Abstract
The human genomic instability syndrome ataxia telangiectasia (A-T), caused by mutations in the gene encoding the DNA damage checkpoint kinase ATM, is characterized by multisystem defects including neurodegeneration, immunodeficiency and increased cancer predisposition. ATM is central to a pathway that responds to double-strand DNA breaks, whereas the related kinase ATR leads a parallel signaling cascade that is activated by replication stress. To dissect the physiological relationship between the ATM and ATR pathways, we generated mice defective for both. Because complete ATR pathway inactivation causes embryonic lethality, we weakened the ATR mechanism to different degrees by impairing HUS1, a member of the 911 complex that is required for efficient ATR signaling. Notably, simultaneous ATM and HUS1 defects caused synthetic lethality. Atm/Hus1 double-mutant embryos showed widespread apoptosis and died mid-gestationally. Despite the underlying DNA damage checkpoint defects, increased DNA damage signaling was observed, as evidenced by H2AX phosphorylation and p53 accumulation. A less severe Hus1 defect together with Atm loss resulted in partial embryonic lethality, with the surviving double-mutant mice showing synergistic increases in genomic instability and specific developmental defects, including dwarfism, craniofacial abnormalities and brachymesophalangy, phenotypes that are observed in several human genomic instability disorders. In addition to identifying tissue-specific consequences of checkpoint dysfunction, these data highlight a robust, cooperative configuration for the mammalian DNA damage response network and further suggest HUS1 and related genes in the ATR pathway as candidate modifiers of disease severity in A-T patients.
INTRODUCTION
Ataxia telangiectasia (A-T, OMIM #208900) is a hereditary genomic instability syndrome caused by loss of function mutations in ATM, which encodes a protein kinase involved in the cellular DNA damage response (DDR) (1). A-T is characterized by progressive cerebellar ataxia, oculocutaneous telangiectasias, immunodeficiency, infertility, increased cancer incidence and radiation hypersensitivity. These clinical manifestations are the consequence of failed responses to DNA double-strand breaks (DSB) that arise spontaneously or as programmed developmental events. Rare patients with atypical manifestation of the disease have been described and referred to as ‘A-T variants’. One such variant, termed A-TFresno, combines typical A-T phenotypes with microcephaly and mental retardation, making it among the most severe A-T forms known (2).
ATM normally functions as the main component of one of the cellular DNA damage checkpoint pathways that detect aberrant DNA structures and coordinate DNA repair with cell cycle progression. Checkpoint mechanisms are part of a broader DDR that protects genomic integrity and ensures normal development, tissue homeostasis and cancer-free survival (3). In response to DSB, the ATM kinase undergoes a transition from inactive dimer to active, auto-phosphorylated monomer and then phosphorylates numerous substrates involved in signaling transduction and downstream effector functions, including H2AX, CHK2 and p53. A second checkpoint pathway, organized around the related kinase ATR, responds to a wide variety of lesions, including bulky DNA adducts and DNA crosslinks, as well as processed DSB. Activated ATR phosphorylates CHK1 and other targets, promoting fork stabilization, inhibition of late origin firing and stimulation of DNA repair (4). Critical for ATR activation is the RAD9–RAD1–HUS1 (911) complex, a PCNA-like clamp that is loaded onto damage sites and recruits the ATR activator TOPBP1 (5). Importantly, the 911 complex additionally has direct roles in regulating base excision repair, translesion DNA synthesis and apoptotic signaling (6). The genes encoding the 911 complex, like all ATR pathway core components, are essential for organismal survival (7–9). Partial ATR impairment in humans leads to Seckel syndrome (SCKL1, OMIM #210600), an autosomal recessive disorder characterized by proportional dwarfism, microcephaly, craniofacial abnormalities and mental retardation (10).
Owing to their distinct componentry and activation by different types of genome damage, the ATM and ATR pathways initially were thought to function as independent, separable mechanisms. However, aberrant DNA structures often engage both pathways to some extent, and there is substantial biochemical crosstalk between the pathways (4). A full appreciation of these interactions in vertebrates has been hampered by a lack of suitable genetic tools. Although Atm-deficient mice are viable and show many A-T phenotypes (11–13), the severe phenotypes associated with ATR pathway impairment have been an impediment to understanding the biological significance of the interplay between the ATM and ATR pathways. In this study, we overcome this limitation with an Hus1 allelic series in which null (Hus1Δ1n) or wild-type (Hus1+) alleles are used in combination with a hypomorphic (Hus1neo) allele that expresses Hus1 at ∼40% of the wild-type level (14). Hus1neo/Δ1n mice, which have the lowest Hus1 expression in the series, are born at expected frequencies and appear grossly normal but are incapable of proper genome maintenance as evidenced by increased spontaneous GIN and hypersensitivity to exogenous genotoxins (14). By combining the Hus1 allelic series with targeted Atm deletion, we identify an essential cooperative relationship between ATM and HUS1 during embryonic development and in specific adult tissues.
RESULTS
Combined inactivation of Atm and Hus1 results in synthetic lethality
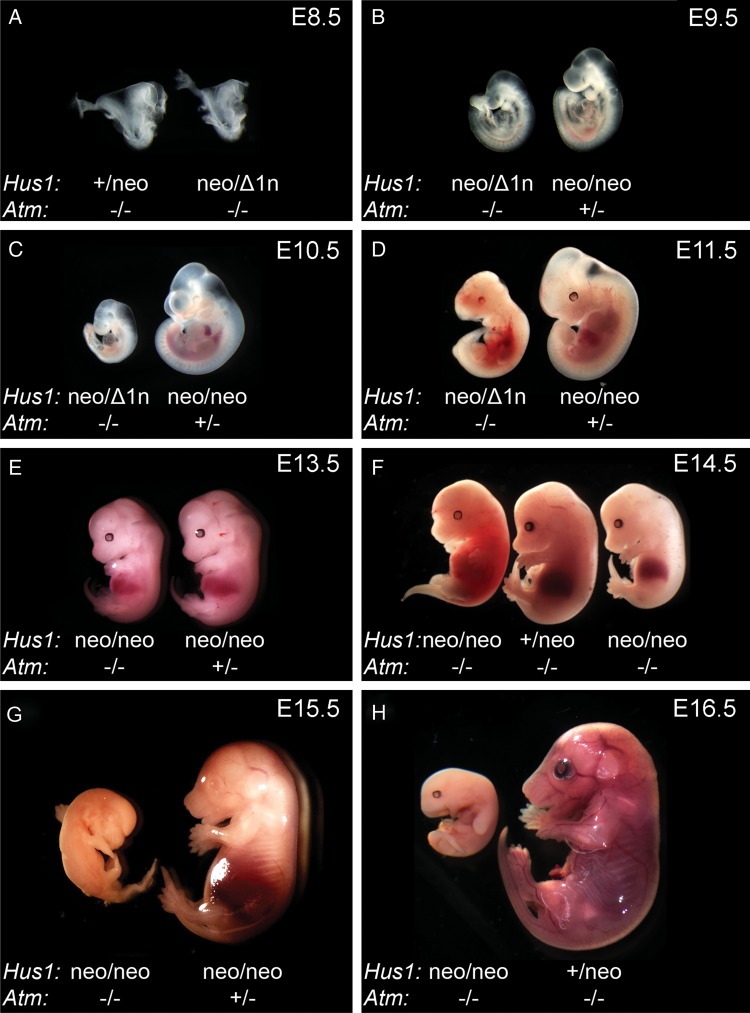
To elucidate the genetic relationships between the ATM- and ATR-mediated DNA damage checkpoint pathways, we generated mice defective for both. Interbreeding of mice bearing targeted Atm and Hus1 alleles was performed in order to produce Atm-deficient mice with incremental reductions in Hus1 gene dosage (Hus1+/+> Hus1+/neo> Hus1+/Δ1n> Hus1neo/neo> Hus1neo/Δ1n). Hus1+/neo cells and mice are phenotypically normal and frequently used as wild-type equivalents in this study (14). Of the 1753 weaned offspring analyzed, no Hus1neo/Δ1nAtm−/− mice were obtained, and only 84 Hus1neo/neoAtm−/− mice were obtained, although 259 were expected (Table 1; P<0.001, χ2 test). The nature of the lethal phenotype associated with combined Atm and Hus1 inactivation was determined by analyzing double-mutant and control embryos from embryonic day E8.5 to birth (Supplementary Material, Table S1). Although Hus1neo/Δ1nAtm−/− embryos appeared normal at E8.5, they were smaller relative to their littermates at E9.5 and all died between E10.5 and E11.5 (Fig. 1A–D). A less severe Hus1 defect in Hus1neo/neoAtm−/− embryos resulted in no gross abnormalities prior to E13.5, at which point the double-mutant embryos were smaller than their littermates. Approximately 30% of Hus1neo/neoAtm−/− embryos were abnormal at E14.5 and died by E15.5 (Fig. 1E–H). Hus1neo/neoAtm−/− mice that survived beyond this mid-gestational period were significantly smaller than control littermates (Fig. 3A and B; P<0.05, Student's t-test), and 30–40% of mice of this genotype died on the first day of life (P1) (Supplementary Material, Table S1), whereas the remainder survived beyond weaning (Table 1). Together, these data indicate that although defects in Atm or Hus1 individually are compatible with grossly normal development and organismal viability, simultaneous defects in Atm and Hus1 cause synthetic lethality in an Hus1 gene dosage dependent manner.
