Abstract
Spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS) are among the most common motor neuron diseases to afflict the human population. A deficiency of the survival of motor neuron (SMN) protein causes SMA and is also reported to be an exacerbating factor in the development of ALS. However, pathways linking the two diseases have yet to be defined and it is not clear precisely how the pathology of ALS is aggravated by reduced SMN or whether mutant proteins underlying familial forms of ALS interfere with SMN-related biochemical pathways to exacerbate the neurodegenerative process. In this study, we show that mutant superoxide dismutase-1 (SOD1), a cause of familial ALS, profoundly alters the sub-cellular localization of the SMN protein, preventing the formation of nuclear ‘gems’ by disrupting the recruitment of the protein to Cajal bodies. Overexpressing the SMN protein in mutant SOD1 mice, a model of familial ALS, alleviates this phenomenon, most likely in a cell-autonomous manner, and significantly mitigates the loss of motor neurons in the spinal cord and in culture dishes. In the mice, the onset of the neuromuscular phenotype is delayed and motor function enhanced, suggestive of a therapeutic benefit for ALS patients treated with agents that augment the SMN protein. Nevertheless, this finding is tempered by an inability to prolong survival, a limitation most likely imposed by the inexorable denervation that characterizes ALS and eventually disrupts the neuromuscular synapses even in the presence of increased SMN.
INTRODUCTION
Muscle wasting and weakness, due to the selective degeneration of spinal motor neurons, are hallmarks of both spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS) (1,2). These phenotypic similarities have prompted investigations into possible links between the two paralytic disorders (3–7). However, SMA is caused by homozygous mutations in the Survival of Motor Neuron 1 (SMN1) gene that lead to reduced levels of the SMN protein (8–10), whereas most instances of ALS are sporadic or, when inherited, caused by mutations in a variety of seemingly disparate genes (reviewed in 11). Molecular mechanisms that may be shared between the two diseases are therefore not readily apparent. In this regard, ALS model mice expressing mutant superoxide dismutase 1 (mSOD1) have been informative. For instance, it was shown that transgenic mSOD1 mice express lower than normal SMN levels and exhibit a more pronounced paralytic phenotype in the absence of a copy of the murine Smn gene (12). Studies on humans have demonstrated a greater susceptibility to ALS when individuals possess genotypes predicted to express lower than normal levels of SMN (4). However, precise SMN levels in ALS patients have never been determined, making it uncertain whether there is indeed a correlation between reduced SMN and increased susceptibility to the disease. In the mSOD1 mice analyzed by Turner et al. (12), considering the advanced stage of the disease at which low SMN levels became apparent, the relatively modest (∼25%) reduction in protein and an early rather than late requirement for it in causing disease (13,14), it is not clear precisely how reduced SMN might exacerbate the ALS phenotype.
The SMN protein is ubiquitously expressed and partitions to both the cytoplasm and the nucleus—observations consistent with a housekeeping role in uridine rich small nuclear ribonucleoprotein (U snRNP) biogenesis (15 and references therein). In the nucleus, it concentrates in particles termed gems that frequently co-localize with Cajal bodies, sites of RNP maturation and small nuclear RNA modification (16–20). Co-localization of the two structures is reportedly mediated by coilin, a marker of Cajal bodies (21). Reduced SMN in SMA patients and mouse models generally results in a loss of gems, but not necessarily of Cajal bodies (9,10,17).
In this study, we extend the findings of reduced SMN accompanying mSOD1 expression and report that the sub-cellular localization of the protein is profoundly altered. In the presence of mSOD1, nuclear gems containing SMN are dramatically reduced, even in pre-symptomatic ALS model mice. In contrast, coilin-positive Cajal bodies within which SMN normally resides persist. The loss of SMN within these nuclear structures is explained, at least in part, by a cell-autonomous, mSOD1-mediated disruption of the interaction between coilin and SMN. Mutant SOD1 expressing astrocytes which were previously reported to cause neurodegeneration (22–24) did not contribute to the loss of motor neuronal gems. However, we observed that overexpressing the SMN protein in spinal motor neurons conferred greater resistance to the degenerative effects of mSOD1 astrocytes. Overexpressing the protein in mSOD1 mice delayed the loss of gems, protected against the characteristic, aggressive loss of spinal motor neurons and extended the disease-free period. Yet, neither the eventual denervation of skeletal muscles nor the survival of mSOD1 mice was prolonged by enhanced SMN expression. Given our collective results, we propose that (i) a combination of mSOD1 and SMN paucity exacerbates motor neuron degeneration, perhaps through disruptions in the proper localization of SMN, (ii) overexpressing SMN appears to preferentially affect the molecular machinery underlying the destruction and the loss of motor neuron cell bodies rather than nerve terminals and (iii) therapeutic agents capable of simultaneously augmenting motor neuronal SMN expression and preserving the neuromuscular synapse will be effective in treating the ALS phenotype in model mice and human patients.
RESULTS
Mutant SOD1 disrupts the formation of nuclear gems
Motor neurons are especially vulnerable to the disease processes in SMA and ALS, raising the possibility of common mechanisms underlying the pathology of the two diseases. Furthermore, reduced SMN is reported to increase susceptibility to ALS, perhaps a consequence of subtle disruptions in SMN function brought about by the pathology of the disease even in the presence of a normal complement of SMN genes. To explore this possibility, we began by assessing the levels and localization of the SMN protein in the motor neurons of a transgenic mouse model of familial ALS that harbors an SOD1G86R mutation. Such mutants exhibit severe spinal motor neuron degeneration accompanied by progressive weakness of the skeletal muscles that culminates in premature death at ∼4 months of age (25). The total amount of SMN protein in the spinal motor neurons of symptomatic [post-natal day 102-127 (PND102-127)] mice, assessed immunohistochemically, appeared unchanged and significantly greater than that in motor neurons of Δ7 SMA mutants (26) which represent a mouse model of severe SMA (average relative SMN intensity in arbitrary units ± SEM—Ntg: 100 ± 6.01; SOD1G86R: 79.3 ± 7.9; Δ7 SMA: 30.73 ± 13.03; n > 30 cells; SOD1G86R versus Δ7 SMA, P < 0.01; SOD1G86R versus Ntg, P > 0.05; one-way ANOVA; also see Fig. 1A). However, the nuclei of the motor neurons were virtually devoid (>95% loss) of SMN staining gems, similar to those of Δ7 SMA mutants (Fig. 1A and B). To further investigate this result, we examined the spinal motor neurons of pre-symptomatic (PND44–50) SOD1G86R mice. Interestingly, gem numbers were significantly greater than those of symptomatic cohorts, but nonetheless fewer (∼70% of wild-type) than those in the motor neurons of age-matched non-transgenic controls (Fig. 1B). In PND14 SOD1G86R mutants, gem numbers were no different from those of age-matched controls, suggesting a progressive depletion of these structures as the disease evolves. To determine whether the depletion of gems was specific to the SOD1G86R transgene, spinal motor neurons from 3–4-month-old transgenic animals expressing either an SOD1G93A mutation or a wild-type SOD1 cDNA construct (27) were examined. The mutant but not wild-type SOD1 transgene caused a depletion of gems (Fig. 1B), indicating that the effect is neither unique to the G86R transgene nor simply a consequence of overexpressing SOD1. Additionally, the result suggests that enzymatically active (G93A) as well as inactive (G86R) mutant SOD1 protein can bring about a loss of gems.
