Abstract
Members of the aldehyde dehydrogenase gene (ALDH) superfamily play an important role in the enzymic detoxification of endogenous and exogenous aldehydes and in the formation of molecules that are important in cellular processes, like retinoic acid, betaine and gamma-aminobutyric acid. ALDHs exhibit additional, non-enzymic functions, including the capacity to bind to some hormones and other small molecules and to diminish the effects of ultraviolet irradiation in the cornea. Mutations in ALDH genes leading to defective aldehyde metabolism are the molecular basis of several diseases, including gamma-hydroxybutyric aciduria, pyridoxine-dependent seizures, Sjögren-Larsson syndrome and type II hyperprolinaemia. Interestingly, several ALDH enzymes appear to be markers for normal and cancer stem cells. The superfamily is evolutionarily ancient and is represented within Archaea, Eubacteria and Eukarya taxa. Recent improvements in DNA and protein sequencing have led to the identification of many new ALDH family members. To date, the human genome contains 19 known ALDH genes, as well as many pseudogenes. Whole-genome sequencing allows for comparison of the entire complement of ALDH family members among organisms. This paper provides an update of ALDH genes in several recently sequenced vertebrates and aims to clarify the associated records found in the National Center for Biotechnology Information (NCBI) gene database. It also highlights where and when likely gene-duplication and gene-loss events have occurred. This information should be useful to future studies that might wish to compare the role of ALDH members among species and how the gene superfamily as a whole has changed throughout evolution.
Keywords: ALDH, aldehyde dehydrogenase, nomenclature, carbonyl metabolism, evolution, gene family
Introduction
The aldehyde dehydrogenase gene (ALDH) superfamily is represented in all three taxonomic domains (Archaea, Eubacteria and Eukarya), suggesting a vital role throughout evolutionary history. Our understanding of the biological roles of this superfamily continues to expand in ways that are often unexpected and, perhaps, unprecedented for an enzyme family. As implied by their name, members of this superfamily serve to metabolise both physiologically and pathophysiologically relevant aldehydes. This capacity prevents the accumulation of toxic aldehydes derived from endogenous production and/or exogenous exposures, which, if left unchecked, adversely affect cellular homeostasis and organismal functions [1].
ALDH activity is also required for the synthesis of vital biomolecules through the metabolism of aldehyde intermediates, such as retinoic acid, folate and betaine, to name a few [2-4]. Whereas the ability of the ALDH family members to metabolise reactive aldehydes represents a major underlying cytoprotective mechanism, it is important to recognise that ALDHs demonstrate functions that extend beyond detoxification. Accumulating evidence supports roles for ALDHs in the modulation of cell proliferation, differentiation and survival, especially through participation in retinoic acid synthesis [2]. Members of this superfamily also exhibit functions that appear to be independent of their enzyme activity, including absorption of ultraviolet (UV) irradiation in the cornea by acting as a crystallin and binding to hormones and other small molecules, including androgens, cholesterol, thyroid hormone and acetaminophen [2,5,6].
Sequencing of the human genome and subsequent identification of mutations in ALDH genes associated with loss of ALDH enzyme activity have led to the identification of many disease associations, such as cataracts (ALDH1A1, ALDH3A1, ALDH18A1), seizures (ALDH7A1), hyperprolinaemia (ALDH4A1), heart disease (ALDH2), alcohol sensitivity (ALDH1A1, ALDH1B1, ALDH2), certain cancers (ALDH2) and a broad array of other metabolic and developmental abnormalities [2]. Recently, a role for ALDHs in normal and cancer stem cells has also been identified. For example, ALDH1A1 is differentially expressed in human haematopoietic stem cells (HSCs) and can be used as a stem cell marker for multiple cancers [2]. Similarly, ALDH1B1 is primarily expressed in stem cells in the normal colon and is strongly upregulated in human colonic adenocarcinomas [7,8]. As described by Nelson and colleagues,[9] genomic gene artefact identification becomes very important when using genotyping techniques to identify disease-causing alleles. Gene-duplication events, leading to multiple functional and/or non-functional genetic copies in the genome, can significantly complicate polymerase chain reaction (PCR)-based genotyping assays. Transgenic animal models have permitted the exploration of the functions of ALDHs under in vivo physiological and pathophysiological conditions [2]. These invaluable studies are heavily dependent upon our understanding of the mouse and human genomes. In addition to mutations in ALDH genes within populations, there is a large variation in the number of ALDH genes between species.
