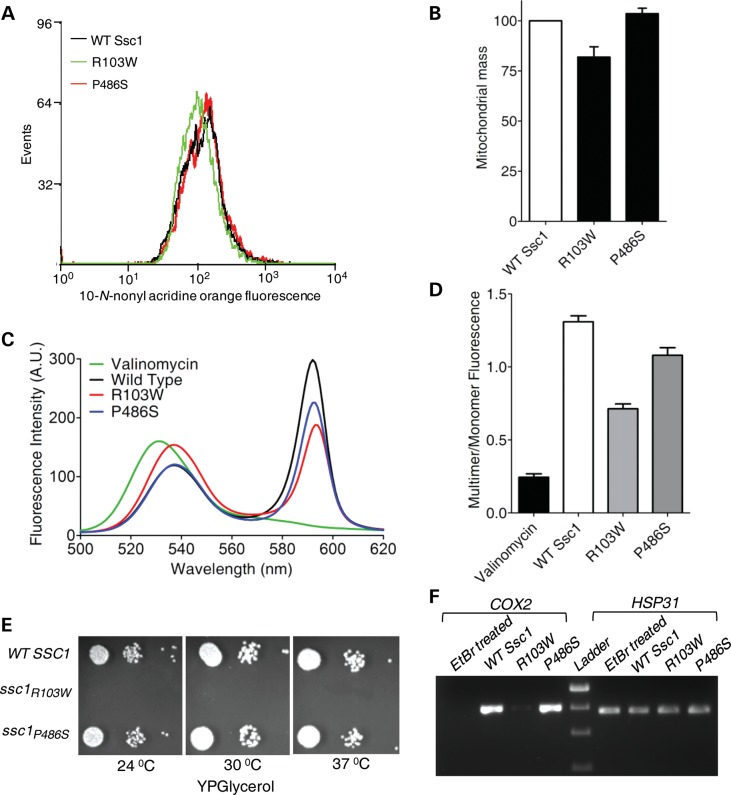
PD pathogenesis has been closely associated with mitochondrial dysfunction at the cellular level. In order to test whether these PD mutants of Ssc1 can also contribute to the overall loss in mitochondrial function, we analyzed the aforementioned using multiple approaches. First, we checked for the overall mitochondrial mass in the yeast PD mutants by using the fluorescent dye 10-N-nonyl acridine orange (NAO), which binds to the cardiolipin of mitochondrial membrane, thereby giving an estimate of the total mitochondrial mass present inside the cell (38). Surprisingly, the R103W mutant showed a shift in the fluorescence signal toward the lower side (highlighted in green) when compared with wild-type and the P486S mutant (Fig. 4A). Upon quantitation of mean fluorescence intensity (MFI), a 20% reduction in the overall mitochondrial mass was reproducibly observed in the R103W mutant in comparison with the wild-type and P486S (P < 0.001; Fig. 4B). Second, we assessed for the relative amount of functional mitochondria, using the JC-1 dye (39). The JC-1 dye uptake is dependent on intact membrane potential which results in an aggregate form of the dye yielding red fluorescence. In contrast, the extra-mitochondrial localization of monomeric form of the dye, as a result of disrupted membrane potential, generates a signal of green fluorescence (39). The ratio of red to green fluorescence is used as an indicator for functional mitochondria with an intact membrane potential (39). Valinomycin is known to disrupt the membrane potential of intact mitochondria and was used as a positive control where minimal emission was detected at 590 nm (40). Strikingly, the R103W mutant showed a significant reduction in the fluorescence intensity at 590 nm maxima when compared with wild-type mitochondria and more closely resembled the valinomycin-treated mitochondria (Fig. 4C). As indicated in Figure 4D, upon quantification, a 40% reduction in the levels of functional mitochondria was estimated for the R103W mutant. Similarly, a 10–15% reduction in the functional mitochondria was observed for the P486S mutant when compared with wild-type, further highlighting the essential role played by Ssc1 in the normal mitochondrial biogenesis (P < 0.05; Fig. 4D).
Figure 4.
Assessment of mitochondria-specific cellular defects in the PD mutants of yeast Ssc1. (A) Mitochondrial mass estimation. The MFI histogram for 10 000 cells stained using NAO, as analyzed by flow cytometry, was plotted to highlight the difference in the overall mitochondrial mass. (B) The total mitochondrial mass associated with each yeast strain as obtained from FACS analysis, represented as a normalized bar chart (P < 0.001). (C) Measurement of mitochondrial membrane potential. Purified mitochondria stained with the JC-1 dye were subjected to an emission wavelength scan ranging from 500 to 620 nm. The fluorescence intensity values obtained were plotted against the wavelengths to calculate the relative distribution of polarized versus depolarized mitochondria in each case. (D) Bar chart representing the fluorescence emission of the JC-1 dye in terms of multimer (590 nm) to monomer (530 nm) ratio, for mitochondria isolated from wild-type (WT) and PD-mutant yeast strains (P < 0.05). (E) Respiratory competence of the yeast strains. Serially diluted yeast cells of WT and PD-mutant strains were spotted on non-fermentable YPG media and incubated for 96 h at indicated temperatures to assess the respiratory efficiency. (F) Estimation of mtDNA abundance. PCR amplification of the COX2 gene was used to assess mtDNA loss using genomic DNA isolated from WT and PD-mutant strains incubated in YPG media for 12 h. HSP31 amplification served as the control for nuclear DNA. Yeast cells treated with EtBr (exposed to 20 ng/ml for multiple generations) to induce mtDNA loss were used as a positive control.
As an additional parameter, we tested for the growth of mutants on YP glycerol (YPG) medium by drop test analysis. Efficient utilization of non-fermentable carbon source such as glycerol requires elevated levels of functional mitochondria. Most notably, the R103W mutant showed a lethal phenotype when grown on media containing glycerol as a sole carbon source (Fig. 4E). However, the P486S mutant grew normally in YPG media. In yeast, a lack of growth on non-fermentable carbon source is often associated with the loss of mtDNA over successive generations of cell growth (41). To demonstrate mtDNA loss in mutants, we employed a PCR-based method by monitoring the level of amplification from a mitochondrial genome-encoded reporter gene, COX2, as described previously (42). Interestingly, the R103W mutant showed significant loss of mitochondria-encoded COX2 levels when compared with wild-type and P486S mutant (Fig. 4F). However, the amplicon levels of the nuclear encoded HSP31 gene remained unaltered in wild-type, P486S and the R103W mutant (Fig. 4F). As a positive control, wild-type yeast cells were exposed to ethidium bromide (EtBr) treatment, which is well documented to promote the loss of mtDNA (43). Notably, a similar pattern of reduction in COX2 levels was observed in the EtBr-treated wild-type yeast cells, thus indicating that R103W is more susceptible to mtDNA loss when compared with the P486S mutant (Fig. 4F). Overall, we conclude that PD mutations in Ssc1 lead to impairment in the mitochondrial function and organelle biogenesis.