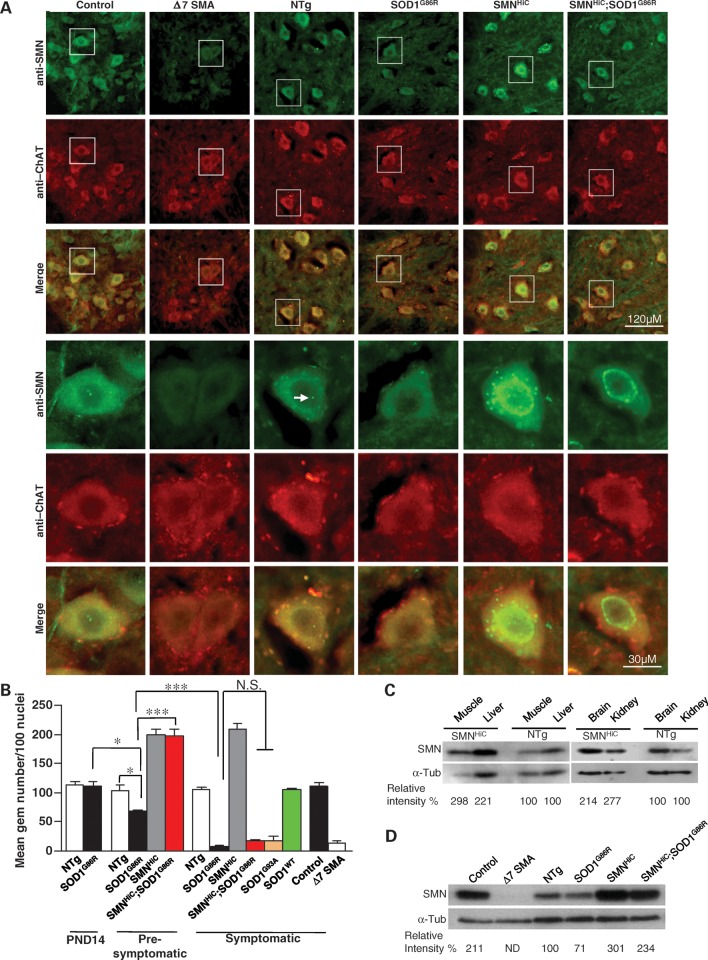
Figure 1.
Mutant SOD1 disrupts the localization of SMN to nuclear gems. (A) Immunostaining of thoracic motor neurons in the anterior horns of the spinal cords of end-stage mSOD1 mice and age-matched controls depicting cells devoid of nuclear gems (arrow) in the mutants. Included is a section from a Δ7 SMA mouse known to express very low SMN and harbor few, if any, nuclear gems. Motor neurons were identified by co-staining with an antibody against ChAT. (B) Quantification of gems in mSOD1 mice and relevant controls at pre- and post-symptomatic stages, demonstrating a delay in the loss of these structures in the presence of high SMN protein. *P < 0.05 (one-way ANOVA); ***P < 0.001 (t-test); N.S., not significant (one-way ANOVA); n ≥ 3 mice in each case. (C) SMNHiC mice harboring eight SMN2 genes in addition to the endogenous murine Smn allele express 2–3-fold as much SMN protein as non-transgenic littermates as assessed on western blots. (D) Spinal cord tissue from end-stage mSOD1 mice exhibits a modest reduction of SMN protein in a western blot assay. ND, not determined. Note: The genotype of the age-matched control for the Δ7 SMA mutant is SMN2;Δ7;Smn+/+.
Gem numbers generally correlate with SMN levels (9). We therefore asked whether overexpressing the SMN protein in the SOD1G86R mutants might restore gems to their motor neurons. To do so, we bred mice harboring eight copies of a human SMN2 transgene to the SOD1G86R mutants. Eight copy SMN2 transgenic mice have been backcrossed over six successive generations to the same strain, FVB/N, on which the SOD1G86R mutation resides, and the mice express ∼2.5 times as much SMN protein as non-transgenic mice in all tissues examined (Fig. 1C; 28). Phenotypically, the mice are no different from wild-type animals and will be referred to henceforth as SMNHigh Copy (SMNHiC) animals. Quantification of gems in the spinal motor neurons of the animals revealed twice as many as those in non-transgenic animals (Fig. 1A and B), an observation consistent with high levels of total SMN proteins which are also known to promote the appearance of supernumerary bodies within and outside the nucleus (29). Expression of the SMNHiC transgene in SOD1G86R mutants appeared to restore the number of gems to those observed in mice expressing SMNHiC alone, when the animals were examined at PND44–50. However, an examination of motor neurons in PND102–127 double-transgenic mice once again revealed an almost complete loss of gems, resulting in numbers similar to those in end-stage disease SOD1G86R mutants (Fig. 1A and B).
Since a depletion of motor neuronal gems generally reflects a loss of total SMN protein (9,10), we quantified the protein in spinal cord tissue of end-stage disease SOD1G86R mutants and relevant controls, including SMA mice. As previously reported (26,30), spinal cord tissue from Δ7 SMA mice contained very little SMN protein (Fig. 1D), reflective of the reduced immunostaining of their spinal motor neurons. On the other hand, despite an almost complete absence of motor neuronal gems, spinal cord tissue from end-stage SOD1G86R mice displayed only a modest ∼30% reduction of the SMN protein. A similar drop in total protein level was observed in SMNHiC;SOD1G86R spinal cords compared with levels in mice expressing SMNHiC alone. These results are similar to those reported by Turner et al. (12) in a study that utilized SOD1G93A mutants. Nevertheless, the reduction in total SMN protein in end-stage SOD1G86R mice still exceeded that observed in heterozygous Smn+/− mice which do not exhibit the loss of motor neuronal gems (31). Viewed in light of these data, our results suggest that the dramatic loss of nuclear gems in SOD1 mutants is likely to be the result of a targeting defect rather than an overall depletion of the SMN protein. Such a defect applies to other cell types too, although to a lesser extent, e.g. Purkinje cells of SOD1G86R mutants displayed half as many gems as those of wild-type littermates (Supplementary Material, Fig. S1A). Cultured fibroblasts from SOD1G86R and SMNHiC;SOD1G86R mice also contained significantly fewer gems than their wild-type and SMNHiC littermates, respectively (Supplementary Material, Fig. S1B and C). Viewed in aggregate, the results suggest that mSOD1 protein profoundly affects nuclear localization of the SMN protein. The depletion of gems is progressive, akin to the evolution of the disease but preceding the onset of neuromuscular symptoms. Overexpressing SMN in the mSOD1 mice delays but does not halt the loss of gems, implying a dominant, deleterious effect of the mutant protein and disease process on proper SMN localization.
Reduced binding of SMN to coilin in the presence of mSOD1 protein
To investigate the cause of reduced gems in the SOD1 mouse mutants, we began by determining whether mSOD1 also depleted these structures in cultured cells. Accordingly, we transfected an NSC-34 motor neuron-like cell line with either an SOD1G86R construct or its wild-type counterpart (SOD1WT) cloned into vectors containing an enhanced yellow fluorescent protein (eYFP). Whereas gems were easily detected in cells 2 and 4 days following transfection with the wild-type SOD1 construct, a marked depletion of these nuclear structures was observed in cells expressing the mutant protein (Fig. 2A and B). Mutant SOD1 is wont to form inclusion bodies in vitro and in the motor neurons of model mice (32,33). We observed such inclusions in the SOD1G86R transfected cells. Importantly, cells containing the inclusions were no more likely to be depleted of gems than cells without them, suggesting that loss is not merely a consequence of aggregate formation (Fig. 2B).