Table 1.
Synthetic lethality following combined impairment of Hus1 and Atm in micea
| Hus1 genotype | Atm genotype | Number expected | Number observed |
|---|---|---|---|
| +/+ | 3 | 5 | |
| +/neo | 95 | 108 | |
| +/Δ1n | +/+ | 12 | 9 |
| neo/neo | 259 | 286 | |
| neo/Δ1n | 69 | 58 | |
| +/+ | 6 | 5 | |
| +/neo | 190 | 208 | |
| +/Δ1n | +/− | 24 | 31 |
| neo/neo | 518 | 659 | |
| neo/Δ1n | 138 | 190 | |
| +/+ | 3 | 4 | |
| +/neo | 95 | 93 | |
| +/Δ1n | −/− | 12 | 13 |
| neo/neo | 259 | 84 | |
| neo/Δ1n | 69 | 0 | |
| TOTAL | 1753 | 1753 |
a Mice were genotyped by PCR analysis of tail DNA at 3 weeks of age. Expected values represent the combined Mendelian ratios from several crosses involving different parental genotypes. The observed and expected genotype frequencies were significantly different (P<0.001; χ2 test).
Figure 1.
Embryonic lethality upon simultaneous deregulation of Atm and Hus1. Embryos from timed matings were isolated at the indicated stage of embryonic development, imaged for morphological assesment and genotyped by PCR. (A–D) Representative images of Hus1neo/Δ1nAtm−/− embryos and control littermates from E8.5 to E11.5. (E–H) Representative images of Hus1neo/neoAtm−/− embryos and control littermates from E13.5 to E16.5. Note that, at E14.5, some Hus1neo/neoAtm−/− embryos are dead, whereas others are smaller than control littermates but otherwise normal.
Figure 3.
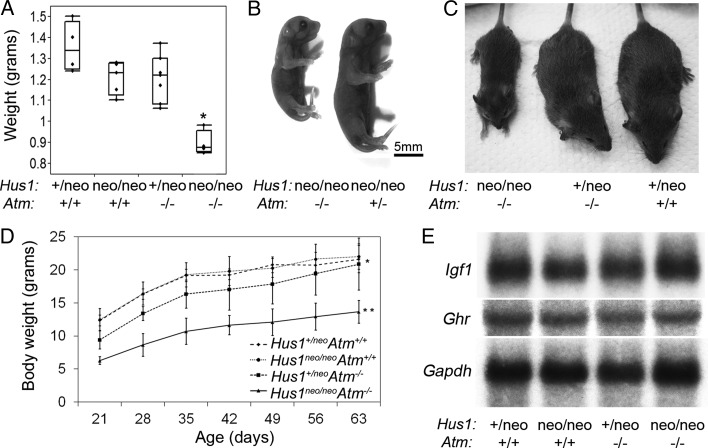
Dwarfism in Hus1neo/neoAtm−/− embryos and adult mice. (A) The box plot indicates the body weight of E18.5 embryos of the indicated genotypes (n≥ 4 per group). Hus1neo/neoAtm−/− embryos were significantly smaller than their littermates (P< 0.001, Student's t-test). (B) A representative image of newborn (P1) littermates of the indicated genotypes is shown. The scale bar represents 5 mm. (C) The photograph of 6-week-old male littermates of the indicated genotypes. (D) Body weight analysis of Hus1neo/neoAtm−/− mice and control littermates. The average body weights of female mice (n≥ 5 per genotype) of the indicated genotypes, from weaning to 9 weeks of age, are shown. The mean body weight was significantly lower for Hus1+/neoAtm−/− mice when compared with the Atm+/+ groups (*P< 0.05, mixed model analysis); Hus1neo/neoAtm−/− mice had significantly lower mean body weight when compared with all other groups (**P<0.01, mixed model analysis). (E) Northern blot analysis of transcript levels of Igf1 and Ghr in total RNA from the liver. Gapdh was used as a loading control.
Increased genomic instability and cell death in Atm/Hus1-deficient embryos
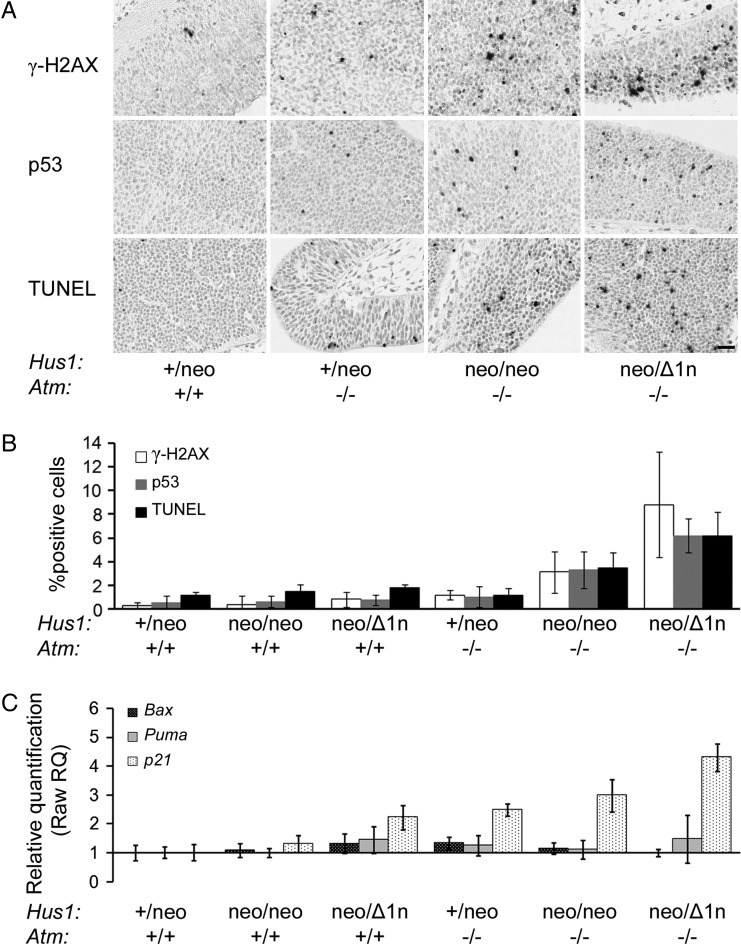
The failure of cells to deal with replication stress and spontaneous DNA lesions can lead to impaired cell proliferation and/or survival (15). To evaluate these outcomes in Atm/Hus1 mutant embryos, we initially performed Feulgen–Schiff staining on E10.5 embryos and quantified the frequency of pyknotic and mitotic nuclei. There was a significant increase in pyknotic nuclei, indicative of apoptosis in Hus1neo/Δ1nAtm−/− and Hus1neo/neoAtm−/− embryos when compared with the Hus1+/neoAtm+/+ control group (Supplementary Material, Fig. S1; P<0.001, Student's t-test). By contrast, cell proliferation was unaffected. We reasoned that the apparent increase in apoptosis in the Atm/Hus1 double-mutants could be due to checkpoint activation and therefore monitored the accumulation of phosphorylated H2AX (γ-H2AX; Supplementary Material, Fig. S1D) and total p53, markers of an activated DDR (3). Terminal uridine deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining also was performed to directly measure apoptosis. Although almost no γ-H2AX, p53 or TUNEL-positive cells were detected in Atm or Hus1 single-mutant embryos, Hus1neo/neoAtm−/− and, to an even greater extent, Hus1neo/Δ1nAtm−/− littermates showed significantly increased positive staining (Fig. 2A and B; P<0.05, Student's t-test). Because p53 can induce cell cycle arrest through transcriptional activation of p21 or apoptosis by upregulating Bax and Puma, we used real-time PCR to measure the expression level of these genes in E10.5 embryos. Consistent with the observation of p53 accumulation in Atm/Hus1 double-mutant embryos, there was a corresponding significant increase in p21 expression (Fig. 2C; P<0.05, Student's t-test). Interestingly, there were no detectable changes in Bax or Puma expression, raising the possibility of a p53-independent apoptotic mechanism. Similar results were observed by northern blot analysis (data not shown). Altogether, these findings suggest that the embryonic lethality following combined deregulation of Atm and Hus1 is associated with a generalized accumulation of genome damage, activation of a DDR and ultimately widespread apoptosis.