PD mutations in Ssc1 elevate the cellular ROS levels
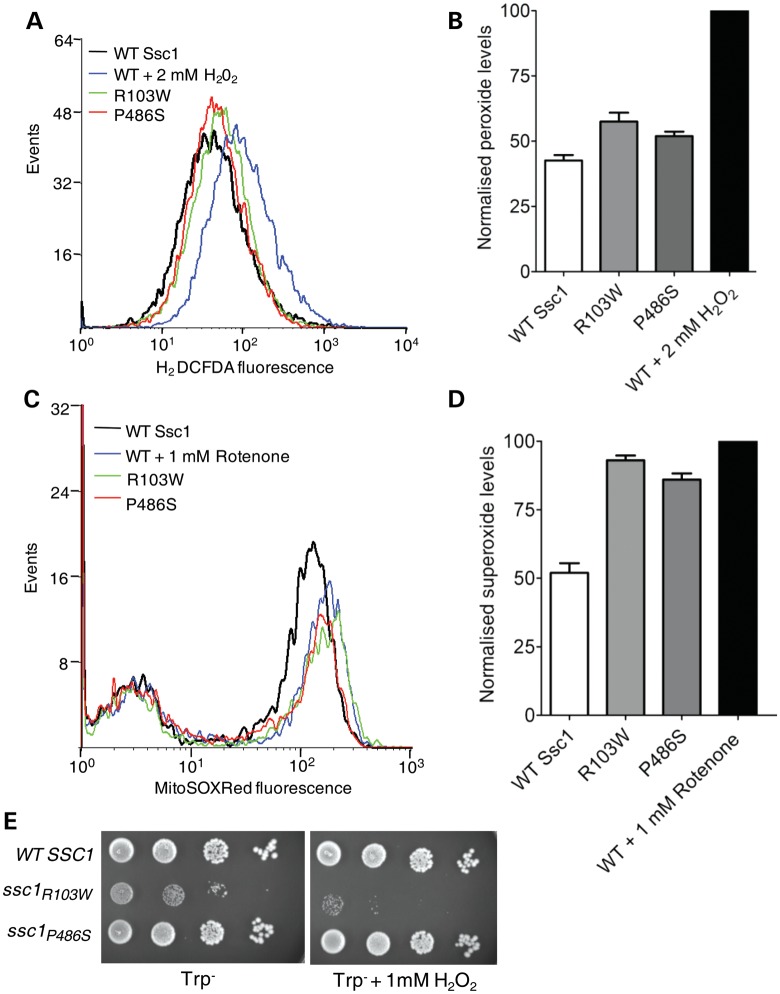
Mitochondria constitute a major source of cellular ROS, and enhanced oxidative stress is frequently associated with mitochondrial dysfunction. To determine whether the PD mutations in Ssc1 lead to an elevation in the ROS levels, we measured the overall ROS in the PD mutant strains using the H2DCF-DA dye, which produces a fluorescence signal upon modification by peroxide species, and was quantitated by FACS-based analysis (44). As an internal control, the wild-type cells were pre-treated with 2 mm H2O2 to record the shift in the fluorescence signal peak, which is an indicator of excess generation of total ROS species (highlighted in blue) (Fig. 5A). Interestingly, both R103W and P486S mutants showed a shift in the fluorescence signal peaks toward higher ROS levels (Fig. 5A). With reference to H2O2-treated and untreated wild-type cells, a 15–20% elevation in the total ROS levels was estimated for R103W and P486S mutants (P < 0.05; Fig. 5B). To investigate the specific contribution of the mitochondria for the observed elevated cellular ROS levels, we subjected PD mutant strains to FACS analysis using the fluorescent dye, MitoSOX Red. The MitoSOX Red dye is a selective indicator of mitochondrial ROS alone wherein its fluorescence emission is dependent upon uptake and its subsequent oxidation within the mitochondrial compartment (45). As an internal control, cells were treated with 1 mm rotenone, which elevates the mitochondrial superoxide levels by inhibiting the respiratory complex-I. Importantly, R103W and P486S mutants exhibited a shift in the fluorescence toward higher levels, and a 35% increment in superoxide ion contents was estimated (P < 0.05; Fig. 5C and D). Furthermore, to demonstrate the enhanced sensitivity of mutant strains to extraneous oxidative stress, cells were subjected to hydrogen peroxide treatment. As shown in Fig. 5E, R103W was extremely sensitive to oxidative stress and was found to have a severely compromised growth phenotype in the minimal media containing 1 mm H2O2. On the other hand, P486S mutant grew normally in the presence of 1 mm H2O2 containing media, similar to wild-type (Fig. 5E). Together, our analysis infers that PD mutations in Ssc1 significantly contribute toward the enhancement of overall cellular ROS levels.
Figure 5.
Estimation of cellular ROS levels and oxidative stress sensitivity of yeast PD mutants. (A and B) Quantification of total cellular ROS levels. (A) Histogram of flow cytometric analysis using the H2DCF-DA dye to detect enhancement in ROS levels where cell numbers are plotted as a function of the fluorescence intensities. (B) Bar chart representing the normalized relative ROS levels in the yeast strains as determined from the same analysis. The MFI of 2 mm H2O2-treated positive control cells was set as 100% (P < 0.05). (C and D) Mitochondrial ROS measurements. (C) Flow cytometric analysis of mitochondrial superoxide levels in the wild-type (WT) and PD-mutant yeast strains as estimated by MitoSOX Red fluorescence. (D) Bar chart representing the relative superoxide levels obtained from the aforementioned analysis. The MFI of positive control cells treated with 1 mm rotenone was set as 100%. For both flow cytometry experiments, 10 000 events were analyzed in each case and the values were plotted based on three independent experiments (P < 0.05). (E) Sensitivity to extraneous oxidative stress. Oxidative stress sensitivity was assessed by performing a drop test analysis on selective medium with and without 1 mm H2O2 as indicated. Equivalent numbers of yeast cells from the WT and PD-mutant strains were serially diluted and spotted to grow at the permissive temperature of 30°C for 96 h.
Compromised stability of mtHsp70 variants: a key determinant for mitochondrial dysfunction in PD progression
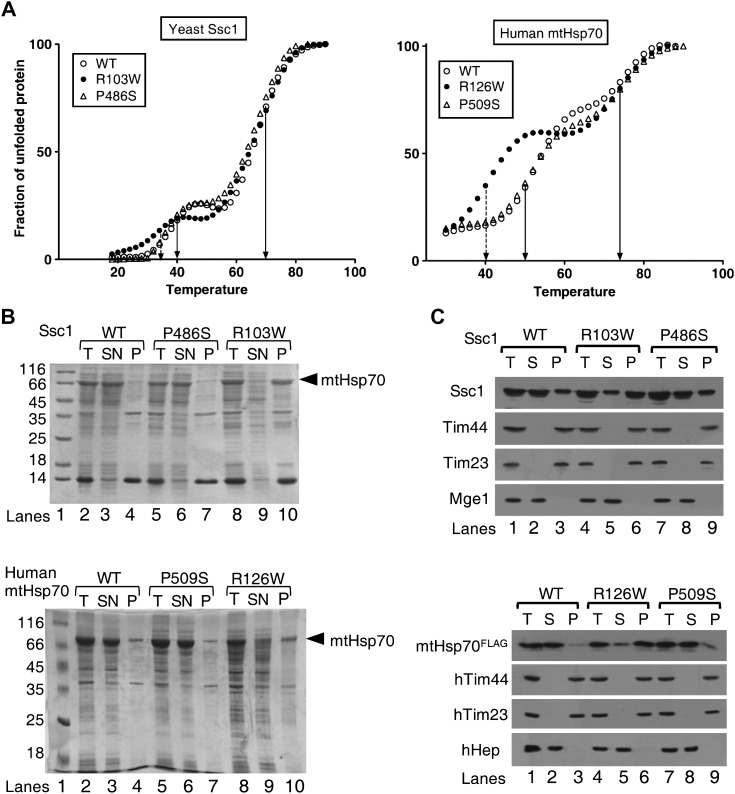
To investigate the differences of yeast and human PD variants at the structural level, the purified proteins were subjected to circular dichroism (CD) analysis. Both Ssc1 and human mtHsp70 PD variants showed a comparable shapes and ellipticity values, suggesting that the mutants harbor similar secondary structural elements identical to their respective wild-type proteins (data not shown). To analyze whether PD mutations alter the stability of mutant proteins, thermal unfolding was monitored by measuring the ellipticity at 222 nm as a function of temperature. Like other Hsp70s, wild-type Ssc1 showed two melting transition temperatures (Tm) for a 50% unfolding of NBD (40 ± 0.5°C) and SBD (70 ± 0.5°C) (Fig. 6A, left panel and Supplementary Material, Table S2). Most strikingly, R103W showed 6°C reduction in the Tm value for the NBD (34 ± 0.5°C), whereas the Tm of SBD remained unchanged (Fig. 6A, left panel and Supplementary Material, Table S2). On the other hand, the Tm for SBD and NBD of P486S was found to be similar to that of wild-type (Fig. 6A, left panel). To address whether the lowered stability of mutants is a common defect associated with the PD mutations in humans as well, the purified human mtHsp70 PD variant proteins were subjected to CD thermal melt analysis. The wild-type human mtHsp70 exhibited Tm of 50 ± 0.5 and 74 ± 0.5°C for NBD and SBD, respectively (Fig. 6A, right panel). Interestingly, a similar 10°C drastic reduction in the Tm (40 ± 0.5°C) for the NBD of the R126W variant was recorded when compared with wild-type and P509S (Fig. 6A, right panel and Supplementary Material, Table S2).