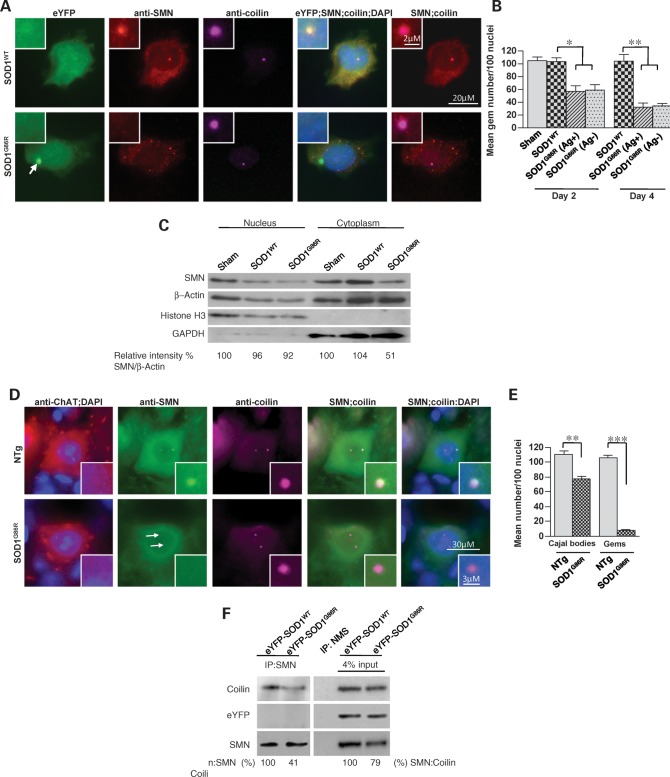
Figure 2.
Mutant SOD1 effects gem loss by weakening the binding affinity of coilin for SMN. (A) Immunohistochemistry of NSC34 neuron-like cells transfected with mutant SOD1 reveals an inability of the cells to localize SMN to coilin-positive Cajal bodies (the arrow denotes mSOD1 inclusion). (B) Quantification of gems in NSC34 cells expressing mutant or wild-type SOD1, 2 or 4 days following transfection. Cells with mSOD1-induced aggregates (Ag+) appear to contain no fewer gems than cells without them (Ag−). (C) Western blot analysis of nuclear and cytoplasmic fractions of NSC34 cells transfected with either a wild-type or mSOD1 construct. (D) Immunostaining of representative spinal motor neurons from SOD1G86R mutant mice and non-transgenic controls demonstrating a loss of SMN within coilin-positive Cajal bodies. Arrows in panels (stained with the SMN antibody) indicate where the gems ought to have localized. (E) Quantification of SMN and coilin-positive nuclear bodies in motor neurons from mutant mice suggests a much greater disruptive effect of mSOD1 on the formation of gems than on Cajal bodies. (F) Western blots following co-immunoprecipitation experiments demonstrate a lower (∼40%) affinity of SMN for coilin in the presence of mSOD1 protein. Neither mutant nor wild-type SOD1 binds SMN as assessed by an inability to detect the eYFP protein. Note: *P < 0.05, **P < 0.01, ***P < 0.001, t-test. Sham refers to transfections without DNA. NMS, normal mouse serum.
We next determined whether the loss of gems was a consequence of a perturbation in the partitioning of SMN between cytoplasm and nucleus. Cells transfected with an empty vector, SOD1G86R or SOD1WT, were lysed, differentially centrifuged to separate nuclear and cytoplasmic fractions, and SMN levels in them were quantified by western blot analysis. In contrast to a previous study (12), mSOD1 did not re-localize nuclear SMN to the cytoplasm, resulting in an increase in cytoplasmic SMN. On the contrary, we found a decrease in the cytoplasmic SMN fraction, whereas nuclear SMN appeared unchanged (Fig. 2C), suggesting that a depletion of gems is more likely a result of changes in the micro-environment of the nucleus. Consistent with this conclusion, nuclear snRNP levels, as assessed by the intensity of Sm protein staining, did not differ between motor neurons of SOD1G86R or SMNHiC;SOD1G86R and age-matched controls (Supplementary Material, Fig. S2A and B). However, the snRNPs failed to localize to Cajal bodies (Supplementary Material, Fig. S2C and D), suggesting that despite entering the nucleus, they are not recruited to these sub-nuclear structures.
Because SMN complexed with snRNPs co-localizes with Cajal bodies, we also determined whether the latter structures were simultaneously lost in the presence of mSOD1. Although there was a decrease in the number of Cajal bodies in SOD1G86R-transfected cells as well as mutant mice, the loss was modest relative to the depletion of gems (Fig. 2D and E and Supplementary Material, Fig. S2E). This suggests that Cajal bodies are able to form in the presence of mSOD1, although not to the same extent as in wild-type cells, but cannot efficiently recruit the SMN–snRNP complex. The protein coilin is reported to bind SMN and mediate its recruitment to Cajal bodies (21). We therefore determined whether this interaction is altered in the presence of mSOD1. NSC-34 cells were transfected with either SOD1G86R or SOD1WT, extracts immunoprecipitated with anti-SMN sera and levels of bound protein assessed. As previously reported, coilin did bind SMN. However, in the presence of mSOD1, this interaction was markedly reduced (Fig. 2F). Since neither wild-type nor mSOD1 bound SMN, we surmise that the mutant protein indirectly disrupts the interaction between coilin and SMN. Collectively, the results explain, at least in part, the persistence of Cajal bodies in conjunction with a depletion of gems in SOD1G86R mice.
A cell-autonomous protective effect of enhanced SMN expression
The neurodegenerative effects of mSOD1 are a combination of cell-autonomous and non-cell-autonomous processes (22–24,34). Given our observation that NSC-34 cells expressing an SOD1G86R mutation bear fewer nuclear gems, we can conclude that the depletion of these structures by mSOD1 involves a cell-autonomous component. To investigate whether non-cell-autonomous mechanisms also contribute to this process, we examined the effects of mSOD1 expressing astrocytes on wild-type or SMNHiC motor neurons. Since the G93A mutation behaved similarly to G86R in depleting motor neuronal gems and because we had previously optimized conditions using astrocytes from SOD1G93A mice, we used the former mutation for these experiments. Spinal motor neurons were derived from E12.5 day embryos that were either wild-type or transgenic for SMNHiC. Both sets of embryos were also transgenic for the HLXB9::eGFP construct, which allows for the identification of spinal motor neurons by virtue of being selectively expressed in these cells (35). Following the isolation of the motor neurons, we cultured them on astrocyte monolayers (AML) that were either mutant (SOD1G93A) or non-transgenic. As previously reported, significantly fewer wild-type motor neurons survived on SOD1G93A astrocytes than on non-transgenic ones when the cells were assessed 7 and 13 days after plating (Fig. 3A and B). We found this to be true of motor neurons overexpressing the SMN protein too. However, relative to motor neurons expressing normal levels of the SMN protein, significantly greater numbers of the SMNHiC motor neurons survived the toxic effects of mSOD1 astrocytes. This difference disappeared when the wild-type and SMNHiC motor neurons were cultured on normal astrocytes, suggesting that the protective effect becomes evident only when the cells are subject to a toxic environment. These results confirm our in vivo findings and serve to emphasize the neuroprotective effects of overexpressing SMN within motor neurons subjected to mSOD1 toxicity.
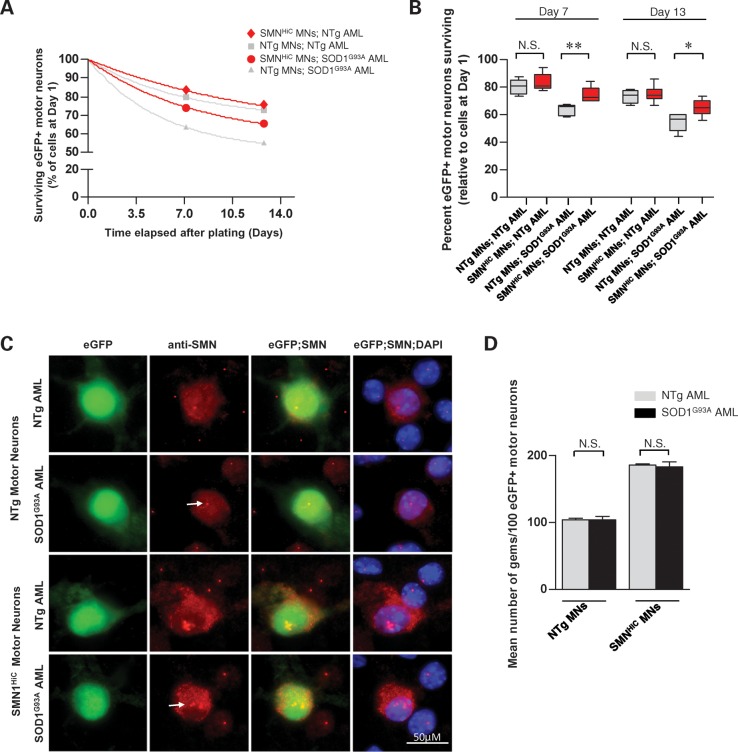
Figure 3.
