Abstract
Here we report an effective method for protein immobilization on a surface plasmon resonance (SPR) gold chip, describing the combination of cysteine- and oligomerization domain-mediated immobilization of enhanced green fluorescent protein (EGFP) as a model protein for the purpose of orientation-controlled surface density packing. In order to facilitate the oligomerization of EGFP, the dimeric and trimeric constructs derived from GCN4- leucine zipper domain were chosen for multimeric EGFP assembly. For orientation-controlled immobilization of the protein, EGFP modified with cysteine residues showing excellent orientation on a gold chip was used as a starting protein, as previously reported in our earlier study (Anal. Chem., 2007, 79, 2680–2687). Constructs of EGFP with oligomerization domains were genetically engineered, and corresponding fusion proteins were purified, applied to a gold chip, and then analyzed under SPR. The immobilized EGFP density on a gold chip increased according to the states of protein oligomerization, as dimeric and trimeric EGFPs displayed better adsorption capability than monomeric and dimeric forms, respectively. Fluorescence measurement corroborated the SPR results. Taken together, our findings indicated that the combination of cysteine- and oligomerization domain-mediated immobilization of protein could be used in SPR biosensor applications, allowing for an excellent orientation and high surface density simultaneously.
Introduction
In proteomics study, much attention has been paid to protein chip technology due to its benefits for simultaneous and high-throughput screening of proteins.1,2 In parallel with this, efforts to develop label-free methods coupled with protein chip technology have been made.3,4 Among them, a promising combination is surface plasmon resonance (SPR) protein chip technology, which is capable of directly detecting biomolecular interactions.5,6
The assembly of oligomeric protein is mediated through an oligomerization domain, which contributes to protein functional properties by enhancing binding, and improving conformational stability.7–9 This feature of protein oligomerization can be genetically mimicked by fusing the gene encoding the oligomerization domain to a target protein gene.10,11 One of the most well-characterized oligomerization domains is the leucine zipper of GCN4, yeast transcriptional activator, comprising 33 amimo acid residues that form parallel dimeric coiled-coils.12,13 The variants of dimeric GCN4 leucine zipper exhibit various multimeric forms upon mutation in a and d potisions.14,15
Many studies have been performed in the area of protein immobilization due to its relevance to the development of sensitive biosensor, particularly the research regarding the orientation-controlled immobilization of proteins.16–19 One of these self-oriented immobilization technologies is the cysteine-mediated immobilization of protein on a gold chip, which is based on the Au–S bond formation, interaction between bare gold and the thiol group (–SH) of cysteine.20–22
In this study, we describe a new method to integrate cysteine with the oligomerization domain-mediated immobilization of protein for the purpose of achieving an orientation-controlled density packing of multimeric protein on a SPR gold chip. Particularly, enhanced green fluorescent protein (EGFP) was chosen as a target protein owing to the feature that allows us to visualize its binding to the chip surface. SPR and fluorescence imaging analyses showed that the oligomeric states of EGFP affect the surface adsorption capability. Dimeric and trimeric EGFPs:Cys proteins bound to the gold surface more densely than monomeric and dimeric form, respectively. Our results showed that well-oriented density packing of a protein on a SPR gold chip surface can be achieved by combining cysteine- and multimerization domain-assisted protein immobilization, thereby allowing for better biosensor performance.
Materials and methods
Cloning of oligomeric EGFP:Cys genes
In order to clone the monomeric, dimeric, and trimeric EGFPs with cysteine residues, the full length gene encoding for the EGFP with a couple of cysteines at its C-terminal end was first amplified using the 5′ primer (GGA ATT CAT GGT GAG CAA GGG CGA G) and the 3′ primer (CCC AAG CTT TCA GCA GCA CTT GTA CAG CTC GTC) via the polymerase chain reaction (PCR). The 5′ and 3′ termini were designed to harbor the EcoRI and HindIII restriction enzyme cleavage sites, respectively. The PCR product was then purified using a DNA purification kit (Qiagen, CA), and digested with the EcoRI and HindIII restriction enzymes. And then, nucleotides coding for dimerization domain (DD) and trimerization domain (TD) with the BamHI and EcoRI enzyme cleavage sites at the 5′ and 3′ termini, respectively, were obtained from chemical synthesis, and digested with the indicated enzymes. Following the ligation of EGFP:Cys with DD and TD using a ligation kit (Takara, Japan), the resulting DD:EGFP:Cys and TD:EGFP:Cys cassettes were inserted into the pET 21a vector using BamHI and HindIII sites to generate in-frame fusion of pDD:EGFP:Cys and pTD:EGFP: Cys, respectively. For the construction of monomeric EGFP:Cys (pEGFP:Cys), the EGFP:Cys fragments were introduced into the pET 21a vector using EcoRI and HindIII sites. The fusion genes were all verified by DNA sequencing, and transformed into Escherichia coli BL21 (DE3) for the expression of the recombinant proteins.