Figure 2.
Significantly increased apoptosis and DDR activation in Atm/Hus1 double-mutant embryos. Sections of E10.5 Atm/Hus1 embryos of the indicated genotypes were stained for γ-H2AX, for p53 or by TUNEL assay. (A) Representative images of embryo neural tube sections stained for γ-H2AX, p53 and TUNEL are shown. The scale bar represents 25 µm. (B) The percentage of γ-H2AX, TUNEL or p53-positive cells. Values are the mean of at least three embryos per genotype; error bars indicate standard deviation. Quantification was performed in the neural ectoderm to ensure that equivalent cell populations were compared between genotypes; however, similar staining patterns were observed throughout the embryos, irrespective of the cell type. The percentage of positively stained cells in Hus1neo/Δ1nAtm−/− and Hus1neo/neoAtm−/− embryos was significantly different from each other (P< 0.05, Student's t-test) and when each was compared with all the other genotypes (P< 0.01, Student's t-test). (C) The bar graph representing the fold increase in the expression level of p21, Bax and Puma for the embryos of the indicated genotype relative to Hus1+/neoAtm+/+ as measured by qPCR. p21 expression was significantly increased in Hus1neo/Δ1nAtm+/+, Hus1+/neoAtm−/−, Hus1neo/neoAtm−/− and Hus1neo/Δ1nAtm−/− embryos (P< 0.05, Student's t-test) and was significantly greater in Hus1neo/Δ1nAtm−/− embryos when compared with all other genotypes (P< 0.05, Student's t-test).
Dwarfism in adult Hus1neo/neoAtm−/− mice
Approximately one-third of Hus1neo/neoAtm−/− mice survived to adulthood, and without exception these animals exhibited a striking dwarfism that became apparent at approximately E13.5 and persisted through adulthood (Fig. 3A–D). Consistent with previous reports (11–13), modestly reduced body size was observed in Hus1+/neoAtm−/− mice defective for Atm alone. This phenotype was significantly enhanced in Hus1neo/neoAtm−/− animals, which on average were 40% smaller by weight than control littermates at 6 weeks of age (Fig. 3D; P<0.01, mixed model analysis). Most organs were proportionally smaller in both Hus1+/neoAtm−/− and Hus1neo/neoAtm−/− mice when compared with Hus1+/neo Atm+/+ controls (Supplementary Material, Table S2). Hus1neo/neoAtm−/− mice showed a slight increase in relative brain size when compared with mice of all other genotypes, but this was not associated with any apparent histopathological abnormalities (Supplementary Material, Fig. S2).
It is well established that the somatotroph axis that includes growth hormone (GH) and insulin-like growth factor 1 (IGF1) plays important roles in somatic growth and aging (16). Deficiencies in the GH/IGF1 somatotroph axis are observed in some mouse mutants that display dwarfism in conjunction with underlying DDR defects (17). In order to understand the molecular basis for the dwarfism in Atm/Hus1 mutant mice, we analyzed the expression levels of Igf1 and Ghr and found no difference in their transcript levels in livers of 6-week-old mice of representative genotypes (Fig. 3E), findings that were confirmed by qPCR (data not shown). Furthermore, preliminary analyses revealed no differences in circulating IGF1 levels in Hus1neo/neoAtm−/− mice relative to controls (data not shown). Overall, these results demonstrate that combined Atm and Hus1 deficiency has a severe impact on body size from mid-gestation to adulthood and that this proportional dwarfism arises independently of detectable abnormalities in the somatotroph axis.
Skeletal abnormalities in Hus1neo/neoAtm−/− mice
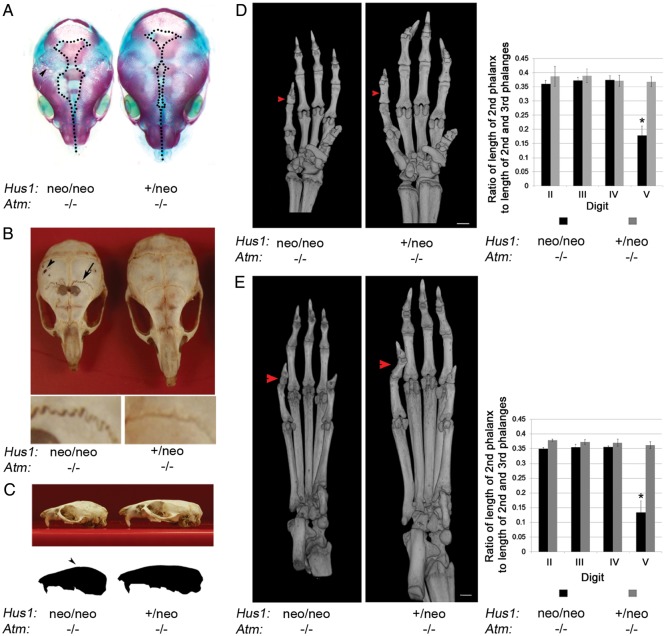
Visual inspection of Hus1neo/neoAtm−/− mice also revealed fronto-parietal alopecia and an abnormal facial morphology (Fig. 3C). To understand the origins of these abnormalities, we performed Alizarin red/Alcian blue staining of embryos. At E18.5, Hus1neo/neoAtm−/− embryos had widely open sagittal (midline) sutures and presented an array of fenestrations primarily in the parietal bone (Fig. 4A) that were also apparent as early as E15.5 (Supplementary Material, Fig. S3A). In order to characterize the craniofacial defects in adult animals, we first produced isolated skull preparations from mice at 6 weeks of age. Grossly, Hus1neo/neoAtm−/− skulls showed a domed morphology with an incomplete closure of the sagittal sutures and reduced bone thickness in the fronto-parietal region, and multiple fenestrations in the parietal bone and jagged coronal (fronto-parietal) sutures (Fig. 4B and C). To identify and quantify patterns in skull morphology, we analyzed high-resolution three-dimensional images generated by micro-computed tomography (micro-CT) (Supplementary Material, Fig. S3D). A series of measurements were then subjected to principal component (PC) analysis to identify suites of changes in skull morphology. The first PC explained 83.6% of the total variation in the 23 skull measurements. Therefore, in a single value PC1 quantified the great majority of the skeletal variation between Hus1neo/neoAtm−/− samples and those of Hus1+/neoAtm+/+, Hus1neo/neoAtm+/+ and Hus1+/neoAtm−/− genotypes (Supplementary Material, Fig. S3E). The correlations or factor loadings between the PC1 score and each distance measured for each mouse further resolved the pattern of variation: the skulls of Hus1neo/neoAtm−/− mice were smaller overall and differed in shape, with an elongated inter-parietal bone and a wider frontal bone, which increased the inter-orbital distance (Supplementary Material, Fig. S3F).
Figure 4.
Skeletal abnormalities in Hus1neo/neoAtm−/− embryos and adult mice. (A) Shown is a representative image of Alizarin red–Alcian blue-stained skulls from E18.5 littermates of the indicated genotypes. Dotted lines outline the region where the skull plates have yet to fill in and fuse. The arrowhead indicates the fenestrations present in the Hus1neo/neoAtm−/− skull. (B) Skulls were prepared from 6-week-old littermates of the indicated genotypes and photographed. The arrowhead indicates the fenestrations present in the parietal bone. The arrow indicates abnormal sutures of the mutant. Lower panels: higher magnification view of sutures in the Hus1neo/neoAtm−/− (left) and Hus1+/neoAtm−/− (right) skulls. (C) A representative image of the skulls from 6-week-old littermates of the indicated genotypes is shown. The outline was generated in Adobe Photoshop to illustrate the doming of the skull. (D and E) 3D reconstructions based on micro-CT data for forelimbs (D) or hindlimbs (E) from Hus1neo/neoAtm−/− and Hus1+/neoAtm−/− mice, and the bar graphs of the corresponding measurement data (n=3 per genotype). Red arrowheads point to the mesophalanx of digit V. The relative length of mesophalanx V in both forelimbs and hindlimbs was significantly shorter in Hus1neo/neoAtm−/− mice when compared with all other genotypes (*P< 0.001, Student's t-test).