During the past decade, the availability of gene and protein information has grown rapidly, primarily due to advances in gene-sequencing technologies. In the 2002 update of ALDH superfamily members,[10] 555 ALDH genes were listed, including 32 from Archaea, 351 from Eubacteria and 172 from Eukarya. Characteristic ALDH motifs were searched in 74 genomes: 16 in Archaea, 51 in Eubacteria and seven in Eukarya. A recent download from the current Pfam database (build version 24.0) includes 16,765 ALDH entries (listed as aldedh in the Pfam database) [11]. This update focuses on 11 representative vertebrate species in which the full genome has been sequenced: five primates, the cow, two rodents, two birds and one fish. Many of these genomes have been annotated automatically; generous algorithms list pseudogenes as protein-coding genes. This update attempts to describe the ALDH complement within these organisms and identify pseudogenes and gene-duplication events, when possible.
Methods
Fully sequenced genomes from 11 representative species: primates (human, Homo sapiens; common chimpanzee, Pan troglodytes; common marmoset, Callithrix jacchus; Sumatran orangutan, Pongo abelii; Rhesus macaque, Macaca mulatta), the cow (Bos taurus), rodents (mouse, Mus musculus; rat, Rattus norvegicus), birds (zebra finch, Taeniopygia guttata; domestic chicken, Gallus gallus) and one fish (zebrafish, Danio rerio were analysed).
ALDH genes were retrieved from Entrez Gene [12] using the terms 'ALDH' or 'aldehyde dehydrogenase'. Peptide sequences for each ALDH gene were retrieved from Entrez Protein [12] and aligned against a reference list of ALDH family members, including known human ALDHs and sequences from the NCBI's HomoloGene [12] using ClustalW [13]. To be included for description, a gene record was required to meet three criteria: 1) the protein product of the gene must be 'full-length' (ie excludes known fragments and partial records); 2) the gene must have a known unique chromosomal location on the annotated genome; and 3) the gene must be listed as protein-coding (ie excludes known pseudogenes).
Parent genes were designated based on highest homology to the known human protein. Identified gene duplications were sequentially named according to nomenclature guidelines, based on decreasing sequence homology to the parent gene. Duplicated genes were further analysed to determine if they represented potentially new protein-coding genes or non-functional pseudogenes. Pseudogenes were identified according to criteria outlined previously [9] and assigned to the following categories: detritus pseudogenes (those which are fragments missing exons) and reverse-transcriptase events (those which resemble mRNA sequences and lack introns). If data suggested that a duplicated gene was protein coding, it was considered to be a new gene family member and named according to the previously established ALDH nomenclature system [14]. Zebrafish aldh genes were named according to the guidelines set out by the zebrafish nomenclature committee (http://www.zfin.org) [15]. Pseudogenes in rodent (or fish) and non-rodent/non-fish genomes were appended with the suffix 'p' or 'P', respectively, and followed by a number designating multiple pseudogenes for a given gene family within each individual species.
It is, again, important to underscore that this initial analysis should be considered preliminary and subject to change as experimental evidence sheds light on actual protein function. Alignment and clustering of protein sequences were used as a basis for assigning homology. Sequences were aligned, and dendrograms based on neighbour-joining distances were created using a ClustalW webserver at (http://align.genome.jp). Percentage amino acid (AA) identities were determined using the Needle webserver at (http://www.ebi.ac.uk/Tools/emboss/align/) [16].
To assess whether protein sequences were actively transcribed, we employed several methods. Numerous promoter-prediction programs were used, but none was sufficiently consistent across species or discriminatory to be useful in the prediction of pseudogenes. The ratio of non-synonymous to synonymous (Ka/Ks) nucleotidesubstitution rates was used as a measure of selective pressure on each individual gene. Rates were calculated using homologous genes for all species in the current analysis, in order to determine ancestral states using the Bergen Center Ka/Ks Calculation Tool (http://services.cbu.uib.no/tools/kaks/) and default values, with the exception that the tree method was set to maximum likelihood [17].
Copy number variants (CNV; defined here as gains and losses of DNA sequences > 1 kiIobase [kb]), insertions and deletions (InDels; gains and losses of DNA sequences of 100-999 base pairs [bp]), and inversions in human ALDH genes were retrieved from the Database of Genomic Variants [18].