Expression and purification of the oligomeric OD:EGFP:Cys proteins
The pEGFP, pEGFP:Cys, pDD:EGFP:Cys, pTD:EGFP:Cys plasmids were transferred to the expression host, E. coli BL21 (DE3) (Stratagene, CA). The transformed cells were grown at 37 °C with shaking to an OD600 of 0.6. The cells were induced with 1 mM isopropyl-2-D-thiogalactopyranoside (IPTG) (GibcoBRL, MD), and grown for an additional 4 h. The cells were then harvested after 10 min of centrifugation at 6,000 g at 4 °C. In order to purify the 6 × His: EGFP, EGFP:Cys, DD:EGFP:Cys, TD: EGFP:Cys proteins, one liter of crude cell lysate was loaded onto a IDA-miniexcellose affinity column (Bioprogen Co., Korea) and washed three times with 10 ml of equilibration buffer (50 mM phosphate, 0.5 N NaCl, pH 8.0), respectively. The recombinant proteins were then eluted with 5 ml of 0.5 M imidazole in the same buffer (50 mM phosphate, 0.5 N NaCl, pH 8.0).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and native gel analyses
The E. coli cells were harvested, and then suspended in 50 mM Tris-HCl buffer (pH 8.0). The cells were disrupted via sonication, after which the soluble and insoluble fractions were separated by centrifugation for 10 min at 10 000g. The soluble fractions were then analyzed using 10% SDS-PAGE and native or non-denaturing gel without SDS. The gels were stained with Coomassie’s Brilliant Blue R250. Protein concentrations were determined by the Bradford method, using bovine serum albumin as a standard.
SPR analysis
The bare gold chip surface was cleaned with piranha solution (70% (v/v) H2SO4, 30% (v/v) H2O2), and the chip was subsequently washed with ethanol and distilled water. The SPR measurements were performed with the pre-cleaned gold chip on a Biacore 3000 device (Biacore AB, Sweden) at 25 °C. A 10 mM PBS (pH 7.4) was used as running buffer for all SPR analysis. A series of oligomeric EGFP proteins with cysteines such as monomeric EGFP:Cys, dimeric EGFP:Cys, and trimeric EGFP: Cys were immobilized onto the bare gold chip by introducing the solution (150 μl, 200 μg ml−1, in PBS, pH 7.4) through the instrument channel at a flow rate of 5 μl min−1. And then, the real-time profile of SPR response was presented in a sensorgram. Experiment for antibody binding to EGFP was performed using a monoclonal anti-EGFP antibody (G1546, Sigma-Aldrich, MO).
Fluorescence measurement
A bare slide glass was cleaned with piranha solution (70% (v/v) H2SO4, 30% (v/v) H2O2) and was thoroughly washed with distilled water and ethanol. The glass slide was immersed in ethanol containing 1.0% APTMS for 20 min, and the slide was subsequently washed with ethanol. The resulting amine glass slide was stored in an oven at 100 °C prior to use. To modify the amine glass surface with maleimide group, the glass slide was incubated in a 10 mM PBS (pH 7.4) containing 50 μg ml−1 BMPS for 30 min. For the fluorescence measurement, the EGFP, EGFP:Cys, DD:EGFP:Cys, TD:EGFP:Cys solutions in PBS (pH 7.4) were spread onto the maleimde functionalized slide glass stuck to sticky tape with wells of approximately 3 mm in diameter and 100 μm in thickness in order to compartmentalize to protein samples. The sticker chip was washed three times with PBST, followed by rinsing with distilled water, and drying with N2 gas. The fluorescence was analyzed by using a GenPix 4200 (Axon Instruments, CA).