Detailed analysis of the embryonic skeletal preparations also revealed brachymesophalangy of the fifth digit (Supplementary Material, Fig. S3B and C). Interestingly, this and the related phenotype of clinodactyly are associated with several hereditary GIN syndromes, other congenital disorders and fetal teratogen exposure (18). Further inspection of micro-CT images indicated that the phalangeal defect was fully penetrant in adult Hus1neo/neoAtm−/− mice and affected both the forelimbs and hindlimbs (Fig. 4D and E). Quantitative measurements showed that the mesophalanx of the fifth digit was less than one-half of the expected size in Hus1neo/neoAtm−/− mice relative to controls, whereas the remaining digits were unaffected (P<0.001, Student's t-test). Considered together with the craniofacial defects described above, these results demonstrate an essential dual function for Atm and Hus1 in the development of the skeleton and suggest a potential elevated sensitivity of particular bone progenitor cells to checkpoint impairment and genomic instability.
Increased genomic instability in adult mice defective for both Atm and Hus1
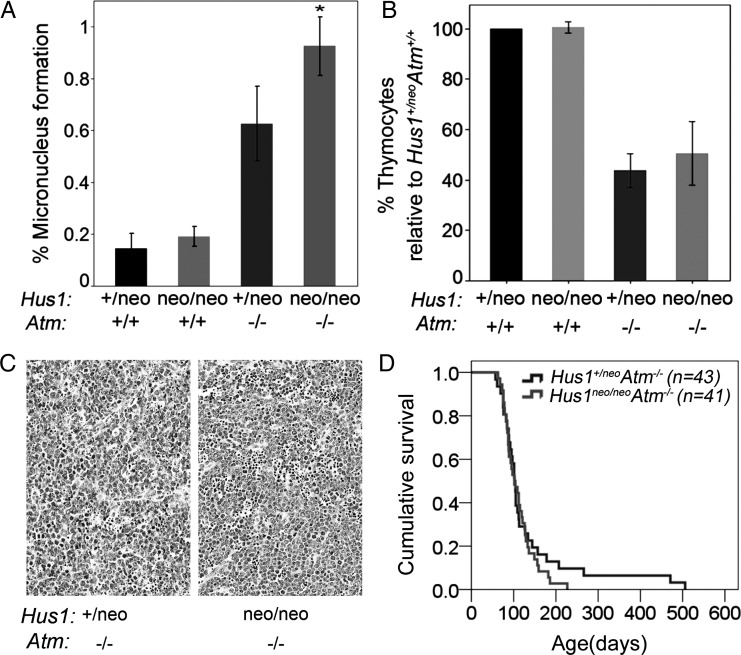
HUS1 and ATM are integral components of DNA damage signaling pathways that function to protect genomic integrity. We therefore tested the impact of combined Atm and Hus1 defects on GIN in adult mice by measuring micronucleus (MN) formation in peripheral blood cells, using a flow cytometry-based assay. MN are entire chromosomes or fragments of chromosomes that have not been incorporated in the main nuclei at cell division and are a hallmark of GIN. Consistent with previous findings (14,19), a moderate Hus1 defect alone in Hus1neo/neoAtm+/+ mice caused no significant change in MN formation relative to Hus1+/neoAtm+/+ littermate controls, whereas Atm deficiency alone was associated with significantly increased MN formation in Hus1+/neoAtm−/− mice (Fig. 5A; P<0.001, Student's t-test). Notably, Hus1neo/neoAtm−/− mice had more than an additive increase in MN formation when compared with Hus1neo/neoAtm+/+ and Hus1+/neoAtm−/− animals (P=0.002, univariate analysis of variance). In contrast, expression of damage-inducible genes such as p21 was unaltered in the adult liver, a relatively quiescent tissue (Supplementary Material, Fig. S4). These observations suggest that phenotypes associated with combined Atm and Hus1 defects in adult mice may be more pronounced in proliferating cell populations such as blood cell precursors and highlight a potential role for replication stress as a key factor in the observed genetic interaction.
Figure 5.
Increased genomic instability with no change in tumor predisposition in Hus1neo/neoAtm−/− mice. (A) The bar graph shows the average percentage of peripheral blood cells with MN in mice of the indicated genotypes. Hus1neo/neoAtm+/+ and Hus1+/neoAtm+/+ mice showed similar levels of GIN, whereas Hus1+/neoAtm−/− mice had a significant increase in MN formation (P< 0.001, Student's t-test). The combined effect of Hus1 impairment in an Atm null background (Hus1neo/neoAtm−/−) resulted in a greater than additive increase in GIN (P=0.002, univariate analysis of variance). (B) The bar graph shows the relative number of thymocytes in 6-week-old mice of the indicated genotypes (n≥3 per group) expressed as a percentage of the value for Hus1+/neoAtm+/+ control littermates. Error bars indicate standard deviation. The entire thymus from each mouse was harvested and mechanically disrupted, and viable trypan-blue-negative thymocytes were counted using a hemocytometer. The relative number of thymocytes was significantly lower for both Hus1+/neoAtm−/− and Hus1neo/neoAtm−/− mice (*P<0.001, Student's t-test). (C) Representative hematoxylin and eosin-stained sections of thymic lymphomas from Hus1+/neoAtm−/− (left) and Hus1neo/neoAtm−/− (right) mice. The scale bar represents 50 µm. (D) Cohorts of Hus1neo/neoAtm−/− (n=41) and Hus1+/neoAtm−/− (n=43) mice were monitored for tumor development as described in Materials and Methods. A Kaplan–Meier survival curve is shown. Overall survival was not significantly different between genotypes (P=0.644, log-rank test).
Increased GIN may influence different steps en route to carcinogenesis, including tumor initiation and progression, and in Atm-null mice contributes to the development of thymic lymphoma with near-complete penetrance (11–13). Prior to tumor development, Hus1neo/neoAtm−/− and Hus1+/neo Atm−/− mice contained similar numbers of lymphocytes in the thymus (Fig. 5B) and circulating blood (Supplementary Material, Table S3) and subsequently showed no significant difference in tumor-free survival as determined by Kaplan–Meier survival analysis (Fig. 5D). Furthermore, the resulting neoplasms were indistinguishable histopathologically (Fig. 5C). Together, these results reveal that combined defects in Atm and Hus1 result in synergistic increases in GIN in adult mice but without major consequences on thymic tumorigenesis.
Partial Hus1 impairment synergizes with Atm deficiency to accelerate senescence without detectably altering key DNA damage checkpoint signaling events or radiation sensitivity
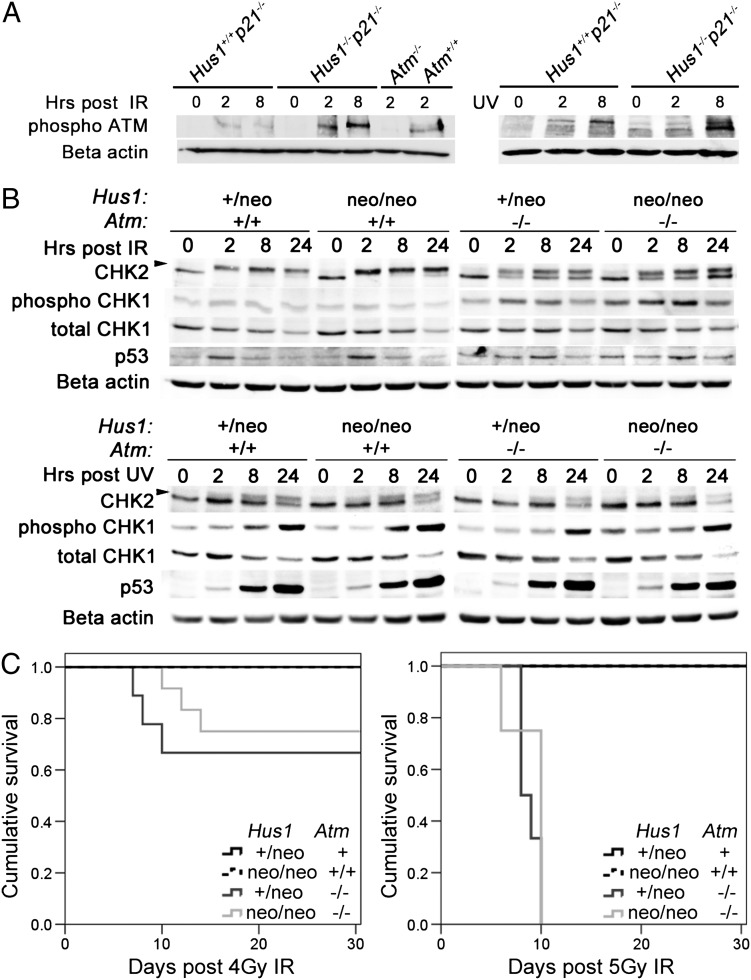
The strong genetic interaction observed between Atm and Hus1 raised the possibility of overlapping roles for these checkpoint proteins in DNA damage signaling. Complete Hus1 inactivation results in impaired activation of the ATR substrate CHK1 but does not diminish the induction of the ATM substrates CHK2 or p53 (20). Consistent with the prevailing view that ATM activation occurs independently of the 911 complex, we observed that IR- or UV-induced ATM phosphorylation occurred to a similar or greater extent in Hus1-null cells when compared with Hus1-proficient controls (Fig. 6A). To better understand the relationship between Atm and Hus1 at the cell and molecular levels, we isolated primary mouse embryonic fibroblasts (MEFs) from the Atm/Hus1 mouse model and analyzed their behavior in culture. Metaphase chromosome analysis of the MEF cultures at passage 1 revealed an increased frequency of chromosomal abnormalities in Hus1neo/neoAtm−/− double-mutant cells when compared with Atm or Hus1 single mutants (Supplementary Material, Fig. S5). In particular, Hus1neo/neoAtm−/− cells exhibited a significant increase in the occurrence of extensive chromosomal damage (P<0.02, Student's t-test) as well as an increased frequency of radial figures (P<0.03, Student's t-test) when compared with cells of all other genotypes analyzed.