Figure 6.
Assessment of protein stability and aggregation propensity of yeast Ssc1 and human mtHsp70 PD variants. (A) Analysis of protein stability. The thermal melting curves for the purified proteins of yeast Ssc1 (WT, R103W, P486S) (left panel) and of human mtHsp70 (WT, R126W, P509S) (right panel) were obtained by monitoring the ellipticity at 222 nm as a function of temperature in 10 mm phosphate buffer (pH 7.5). (B) Protein solubility analysis. The yeast (upper panel) and human (lower panel) mtHsp70 proteins were coexpressed with Zim17 in E. coli cells grown at 37 °C, for 5 h post-induction. The crude cell lysates were centrifuged to separate the soluble supernatant fraction (SN) and insoluble pellet fraction (P). The samples were dissolved in SDS sample buffer and analyzed on SDS–PAGE followed by Coomassie dye staining. The crude lysates prior to centrifugation were used as total loading control (T). The bands corresponding to mtHsp70 are indicated by an arrow. (C) Intra-mitochondrial aggregation analysis. Equivalent amounts of isolated mitochondria corresponding to yeast (upper panel) and human PD variants (lower panel) were fractionated by sonication-based lysis, followed by ultracentrifugation. Samples corresponding to supernatant (S), insoluble pellet (P) and un-fractionated extract (T) were analyzed by SDS–PAGE and subjected to immunoblotting using appropriate yeast and human antibodies. Tim44, Tim23 (insoluble) and Mge1 (soluble) (for yeast proteins) and hTim44, hTim23 (insoluble) and hHep (soluble) (for human proteins) were used as controls for the fractionation. Anti-FLAG antibody was used for probing the wild-type and PD variants of mtHsp70 expressed in HEK293T cells.
To validate the findings of lowered stability of mtHsp70 due to PD mutations, Ssc1 and human mtHsp70 PD variants were co-expressed with Hep1 in E. coli BL21 (DE3) bacterial strain. Mitochondrial Hsp70 members are prone to aggregate when overexpressed alone in E. coli cells. However, upon coexpression with Hep1, the major portion of wild-type Ssc1 and human mtHsp70 was recovered in the soluble fraction at 37°C (Fig. 6B, upper and lower panels). Most strikingly, >95% of Ssc1 R103W and human mtHsp70 R126W mutants were recovered in the pellet fraction at 37 °C (Fig. 6B, upper and lower panels). At the same time, the mutant proteins remained soluble upon overexpression at lower temperature conditions (data not shown). An increased tendency for aggregation in E. coli suggests that arginine-to-tryptophan mutation in mtHsp70s reduces the overall in vivo stability, which is in agreement with the CD analysis. On the other hand, Ssc1 P486S and human mtHsp70 P509S mutants stably expressed and were recovered in the supernatant fraction similar to their respective wild-type proteins.
To demonstrate further whether the reduced stability of the mutant proteins leads to aggregation in the yeast and human mitochondrial matrix, the purified yeast mitochondria were subjected to fractionation analysis. After hypotonic swelling, the mitoplast was gently lysed by sonication followed by fractionation, using ultracentrifugation as described previously (46). The samples were separated on SDS–PAGE and subjected to immunodetection using Ssc1-specific antibodies. The Tim23, Tim 44 (insoluble) and Mge1 (soluble) marker proteins were used as positive controls. Most notably, R103W showed a higher propensity for aggregation in the mitochondrial matrix and >70% of the R103W protein was recovered in the pellet fraction (Fig. 6C, upper panel). However, the P486S mutant remained in the soluble fraction like the wild-type (Fig. 6C, upper panel). To understand the relative distribution of mtHsp70 in soluble and insoluble fractions in mammalian cells, we generated FLAG-tagged mammalian constructs of wild-type and human mtHsp70 PD variants. Using a Lipofectamine 2000 (Invitrogen)-based transfection method, the aforementioned constructs were transiently transfected into HEK 293T cells. Subsequently, 48 h post-transfection, the cells were harvested, and isolated mitochondria were subjected to fractionation. To detect the exogenously expressed mtHsp70 and its PD variants, anti-FLAG antibodies were used to study the distribution between the supernatant and the pellet fractions. A similar pattern of aggregation was observed for R126W upon fractionation of human mitochondria (Fig. 6C, lower panel). On the other hand, P509S and wild-type remained in the soluble fraction in comparison with positive control proteins, hTim23 and hTim44 (insoluble) and hHep (soluble) (Fig. 6C, lower panel). In conclusion, our in vitro CD analysis and in vivo aggregation studies provide direct experimental evidence to indicate that the compromised protein stability associated with the R126W mutant is the primary cause for mitochondrial dysfunction in PD.