Results and discussion
Research concept for on-chip density packing of protein in a good orientation
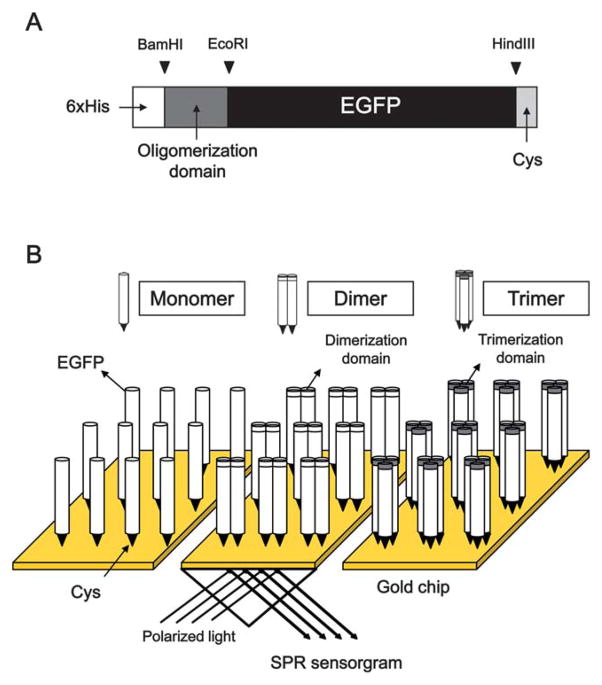
The cysteine residue holds the thiol group (–SH) which plays a role in folding and stabilizing protein via disulfide lingkages. It is also well known that thiol adsorption occurs on the gold surface. Using genetic engineering technique, cysteine residues can be introduced at a specific location on a protein. Thus, cysteine-mediated protein immobilization on a gold surface has been widely employed in biosensor applications. In our previous study, we showed the site-specific immobilization of protein G linked by cysteine residues on a SPR gold chip, illustrating an easy, one-step, and oriented immobilization of protein G without necessitating any surface modification and reagents to cross-link.22 The orientation of proteins on a chip is essential for optimal function of biosensors. Besides, high density packing of protein on a chip surface is related to biosensor efficiency including high sensitivity and minimum amount of analyte samples. However, little research has been conducted to advance the adsorption capability of protein without sacrificing its functional properties. Along this line, we hypothesized that well-oriented cysteine-tagged protein with oligomerization domains would display high surface density on a SPR gold chip. To test this hypothesis, GCN4-derived dimeric and trimeric mutants were adopted for mutimerization of EGFP which is capable of facilitating visualization of its adsorption (Fig. 1A and 1B). Nucleotide and amino acid sequences of the dimerization domain (DD) and trimerization domain (TD) are listed in Table 1.
Fig. 1.
Schematic representation of the immobilization of the oligomeric EGFP:Cys proteins. The oligomeric EGFP constructs attached by a couple of cysteine residues were cloned by inserting the genes encoding GCN4-derived dimerization domain (DD) and trimerization (TD) into the C-terminal region of cysteine-tagged EGFP gene. The 33 amino acid sequences of DD and TD were described in the box (A). The cysteine residues and the oligomerization domains are responsible for the orientation and the density packing of the oligomeric EGFP:Cys proteins onto the SPR gold chip, respectively (B).
Table 1.