Enhanced SMN expression protects against the toxic effects of astrocytes expressing mSOD1 protein. (A) The decay in relative numbers of normal (NTg) or SMN overexpressing (SMNHiC) primary motor neurons plated on AML derived from either mutant SOD1G93A mice or wild-type controls. Neuron numbers are relative to those present 1 day after plating the respective cells. (B) Quantification of motor neuron numbers in the various co-cultures 7 and 13 days after plating the cells reveals a protective effect of overexpressing SMN in neurons exposed to mSOD1-bearing astrocytes. Note: *P< 0.05, **P < 0.01, one-way ANOVA. Values were obtained from at least three independent experiments performed in triplicate. (C) Representative HLXB9:eGFP-positive motor neurons (MN) examined 7 days after plating continue to form gems (arrows) despite being exposed to astrocytes expressing mSOD1 protein. (D) Quantification of gems in HLXB9:eGFP-positive motor neurons reveals equivalent numbers of these structures in the presence or absence of astrocytes bearing a mutant SOD1 transgene. P-values were calculated by the t-test and found to be >0.5 in each case. N.S., not significant.
Is it possible that augmenting SMN in motor neurons provides a greater generic resistance to these neurons irrespective of the nature of the insults? To address this question, we assessed whether enhanced SMN expression might also protect against the degenerative effects of withdrawing neurotrophic factor support from cultured motor neurons. We found that the SMNHiC transgene conferred no protection to motor neurons against the removal of neurotrophic factors (Supplementary Material, Fig. S3). Neurotrophic factor deprivation also failed to alter the nuclear localization of gems (data not shown). Collectively, these results suggest that the benefit we observe appears to be specific to mSOD1 protein.
We next determined whether the toxic effects of mutant astrocytes also caused a depletion of motor neuronal gems. Considering their higher SMN levels, we were not surprised to see about twice the number of gems in the SMNHiC-expressing motor neurons as in their wild-type counterparts. However, neither group exhibited a loss of gems on mutant versus wild-type astrocytes (Fig. 3C and D). Culturing mSOD1 motor neurons on mSOD1 or wild-type astrocytes produced similar results (Supplementary Material, Fig. S4). This suggests that the loss of gems in the motor neurons of mSOD1 mice is unlikely to be mediated by surrounding glia but is instead entirely a consequence of mutant protein within the neurons. The results also suggest that a depletion of SMN in gems does not on its own cause cell death, but could exacerbate the toxic effect of mSOD1.
Enhanced SMN expression extends the disease-free period in transgenic mSOD1 mice
Considering reports of an exacerbated disease phenotype in mSOD1 mice expressing reduced SMN (12) and our own results demonstrating (i) a perturbation of nuclear SMN by mSOD1, and (ii) at least a partial mitigation of this effect by overexpressing the SMN protein, we sought to determine the phenotypic consequence of overexpressing SMN on the ALS-like phenotype of SOD1G86R mutant mice. We therefore examined the onset of disease symptoms, motor behavior and survival outcome of SOD1G86R mice with or without the SMNHiC transgene. To compare symptom onset, animals were assessed for their motor performance in the vertical pole and rotarod tests. Whereas the average age of SOD1G86R mice at which the disease phenotype, based upon the vertical pole test, became apparent was 56 days, the average age at which the disease appeared in SMNHiC;SOD1G86R was significantly delayed by 26% to ∼70 days (Fig. 4A). Similarly, the onset of motor deficits based on the rotarod test was delayed by 20% from 61 days in SOD1G86R mice to 73 days in SMNHiC;SOD1G86R mutants. However, there was a difference in neither the average age at which we recorded a ≥5% decrease in body weight as the disease progressed nor the survival of the two cohorts of mice (Fig. 4B and C). Indeed, end-stage disease SMNHiC;SOD1G86R mutants exhibited many of the phenotypic characteristics of previously described SOD1G86R mice and appeared readily distinguishable from mice expressing the SMNHiC transgene alone (Fig. 4D). The results described above are summarized in Table 1 and demonstrate that although a high level of the SMN protein delays the onset of the ALS-like phenotype caused by mSOD1, it does not prolong the survival of mSOD1 mice.
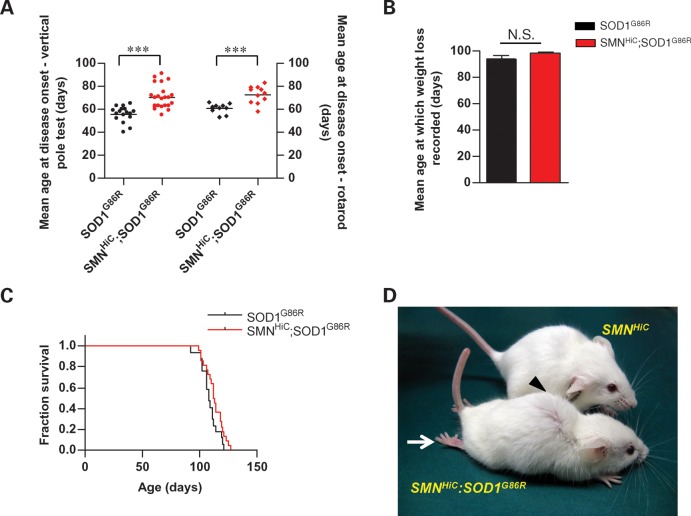
Figure 4.
A delay in the onset of the ALS phenotype in mutant SOD1 mice overexpressing the SMN protein. (A) Mutant SOD1G86R mice co-expressing the SMNHiC transgene exhibit a delay in disease onset measured by the vertical pole and rotarod tests, but (B) lose weight equally rapidly and (C) succumb to the disease at the same time as animals expressing the mutant SOD1 transgene alone. (D) SOD1G86R mice develop a severe neuromuscular disease phenotype (the arrow indicates hindlimb paralysis; the arrowhead indicates kyphosis of spine) by ∼3.5 months of age despite expressing higher than normal levels of SMN protein. Note: 17 or more mice were used in the phenotypic studies. ***P < 0.001, t-test; differences in survival were determined to be insignificant by the log-rank test.
Table 1.