P486S Ssc1 PD mutant exhibits a similar enhanced interaction with J-protein cochaperones and reduction in chaperone activity
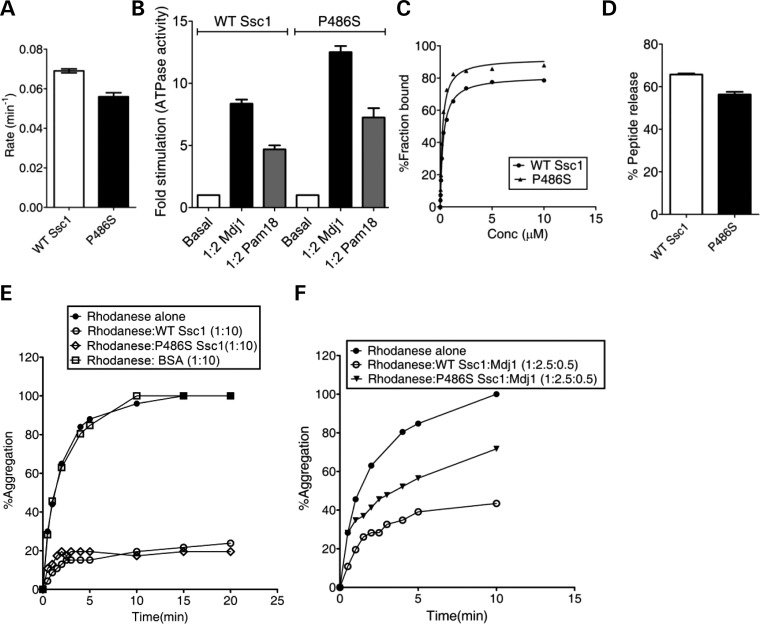
The human mtHsp70 P509S PD variant showed elevated ATPase activity stimulation by J-proteins. To investigate whether a similar level of enhanced J-protein stimulation is observed in yeast counterpart, purified P486S PD mutant protein was subjected to ATP hydrolysis analysis using single-turnover ATPase assays. P486S and wild-type showed a comparable basal catalytic rate constant of 0.056 and 0.075 min−1, respectively (P < 0.01; Fig. 7A). For estimating J-protein stimulation, we utilized two J-proteins: (i) Mdj1, an ortholog of E. coli DnaJ required for the folding of matrix proteins, and (ii) Pam18, an inner mitochondrial membrane-bound J-protein, which is essential for the protein translocation function. At 1:2 Ssc1-to-Mdj1 ratio, an 8-fold stimulation was observed for wild-type (P < 0.05; Fig. 7B). Under identical conditions, a 13-fold stimulation was observed for the P486S mutant (P < 0.05; Fig. 7B). An ∼2-fold enhancement in the J-protein stimulation is indicative of ‘gain of function’ for the P486S PD mutant similar to its human counterpart, P509S. Similarly, at 1:2 Ssc1-to-Pam18 ratio conditions, a 4-fold stimulation was observed for the wild-type when compared with a 7-fold for P486S (P < 0.05; Fig. 7B). A similar enhancement in the stimulation was recorded for human Pam18 ortholog—DnaJC19, which suggests that the C-terminal mutation leads to an overall gain of function with J-protein cochaperones for the PD variants. To ascertain that P486S possesses a normal client–protein interaction, we analyzed for the peptide-binding activity, using fluorescein-labeled P5 by anisotropic measurements. As indicated in Figure 7C, P486S showed a similar affinity for P5 binding when compared with wild-type, suggesting that the mutation does not compromise the client–protein interaction like its human counterpart, the P509S PD variant (Supplementary Material, Table S1). Also, upon addition of ATP to prebound F-P5-mtHsp70 complex, the P486S mutant exhibited the release of bound F-P5 to comparable level as the wild-type (P < 0.05; Fig. 7D). To demonstrate whether enhanced stimulation of the ATPase activity of P486S by J-proteins influences its overall chaperoning activity like its human counterpart, we tested for its ability to prevent the aggregation of the denatured rhodanese substrate. Similar to wild-type, the P486S mutant solely displayed a robust activity for the prevention of rhodanese substrate aggregation, which is consistent with the human PD variants (Fig. 7E). However, a 35% reduction in the prevention of rhodanese aggregation was observed in the presence of a preformed complex of P486S and J-protein (Mdj-1) when compared with wild-type Ssc1 and BSA-alone controls (Fig. 7E and F). These observations are consistent with the human PD variants, where a similar reduction in rhodanese aggregation was observed in the presence of Hsp70/J-protein bichaperone complex. Overall, we conclude that the PD mutation (P486S/P509S) in the C-terminal domains of mtHsp70s leads to an elevation in the J-protein-dependent stimulation and as a consequence, results in reduced ability to prevent aggregate formation of unfolded substrates.
Figure 7.
Biochemical defect analyses of yeast P486S PD mutant. (A and B) Measurement of the ATPase activity and stimulation. 1 µm of prebound mtHsp70-radiolabeled ATP complex was incubated with (A) buffer alone (P < 0.01), (B) 2 µm of Mdj1(P < 0.05) and Pam18 (P < 0.05) each and the rate of ATP hydrolysis was monitored under single-turn-over conditions at different time intervals, and the percentage of ATP-to-Pi conversion was plotted. Fold stimulation was calculated by setting the basal rate as 1. (C and D) Substrate interaction studies. (C) 25 nm fluorescein-labeled P5 peptide (F-P5) was incubated with increasing concentrations of the purified proteins and allowed to reach equilibrium. The observed anisotropy values were fitted to a one-site binding equation using the GraphPad Prism 5 software to obtain the Kd values. (D) 1000-fold excess ATP was added to saturated protein-F-P5 complexes in the ADP-bound state. The resultant maximal decrease in fluorescence anisotropy of the bound F-P5 was noted to determine the percentage of bound F-P5 released due to ATP binding. (E and F) Prevention of rhodanese aggregation. Denatured rhodanese (0.46 µm) was added to (E) mtHsp70 protein (4.6 µm) alone, and (F) a preformed complex of 1.15 µm of mtHsp70 (WT and P486S Ssc1) in combination with the J-protein Mdj1 (0.23 µm). BSA was used as a negative control for chaperoning activity. The aggregation of rhodanese was measured at a wavelength of 320 nm and the normalized values were plotted setting the aggregation pattern of rhodanese alone as 100%.
DISCUSSION
Besides selective death of dopaminergic neurons and accumulation of Lewy bodies, mitochondrial dysfunction has been one of the major hallmarks of PD (47,48). Until now, several gene loci have been identified in multifactorial familial PD that is implicated in the disease progression. Importantly, a gene-specific screen conducted on PD patients identified two novel variants in mtHSP70, namely R126W and P509S, which are suggested to be involved in PD pathogenesis. Recently, another separate large-scale genomic screen involving PD patients revealed a strong association of genes present on the long arm of chromosome 5 to be involved in the overall PD pathogenesis (49,50). This region includes the gene locus of HSPA9 (mtHSP70), thus supporting a likely involvement of mtHsp70 in PD pathology (49,50). In addition to that, in an independent screen, a third novel variant (A476T) has been identified in the SBD of mtHsp70 (7). All the three PD variants are substitutions found in highly conserved regions of mtHsp70, across species. Moreover, accumulation of these mutations in mtHsp70 in heterozygous condition results in mitochondrial dysfunction in neuronal as well as non-neuronal cell lines (7). At the same time, mtHsp70 is also known to interact with multiple PD-causing players at the cellular level, such as α-synuclein, Parkin and DJ-1. However, the molecular mechanism behind how mtHsp70 variants are centrally involved in conjunction with other known PD-causing proteins toward the PD progression is still unclear. Owing to the essential role played by mtHsp70 in the mammalian system, only up to a 50% downregulation of the endogenous wild-type protein has been reported (7). As a result, it is difficult to assess the impact of mutations in mtHsp70 PD variants in the heterozygous state using cell lines. Therefore, we established ‘yeast’ as a suitable model system by creating analogous mutations in the orthologous SSC1 gene and in parallel compared the phenotypic evidence observed in cell lines, enabling us to address the possible mechanisms by which the disruption of mtHsp70 function leads to PD-like manifestations at the cellular level.
Our previous findings highlighted that human mtHsp70 interacts with multiple components of the folding machinery such as J-domain proteins (hTid-1S and hTid-1L), nucleotide exchange factor (GrpEL1) and ‘Hep’ protein for maintaining the protein quality control in the mitochondrial matrix (29). Therefore, as a foremost approach, we tested interactions of human mtHsp70 PD variants with the components of the folding machinery. Both R126W and P509S variants displayed a similar level of ability to bind substrates when compared with wild-type, indicating that mitochondrial client protein sequestration is not compromised in the folding cycle. Biochemically, R126W exhibits slightly elevated basal ATP-hydrolysis activity as well as reduced ATP-dependent release of bound substrates, suggesting a minor allosteric communication defect. On the other hand, both the variants possess a similar wild-type level of nucleotide exchange activity and escort protein-dependent activity, thus making it unlikely that this part of the chaperone cycle is affected during the folding process. Most strikingly, PD variants showed enhanced interaction with J-proteins, which are involved in the folding function (hTid-1S and hTid-1L) as well as import reaction (DnaJC19) as indicated by their ability to get the ATPase activity stimulated through J-domain. In general, J-class protein family members are known to interact transiently with the ATPase domain of Hsp70s in ATP-bound conformation via their conserved J-domain (51). In vivo, the folding cycle of Hsp70 is initiated in ATP-bound conformation (21). J-proteins accelerate the conversion of ATP-bound conformation of Hsp70 to ADP state by catalytically stimulating the ATP hydrolysis, thereby stabilizing substrate interaction in the chaperone-mediated folding cycle (21).