Nucleotide and amino acid sequences of GCN4-derived dimerization domain (DD) and trimerization domain (TD). Amino acid symbols are enclosed in parentheses
| Domain | Nucleotide and amino acid sequences |
|---|---|
| Dimerization | AGA(R) ATG(M) AAA(K) CAA(Q) CTT(L) GAA(E) GAC(D) AAG(K) GTT(V) GAA(E) GAA(E) TTG(L) CTT(L) TCG(S) AAA(K) AAT(N) TAT(Y) CAC(H) TTG(L) GAA(E) AAT(N) GAG(E) GTT(V) GCC(A) AGA(R) TTA(L) AAG(K) AAA(K) TTA(L) GTT(V) GGC(G) GAA(E) CGC(R) |
| Trimerization | AGA(R) ATG(M) AAA(K) CAA(Q) ATT(I) GAA(E) GAC(D) AAG(K) ATT(I) GAA(E) GAA(E) ATC(I) CTT(L) TCG(S) AAA(K) ATT(I) TAT(Y) CAC(H) ATC(I) GAA(E) AAT(N) GAG(E) ATT(I) GCC(A) AGA(R) ATT(I) AAG(K) AAA(K) TTA(L) ATT(I) GGC(G) GAA(E) CGC(R) |
Analysis of oligomeric EGFP:Cys on a native gel electrophoresis
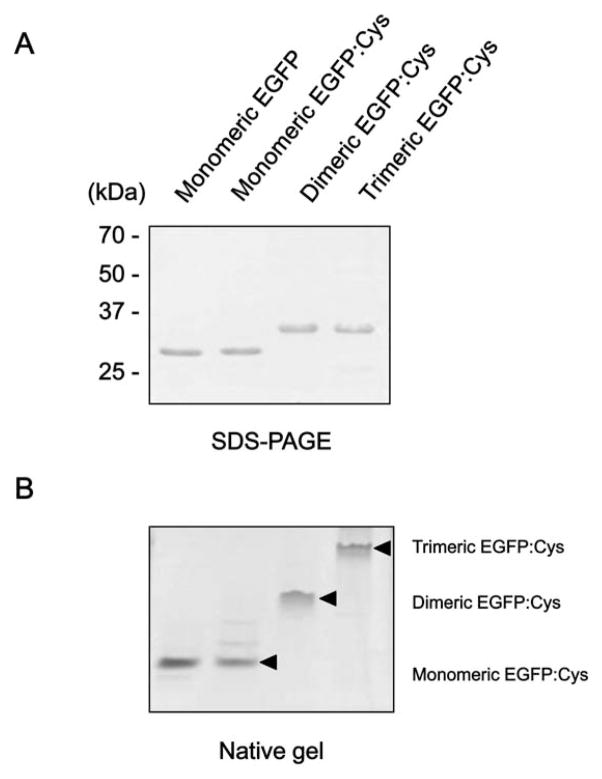
To generate the oligomeric EGFPs with cysteine residues, we cloned two constructs; (a) pDD:EGFP:Cys and (b) pTD:EGFP: Cys by introducing the genes encoding the dimerization domain (DD) and trimerization domain (TD) into the N-terminal region of EGFP. Both constructs contain two cysteines at its C-terninal end. The oligomerization domains were originally derived from GCN4 leucine zipper, which is characterized by a repeat of leucines regularly spaced every seven residues. The 33 amino acid sequences of the oligomerization domains are described in Fig. 1A. Additional constructs, pEGFP alone as negative control and pEGFP:Cys, were PCR-amplified. Procedures for the expression and purification of chimeric EGFPs are described in the Materials and Methods section. After elution, the resulting proteins were subject to SDS-PAGE, which exhibited approximately 27 and 31 kDa of single protein bands corresponding to the molecular weights of EGFP only, and OD:EGFP:Cys, respectively (Fig. 2A). SDS-PAGE, a denaturing gel electrophoresis, can be used to separate protein molecule according to its molecular weight. This is suitable for resolving the molecular weights of non-covalently bound subunits of protein complexes. When denatured with the denaturing detergent SDS, oligomeric proteins will be divided into smaller monomeric fractions, meaning that it is impossible to separate the proteins in the native oligomeric forms by using the SDS-PAGE. For the reason, in order to evaluate the oligomeric states of EGFP:Cys, the proteins were analyzed in a non-denaturing condition. As shown in Fig. 2B, EGFP migrated as monomer, dimer, and trimer in a native or non-denaturing gel without SDS, which clearly evidenced that EGFP with oligomeric domains formed the corresponding mutimeric proteins. These purified oligomeric EGFP: Cys proteins were then analyzed for testing their adsorption capability on a gold chip by surface plasmon resonance (SPR) system.
Fig. 2.
SDS-PAGE and native gel analyses of purified OD:EGFP:Cys proteins. Purified recombinant proteins were analyzed via 10% SDS PAGE (A), and non-denaturing gel without SDS (B), and then visualized via Coomassie staining. The arrowhead indicates the expressed monomeric, dimeric, and trimeric EGFP:Cys.