Results
Records for ALDH genes were retrieved and sorted for all 11 species analysed (Table 1). The number of records that met the above-mentioned criteria is provided (ie the number of genes excluding nonfunctional pseudogenes). The number of ALDH genes per species varied from 14 in chicken to 25 in zebrafish. There are currently 207 distinct genes present within the database for these 11 species; this is a greater than fourfold increase from 2002, when only 51 were annotated [10]. This allows for a much more comprehensive comparison of ALDH superfamily members throughout vertebrate evolution during the past 450 million years. It is important to keep in mind that, for many species, some genes have yet to be identified. Further, many annotated genes may reflect gene-duplication events that represent non-functional pseudogenes. These situations will be explored in greater depth below.
Table 1.
List of all species examined in the current study, including the Latin name and common name and the number of unique ALDH genes found in each species
| Latin name | Common name | # ALDH genes |
|---|---|---|
| Homo sapiens | Human | 19 |
| Pan troglodytes | Common chimpanzee | 18 |
| Callithrix jacchus | Common marmoset | 16 |
| Pongo abelii | Sumatran orangutan | 18 |
| Macaca mulatta | Rhesus macaque | 20 |
| Bos taurus | Cow | 20 |
| Rattus norvegicus | Norway rat | 21 |
| Mus musculus | House mouse | 21 |
| Taeniopygia guttata | Zebra finch | 15 |
| Gallus gallus | Chicken | 14 |
| Danio rerio | Zebrafish | 25 |
The data reflect the number of gene records found in the NCBI Gene Entrez database for each species, as of 13th March 2011
The total number of human annotations has remained unchanged since 2005, with 19 functional protein-coding genes [19]. The chimpanzee and the orangutan genomes diverged from humans ~5 and ~14 million years ago (MYA), respectively [20,21]. Both the chimpanzee and orangutan genomes contain 18 ALDH genes, each corresponding to a known human orthologue. The macaque and common marmoset genomes are more distantly related. They diverged ~25 and 35-40 MYA [22] and contain 20 and 16 ALDH members, respectively. Orthologues for all 19 human genes were identified in mouse and rat. In addition, rodent genomes contain an Aldh1a1 paralogue (Aldh1a7) and an Aldh3b2 gene duplication, resulting in a total of 21 Aldh genes. The most recent common ancestor of humans and rodents lived 75-90 MYA.
The cow genome, which diverged from that of the human 80-100 MYA, has 20 annotated ALDH entries which, again, closely parallel human members. Variations include two gene duplications and one possible deletion. Both avian genomes currently lack orthologous entries for ALDH1A1, ALDH1B1, ALDH1L1, ALDH3A1, ALDH3B2 and ALDH16A1. Moreover, the zebra finch genome is also missing annotated sequences for ALDH18A1 and includes two apparent gene duplications.
Table 2 summarises these ALDH orthologues, their chromosomal locations and the associated NCBI Entrez gene identification (ID) number for each of the 11 species. For zebrafish, Entrez gene ID 100334142 was listed as 'aldehyde dehydrogenase 1A1-like [D. rerio]'. This gene record appears to be derived from an unplaced chromosomal fragment, however, because no genome location could be determined. In addition, alignment of the peptide sequence for this gene ID to other mammalian ALDH1A1 protein sequences was poor. Specifically, sequence homology with human, mouse and rat ALDH1A1 was only 26.2 per cent, 26.4 per cent and 26.8 per cent, respectively. NCBI BlastP analysis indicated that it most closely resembles bacterial ALDH proteins. Together, this evidence suggests that this record may represent bacterial contamination, rather than a true zebrafish gene; thus, we have not included this gene. This also makes the zebrafish the only species among the 11 analysed that lacks a record for ALDH1A1. Interestingly, a protein blast (blastp) search using human ALDH1A1 and limiting results to fish species only (NCBI taxid: 7898) revealed ALDH1A2 homologues in multiple species (including salmon, pufferfish, ricefish and bichir), but no records for ALDH1A1 in any fish species. This is consistent with previous findings that indicate that ALDH1A1 is not present in the teleost lineage [23].
Table 2.