Although Hsp70 can bind substrates independently in the ATP-bound state, the J-proteins significantly influence the regulation of the chaperone cycle and folding process (21,22,31,51). Retrospectively, the enhanced J-protein interaction associated with the PD variants may therefore impede the regulation of the chaperone cycle by altering the equilibrium between ATP- and ADP-bound states of Hsp70, thus directly influencing the overall efficiency of the folding reaction. Consequently, this would generate an imbalance in general ‘quality control’ inside the matrix compartment and the overall ‘protein homeostasis’, leading to mitochondrial dysfunction. Particularly, P509S showed a robust interaction with all J-proteins (hTid-1S, hTid-1L, DnaJC19) and as a result significantly imparting the folding phenotype. Critically, our in vitro aggregation prevention experiments using denatured rhodanese as a model substrate provide evidence in favor of this hypothesis. Notably, both P509S and R126W variants were found inefficient in preventing the aggregation of unfolded rhodanese as a bichaperone complex with J-proteins, compared with wild-type. Thus, the abnormal J-protein interaction associated with PD variants significantly reduces the overall chaperoning activity, resulting in protein quality control defect in the mitochondrial matrix. The imbalance in quality control may be restricted to either a generic set of proteins or a subset of neuronal-specific proteins, which has higher tendency for aggregation due to oxidative modification of proteins by abnormally accumulated reactive dopamine adducts (52). Importantly, as recently reported, the Hsp70-Hsp40 system plays a major role in disaggregating α-synuclein oligomers in neurons and thus reducing their toxicity at the cellular level (53). Therefore, we predict that an enhanced interaction of J-proteins with Hsp70 could lead to inefficient solubilization of intra-mitochondrial α-synuclein oligomers, resulting in their accumulation in the organelle and contributing to mitochondrial dysfunction (54).
Our biochemical analysis reveals that the R126W mutant exhibits severely compromised stability in vivo when compared with the P509S variant. Although, both the mutants possess a similar secondary structural content, the stability of ATPase domain of the R126W variant is significantly altered. The human mtHsp70 showed a higher thermal melting transition of 50°C for the ATPase domain when compared with the yeast ortholog, Ssc1 (40°C), which is concurrent with their permissive growth temperatures. Remarkably, R126W exhibits a 10°C lower unfolding transition for the ATPase domain, which is closer to mammalian physiological temperature. Therefore, the R126W variant is highly susceptible to aggregation at human physiological conditions. The property of in vivo aggregation is further demonstrated by overexpressing the protein in E. coli, wherein R126W is recovered in the insoluble pellet faction at 37°C. Similarly, in case of overexpressed FLAG-tagged PD variants in human mitochondria, a significant level of the R126W protein was recovered in the aggregated pellet upon fractionation analysis. These findings clearly highlight that, due to the inherent aggregation property of R126W, the cellular levels of functional protein might vary considerably in the PD condition. This observation is in agreement with the previous findings where the mtHsp70 levels were found to be reduced in the affected brain regions of PD patients as well as in the cellular disease model system (8,9). Based on our results, it is reasonable to believe that the mitochondrial dysfunction phenotype of the R126W variant in PD patients is attributable to an overall reduced stability associated with the protein due to a high degree of conformational plasticity. In retrospect, the reduced levels of functional R126W protein may directly influence the mitochondrial protein homeostasis and its biogenesis in PD patients.
To gain further insights into human PD variants at the cellular level in the homozygous state, we developed S. cerevisiae as an elegant model system for genetic and biochemical analyses. Owing to a high degree of conservation in basic cellular processes with higher eukaryotes, the budding yeast has been successfully utilized as a model system to demonstrate the conserved mechanisms underlying many neurodegenerative diseases (55). This model confers an added advantage as it allows flexible genetic manipulations which ensure complete elimination of the background effect of endogenous wild-type protein. Furthermore, recent studies have demonstrated that other PD susceptibility factors such as Parkin and DJ-1 associate with mtHsp70, hence adding another layer of complexity in uncovering the specific role of mtHsp70 alone in PD pathogenesis (7,56). Most importantly, the variants of mtHsp70 are found to be present in highly conserved regions of the protein, emphasizing the possibility of an intrinsic defect associated with the protein affecting the overall mtHsp70 function to a similar extent across species (7).
An analogous PD mutation, R103W in yeast Ssc1, results in conditional phenotype with highly compromised growth at permissive and inviability at non-permissive temperatures. A severe growth phenotype of R103W amino acid substitution highlights the critical nature of the mutation in eukaryotes in homozygous conditions. The yeast cells harboring R103W mutation showed multiple phenotypes at the cellular levels, namely (i) defect in translocation kinetics of pre-proteins across mitochondrial inner membranes that lead to the accumulation of precursor proteins in the cytosol; (ii) increased total cellular and mitochondrial ROS levels; (iii) enhanced susceptibility toward exogenous oxidative stress such as H2O2; (iv) almost 40% reduction of functional mitochondria due to the loss of inner mitochondrial membrane potential. In addition to that, R103W showed an increased tendency to lose mtDNA, thus imparting respiration defects, which is a unique property of this yeast PD mutant. At the molecular level, the purified protein showed lower stability as indicated by a 6°C reduction in the thermal melting transition for ATPase domain when compared with wild-type. Besides, the protein showed an increased tendency to aggregate at a non-permissive temperature as observed upon overexpression in the E. coli system at 37°C. Furthermore, a similar aggregation pattern was observed in the yeast mitochondrial lysates as demonstrated by fractionation analysis. These results clearly indicate that due to compromised stability of the R103W mutant, the overall protein quality control is altered, leading to multiple physiological defects, which are implicated in the mitochondrial dysfunction under PD conditions.
On the other hand, the P486S yeast PD variant was found viable and showed less drastic cellular phenotypes. This includes a 15% reduction in the functional mitochondria and elevated mitochondrial specific ROS levels. Biochemically, the purified P486S mutant displayed a wild-type level of stability as well as an ability to bind mitochondrial client proteins. Strikingly, the P486S variant also exhibited enhanced J-protein-dependent stimulation similar to the analogous human PD variant, P509S. P486S showed enhancement in the ATPase stimulation with the J-protein, Pam18, which is a part of the import motor complex involved in protein translocation across the inner mitochondrial membrane. The enhanced stimulation by Pam18 favors the translocation process as the mutant showed an increased kinetics of protein import when compared with wild-type. A similar enhancement in stimulation was also observed with yeast Mdj1, although to a much lesser extent than human hTid-1S and hTid-1L. A tighter interaction with Mdj1 also inhibits the chaperoning activity of P486S/J-protein bichaperone complex, thus leading to reduction in protein quality control in the matrix compartment. Therefore, it is reasonable to believe that mitochondrial dysfunction associated with the P486S is due to a consequence of J-protein-dependent gain of function. In case of yeast cells, a minor reduction in the levels of ‘functional mitochondria’ is tolerable based on growth conditions and hence may not display a drastic growth phenotype. Moreover, the yeast mitochondria possess other built-in compensatory mechanisms like presence of additional paralogs such as Ssq1, which might directly contribute for the functional overlap at least in synthesizing Fe/S cluster proteins. Unlike yeast, the human mitochondria consist of single mtHsp70 for all constitutive functions, including folding twice the amount of proteome size when compared with lower eukaryotes. Owing to the absence of well-defined additional compensatory mechanisms, mammalian mitochondria are heavily dependent on the functional integrity of the mtHsp70 protein. In that respect, the severity of the cellular defects may be even significantly higher for the neurons which require high-energy demands for their constitutive functions. Therefore, we hypothesize that a loss of regulation in the chaperone folding cycle due to enhanced J-protein interaction with mtHsp70 variants may generate an elevated level of neuronal-specific protein toxicity, leading to mitochondrial dysfunction in PD patients.