SPR analysis to evaluate the site-directed density packing of oligomeric EGFP:Cys
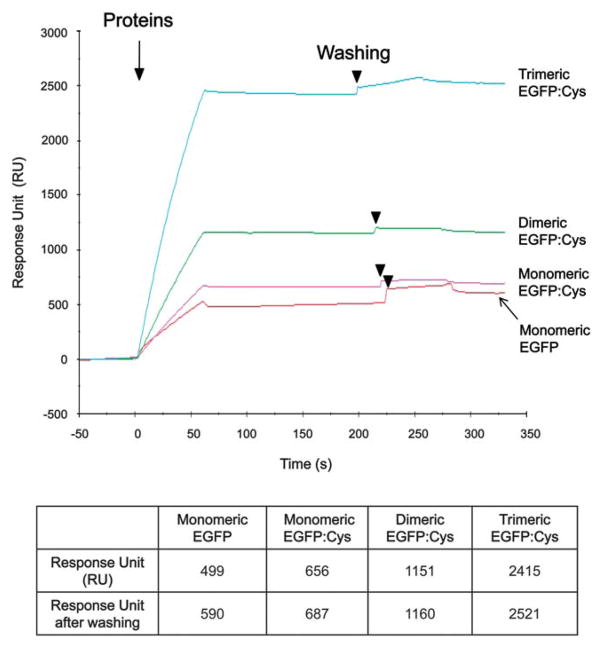
The adsorption capability of oligomeric EGFP:Cys proteins were investigated by means of mass change via real-time SPR measurement. The SPR sensorgram (Fig. 3) shows the changes of RU values in response to OD:EGFP:Cys proteins on the gold chip surface. Upon the addition of monomeric EGFP:Cys onto the gold film, the RU value was approximately 687 RU after washing with Tween-20 buffer (Fig. 3). After treatment with dimeric EGFP:Cys and trimeric EGFP:Cys proteins, the SPR responses were shifted up to about 1,160 and 2,521, respectively, in the plateau region after washing (Fig. 3). Based on the SPR signal expressed in response unit (RU), the RU value of 1,000 corresponds to the binding of approximately 1 ng per square mm of protein on the gold surface. Thus, the surface density of the bound monomeric EGFP:Cys, dimeric EGFP:Cys, and trimeric EGFP:Cys proteins can be calculated as 6.87 × 10−10 g mm−2 (or 22.9 fM, 1.4 × 1010 number of EGFP units mm−2), 11.6 × 10−10 g mm−2 (or 19.3 fM, 1.2 × 1010 number of EGFP units mm−2), and 25.2 × 10−10 g mm−2 (or 28.0 fM, 1.7 × 1010 number of EGFP units mm−2), respectively. Since a dimer or a trimer consists of double or triple molecules, respectively, the bound protein amounts of dimeric EGFP:Cys and trimeric EGFP:Cys were 1.7 and 3.6 times higher than that of monomeric EGFP:Cys, respectively. The data indicate that the oligomerized proteins modified with cysteines can pack more densely according to their oligomeric states onto the gold chip surface. The surface coverage was estimated by the means that experimentally obtained number of protein units mm−2 is compared with the theoretical value calculated in the saturation adsorption of proteins. Considering that the dimension of trimeric EGFP:Cys on a gold surface comes to 4 nm × 12 nm, the theoretical number of trimeric EGFP units mm−2 can be estimated as 2.7 × 1010 number of EGFP units mm−2. Based on this value, the surface coverage for trimeric EGFP:Cys on the gold surface can be estimated approximately 63% (= 1.7 × 1010/2.7 × 1010 × 100 (%)). As shown in Fig. 3, the construct without cysteines seemed to bind to the gold surface, as observed to be 590 RU (or 19.6 fM, 1.2 × 1010 number of EGFP units mm−2). It is thought that such binding is random as non-specific binding, and the construct does not retain orientation. Non-specific binding of protein on a sensing layer is one of the major bottleneck to biosensor applications. The EGFP used in this study was tagged by six histidine residues. The hexahistidine (His×6) tag is commonly used to facilitate the purification of recombinant protein in biology. Recently, Kogot et al. reported the binding of histidine tag to Au surface.23 Considering this notion, the N-terminal histidine residues may cause the nonspecific adsorption of cysteine-free EGFP on the gold surface. Thus, in order to assess this issue, the effect of histidine residues on the immobilization of His×6–tagged proteins on a gold surface requires further study.