Figure 6.
DNA damage signaling in Atm/Hus1 double-mutant MEFs. (A) Immortalized MEFs of the indicated genotypes were treated with 20 Gy IR or 65 J/m2 UV, and total protein lysates were prepared at 0, 2 and 8 h post-treatment. Samples were immunoblotted using antibodies specific for phospho-Ser1981-ATM, or beta-actin as a loading control. Similar results were obtained with an independent set of Hus1−/−p21−/− and matched control Hus1+/+p21−/− MEFs (data not shown). (B) Primary MEFs at passage 1 or 2 were treated with 30 Gy IR or 65 J/m2 UV, and total cell protein lysates were prepared at 0, 2, 8 and 24 h post-treatment and immunoblotted for CHK1, phosphoSer345-CHK1, CHK2 and p53. The arrow indicates the position of phosphorylated CHK2. Beta-actin was used as a loading control. (C) Kaplan–Meier survival curves show the radiation sensitivity of mice of the indicated genotypes, following exposure to 4 Gy (left) or 5 Gy (right) IR. The cohort treated with 4 Gy included Hus1+/neoAtm+ (n=7; Atm+ is a combination of Atm+/+ and Atm+/−), Hus1neo/neoAtm+/+ (n=6), Hus1+/neoAtm−/− (n=9) and Hus1neo/neoAtm−/− (n=12) mice. The cohort treated with 5 Gy included Hus1+/neoAtm+ (n= 4), Hus1neo/neoAtm+/+ (n=6), Hus1+/neoAtm−/− (n=6) and Hus1neo/neoAtm−/− (n=4) mice. There were no significant differences in survival between Hus1+/neoAtm−/− and Hus1neo/neoAtm−/− mice after 4 Gy (P=0.295) or 5 Gy (P=0.366) as determined by the log-rank test.
Consistent with previous results (14,21), Atm-deficient Hus1+/neoAtm−/− MEFs underwent senescence prematurely, whereas Hus1neo/neoAtm+/+ MEFs doubled in a manner similar to Hus1+/neoAtm+/+ control cells. The combination of Atm and Hus1 defects in Hus1neo/neoAtm−/− MEFs resulted in extremely rapid senescence after just a few doublings (Supplementary Material, Fig. S6A). Nevertheless, cells of all genotypes were capable of undergoing spontaneous immortalization, after which they doubled at similar rates when maintained under standard 3T3 culture conditions, both in low (3%) (data not shown) and normal (20%) O2 levels (Supplementary Material, Fig. S6A). The proliferative capacity of the spontaneously immortalized Atm/Hus1 MEFs was analyzed under more stringent conditions by plating cells at low density and assessing the colony-forming potential of each cell line. Colony formation was reduced in immortalized Hus1neo/neoAtm+/+ cells and impaired to an even greater degree in Hus1+/neoAtm−/− MEFs. Remarkably, Hus1neo/neoAtm−/− MEFs were completely defective for colony formation, under either atmospheric (Supplementary Material, Fig. S6B) or low-oxygen conditions (data not shown), despite the fact that these cells showed normal proliferation when cultured at higher densities. Thus, simultaneous defects in Atm and Hus1 result in accelerated senescence and, despite being compatible with spontaneous immortalization, cause profound impairment of the capacity of single cells to grow out as colonies.
To further understand the cooperative relationship between ATM and HUS1 in DNA damage signaling, we next treated the Atm/Hus1 primary MEFs with UV or IR and assessed the activation of canonical ATM and ATR downstream targets. Untreated Hus1neo/neoAtm−/− MEFs had slightly higher basal levels of p53 and phospho-CHK1 (Fig. 6B), suggestive of increased spontaneous DNA damage accumulation. Moderate Hus1 impairment alone had no detectable effect on DNA damage signaling upon IR treatment, with comparable levels of CHK2 phosphorylation and p53 accumulation occurring in Hus1+/neoAtm+/+ and Hus1neo/neoAtm+/+ cells. The slight accumulation of phosphorylated CHK1 following IR also was unchanged in Hus1neo/neoAtm+/+ cells. Lacking ATM alone, Hus1+/neoAtm−/− cells showed delayed induction of both CHK2 and p53 following IR and a slight increase in CHK1 phosphorylation. Interestingly, Hus1neo/neoAtm−/− MEFs showed the same IR-induced DDR kinetics as Hus1+/neoAtm−/− cells, and the magnitude of the response was similar with the exception that CHK1 activation after IR was even greater in cells defective for both Atm and Hus1. In response to UV, CHK2 and CHK1 phosphorylation as well as p53 accumulation were unaffected by Atm inactivation, partial Hus1 impairment, or both (Fig. 6B). Although defects in other signaling events not analyzed here cannot be ruled out, these data suggest that primary Atm/Hus1 double-mutant cells can trigger key DNA damage checkpoint signaling events as well as the single mutants. As a final measure of how interactions between Atm and Hus1 affect the DDR, we compared the radiation sensitivity of mice with individual or combined defects in Atm and Hus1. Consistent with the lack of detectable synergistic effects on IR-induced checkpoint signaling in MEFs, Atm/Hus1 double-mutant mice showed an equivalent sensitivity to whole-body IR exposure as Atm single-mutant mice, which are hypersensitive to IR (Fig. 6C). Thus, the defective DSB response in Atm-deficient mice is not substantially worsened by additional partial Hus1 impairment.
DISCUSSION
The ATM and ATR DNA damage checkpoint pathways were initially viewed as separable, parallel signaling cascades that respond to distinct DNA lesions. However, recent biochemical evidence indicating significant crosstalk between the two pathways has led to the concept of a broad, integrated DDR network (22). Nevertheless, the genetic relationship between these pathways in vertebrates has been difficult to investigate due to the lethality associated with a complete inactivation of ATR pathway components. Here we used a system for partial Hus1 inactivation to show for the first time that ATM and HUS1 cooperate in genome maintenance in vivo. Whereas mice defective for either Atm or Hus1 alone are viable, simultaneous dysfunction of both genes resulted in synthetic lethality. The ability to fine-tune Hus1 expression levels further revealed tissue-specific phenotypes, such as craniofacial and limb defects, associated with moderate Hus1 impairment in an Atm-deficient background. This was unexpected, as the two main mammalian DDR pathways are often considered ubiquitously important, and suggests that there is significant tissue-specific variation in the level of endogenous DNA damage and/or the activity of additional pathways that respond to DNA lesions. Mapping out these specificities will be critical for understanding the etiology of many inborn diseases as well as the nature of organismal responses to chemotherapeutics and other genotoxic agents.