Effect of overexpressing SMN on the mSOD1 phenotype
| Outcome measure | Mouse genotype | Survival/day of symptom onset |
|---|---|---|
| Vertical pole test | SOD1G86R | 55.7 ± 6.8 |
| SMNHiC;SOD1G86R | 70.3 ± 10.1 | |
| Rotarod test | SOD1G86R | 60.7 ± 4.1 |
| SMNHiC;SOD1G86R | 72.6 ± 7.5 | |
| Loss of body weight | SOD1G86R | 94.0 ± 8.2 |
| SMNHiC;SOD1G86R | 97.8 ± 7.2 | |
| Survival | SOD1G86R | 108.7 ± 7.4 |
| SMNHiC;SOD1G86R | 112.9 ± 8.0 |
Enhanced SMN protein expression protects against mSOD1-mediated damage to spinal motor neurons
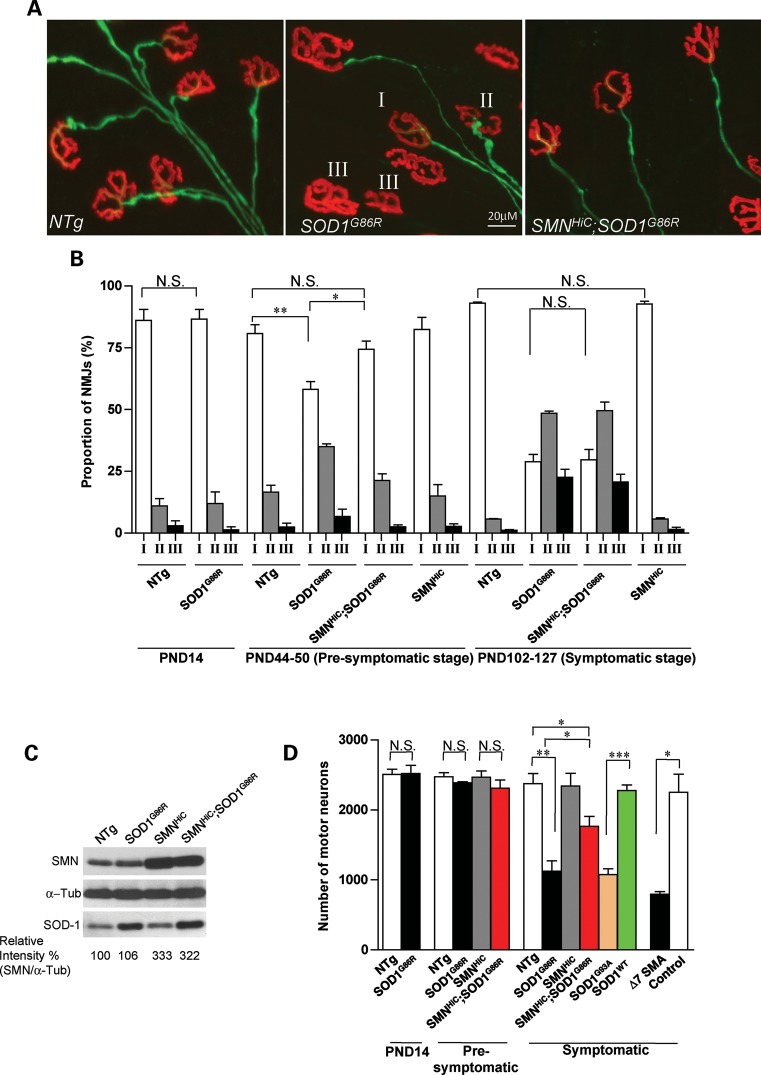
To investigate the cellular basis of a delayed onset of disease symptoms in SOD1 mutants overexpressing SMN, we examined the pathology of motor neuronal cell bodies and their neuromuscular junctions (NMJs) in SOD1G86R and SMNHiC;SOD1G86R mice. Since the neuromuscular disease in ALS model mice is reported to first affect the distal axon (36), we began by inspecting the NMJs of PND14, adult pre-symptomatic (PND44–50) and symptomatic (PND102–127) animals. We chose to focus on the intercostals, paralysis of which leads to respiratory failure and death in ALS. Tissue from age-matched non-transgenic and SMNHiC animals served as controls. Synapses were defined as fully innervated, partially innervated or fully denervated (Fig. 5A and see Materials and Methods). PND14 SOD1G86R and non-transgenic controls displayed no difference in NMJ profiles. However, a quantification of NMJ types in PND44–50 pre-symptomatic SOD1G86R mice revealed greater numbers of denervated NMJs and concomitantly fewer innervated synapses (Fig. 5B). Overexpressing SMN in SOD1G86R mutants at this age protected the synapses such that the number of fully innervated NMJs did not differ between SMNHiC;SOD1G86R and non-transgenic animals. Importantly, no difference in NMJ profile between SMNHiC mice and non-transgenic littermates was observed, suggesting that rescue was not simply due to an SMNHiC-specific transgene effect that could endow animals harboring it with greater than normal numbers of innervated NMJs. A rescue could also be accounted for by an SMNHiC transgene-mediated down-regulation of mSOD1 or, conversely, an mSOD1-mediated increase in SMN expression. Western blot analysis of SOD1 and SMN in spinal cord tissue of PND50 SOD1G86R, SMNHiC;SOD1G86R and age-matched control mice indicated that this was not the case (Fig. 5C). In contrast to the NMJs of pre-symptomatic PND44–50 SOD1G86R mice which were protected by SMN overexpression and were morphologically indistinguishable from those of non-transgenic controls, the NMJs of symptomatic PND102–127 SMNHiC;SOD1G86R mutants were denervated to the same extent as those of age-matched SOD1G86R mice. Together, these results suggest that enhanced SMN expression delays axonal retraction from the skeletal muscle and provides a cellular basis for the delay in the onset of the disease phenotype observed in SMNHiC;SOD1G86R mice.
Figure 5.
SMN protects the motor unit in mice against mutant SOD1-mediated neurodegeneration. (A) The three classes (fully innervated—class I, partially innervated—class II and fully denervated—class III) of neuromuscular synapses were used to construct and compare distal motor unit profiles of mSOD1 mice and control littermates. Also depicted are fully inervated NMJs from SMNHiC;SOD1G86R mice. (B) Quantification of synapses and a comparison of their profiles reveal a protective effect of overexpressing SMN in early, pre-symptomatic mSOD1 mice. In contrast, the protective effect was lost in symptomatic mice. As expected, no difference was detected between post-natal day 14 (PND14) mutants and controls. (C) Western blot analysis of spinal cord tissue from pre-symptomatic mice indicates that SMN overexpression does not suppress mSOD1 levels or vice versa. (D) Quantification of spinal motor neurons attesting to the protective effect of enhanced SMN expression on the motor units of mSOD1 mice. Quantification was carried out on at least three animals of each genotype; *P< 0.05, **P< 0.01, ***P < 0.001, t-test. Note: Genotype for the control (D) is SMN2;Δ7;Smn+/+. N.S., not significant.
Having discovered a protective effect of enhanced SMN expression on the NMJs of the motor neurons of ALS mice, we were interested in determining whether the cell bodies within the spinal cord were similarly protected. We therefore examined spinal motor neuron cell bodies innervating the intercostals of the mutants and relevant controls. We found that the number of motor neurons in PND14 SOD1G86R mice was similar to those in control mice (Fig. 5D). Additionally and consistent with a degenerative process that begins in the distal axon, no cell loss was apparent in either pre-symptomatic (PND44–50) SMNHiC;SOD1G86Ror SOD1G86R animals. However, symptomatic SOD1G86R mice were found to have lost more than half (53 ± 6%) the number of motor neurons present in age-matched non-transgenic controls, similar to the loss observed in end-stage Δ7 SMA mice (26). Remarkably, but in keeping with the protective effect of the SMN protein, SMNHiC;SOD1G86R mice lost significantly fewer (25 ± 6%) choline acetyltransferase (ChAT)-positive motor neurons than mutants without the SMNHiC transgene. This was determined to not simply be because of increased numbers of these cells within the spinal cords of SMNHiC. To rule out the possibility that cell loss in SOD1G86R and SMNHiC;SOD1G86R mice was merely because of abnormally high levels of SOD1 expression, counts were also carried out on mice overexpressing wild-type and a G93A mutation in the SOD1 gene (27). As expected, only the latter exhibited obvious evidence of neurodegeneration. Collectively, these results are indicative of a marked neuroprotective effect of enhanced SMN expression.