Fig. 3.
SPR measurement. The SPR sensorgram shows the binding of cysteine-tagged EGFP with oligomerization domain onto a gold chip surface. Surface densities of immobilized proteins are shown in the lower panel. The SPR signal intensities were expressed in response units (RU).
Another shortcoming in the immobilization of protein onto a gold chip is weak attachment, which may result in the elimination of the protein from the chip surface when washed by certain buffers with detergents, possibly leading to a poor performance of SPR measurement. Thus, we also examined the binding strength of immobilized proteins with buffer containing the detergent Tween-20. Tween-20 is commonly added to PBS buffer to remove unbound proteins for ELISA, Western blotting, immunohistochemisty, etc. As shown in Fig. 3, no considerable change of SPR response before and after treatment with Tween-20 wash buffer was observed among the monomeric EGFP:Cys, dimeric EGFP:Cys, and trimeric EGFP:Cys proteins. Likewise, similar SPR result was also obtained with CHAPS buffer widely used for cell lysis (data not shown). It is assumed that the cysteine-tagged EGFP monomer possesses high binding affinity that when bound to the chip imparts sufficient strength similar to the dimer and trimer. Thus, no significant difference exists between the three configurations used (Fig. 3).
Fluorescence analysis of the oriented surface density packing of oligomeric EGFP:Cys onto the maleimide-modified glass slide
Our constructs with cysteine residues were designed to provide a controlled orientation of protein on a gold surface. To assess the controlled protein immobilization, the SPR experiment of antibody binding onto EGFP-modified gold surface was carried out. However, the results of antibody binding analysis were not satisfactory under SPR (data not shown). Proper orientation of the immobilized proteins on a solid support improves the binding activity, because the recognition site of its cognate antibody remains accessible.24 In our experimental scheme, the N-terminal fragment of EGFP is away from the gold surface. Therefore, it is very likely that the spacial proximity between the C-terminal half of EGFP and the gold surface may interfere with the binding of antibody, since the EGFP antibody used herein epitopes the C-terminal lobe of EGFP.
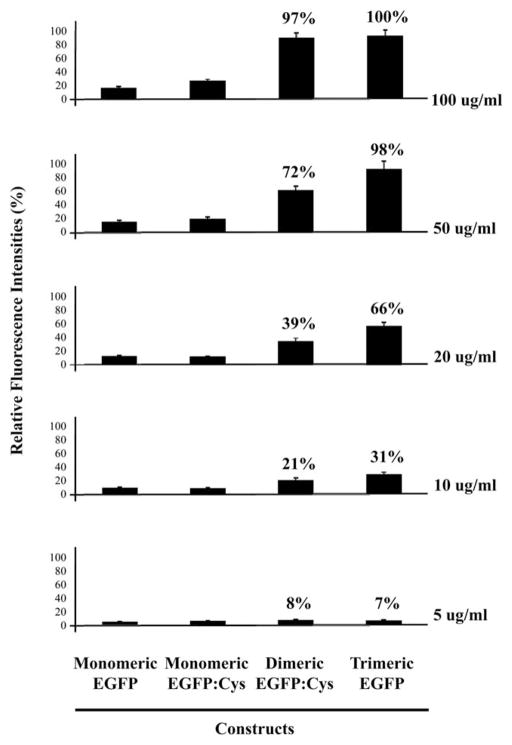
Thus, in order to show the evidence of specific binding of cysteine-modified EGFP onto the cognate surface via thio-bearing cysteine residues, the direct fluorescence measurement was employed, as our constructs tested in this work were basically designed to be detectable using a simple fluorescent readout. Prior to fluorescence measurement, the glass surface was modified with maleimide-functional groups for capturing cysteine-tagged EGFP with oligomerization domain. A series of oligomeric EGFP:Cys proteins ranging from 100 μg ml−1 to 5 μg ml−1 were spread on a maleimide-coated glass slide stuck to sticky tape with wells of approximately 3 mm in diameter and 100 μm thickness in order to compartmentalize the protein solutions. As shown in Fig. 4, the fluorescence intensities of the protein binding showed approximately direct proportion according to the states of oligomeric EGFP among those proteins, which is consistent with the SPR measurement. The relative fluorescence signal intensity of trimeric EGFP:Cys showed 100 μg ml−1 as a saturation concentration, while cysteine-unmodified oligomeric EGFP as a negative control did not bind to the maleimide-modified glass slide, if any, it is only a minimally detectable signal. The differences in fluorescence intensities were seldom observed with both monomeric EGFP and monomeric EGFP:Cys at 5, 10, 20, and 50 μg ml−1 protein concentrations on the maleimide-coated glass slide (Fig. 4). However, a meaningful increase in the adsorbed protein mass with monomeric EGFP:Cys was detected in response to 100 μg ml−1 protein via a fluorescence enhancement (Fig. 4). The fluorescence intensity of monomeric EGFP:Cys was approximately 1.8 times higher than that observed in cysteine-free monomeric EGFP at the concentration of 100 μg ml−1 protein. The data indicate that monomeric EGFP without cysteines may physically bind to the maleimide-modified surface regardless of orientation, whereas the protein with cysteine residues binds to the surface via the direct and specific bond between thiol-bearing cysteine residues and the cognate surface.