In summary, our study delineates the possible molecular mechanisms of mtHsp70 participation in the manifestation of pathological symptoms found in dopaminergic neurons in cases of PD. Additionally, our findings unravel the cellular mechanisms behind the mitochondrial dysfunction associated with mtHsp70 PD variants in humans as well as in yeast. The multiple in vivo phenotypes and biochemical defects associated with the human mtHsp70 PD variants were remarkably recapitulated using yeast as a model system. Furthermore, our findings provide a molecular basis for reduced levels of mtHsp70 found in affected brain regions of PD patients based on our observation of aggregation susceptibility of R126W, a single mtHsp70 mutant. Based on the severity of the phenotypes observed in the yeast haploid state, we propose that homozygous R126W PD mutation may by itself be capable of contributing to the overall disease outcome. On the other hand, accumulation of the P509S variant might contribute toward the PD progression by altering the mitochondrial quality control, in addition to mutations at other genetic loci. These initial findings will therefore provide invaluable insights for the future in uncovering the genetic and environmental factors that influence the development of familial PD in connection with mutations in mtHsp70 loci, using well-established yeast as a model system.
MATERIALS AND METHODS
Yeast strains, plasmids construction and genetic analysis
For genetic analysis in yeast, wild-type SSC1 was cloned under the control of its native promoter in the yeast centromeric plasmid pRS314 containing a Trp marker for selection. The yeast haploid strain, DG252 (trp1–1 ura3–1 leu2–3, 112 his3–11, 15 ade2–1 can1–100 GAL2+ met2-Δ1 lys2-Δ2 ssc1ΔClaI::LEU2), containing the plasmid pRS316-SSC1 for the maintenance of viability was a kind gift from Professor E.A. Craig. For in vivo phenotype analysis, the haploid Δssc1 strain was transformed with the plasmid carrying the point mutants of SSC1, and the transformants were selected on tryptophan omission plates. The transformants (Trp+) were then counter-selected on medium containing 5-FOA to eliminate the wild-type containing the plasmid. The viable yeast cells were recovered on rich media and subjected to spot test analysis using serial dilutions of the cells on various media like Trp−, YPG and Trp− containing H2O2 to confirm the growth phenotypes.
The ORF of human mtHsp70 was amplified using a cDNA library from HeLa cells, using suitable primers. The PD variants of Ssc1 and human mtHsp70 were generated by QuikChange site-directed mutagenesis, using high-fidelity PfuTurbo DNA Polymerase (Stratagene). All the clones were verified by DNA sequencing carried out at Eurofins, Inc. For solubilization and purification using the bacterial expression system, the ORFs of wild-type human mtHsp70 and Ssc1 were cloned in pRSFDuet-1vector along with the yeast Zim17 as previously described (29,57). Similarly, the corresponding coding nucleotide sequences of GrpEL1, Hep, hTid-1S and hTid-1L proteins were cloned in pET vectors as described earlier (29). All the clones used for purification and in vitro experimental analysis were devoid of mitochondrial leader sequence based on the reported mature forms. For transient mammalian transfection experiments, the full-length human mtHsp70 and its PD variants were cloned into a CMV-based promoter, pCI-neo vector (Promega) with a FLAG-tag at the C-terminus of the gene.
Protein purification analysis
The hexahistidine-tagged human mtHsp70, Ssc1 and PD variants were coexpressed with yeast Zim17 in E. coli BL21 (DE3) strain to ensure enhanced solubility by allowing growth at 25°C. Briefly, the cultures were allowed to reach an A600 of 0.5, followed by induction using 1 mm isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 6 h. Cells were harvested by centrifugation and then lysed in buffer C (20 mm Hepes-KOH, pH 7.5, 150 mm KCl, 20 mm imidazole, 10% glycerol) containing 2.5 mm magnesium acetate, 0.2 mg/ml lysozyme and protease inhibitor mixture, followed by incubation at 4°C for 1 h. The samples were gently lysed with 0.2% deoxycholate, followed by DNase I (10 μg/ml) treatment for 15 min at 4°C. The mixtures were further lysed by sonicating three times (15 s) at a 25% amplitude using ultrasonic processor with intervals of 2 min on ice. The cell lysates were clarified by centrifuging at 28 000g for 30 min at 4°C. The supernatant and pellet fractions were dissolved in SDS sample buffer and analyzed on 12.5% SDS–PAGE. After centrifugation, the soluble supernatant was incubated with Ni-NTA Sepharose (GE Healthcare) for 2 h at 4°C to allow binding. Unbound proteins and non-specific contaminants were removed by multiple washes of buffer C alone followed by sequential single washes of 1 m KCl, 0.05% Triton X-100, 1 mm ATP and 40 mm imidazole contained in buffer C. Finally, the bound-proteins were eluted with buffer C containing 250 mm imidazole, and the samples were dialyzed against appropriate buffers for use, in particular experiments. The purification of His-tagged proteins such as GrpEL1, Hep, hTid-1S and hTid-1L were carried out using our previously published protocols (29). Greater than 95% purity was obtained for all the protein preparations of mtHsp70, Ssc1 and PD variants.
ELISA for mtHsp70 and J-protein interaction analysis
The ELISA plate (Nunc, F8 Maxisorp Loose) wells were coated with hTid-1S (3 µg/well) diluted in phosphate-buffered saline (PBS, pH 7.4) for 1 h. Subsequently, the wells were blocked with BSA (0.01%) in PBS for 1 h. The plates were then incubated with increasing concentrations of wild-type or human mtHsp70 PD variants for 1 h in a buffer containing 0.005% BSA, 10 mm Mg(OAc)2 and 1 mm AMP-PNP (non-hydrolyzable ATP-analog). BSA was used as a negative control. The wells were washed for 5 min with PBS. Polyclonal antibody specific to human mtHsp70 was added and incubated for 1 h. The wells were washed with PBS and further incubated with goat anti-rabbit secondary antibody conjugated to HRP, in PBS for 1 h. The wells were washed and incubated with the addition of tetramethylbenzidine/H2O2 substrate and monitored for blue color development. The reaction was terminated by the addition of 2 N H2SO4, and the absorbance was recorded at 450 nm. All steps of the assay were performed at room temperature (25°C).
Analysis of mitochondrial mass, membrane potential and mtDNA loss
The total mitochondrial masses of the transformed PD-mutant yeast strains were determined using NAO (Molecular Probes). Briefly, 0.2 OD of log-phase yeast cells were harvested, washed with PBS and incubated with 10 µm NAO for 30 min at room temperature. Subsequently, the cells were washed and resuspended in PBS, and FACS analysis was carried out using the Becton Dickinson (BD) FACSCanto II flow cytometer. An argon laser was used for the excitation at wavelength 488 nm, while the emission was recorded at 520 nm. For each analysis, 10 000 events were recorded and the data were analyzed based on three independent experiments, using the WinMDI 2.9 software. In order to detect changes in mitochondrial membrane potential, isolated mitochondria (35 µg) from PD-variant strains were incubated with the JC-1 dye (5 µg/ml) for 5–10 min in dark. Subsequently, they were excited at 490 nm and subjected to an emission scan between 500 and 620 nm in a JASCO FP-6300 spectrofluorometer. The ratios of the peaks at 590 (aggregate form of the JC-1 dye) and 530 nm (monomeric form) were used as an indicator of membrane polarization. The wild-type mitochondria were incubated in 100 µm valinomycin for 15 min prior to dye staining as a control for complete depolarization. To determine mtDNA loss, a PCR-based method was used as described previously (42). Briefly, yeast cells grown in YPG media were harvested during the log phase and subjected to genomic DNA isolation. The genes encoding COX2 (mtDNA marker) and HSP31 (nuclear DNA marker) were PCR-amplified in separate reactions to determine the relative abundance of mtDNA in each of the yeast PD strains. EtBr-treated cells induced to lose mtDNA were used as a positive control as reported earlier (43).