DISCUSSION
The selective degeneration of a cell type due to the loss or dysfunction of a protein reported to play a housekeeping function inevitably raises questions about the precise biochemical pathways underlying the degeneration. A similar degeneration of that cell type triggered by mutant forms of one or more additional proteins could be indicative of common, cell-specific pathways shared by the different molecules in maintaining cellular viability and function. In this study, we have probed possible links between two proteins, SMN and SOD1, both of which underlie motor neuron disorders. The study stems, in part, from previous reports of the exacerbation of the neurodegenerative ALS phenotype by a relative paucity of the SMN protein (4,12), and suggestions from association studies that one or both SMN genes may be linked to sporadic ALS (3,5–7,37). However, a drawback of the association studies, given the limited number of polymorphic markers used, is the inability to conclude precisely whether either of the SMN genes or a linked locus within the proximity of the genes is a risk factor for ALS. Our study addresses this question directly by investigating, through the use of a high copy number SMN2 transgene, the effects of SMN on the ALS phenotype. Our main findings are summarized as follows. First, we have demonstrated that mSOD1 strikingly alters the sub-cellular localization of the SMN protein. Nuclear gems are virtually eliminated in the presence of the mutant protein, not as a consequence of a dramatic reduction of SMN or disruption of Cajal body formation, but more likely because of the reduced capacity for coilin to bind and recruit SMN to Cajal bodies. Second, we have shown that overexpressing SMN not only delays the loss of gems but also protects against an ALS-like phenotype observed in a mouse model of the disease. However, the mitigation of the phenotype is limited to the onset of disease symptoms and does not prolong survival. Third, we have shown that the protection observed is due, at least in part, to a delay in the onset of neuromuscular synapse loss and preservation of motor neuron cell bodies. An inability to extend survival is most likely a result of a failure to prevent denervation in late stages of the disease. Consistent with these in vivo findings, we found SMN to protect motor neurons in culture against mSOD1-mediated toxicity. Finally, we have demonstrated that the disruption of gems by mSOD1 occurs cell-autonomously, and is not brought about by the toxicity of neighboring glial cells such as astrocytes.
An increased susceptibility to ALS in humans and enhanced severity of the neuromuscular phenotype in model mice predicted or shown, respectively, to express reduced SMN protein is indicative of a protective effect of the protein against processes that adversely affect the motor neuron. From studies of SMA mice, the protective effect of SMN operates not only in neonates but also in adults (31) when motor neuron loss due to ALS-causing mutations becomes evident. Taking advantage of transgenic mice that overexpress SMN, we showed that the protein is indeed protective against mSOD1 protein, a known cause of motor neuron disease. However, phenotypically the effect is restricted to symptom onset and the initial manifestations of the disease. In light of our findings, this result is not overly surprising. If eventual death of ALS patients and model mice is in fact a consequence of paralysis due to denervation, then the inability to prevent this as the disease progresses would indeed preclude extending survival. The marginal, ∼15% delay in disease onset and extension of life of SOD1G93A;Bax−/− mutants in which motor neuron loss is completely blocked but muscle denervation readily apparent (38) is consistent with this line of reasoning. Our result apparently contrasts with a prior report in which reducing SMN protein by half in an SOD1G93A ALS mouse model, by generating mutants heterozygous for the murine Smn gene (SOD1G93A;Smn+/−), decreased lifespan by ∼10% relative to that of SOD1G93A;Smn+/+ littermates. The 10% difference is decidedly modest and, even if real, is not necessarily inconsistent with our result if haploinsufficiency of the Smn gene, as previously suggested, is sufficient to cause denervation at the neuromuscular synapse (30). Viewed in aggregate, the various results suggest that even if a robust increase in SMN cannot alter survival as an outcome in ALS, it does protect the motor unit albeit temporarily at the neuromuscular synapse. The protective effect on the motor neuron cell body relative to that on the NMJs is noteworthy and is an indication of the sub-cellular compartment in which the molecular mechanisms underlying mSOD1-mediated ALS begin and on which SMN preferentially acts. One caveat of this conclusion is the development of functional defects within the cell body that we did not analyze but manifest by initially affecting the morphology of distal processes such as the axon terminal. Still, our results suggest that future therapies that effectively protect the neuromuscular synapse may be combined with SMN-augmenting approaches and expected to offer real benefit to at least one class of ALS patients, provided they are identified in a timely manner.
Perhaps the most intriguing finding from our work but one whose dynamics can be correlated with the progression of the ALS phenotype is the dramatic loss, particularly in spinal motor neurons, of gems in the presence of mSOD1 protein, a finding consistent with a recent independent study (39). The precise function of nuclear gems is uncertain and they are unlikely to be required for cell survival given their absence in certain cell types (17). However, spinal motor neurons contain a particularly prominent set of these structures (9,10,17) and it is possible that their formation and presence enhances the efficiency of a yet-to-be defined function whose requirement is especially stringent in these cells. The loss of gems in the presence of mSOD1 proceeds rapidly and appears to be much less disruptive to Cajal bodies, which, for the most part, persist as foci lacking SMN and snRNPs. Moreover, gem loss seems to be an early event in the progression of the disease as it mirrors the onset of denervation and appears in pre-symptomatic mice. Interestingly, overexpressing SMN delayed both the loss of gems as well as the onset of denervation. The precise timing of these two cellular events is not clear, but it is tempting to speculate that inefficient recruitment of the SMN–snRNP complex to Cajal bodies further perturbs the health of an already fragile motor neuron, thereby accelerating its eventual demise. One possibility therefore is that overexpressing SMN on an ALS background enhances SMN-related functions, including those involving nuclear gems that may be disrupted by mutations in SOD1, thus delaying motor neuron loss and muscle denervation. A boost in overall SMN as observed in the SMNHiC mice therefore mediates the correlation between an increase in the number of gems and delayed motor neuron pathology. Whether protein increase in the SMNHiC mice enhances snRNP biogenesis, or conversely, whether the decline in SMN in mSOD1 mice reduces it is unclear from our study. However, given the modest reduction of SMN in the mSOD1 mice, the efficiency of snRNP assembly is unlikely to be lower than that of Smn+/− mice which express half the normal amounts of SMN protein. Our results indicate that mSOD1 interferes with the ability of SMN to bind coilin. A second, parallel possibility is a disruption in the interaction between SMN and WRAP53, a protein also thought to target SMN to Cajal bodies (40). However, this is unlikely as a disruption of this interaction depletes not only gems but also overall nuclear SMN, which was not observed in our experiments. Forced expression of SMN from SMNHiC promotes gem formation and therefore temporarily retards their loss even in the presence of mSOD1. Their eventual loss coupled with an inability to permanently halt the denervation or prolong the survival of ALS mice expressing the SMNHiC transgene suggests first, that even enhanced SMN expression is unable to overcome the accumulating toxic effects of mSOD1 and, second, that the disease process is very likely accelerated between pre-symptomatic and symptomatic stages in these mutants. Whether the increased expression of SMN in the SOD1 mutants also enhances a reported chaperone activity of the protein (41), thereby retarding disease onset is unclear from our experiments. However, it is quite possible that multiple mechanisms involving gem restoration as well as an increase in cytoplasmic SMN contribute to the phenotypic and cellular rescue we observe in our model systems.
The fact that mSOD1 glia do not perturb motor neuronal gems is noteworthy. On the other hand, their role in the degeneration of motor neurons via a non-cell-autonomous pathway ostensibly involving diffusible factors has been well established (22–24). Viewed together the two observations might form the basis of an argument questioning any significance ascribed to gem loss in motor neuronal death. However, it is to be noted that selective expression of mSOD1 within neurons is sufficient to cause dysfunction and disease when the protein is at sufficiently high levels (34). If so, it is likely that the mitigating effects of enhanced SMN expression we observe are predominantly if not entirely cell-autonomous and future treatments for familial and/or sporadic forms of ALS will require targeting multiple cell types. In conclusion, we have found that the mSOD1 protein and its associated pathology have a remarkably disruptive effect on the sub-cellular localization of the SMN protein. Such a result in which one component, SMN, of the Cajal body is lost without a dramatic reduction in the protein highlights an experimental system that may be exploited in future studies using model systems and human tissue to probe the precise function of these nuclear structures.