Fig. 4.
Fluorescence measurement of the oligomeric EGFP:Cys proteins. Fluorescence of the proteins were quantified using the GenePix Pro 6.0 system (Axon Instruments, CA). Relative fluorescence signal intensities (%) are graphed.
Next, the dissociation constants (KD values) of oligomeric EGFP:Cys to their cognate glass slide under the conditions used in fluorescence measurement were determined. Now that the dimeric and trimeric EGFP:Cys proteins showed excellent performance of the protein binding affinity in our experimental settings, the KD values of the two proteins were measured. Fractional saturation sites were calculated as V = B/Bmax, where B was the fluorescence intensity change due to the protein binding (Pb) to the maleimide-modified surface at a given point of titration, and Bmax was the fluorescence intensity change when all glass surfaces were occupied by the protein. Bmax was estimated by fitting B data versus total protein concentration (Pt) with an empirical rectangular hyperbola. The equilibrium dissociation constant (KD) of the protein to the cognate surface was calculated by nonlinear least squares using the following equation: V = [Pf]/(KD + [Pf]), where Pf is the free protein concentration which was calculated at each equilibrium point as [Pf] = [Pt] − [VMt], where Mt is the total maleimide concentration. From the relationship between fluorescence signal intensity and protein concentration profile, the dissociation constants for dimeric EGFP:Cys and trimeric EGFP:Cys binding to the maleimide-functionalized surface were calculated by Kaleida Graph ver. 4.0 program. The dissociation constants of dimeric and trimeric EGFP:Cys were estimated approximately 29.5 (±5.2) and 15.4 (±1.2) μg ml−1, respectively, indicating that the binding affinity of trimeric EGFP:Cys is higher in fluorescence measurement than dimeric EGFP:Cys construct onto the maleimide-modified glass slide. The data suggest that trimeric EGFP:Cys displays better directed density packing of the protein on the cognate surface than dimeric form.
The goal of this study was to develop a suitable method for immobilizing a number of proteins in a high density. The assembly of oligomeic protein is advantageous in terms of its functionality such as binding enhancement and conformational stability. Besides the positive effects of the oligomeric structure, we need to take the unwanted steric hindrance effect interfering with active site due to the oligomerization of protein into consideration. In this regard, Pack et al. reported a method to produce multivalent miniantibodies using GCN4-based sequences.25 In that study, it has been shown that the GCN4-derived tetravalent miniantibody exhibited the enhanced functional affinity compared to the bivalent construct, implying that the probable steric hindrance effect coming from the multimerization of a receptor protein is negligible. In the case of steric problem, the steric exclusion may be possibly resolved by introducing some flexible linker residues such as glycine and serine, resulting in a spacing of the active site due to flexiblilty of the oligomeric protein.
Here we showed that cysteine-tagged EGFP with oligomerization domains binds tightly to a gold chip layer in high density. The current technology has potential applications for on-chip immobilization of antibody-capturing proteins for the disease diagnosis, environmental monitoring or food pathogen detection, as well as target proteins for high-throughput drug screening. Results described herein are novel as far as immobilization of protein to an on-chip density packing of protein in a good orientation is concerned. The results also suggest that density packing coupled with site-specific adsorption of protein on a chip surface contributes to the sensing performance of biosensors. The current technology maybe of potential importance in the area of protein immobilization for biosensor applications. However, since we assessed the feasibility of orientation-controlled density packing of EGFP as a model protein on the SPR gold chip, additional evaluations with other proteins are needed.