The most striking phenotype in Hus1neo/neoAtm−/− mice was their significantly reduced overall size, which was apparent prior to birth. In mice and humans, there are several reports of dwarfism associated with DDR defects, but the underlying mechanisms are still poorly understood in many cases. Most theories on how DDR dysfunction causes dwarfism center on either cell-autonomous defects or systemic growth axis perturbations. In particular, the impairment of nucleotide excision repair in mouse models of Cockayne syndrome results in dwarfism, which is associated with the suppression of the GH/IGF1 axis in what is believed to represent a homeostatic feedback response to GIN (17). Likewise, SIRT6 deficiency in mice results in genomic instability, reduced Gh/Igf1 expression and dwarfism (23). Conversely, other mouse models in which inactivation of genome maintenance mechanisms leads to dwarfism maintain an intact somatotroph axis. For instance, Atm-null mice are slightly smaller than their littermates (11–13) but show no differences in the circulating levels of GH or free IGF1 (24). Similarly, mice lacking KU80, a subunit of the DNA–PK complex, are smaller than their littermates but have an intact somatotroph axis (25,26).
Notably, proportional dwarfism is a characteristic of humans with Seckel syndrome. Clinical reports indicate that the GH/IGF1 axis is normal in Seckel patients, although an Atr-Seckel mouse model shows reduced Igf1 and Ghr expression (27,28). Interestingly, Seckel syndrome also can be caused by mutations in genes encoding centrosomal proteins, including PCNT, CEP152 and CENPJ (29–32). In these cases, defective cell division has been hypothesized to drive the cell loss that underlies the reduced body size. The apparently normal GH/IGF1 axis in Atm/Hus1 mice suggests that a cell-autonomous defect also might be responsible for the proportional dwarfism reported here and raises the possibility that DNA damage accumulation upon combined Atm and Hus1 impairment leads to small size through defective expansion of key cell populations, either throughout the body or in specific compartments that impact body size, such as stem cell pools or developing bone growth plates (33,34).
In addition to their small size, Atm/Hus1 double-mutant animals showed skeletal abnormalities that included incomplete ossification and aberrant architecture of the skull, as well as significant shortening of the mesophalanx of the fifth digit (brachymesophalangy V) in both forelimbs and hindlimbs. Craniofacial defects are common in patients suffering from GIN-related disorders, including Seckel syndrome (35), Nijmegen breakage syndrome (due to NBS1 mutations) (36) and Williams–Beuren syndrome (caused by a micro-deletion at 7q11.23 that includes RFC2) (37). Digit abnormalities, including brachymesophalangy and clinodactyly, are frequently observed in patients with Seckel syndrome as well as another GIN disorder, Fanconi anemia (38). In addition to their association with hereditary disease, craniofacial defects and brachymesophalangy V also are present following fetal exposure to teratogens, such as alcohol (39), and may be hallmarks of excessive DNA damage during development (40). Why GIN is frequently associated with these particular developmental phenotypes is largely unknown, but may reflect a heightened stress sensitivity in certain cell types. Such a mechanism has been identified in Treacher Collins syndrome, in which the occurrence of craniofacial abnormalities has been attributed to a lower threshold of neural crest cells for stress-induced p53-dependent apoptosis (41). In Atm/Hus1 mice, cell death triggered by increased genomic instability could potentially impact crest-derived cells required for suture maintenance, possibly explaining the retarded dorsal expansion of these roofing bones. The lateral fenestrations in Atm/Hus1 skull bones beginning at the initial stages of bone formation suggest that early ossification is disrupted, but do not provide clues as to why these specific sites are affected. The mouse model described here will be a powerful tool for understanding cell-type-specific responses to GIN and their roles in the developmental defects observed in patients with instability syndromes.
Increased GIN in Atm/Hus1 double-mutant mice was readily apparent during embryogenesis as well as in a tissue-specific pattern in adults, as evidenced by increased MN formation in hematopoietic cells without detectable upregulation of DNA damage-responsive genes in the liver. HUS1 loss causes chromosomal instability specifically during S-phase (42), and the observations reported here are consistent with a model in which the phenotypes associated with combined Atm and Hus1 defects are due primarily to the failure of actively dividing cells to deal with replication stress. Surprisingly, the elevated GIN in Atm/Hus1 mice did not significantly change the onset or histological characteristics of thymic lymphomas relative to those arising in mice lacking Atm alone. It may be that the rapid onset of lymphomas in Atm-null mice obscures effects associated with partial Hus1 impairment. Alternatively, a greater level of genomic instability in Atm/Hus1 double-mutant mice may have opposing consequences that neutralize each other, both fueling the accumulation of cancer-promoting mutations and interfering with the ability of cancer cells to withstand stress-associated malignant transformation. Finally, the absence of a difference in tumor-free survival may reflect a limited requirement for Hus1 in this compartment. Previous studies have shown that thymocytes rely more on ATM and DNA-PK than on ATR (43), and our own work suggests that lymphocytes can tolerate Hus1 loss better than other cell types (44–46).
Unexpectedly, although we deregulated the two primary DNA damage checkpoint pathways in the Atm/Hus1 mouse model and observed clear phenotypic consequences, we detected only limited alterations in canonical ATM/ATR-mediated DNA damage signaling events when compared with those in Atm mutants alone. Indeed, for some of the factors analyzed, DNA damage signaling was increased in Atm/Hus1 embryos and MEFs. There are several possible explanations for these observations. First, upon deregulation of the ATM and ATR pathways, an overlapping DDR kinase could become activated. Among the several possible factors that could serve such a role, the best candidate may be the PIKK DNA-PK, which is known to cooperate with ATM and ATR in H2AX phosphorylation and is capable of phosphorylating an array of DDR proteins in vitro, including CHK2 and p53 (47). However, CHK1 activation was readily apparent in Atm/Hus1 double-mutants and there is limited evidence that DNA-PK phosphorylates CHK1 (48). Alternatively, ATR could mediate the checkpoint signaling either in a 911 independent manner (49) or via residual HUS1 expression from the hypomorphic Hus1 allele. Although Atm/Hus1-defective cells can sense and mount signaling responses to DNA damage, this response is clearly ineffective, resulting in GIN, followed by massive apoptosis and embryonic lethality. The observed accumulation of genome damage could be due to a repair defect, an elevated damage load that overwhelms the repair machinery, or both. Hus1 impairment did not further worsen the radiation sensitivity of Atm-null mice, indicating that the synthetic phenotypes in Hus1/Atm double-mutant mice are unlikely to originate from more severe DSB response defects relative to those in Atm single-mutants. Given that the 911 complex has been directly implicated in several DNA repair processes, including base excision repair and translesion synthesis, we favor the model that defective DNA repair following HUS1 deregulation leads to an increased number of DNA lesions that are normally resolved efficiently in an ATM-dependent manner, resulting in the observed synthetic lethality when both pathways are defective simultaneously (Supplementary Material, Fig. S7).
The strong genetic interaction between Atm and Hus1 in mice also has implications for human disease. First, the synergistic increases in genome damage and apoptosis observed here following combined Atm and Hus1 impairment suggest that therapeutic targeting of HUS1 or the broader ATR pathway may be effective for some malignancies that arise due to ATM pathway mutations, which may be as many as 70% of all cancers (50,51). However, the efficacy of such a strategy may depend on the cell of origin or other factors like the particular initiating oncogenic events in the tumor, as the impact of simultaneous Atm and Hus1 defects varied between tissues. Another implication of this work relates to the basis for variant forms of A-T, like A-TFresno, which is associated with ATM mutations but combines typical A-T phenotypes with microcephaly and mental retardation (2). This variation has been attributed to genetic modifiers outside of ATM. The findings presented here suggest that polymorphisms in HUS1 or other ATR pathway genes could significantly impact the onset and severity of A-T phenotypes.
MATERIALS AND METHODS
Animals
All animals used in this study were handled in accordance with federal and institutional guidelines, under a protocol approved by the Cornell University Institutional Animal Care and Use Committee (IACUC). Hus1neo and Hus1Δ1n mice were maintained on the 129S6 background (7,14). Atm knockout mice were maintained on the FVB background (12). These strains were interbred to create Atm/Hus1 double-mutant mice. Mice were genotyped by PCR analysis of genomic DNA isolated from tail snip biopsies as previously described (7). Images of live mice and skulls were taken using a Canon EOS Digital Rebel XTi digital camera equipped with a Canon EF-S 18–55 mm lens. For analysis of embryonic development, timed matings were performed. Noon of the day of vaginal plug detection was defined as embryonic day E0.5. For analysis of tumor-free survival, a cohort of mice was maintained for up to 18 months of age or until the appearance of visible neoplasms or signs of clinical disease, including weight loss, hunched posture, labored breathing, poor grooming or wasting, at which point they were euthanized by carbon dioxide asphyxiation and necropsied. For analysis of radiation sensitivity, mice were subjected to 4 or 5 Gy doses of γ-irradiation, using a Mark I Model 68 sealed 137Cs source gamma irradiator (JL Shepherd & Associates), and monitored for up to 30 days or until reaching humane endpoint criteria based on the clinical disease signs noted above.