MATERIALS AND METHODS
Mice
Mutant SOD1G86R mice were obtained from the Jackson Laboratory (#05110) and genotyped using primers SOD1F2: 5′-CGGTCTGGAAAAGCCATCAT-3′ and E4A-SOD1: 5′-AATGATGGAATGCACTCCTGA-3′. The 256 bp PCR product was digested with Fsp1, which recognizes the G256C mutation in the SOD1 gene to generate 170 and 86 bp fragments. Mice overexpressing the SMN protein (SMNHiC; Jax #008206) were generated at the Ohio State University (28) and transferred to Columbia University. Such animals bear eight SMN2 genomic copies. Mice hemizygous for the transgene (SMN-HiC+/−;Smn+/+) express ∼2.5-fold wild-type levels of the SMN protein and were used for the experiments described in the manuscript. They were genotyped as previously described (28). To generate mice with spinal motor neurons fluorescently marked with the eGFP protein, SMNHiC animals were bred to HLXB9::GFP mice (Jax #005029) and genotyped for eGFP, using standard GFP primers. All animal procedures were performed in accordance with institutional guidelines.
Phenotypic assessment of mice
Motor behavior and body weights of the mice were measured beginning at PND30 every 2–3 days to assess the phenotypic effects of overexpressing SMN in SOD1 mutants. The vertical pole test was a modified version of the protocol described by Matsuura et al. (42). Briefly, mice were placed in the center of a vertically positioned metal pole (60 cm long, 1 cm diameter) covered with a mesh tape, with their snouts oriented toward the ceiling. Motor performance was measured by the latency to turn around in order to descend the pole. Each animal was subjected to 10 trials and a motor defect noted when a mouse was unable to turn around within 30 s in any of the trials. The disease was defined as phenotypically discernible if a mouse performed in a similar manner on 2 consecutive days. The 30 s cut-off time was chosen based on a 100% success rate in age-matched control mice (n = 50). The rotarod test was performed using a machine from Columbus Instruments (Columbus, OH, USA) under conditions of constant (20 r.p.m.) velocity. Mice were subjected to 10 trials following 3 training periods of 1 min each. An inability to remain on the rotating rod for at least 30 s in each of the trials constituted a motor defect. A similar performance the following day was required to confirm disease onset. The onset of the disease based on body weight loss was defined as the day on which loss exceeded 5% of peak body weight.
Immunoprecipitation and western blot analysis
SMN and SOD1 protein levels were determined by western blot analysis using standard procedures as previously described (10,28). SMN (1:4000, BD Biosciences, San Jose, CA, USA), SOD1 (1:1600, Calbiochem, Rockland, MA, USA), α-tubulin (1:1000, Sigma-Aldrich, St Louis, MO, USA), β-actin (1:1000, Millipore), coilin (1:900, Santa Cruz), GFP (1:1200, Invitrogen, Carlsbad, CA, USA), Histone H3 (1:2000, Cell Signaling Technology, Danvers, MA, USA), GAPDH (1: 2000, Cell Signaling Technology) were visualized using the ECL Detection Kit (RPN2109, GE Healthcare, Piscataway, NJ, USA). Quantification of band intensities was performed using the ImageJ software (NIH, Bethesda, MD, USA). To determine the effect of mSOD1 on SMN levels in the cytoplasmic and nuclear compartments, NSC34 cells were washed twice on ice in RSB100 buffer (20 mm Tris–Cl, pH 7.4, 150 mm NaCl, 2.5 mm MgCl2) and then permeabilized for 13 min at 37°Cwith RSB100 buffer containing 260 μg/ml digitonin (Sigma-Aldrich) and 1% (v/v) protease inhibitors. Lysates were clarified by centrifugation (3000g, 2 min) to collect the cytoplasmic fraction in the supernatant. The pellets were washed twice in RSB100 buffer, solubilized by sonication in RIPA lysis buffer and then centrifuged to obtain the nuclear fraction in the supernatant. To immunoprecipitate proteins bound to SMN, G-Sepharose beads (Sigma-Aldrich) were incubated with anti-SMN 7F3 for 2 h at 4°C, washed with RSB100 buffer to remove excess antibody and then further incubated with 100 mg of protein sample from either cellular compartment of the NSC34 cells. The beads were then washed with RSB100 buffer to remove unbound proteins, boiled for 10 min at 95°C and then centrifuged before collecting the supernatant for western blot analysis.
Immunohistochemistry
NMJs of the intercostal muscle were stained as previously described (30). NMJ types (I, II and III) were defined by the extent of overlap between AChRs stained with labeled α-bungarotoxin and nerve terminals stained with anti-NF(M). To examine the spinal motor neurons, the thoracic spinal cord was dissected from animals and treated as previously described (14). Primary antibodies used were anti-SMN 7F3 (1:200, a generous gift from L. Pellizzoni), anti-ChAT (1:100, Millipore, Temecula, CA, USA), anti-coilin (1:250, Santa Cruz, Santa Cruz, CA, USA), SOD1 (1:180, Calbiochem, San Diego, CA, USA) and anti-Sm 18F6 (1:200, a gift from L. Pellizzoni). To ensure we were quantifying gems as defined by Liu and Dreyfuss ( 16), initial immunohistochemistry experiments were carried out by co-labeling motor neurons with antibodies against SMN and coilin. Since the two proteins always co-localized as depicted in Supplementary Material, Figure S5, subsequent experiments were carried out with anti-SMN alone, and reactive nuclear foci assumed to be gems. SMN intensity within the spinal motor neurons was assessed after images were captured under identical gain and exposure and as previously described (30). Immunostaining of cultured cells was carried out following fixation with 4% PFA for 10 min at 4°C. The cells were then blocked in PBS solution containing 3% BSA and 1% Triton-X (PBS-BT) for 30 min at room temperature and incubated with a primary antibody in PBS-BT for 2 days at 4°C. They were then rinsed in PBS, incubated with a labeled secondary antibody for a day at 4°C and rinsed an additional three times before mounting in Vectashield (Vector Labs, Burlington, VT, USA) containing DAPI for image analysis. Images were acquired on a Nikon Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan) equipped with a Spot Flex digital camera (Diagnostic Instruments, Starling Heights, MI, USA). Confocal images were obtained on a laser scanning confocal LSM5 PASCAL microscope (Zeiss, Germany).
Primary cell cultures
Fibroblasts were obtained from the tails of PND0–1 mice. The tissue was minced and then seeded onto 0.1% gelatin-coated cover slips immersed in DMEM media supplemented with 10% fetal bovine serum (FBS). Cells were harvested 12 days after plating for immunostaining experiments. Primary motor neurons were extracted from the spinal cords of eGFP+ E12.5-day-old embryos. The tissue was mechanically dissociated and then incubated for 8 min at 37°C in 0.025% trypsin. The resulting cells were washed with motor neuron medium [Neurobasal medium (Invitrogen), containing 2% heat-inactivated horse serum, B27 supplement, 0.5 mm glutamine, 25 m β-mercaptoethanol, penicillin/streptomycin and trophic factor cocktail (0.5 ng/ml glial-derived neurotrophic factor, 1 ng/ml brain-derived neurotrophic factor and 10 ng/ml ciliary neurotrophic factor), R&D Systems] three times and plated on astrocyte monolayers at a density of 1500 GFP+ cells/cm2. Mouse astrocytes were extracted from spinal cords of PND3–4 animals by mechanically dissociating the tissue and then transferring the resulting cells to culture flasks containing DMEM medium supplemented with 10% FBS. Following a 15 day incubation period, the flasks were placed on a shaker (200 r.p.m. for 8h) to detach microglia. The astrocytes were then re-plated on cover slips placed in 24-well plates until 80% confluent at which point they were co-cultured with the neurons. Motor neurons for experiments involving neurotrophic factor deprivation were plated on poly-d-lysine/laminin-coated cover slips with motor neuron media with or without the trophic factor cocktail.