Measurement of ROS levels and oxidative stress sensitivity in yeast
To estimate the cellular basal ROS levels, we used the fluorescein derivative H2DCF-DA (Calbiochem). Subsequent to cleavage by cellular esterases, the lipophilic groups of the dye are oxidized by the reactive free oxygen radicals and the emitted fluorescence can be measured using flow cytometry. The yeast cells from the mid-log phase were incubated with 50 µm dye for 20 min. The cells were washed twice with PBS and analyzed on a BD FACSCanto II flow cytometer using the standard excitation/emission parameters for the dye (510/530 nm). Cells pre-treated with 2 mm H2O2 for 30 min were used as a positive control for indicating higher ROS levels. In order to analyze the extent of superoxide generation by the mitochondria, we utilized MitoSOX Red (Molecular Probes). This dye is specifically targeted to the mitochondria where it undergoes oxidation by the mitochondrial superoxide ions and fluoresces upon binding to mtDNA with an emission maxima of 580 nm. Yeast cells (0.1 OD) from the early log phase were harvested and incubated with the dye (2.5 µm) for 10 min, following which they were washed with PBS and subjected to FACS analysis immediately using a 488 nm argon laser for excitation (BD FACSCanto II). The respiratory inhibitor rotenone (1 mm) was used as a control for generating higher superoxide levels. The MFI values of 10 000 events were recorded per sample for both H2DCF-DA and MitoSOX and plotted to compare the relative ROS levels using the WinMDI 2.9 software. For analysis of sensitivity to extraneous oxidative stress, the yeast cells were spotted on selective media containing 1 mm H2O2. The plates were incubated at 30°C for 96 h in order to observe the differential sensitivity of the strains toward an oxidizing agent.
Miscellaneous methods
Fluorescence anisotropy-based peptide-binding assays (58), single-turnover ATPase assays using γ-32P ATP (29), thermal CD analysis (46), in vitro aggregation (29,59), in vivo precursor accumulation analysis (12), import kinetics of pre-proteins into purified mitochondria (37), mitochondria isolation and fractionation (30) were done using standard protocols as described previously unless specified. The antisera used for immunodecoration against specific proteins such as human mtHsp70, Ssc1, Hsp60, Mge1, Tim23, Tim44, Ydj1 and hHep were raised in rabbits as previously reported (30). Anti-hTim23 and anti-hTim44 antibodies were obtained from BD Biosciences. Anti-FLAG tag antibody (Sigma) was used for immunodetection of exogenously expressed mtHsp70 and its PD variants in HEK293T cells. Immunoblot analysis was carried out using the ECL system (Perkin Elmer) according to the manufacturer's instructions. The reagents used for the experiments were obtained from Sigma-Aldrich, unless specified.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We thank Dr Elizabeth A. Craig, Dr William Walter and Dr Brenda Schilke for SSC1-deleted yeast strains and yeast-specific antibodies. We also thank the Flow Cytometry Facility of the Indian Institute of Science, Bangalore. The authors also wish to thank Dr Ganesh Nagaraju and his laboratory members for providing the cell culture facility. The authors (A.V.G, M.S. and D.S.) wish to thank the Council of Scientific and Industrial Research for providing senior research fellowship.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by Lady Tata Memorial Trust Young Researcher Award – 2012 (to P.D.S.). Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust International Senior Research Fellowship in Biomedical Science WT081643MA (to P.D.S).
REFERENCES
- 1.Davie C.A. A review of Parkinson's disease. Br. Med. Bull. 2008;86:109–127. doi: 10.1093/bmb/ldn013. doi:10.1093/bmb/ldn013. [DOI] [PubMed] [Google Scholar]
- 2.Sulzer D. Multiple hit hypotheses for dopamine neuron loss in Parkinson's disease. Trends Neurosci. 2007;30:244–250. doi: 10.1016/j.tins.2007.03.009. doi:10.1016/j.tins.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Cookson M.R., Bandmann O. Parkinson's disease: insights from pathways. Hum. Mol. Genet. 2010;19:R21–R27. doi: 10.1093/hmg/ddq167. doi:10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. doi:10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 5.Shen J., Cookson M.R. Mitochondria and dopamine: new insights into recessive parkinsonism. Neuron. 2004;43:301–304. doi: 10.1016/j.neuron.2004.07.012. doi:10.1016/j.neuron.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 6.De Mena L., Coto E., Sanchez-Ferrero E., Ribacoba R., Guisasola L.M., Salvador C., Blazquez M., Alvarez V. Mutational screening of the mortalin gene (HSPA9) in Parkinson's disease. J. Neural Transm. 2009;116:1289–1293. doi: 10.1007/s00702-009-0273-2. doi:10.1007/s00702-009-0273-2. [DOI] [PubMed] [Google Scholar]
- 7.Burbulla L.F., Schelling C., Kato H., Rapaport D., Woitalla D., Schiesling C., Schulte C., Sharma M., Illig T., Bauer P., et al. Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum. Mol. Genet. 2010;19:4437–4452. doi: 10.1093/hmg/ddq370. doi:10.1093/hmg/ddq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J., Hulette C., Wang Y., Zhang T., Pan C., Wadhwa R., Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol. Cell. Proteomics. 2006;5:1193–1204. doi: 10.1074/mcp.M500382-MCP200. doi:10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Shi M., Jin J., Wang Y., Beyer R.P., Kitsou E., Albin R.L., Gearing M., Pan C., Zhang J. Mortalin: a protein associated with progression of Parkinson disease? J. Neuropathol. Exp. Neurol. 2008;67:117–124. doi: 10.1097/nen.0b013e318163354a. doi:10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya T., Karnezis A.N., Murphy S.P., Hoang T., Freeman B.C., Phillips B., Morimoto R.I. Cloning and subcellular localization of human mitochondrial hsp70. J. Biol. Chem. 1995;270:1705–1710. doi: 10.1074/jbc.270.4.1705. doi:10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- 11.Ran Q., Wadhwa R., Kawai R., Kaul S.C., Sifers R.N., Bick R.J., Smith J.R., Pereira-Smith O.M. Extramitochondrial localization of mortalin/mthsp70/PBP74/GRP75. Biochem. Biophys. Res. Commun. 2000;275:174–179. doi: 10.1006/bbrc.2000.3237. doi:10.1006/bbrc.2000.3237. [DOI] [PubMed] [Google Scholar]
- 12.Kang P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. doi:10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 13.Neupert W., Brunner M. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. doi:10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt O., Pfanner N., Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. doi:10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 15.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. doi:10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germaniuk A., Liberek K., Marszalek J. A bichaperone (Hsp70-Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J. Biol. Chem. 2002;277:27801–27808. doi: 10.1074/jbc.M201756200. doi:10.1074/jbc.M201756200. [DOI] [PubMed] [Google Scholar]
- 17.Lill R., Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. doi:10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 18.Lutz T., Westermann B., Neupert W., Herrmann J.M. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol. 2001;307:815–825. doi: 10.1006/jmbi.2001.4527. doi:10.1006/jmbi.2001.4527. [DOI] [PubMed] [Google Scholar]
- 19.Schilke B., Williams B., Knieszner H., Pukszta S., D'Silva P., Craig E.A., Marszalek J. Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr. Biol. 2006;16:1660–1665. doi: 10.1016/j.cub.2006.06.069. doi:10.1016/j.cub.2006.06.069. [DOI] [PubMed] [Google Scholar]
- 20.Kaul S.C., Deocaris C.C., Wadhwa R. Three faces of mortalin: a housekeeper, guardian and killer. Exp. Gerontol. 2007;42:263–274. doi: 10.1016/j.exger.2006.10.020. doi:10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. doi:10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 22.Pukszta S., Schilke B., Dutkiewicz R., Kominek J., Moczulska K., Stepien B., Reitenga K.G., Bujnicki J.M., Williams B., Craig E.A., et al. Co-evolution-driven switch of J-protein specificity towards an Hsp70 partner. EMBO Rep. 2010;11:360–365. doi: 10.1038/embor.2010.29. doi:10.1038/embor.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deloche O., Liberek K., Zylicz M., Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiae Mdj1p, the mitochondrial DnaJ homologue. J. Biol. Chem. 1997;272:28539–28544. doi: 10.1074/jbc.272.45.28539. doi:10.1074/jbc.272.45.28539. [DOI] [PubMed] [Google Scholar]