Histology and immunohistochemistry
Following euthanasia, isolated tissues were weighed and then fixed in 10% formalin. Embryos were fixed in 4% paraformaldehyde. Histological sections of paraffin-embedded tissues were stained with hematoxylin and eosin and subjected to pathological assessment. For the assessment of nuclear morphology, Feulgen–Schiff staining was carried out using a standard protocol (7). For immunohistochemistry, E10.5 embryos were embedded in paraffin, and serial 5 μm sections were collected on Superfrost Plus slides (Fisher) and processed for immunohistochemistry using citric acid for p53 or EDTA for γ-H2AX-based antigen retrieval. Sections were incubated with anti-γ-H2AX (Upstate #05-636) or p53 (Cell Signaling #2524) antibodies, followed by a secondary biotinylated anti-mouse antibody (Invitrogen, Histostain SP 95-6543B). DAB (DAKO) was used as a chromogen, and hematoxylin as a counter-stain. TUNEL staining was performed using the Apoptag (Chemicon International) system according to the manufacturer's instructions. Quantification was done by counting the frequency of DAB-positive cells as depicted in Supplementary Material, Figure S1D. More than 1000 cells from at least three 100× fields were counted per embryo. Histology images were obtained using an Aperio Scanscope (Aperio Technologies, USA).
Skeletal morphology and measurements
Micro-CT scans were done on both living mice and isolated specimens, using GE CT120 micro-CT (GE Healthcare, London, Ontario, Canada) and Xradia VersaXRM-500 (Xradia, Pleasanton, CA, USA) scanners. Standard micro-CT scan parameters were used and are available upon request. For isolated skull preparations, mice were euthanized and decapitated, and heads were de-skinned and boiled in tap water for 20 min. Skulls were then mechanically cleaned using forceps, whereas the brain was removed by flushing with water. Skulls were dried before being stored. For live imaging, mice were anesthetized with isoflurane and scanned. In order to accurately measure the distance between two points of interest, we developed a tool for marking points on the skull surface in three-dimensional (3D) image space. For every marked point, a surface coordinate location was computed in millimeters for x, y and z 3-D coordinates. The distance between any two surface points was computed using the Euclidean distance of the mapped image coordinates. Statistical analysis of the resulting morphometry data was performed by a PC analysis using the princomp() function and a covariance matrix in R 2.6.0. Limbs were isolated after euthanasia and subjected to micro-CT imaging as described above. Analysis of the resulting data and 3D reconstructions were done using the OsiriX software (52). Skeletal staining with Alcian blue and Alizarin red was performed as previously described (53). Briefly, embryos were eviscerated, fixed in 80% ethanol for at least 24 h, dehydrated in 95% ethanol and stained overnight with Alcian blue (Sigma A3157) in 95% ethanol:1% glacial acetic acid solution. The bone was counterstained with Alizarin red (Sigma A5533) in 2% KOH for 3 h. Embryos were cleared in a decreasing 2% KOH/glycerol series (80:20; 60:40; 40:60) for 24 h each and stored indefinitely in 2% KOH:glycerol (20:80). The stained embryos were imaged using a Leica MZ125 inverse microscope equipped with an RT slider camera (Diagnostic Instruments).
MN assay
Analysis of MN formation in peripheral blood cells was performed as previously described (14,19). Briefly, peripheral blood was collected from the mandibular vein, fixed in methanol and incubated in bicarbonate buffer containing RNase A and anti-CD71: FITC antibody (Biodesign International). After washing and staining with propidium iodide, the cells were analyzed on a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, CA, USA).
Cell culture and proliferation measurements
MEFs were prepared from E13.5 embryos, following timed matings between Hus1+/neoAtm+/− and Hus1neo/neoAtm+/− mice. Briefly, embryos were dissected from the deciduum, mechanically disrupted and cultured in DMEM supplemented with 10% fetal bovine serum, 1.0 mm l-glutamine, 0.1 mm minimal essential medium nonessential amino acids, 100 µg/ml streptomycin sulfate and 100 U/ml penicillin. The initial plating was defined as passage zero (p0), and cells were subsequently maintained on a 3T3 protocol (54). Clonogenic potential was assessed by plating MEFs in six-well dishes at 5 × 102 or 3 × 103 cells per well and changing the media every 3 days. After 11 days in culture, cells were washed with PBS and fixed in methanol for 1 h at room temperature, stained overnight with a 5% crystal violet solution in 70% ethanol and washed with tap water. Immortalized Hus1+/+p21−/− and Hus1−/−p21−/− MEFs were described previously (7). For IR treatment, cells at passage 2 or 3 were exposed to γ-irradiation, using a Mark I Model 68 sealed 137Cs source gamma irradiator (JL Shepherd & Associates), at a dose rate of 123.1 rad/min. For UV treatment, the medium was removed from the cells, cells were washed with PBS, the top of the culture dish was removed and the cells were exposed to 254 nm UV light, using a XL-1000 UV Crosslinker (Spectro Linker).
Northern blotting and real-time PCR
Total RNA was prepared from E10.5 embryos and from the liver, using RNA STAT-60 reagent (Tel-Test, Friendswood, TX, USA), and then resolved on a 1% agarose/formaldehyde gel, transferred to a nylon membrane, and hybridized with 32P-labeled cDNA probes. Quantification of signal intensity was done using a Storm 860 phosphorimager (Molecular Dynamics, Sunnyvale, CA, USA). Total RNA was used for reverse transcription, using the SuperScript III kit (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR (q-PCR) was performed on an ABI 7500 System (PE Applied-Biosystems) using cDNA as a template in the presence of SYBR-green (Quanta Biosciences PerfeCTa SYBR Green FastMix). Primer pairs were designed using the Integrated DNA Technologies' RealTime PCR Assay Design Tool (www.idtdna.com) to generate intron-spanning products of 150–200 bp as follows: Ghr: 5′-AGCCTCGATTCACCA AGTG-3′ and 5′-AATTCTTGCAGCTTGTCGTTG-3′; Igf1: 5′-GAGACTGGAGATGTACTGTGC-3′ and 5′-CTCCTTTG CAGCTTCGTTTTC-3′; Gstt2: 5′-TGCCCAAGTCCACGAA TAC-3′ and 5′-CCAGGACCATTCTATCTCTGTTC-3′; Bax: 5′-TTGGAGATGAACTGGACAGC-3′ and 5′-CAGTTGAAG TTGCCATCAGC-3′; Puma: 5′-CTGGAGGGTCATGTACA ATCTC-3′ and 5′-GGTGTCAGAAGGCGGAG-3′; p21: 5′-C TTGCACTCTGGTGTCTGAG-3′ and 5′-GCACTTCAGGGT TTTCTCTTG-3′; beta-actin: 5′-ACCTTCTACAATGAGCT GCG-3′ and 5′-CTGGATGGCTACGTACATGG-3′. The generation of specific PCR products was confirmed by melting curve analysis and gel electrophoresis. Each primer pair was tested with a logarithmic dilution of cDNA mix to generate a linear standard curve, which was used to calculate the primer pair efficiency (55).