Transfection studies
To test the effect of mSOD1 on SMN localization in cultured cells, NSC34 motor neuron-like cells were transfected with SOD1G86R or SOD1WT constructs. Briefly, RNA extracted from mSOD1G86R mouse spinal cord was reverse-transcribed and the cDNA amplified with primers vecSOD-F2: 5′-TA AAGCTTCGGGGAAGCATGGCGATGAAAGCGGTGTGCGTG-3′ and vecSOD-R1: 5′-AAGGATCCTGCGCAATCCCAATCACTCCACAGGCCAAGCGGC-3′ to generate the wild-type and mSOD1 transcripts. The PCR fragments were digested with BamH1 and HindIII and then cloned into the pEYFP-N1 (Invitrogen) vector. Mutant SOD1 was distinguished from its wild-type counterpart by virtue of the Fsp1 site, which is created by the G256C mutation. A total of 2 × 105 NSC34 cells were transfected with 5 μg of plasmid suspended in Lipofectamine (Invitrogen) according to the manufacturer's recommendation, which resulted in a >90% transfection efficiency.
Statistics
Kaplan–Meier survival curves were compared and assessed for differences, using the log-rank test equivalent to the Mantel–Haenszel test. The unpaired, two-tailed Student's t-test or one-way ANOVA followed by Tukey's post hoc comparison, where indicated, were used to compare means for statistical differences. Data are represented as mean ± SEM unless otherwise indicated. P < 0.05 was considered significant. Statistical analyses were performed with GraphPad Prism v4.0 and StatMate v2.0 (GraphPad Software).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We thank Drs D.C. De Vivo and C.E. Henderson for advice and suggestions and Dr N. Shneider for access to essential equipment. We would also like to express our gratitude to Drs L. Pellizzoni and F. Lotti for SMN antibodies and for insightful discussions of our work. K. Brown, K. Malik and C. Caine provided technical help.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the SMA Foundation, Muscular Dystrophy Association of America, AFM-France, SMA-Europe, Department of Defense (W81XWH-09-1-0245) and National Institutes of Health (NS057482 to U.R.M., NS042269 to S.P.). D.B.R. is the recipient of an NIH Career Development Award (ES009089).
REFERENCES
- 1.Kolb S.J., Kissel J.T. Spinal muscular atrophy: a timely review. Arch. Neurol. 2011;68:979–984. doi: 10.1001/archneurol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen P.M. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr. Neurol. Neurosci. Rep. 2006;6:37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- 3.Veldink J.H., van den Berg L.H., Cobben J.M., Stulp R.P., De Jong J.M., Vogels O.J., Baas F., Wokke J.H., Scheffer H. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology. 2001;56:749–752. doi: 10.1212/wnl.56.6.749. [DOI] [PubMed] [Google Scholar]
- 4.Veldink J.H., Kalmijn S., Van der Hout A.H., Lemmink H.H., Groeneveld G.J., Lummen C., Scheffer H., Wokke J.H., Van den Berg L.H. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology. 2005;65:820–825. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- 5.Corcia P., Khoris J., Couratier P., Mayeux-Portas V., Bieth E., De Toffol B., Autret A., Müh J.P., Andres C., Camu W. SMN1 gene study in three families in which ALS and spinal muscular atrophy co-exist. Neurology. 2002;59:1464–1466. doi: 10.1212/01.wnl.0000032500.73621.c5. [DOI] [PubMed] [Google Scholar]
- 6.Corcia P., Camu W., Halimi J.M., Vourc'h P., Antar C., Vedrine S., Giraudeau B., de Toffol B., Andres C.R. French ALS Study Group. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67:1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- 7.Gamez J., Barceló M.J., Muñoz X., Carmona F., Cuscó I., Baiget M., Cervera C., Tizzano E.F. Survival and respiratory decline are not related to homozygous SMN2 deletions in ALS patients. Neurology. 2002;59:1456–1460. doi: 10.1212/01.wnl.0000032496.64510.4e. [DOI] [PubMed] [Google Scholar]
- 8.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 10.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 11.Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 12.Turner B.J., Parkinson N.J., Davies K.E., Talbot K. Survival motor neuron deficiency enhances progression in an amyotrophic lateral sclerosis mouse model. Neurobiol. Dis. 2009;34:511–517. doi: 10.1016/j.nbd.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Le T.T., McGovern V.L., Alwine I.E., Wang X., Massoni-Laporte A., Rich M.M., Burghes A.H. Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet. 2011;20:3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutz C.M., Kariya S., Patruni S., Osborne M.A., Liu D., Henderson C.E., Li D.K., Pellizzoni L., Rojas J., Valenzuela D.M., et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest. 2011;121:3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 17.Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 18.Ogg S.C., Lamond A.I. Cajal bodies and coilin—moving towards function. J. Cell Biol. 2002;159:17–21. doi: 10.1083/jcb.200206111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanek D., Neugebauer K.M. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- 20.Morris G.E. The Cajal body. Biochim. Biophys. Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Hebert M.D., Szymczyk P.W., Shpargel K.B., Matera A.G. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;47:1049–1056. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Giorgio F.P., Carrasco M.A., Siao M.C., Maniatis T., Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripps M.E., Huntley G.W., Hof P.R., Morrison J.H., Gordon J.W. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 27.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1993;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 28.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossol W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 29.Le T.T., Coovert D.D., Monani U.R., Morris G.E., Burghes A.H. The survival motor neuron (SMN) protein: effect of exon loss and mutation on protein localization. Neurogenetics. 2000;3:7–16. doi: 10.1007/s100480000090. [DOI] [PubMed] [Google Scholar]
- 30.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jablonka S., Schrank B., Kralewski M., Rossoll W., Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum. Mol. Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 32.Kato S., Takikawa M., Nakashima K., Hirano A., Cleveland D.W., Kusaka H., Shibata N., Kato M., Nakano I., Ohama E. New consensus research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 (SOD1) gene mutations: inclusions containing SOD1 in neurons and astrocytes. Amyotroph. Lateral. Scler. Other Motor Neuron Disord. 2000;1:251–258. doi: 10.1080/14660820050515160. [DOI] [PubMed] [Google Scholar]
- 33.Meiering E.M. The threat of instability: neurodegeneration predicted by protein destabilization and aggregation propensity. PLoS Biol. 2008;6:e193. doi: 10.1371/journal.pbio.0060193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaarsma D., Teuling E., Haasdijk E.D., De Zeeuw C.I., Hoogenraad C.C. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J. Neurosci. 2008;105:6338–6343. doi: 10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 36.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Blauw H.M., Barnes C.P., van Vught P.W., van Rheenen W., Verheul M., Cuppen E., Veldink J.H., van den Berg L.H. SMN1 gene duplications are associated with sporadic ALS. Neurology. 2012;78:776–780. doi: 10.1212/WNL.0b013e318249f697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould T.W., Buss R.R., Vinsant S., Prevette D., Sun W., Knudson C.M., Milligan C.E., Oppenheim R.W. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J. Neurosci. 2006;26:8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gertz B., Wong M., Martin L.J. Nuclear localization of human SOD1 and mutant SOD1-specific disruption of survival motor neuron protein complex in transgenic amyotrophic lateral sclerosis mice. J. Neuropathol. Exp. Neurol. 2012;71:162–177. doi: 10.1097/NEN.0b013e318244b635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmoudi S., Henriksson S., Weibrecht I., Smith S., Söderberg O., Strömblad S., Wiman K.G., Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010;8:e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou T., Ilangovan R., Yu F., Xu Z., Zhou J. SMN protects cells against mutant SOD1 toxicity by increasing chaperone activity. Biochem. Biophys. Res. Commun. 2007;364:850–855. doi: 10.1016/j.bbrc.2007.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuura K., Kabuto H., Makino H., Ogawa N. Pole test is a useful method for evaluating the mouse movement disorder caused by striatal dopamine depletion. J. Neurosci. Methods. 1997;73:45–48. doi: 10.1016/s0165-0270(96)02211-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.