- 24.D'Silva P.D., Schilke B., Walter W., Andrew A., Craig E.A. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl Acad. Sci. USA. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. doi:10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horst M., Oppliger W., Rospert S., Schonfeld H.J., Schatz G., Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. doi:10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokranjac D., Sichting M., Neupert W., Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. doi:10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truscott K.N., Voos W., Frazier A.E., Lind M., Li Y., Geissler A., Dudek J., Muller H., Sickmann A., Meyer H.E., et al. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. doi:10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westermann B., Gaume B., Herrmann J.M., Neupert W., Schwarz E. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell. Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goswami A.V., Chittoor B., D'Silva P. Understanding the functional interplay between mammalian mitochondrial Hsp70 chaperone machine components. J. Biol. Chem. 2010;285:19472–19482. doi: 10.1074/jbc.M110.105957. doi:10.1074/jbc.M110.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha D., Joshi N., Chittoor B., Samji P., D'Silva P. Role of Magmas in protein transport and human mitochondria biogenesis. Hum. Mol. Genet. 2010;19:1248–1262. doi: 10.1093/hmg/ddq002. doi:10.1093/hmg/ddq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kampinga H.H., Craig E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. doi:10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laloraya S., Gambill B.D., Craig E.A. A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc. Natl Acad. Sci. USA. 1994;91:6481–6485. doi: 10.1073/pnas.91.14.6481. doi:10.1073/pnas.91.14.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franssens V., Boelen E., Anandhakumar J., Vanhelmont T., Buttner S., Winderickx J. Yeast unfolds the road map toward alpha-synuclein-induced cell death. Cell. Death Differ. 2010;17:746–753. doi: 10.1038/cdd.2009.203. doi:10.1038/cdd.2009.203. [DOI] [PubMed] [Google Scholar]
- 34.Outeiro T.F., Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. doi:10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens S.Y., Cai S., Pellecchia M., Zuiderweg E.R. The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393–507) in complex with the peptide NRLLLTG. Protein Sci. 2003;12:2588–2596. doi: 10.1110/ps.03269103. doi:10.1110/ps.03269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han W., Christen P. Interdomain communication in the molecular chaperone DnaK. Biochem. J. 2003;369:627–634. doi: 10.1042/BJ20020943. doi:10.1042/BJ20020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krayl M., Lim J.H., Martin F., Guiard B., Voos W. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol. Cell. Biol. 2007;27:411–425. doi: 10.1128/MCB.01391-06. doi:10.1128/MCB.01391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallet P.F., Maftah A., Petit J.M., Denis-Gay M., Julien R. Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur. J. Biochem. 1995;228:113–119. doi: 10.1111/j.1432-1033.1995.tb20238.x. doi:10.1111/j.1432-1033.1995.tb20238.x. [DOI] [PubMed] [Google Scholar]
- 39.Nowikovsky K., Reipert S., Devenish R.J., Schweyen R.J. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. doi:10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- 40.Johnson L.V., Walsh M.L., Bockus B.J., Chen L.B. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J. Cell Biol. 1981;88:526–535. doi: 10.1083/jcb.88.3.526. doi:10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson R.B., Roof D.M. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 1997;16:352–357. doi: 10.1038/ng0897-352. doi:10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 42.Blank H.M., Li C., Mueller J.E., Bogomolnaya L.M., Bryk M., Polymenis M. An increase in mitochondrial DNA promotes nuclear DNA replication in yeast. PLoS Genet. 2008;4:e1000047. doi: 10.1371/journal.pgen.1000047. doi:10.1371/journal.pgen.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldring E.S., Grossman L.I., Marmur J. Petite mutation in yeast. II. Isolation of mutants containing mitochondrial deoxyribonucleic acid of reduced size. J. Bacteriol. 1971;107:377–381. doi: 10.1128/jb.107.1.377-381.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pozniakovsky A.I., Knorre D.A., Markova O.V., Hyman A.A., Skulachev V.P., Severin F.F. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 2005;168:257–269. doi: 10.1083/jcb.200408145. doi:10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cap M., Vachova L., Palkova Z. Yeast colony survival depends on metabolic adaptation and cell differentiation rather than on stress defense. J. Biol. Chem. 2009;284:32572–32581. doi: 10.1074/jbc.M109.022871. doi:10.1074/jbc.M109.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pareek G., Samaddar M., D'Silva P. Primary sequence that determines the functional overlap between mitochondrial heat shock protein 70 Ssc1 and Ssc3 of Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:19001–19013. doi: 10.1074/jbc.M110.197434. doi:10.1074/jbc.M110.197434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin T.K., Liou C.W., Chen S.D., Chuang Y.C., Tiao M.M., Wang P.W., Chen J.B., Chuang J.H. Mitochondrial dysfunction and biogenesis in the pathogenesis of Parkinson's disease. Chang Gung Med. J. 2009;32:589–599. [PubMed] [Google Scholar]
- 48.Schapira A.H., Gegg M. Mitochondrial contribution to Parkinson's disease pathogenesis. Parkinsons Dis. 2011;2011 doi: 10.4061/2011/159160. 159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez M., Brice A., Vaughan J.R., Zimprich A., Breteler M.M., Meco G., Filla A., Farrer M.J., Betard C., Hardy J., et al. Genome-wide scan linkage analysis for Parkinson's disease: the European genetic study of Parkinson's disease. J. Med. Genet. 2004;41:900–907. doi: 10.1136/jmg.2004.022632. doi:10.1136/jmg.2004.022632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott W.K., Nance M.A., Watts R.L., Hubble J.P., Koller W.C., Lyons K., Pahwa R., Stern M.B., Colcher A., Hiner B.C., et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. doi:10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- 51.Misselwitz B., Staeck O., Rapoport T.A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. doi:10.1016/S1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 52.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. doi:10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 53.Hinault M.P., Cuendet A.F., Mattoo R.U., Mensi M., Dietler G., Lashuel H.A., Goloubinoff P. Stable alpha-synuclein oligomers strongly inhibit chaperone activity of the Hsp70 system by weak interactions with J-domain co-chaperones. J. Biol. Chem. 2010;285:38173–38182. doi: 10.1074/jbc.M110.127753. doi:10.1074/jbc.M110.127753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devi L., Raghavendran V., Prabhu B.M., Avadhani N.G., Anandatheerthavarada H.K. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. doi:10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller-Fleming L., Giorgini F., Outeiro T.F. Yeast as a model for studying human neurodegenerative disorders. Biotechnol. J. 2008;3:325–338. doi: 10.1002/biot.200700217. doi:10.1002/biot.200700217. [DOI] [PubMed] [Google Scholar]
- 56.Yang H., Zhou X., Liu X., Yang L., Chen Q., Zhao D., Zuo J., Liu W. Mitochondrial dysfunction induced by knockdown of mortalin is rescued by Parkin. Biochem. Biophys. Res. Commun. 2011;410:114–120. doi: 10.1016/j.bbrc.2011.05.116. doi:10.1016/j.bbrc.2011.05.116. [DOI] [PubMed] [Google Scholar]
- 57.Zhai P., Stanworth C., Liu S., Silberg J.J. The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J. Biol. Chem. 2008;283:26098–26106. doi: 10.1074/jbc.M803475200. doi:10.1074/jbc.M803475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q., Krzewska J., Liberek K., Craig E.A. Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 2001;276:6112–6118. doi: 10.1074/jbc.M009519200. doi:10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- 59.Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., Hartl F.U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. doi:10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.