Acknowledgments
This research was supported in part by the DHHS/NIH/NCRR/ RCMI Award (2 G12RR03059-21A1, Infrastructure Development in Computational Biology and Bioinformatics), and the Nano/Bio Science & Technology Program from the Ministry of Education, Science and Technology (MEST) of the Korean Government, and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Initiative Research Program.
References
- 1.Stoevesandt O, Taussig MJ, He M. Expert Rev Proteomics. 2009;6:145–157. doi: 10.1586/epr.09.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huels C, Muellner S, Meyer HE, Cahill DJ. Drug Discovery Today. 2002;7:S119–124. doi: 10.1016/s1359-6446(02)02389-9. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Xu D, Cheng Q. Proteomics. 2006;6:5493–5503. doi: 10.1002/pmic.200600216. [DOI] [PubMed] [Google Scholar]
- 4.Boozer C, Kim G, Cong S, Guan H, Londergan T. Curr Opin Biotechnol. 2006;17:400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Canziani G, Zhang W, Cines D, Rux A, Willis S, Cohen G, Eisenberg R, Chaiken I. Methods. 1999;19:253–269. doi: 10.1006/meth.1999.0855. [DOI] [PubMed] [Google Scholar]
- 6.Malmqvist M. Biochem Soc Trans. 1999;27:335–340. doi: 10.1042/bst0270335. [DOI] [PubMed] [Google Scholar]
- 7.Ali MH, Imperiali B. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 8.Engel J, Kammerer RA. Matrix Biol. 2000;19:283–288. doi: 10.1016/s0945-053x(00)00075-5. [DOI] [PubMed] [Google Scholar]
- 9.D’Alessio G. Nat Struct Biol. 1995;2:11–13. doi: 10.1038/nsb0195-11. [DOI] [PubMed] [Google Scholar]
- 10.De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, Vandekerckhove J, Fiers W, Saelens X. J Biol Chem. 2008;283:11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Calder LJ, Dessen A, Laue T, Skehel JJ, Wiley DC. Proc Natl Acad Sci U S A. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 13.O’Shea EK, Rutkowski R, Kim PS. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 14.Eckert DM, Malashkevich VN, Kim PS. J Mol Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 15.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 16.Jung Y, Jeong JY, Chung BH. Analyst. 2008;133:697–701. doi: 10.1039/b800014j. [DOI] [PubMed] [Google Scholar]
- 17.Rusmini F, Zhong Z, Feijen J. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 18.Cretich M, Damin F, Pirri G, Chiari M. Biomol Eng. 2006;23:77–88. doi: 10.1016/j.bioeng.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Anal Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 20.Weng HA, Wu CC, Chen CC, Ho CC, Ding SJ. J Mater Sci: Mater Med. 2010;21:1511–1519. doi: 10.1007/s10856-010-4026-4. [DOI] [PubMed] [Google Scholar]
- 21.Sejwal P, Narasimhan SK, Prashar D, Bandyopadhyay D, Luk YY. J Org Chem. 2009;74:6843–6846. doi: 10.1021/jo901085u. [DOI] [PubMed] [Google Scholar]
- 22.Lee JM, Park HK, Jung Y, Kim JK, Jung SO, Chung BH. Anal Chem. 2007;79:2680–2687. doi: 10.1021/ac0619231. [DOI] [PubMed] [Google Scholar]
- 23.Kogot JM, England HJ, Strouse GF, Logan TM. J Am Chem Soc. 2008;130:16156–16157. doi: 10.1021/ja8064717. [DOI] [PubMed] [Google Scholar]
- 24.Rao SV, Anderson KW, Bachas LG. Microchim Acta. 1998;128:127–143. [Google Scholar]
- 25.Pack P, Muller K, Zahn R, Pluckthun A. J Mol Biol. 1995;246:28–34. doi: 10.1006/jmbi.1994.0062. [DOI] [PubMed] [Google Scholar]