Western blotting
Cells were harvested and solubilized in RIPA buffer as described previously (56), or protein was extracted using a high-salt extraction procedure (57). Total protein was quantified by the Bradford assay, resolved on 6–14% SDS–PAGE gels and subjected to immunoblotting using antibodies from Millipore (anti-γ-H2AX, #05-636 and anti-CHK2, #05-649), Santa Cruz (anti-CHK1, G-4; sc-8408 and anti-p53, FL-393; sc-6243), Cell Signaling (anti-phospho-CHK1, Ser345, #2341), Rockland (anti-ATM, pSer1981, 200-310-400) and Sigma (anti-beta actin, #A5441). Western blot imaging and quantification were performed using a Versa Doc Imaging System (Bio-Rad Laboratories).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
The authors thank Eric Alani, Sylvia Lee and Yolanda Sanchez for helpful discussions and comments on the manuscript; Yves Boisclair for measurements of circulating IGF1 levels; and the staff of the Cornell Lab Animal Services and CARE programs for excellent animal care.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institutes of Health grants (R01 CA108773 and R03 HD058220 to R.S.W.); a Cornell University College of Veterinary Medicine Graduate Research Assistantship to G.B.; National Institutes of Health training grant (T32 HD052471) funding to A.M.L.; a Cornell University College of Veterinary Medicine Clinical Fellowship to K.R.H.; and a National Center for Research Resources grant (S10RR023781) for instrumentation. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
REFERENCES
- 1.McKinnon P.J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 2.Gilad S., Chessa L., Khosravi R., Russell P., Galanty Y., Piane M., Gatti R.A., Jorgensen T.J., Shiloh Y., Bar-Shira A. Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am. J. Hum. Genet. 1998;62:551–561. doi: 10.1086/301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kemp M., Sancar A. DNA distress: just ring 9-1-1. Curr. Biol. 2009;19:R733–R734. doi: 10.1016/j.cub.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Helt C.E., Wang W., Keng P.C., Bambara R.A. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 7.Weiss R.S., Enoch T., Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins K.M., Auerbach W., Wang X.Y., Hande M.P., Hang H., Wolgemuth D.J., Joyner A.L., Lieberman H.B. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Driscoll M. Mouse models for ATR deficiency. DNA Repair (Amst.) 2009;8:1333–1337. doi: 10.1016/j.dnarep.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 10.O'Driscoll M. Haploinsufficiency of DNA damage response genes and their potential influence in human genomic disorders. Curr. Genomics. 2008;9:137–146. doi: 10.2174/138920208784340795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 12.Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl Acad. Sci. USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 14.Levitt P.S., Zhu M., Cassano A., Yazinski S.A., Liu H., Darfler J., Peters R.M., Weiss R.S. Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol. Cell. Biol. 2007;27:2189–2201. doi: 10.1128/MCB.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartek J., Lukas C., Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 16.Lupu F., Terwilliger J.D., Lee K., Segre G.V., Efstratiadis A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 2001;229:141–162. doi: 10.1006/dbio.2000.9975. [DOI] [PubMed] [Google Scholar]
- 17.Niedernhofer L.J. Tissue-specific accelerated aging in nucleotide excision repair deficiency. Mech. Ageing Dev. 2008;129:408–415. doi: 10.1016/j.mad.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rypens F., Dubois J., Garel L., Fournet J.C., Michaud J.L., Grignon A. Obstetric US: watch the fetal hands. Radiographics. 2006;26:811–832. doi: 10.1148/rg.263055113. [DOI] [PubMed] [Google Scholar]
- 19.Shima N., Hartford S.A., Duffy T., Wilson L.A., Schimenti K.J., Schimenti J.C. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss R.S., Matsuoka S., Elledge S.J., Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Curr. Biol. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y., Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes. Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 23.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 24.Schubert R., Schmitz N., Pietzner J., Tandi C., Theisen A., Dresel R., Christmann M., Zielen S. Growth hormone supplementation increased latency to tumourigenesis in Atm-deficient mice. Growth Factors. 2009;27:265–273. doi: 10.1080/08977190903112663. [DOI] [PubMed] [Google Scholar]
- 25.van de Ven M., Andressoo J.O., Holcomb V.B., von Lindern M., Jong W.M., De Zeeuw C.I., Suh Y., Hasty P., Hoeijmakers J.H., van der Horst G. T., et al. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2006;2:e192. doi: 10.1371/journal.pgen.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 27.Kjaer I., Hansen N., Becktor K.B., Birkebaek N., Balslev T. Craniofacial morphology, dentition, and skeletal maturity in four siblings with Seckel syndrome. Cleft Palate Craniofac. J. 2001;38:645–651. doi: 10.1597/1545-1569_2001_038_0645_cmdasm_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 28.Murga M., Bunting S., Montana M.F., Soria R., Mulero F., Canamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Dosari M.S., Shaheen R., Colak D., Alkuraya F.S. Novel CENPJ mutation causes Seckel syndrome. J. Med. Genet. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- 30.Kalay E., Yigit G., Aslan Y., Brown K.E., Pohl E., Bicknell L.S., Kayserili H., Li Y., Tuysuz B., Nurnberg G., et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch A., Thiel C.T., Schindler D., Wick U., Crow Y.J., Ekici A.B., van Essen A.J., Goecke T.O., Al-Gazali L., Chrzanowska K.H., et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 32.Griffith E., Walker S., Martin C.A., Vagnarelli P., Stiff T., Vernay B., Al Sanna N., Saggar A., Hamel B., Earnshaw W.C., et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klingseisen A., Jackson A.P. Mechanisms and pathways of growth failure in primordial dwarfism. Genes. Dev. 2011;25:2011–2024. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lui J.C., Baron J. Mechanisms limiting body growth in mammals. Endocr. Rev. 2011;32:422–440. doi: 10.1210/er.2011-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodship J., Gill H., Carter J., Jackson A., Splitt M., Wright M. Autozygosity mapping of a seckel syndrome locus to chromosome 3q22. 1-q24. Am. J. Hum. Genet. 2000;67:498–503. doi: 10.1086/303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Digweed M., Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst.) 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Osborne L.R. Williams-Beuren syndrome: unraveling the mysteries of a microdeletion disorder. Mol. Genet. Metab. 1999;67:1–10. doi: 10.1006/mgme.1999.2844. [DOI] [PubMed] [Google Scholar]
- 38.De Kerviler E., Guermazi A., Zagdanski A.M., Gluckman E., Frija J. The clinical and radiological features of Fanconi's anaemia. Clin. Radiol. 2000;55:340–345. doi: 10.1053/crad.2000.0445. [DOI] [PubMed] [Google Scholar]
- 39.Manning M.A., Eugene Hoyme H. Fetal alcohol spectrum disorders: a practical clinical approach to diagnosis. Neurosci. Biobehav. Rev. 2007;31:230–238. doi: 10.1016/j.neubiorev.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Holmes L.B., Kleiner B.C., Leppig K.A., Cann C.I., Munoz A., Polk B.F. Predictive value of minor anomalies: II. Use in cohort studies to identify teratogens. Teratology. 1987;36:291–297. doi: 10.1002/tera.1420360304. [DOI] [PubMed] [Google Scholar]
- 41.Jones N.C., Lynn M.L., Gaudenz K., Sakai D., Aoto K., Rey J.P., Glynn E.F., Ellington L., Du C., Dixon J., et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu M., Weiss R.S. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol. Biol. Cell. 2007;18:1044–1055. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callen E., Jankovic M., Wong N., Zha S., Chen H.T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A., et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francia S., Weiss R.S., Hande M.P., Freire R., d'Adda di Fagagna F. Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr. Biol. 2006;16:1551–1558. doi: 10.1016/j.cub.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 45.Levitt P.S., Liu H., Manning C., Weiss R.S. Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics. 2005;86:212–224. doi: 10.1016/j.ygeno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Yazinski S.A., Westcott P.M., Ong K., Pinkas J., Peters R.M., Weiss R.S. Dual inactivation of Hus1 and p53 in the mouse mammary gland results in accumulation of damaged cells and impaired tissue regeneration. Proc. Natl Acad. Sci. USA. 2009;106:21282–21287. doi: 10.1073/pnas.0904965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meek K., Dang V., Lees-Miller S.P. DNA-PK: the means to justify the ends? Adv. Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 48.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 49.Navadgi-Patil V.M., Burgers P.M. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst.) 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 51.Reaper P.M., Griffiths M.R., Long J.M., Charrier J.D., Maccormick S., Charlton P.A., Golec J.M., Pollard J.R. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 52.Rosset A., Spadola L., Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging. 2004;17:205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaufman M.H. The Atlas of Mouse Development. London/San Diego: Academic Press; 1992. [Google Scholar]
- 54.Todaro G.J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dean F., Tea M., Fenster S. Real-time PCR. Curr. Protoc. Essent. Lab. Tech. 2008 UNIT 10.3, 10.3.1-10.3.34. [Google Scholar]
- 56.Harlow E., Lane D. Lysing tissue-culture cells for immunoprecipitation. Cold Spring Harb. Protoc. 2006 doi: 10.1101/pdb.prot4531. doi:10.1101/pdb.prot4531. [DOI] [PubMed] [Google Scholar]
- 57.Achari Y., Lees-Miller S.P. Detection of DNA-dependent protein kinase in extracts from human and rodent cells. Methods Mol. Biol. 2000;99:85–97. doi: 10.1385/1-59259-054-3:85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.