ALDH genes and duplicated genes across species with respective chromosome (Chr) locations
| Gene (by homology) | Primates | Rodents | Birds | Fish Zebrafish* |
||||
|---|---|---|---|---|---|---|---|---|
| Human | Orangutan | Cow | Rat | Mouse | Zebra finch | Chicken | ||
| ALDH1A1 | 9q21.13 (216) | 9 (100174688) | 8 (281615) | 1q51 (24188) | 19 12.0 cM (11668) | Z (100223406) | Z (395264) | |
| ALDH1A2 | 15q21.3 (8854) | 15 (100171834) | 10 (535075) | 8q24 (116676) | 9 42.0 cM (19378) | 10 (751771) | 10 (395884) | 7 (116713) |
| ALDH1A3 | 15q26.3 (220) | 15 (100452276) | 21 (507093) | 1q22 (266603) | 7 (56847) | 10 (100231202) | 10 (395389) | 7 (751785) |
| 28 (534200) | ||||||||
| ALDH1A7 | 1q51 (29651) | 19 20.0 cM (26358) | ||||||
| ALDH1B1 | 9q11.1 (219) | 9 (100174654) | 8 (281618) | 5q22 (298079) | 4 B2 (72535) | |||
| ALDH1L1 | 3q21.3 (10840) | 3 (100172380) | 3 (505677) | 4 (64392) | 6 (107747) | 6 (798292) | ||
| ALDH1L2 | 12q23.3 (160428) | 12 (100459691) | 5 (516864) | 7q13 (299699) | 10 (216188) | 1A (100230131) | 1 (418078) | 4 (100333269) |
| ALDH2 | 12q24.2 (217) | 12 (100171596) | 17 (508629) | 12q16 (29651) | 5 F-G1 (11669) | 15 (100217978) | 15 (416880) | 5 (393462) |
| 5 (368239) | ||||||||
| 5 (100332355) | ||||||||
| ALDH3A1 | 17p11.2 (218) | 17 (100446485) | 19 (281617) | 10q22 (25375) | 11 34.25 cM (11670) | |||
| ALDH3A2 | 17p11.2 (224) | 17 (100171557) | 19 (513967) | 10q22 (65183) | 11 34.3 cM (11671) | 19 (100230924) | 19 (417615) | 15 (323653) |
| 19 (100226132) | 15 (100000026) | |||||||
| 21 (100329417) | ||||||||
| 21 (447920) | ||||||||
| ALDH3B1 | 11q13 (221) | 11 (100450634) | 29 (511469) | 1q42 (309147) | 19 (67689) | 5 (100232483) | 5 (428813) | 5 (557008) |
| 29 (508879) | 5 (100229547) | |||||||
| ALDH3B2 | 11q13 (222) | 1q42 (688800) | 19 (621603) | |||||
| 1q42 (688778) | 19 (73458) | |||||||
| ALDH3D1 | 3 (282559) | |||||||
| ALDH4A1 | 1p36 (8659) | 1 (10072770) | 2 (100126042) | 5q36 (641316) | 4 66.1 cM (212647) | 21 (100228902) | 21 (419467) | 11 (394133) |
| ALDH5A1 | 6p22 (7915) | 6 (100458767) | 23 (532724) | 17p11 (291133) | 13 A3.1 (214579) | 2 (100222151) | 2 (420818) | 16 (565235) |
| 16 (100330723) | ||||||||
| ALDH6A1 | 14q24.3 (4329) | 14 (100171652) | 10 (327692) | 6q31 (81708) | 12 39.0 cM (104776) | 5 (100226750) | 5 (423345) | 17 (436647) |
| ALDH7A1 | 5q31 (501) | 5 (100461726) | 7 (507477) | 18q12.1 (291450) | 18 29.0 cM (110695) | Z (100223716) | Z (426812) | 10 (334197) |
| ALDH8A1 | 6q23.2 (64577) | 6 (100450228) | 9 (513537) | 1p12 (685750) | 10 (237320) | 3 (100222753) | 3 (421695) | 23 (447801) |
| ALDH9A1 | 1q23.1 (223) | 1 (100173126) | 3 (537539) | 13q24 (64040) | 1 H2 (56752) | 8 (100225645) | 8 (424405) | 8 (100005587) |
| 2 (399481) | ||||||||
| 8 (100006238) | ||||||||
| ALDH16A1 | 19q13.33 (126133) | 19 (100434496) | 18 (506329) | 1q22 (361571) | 7 (69748) | 3 (492710) | ||
| ALDH18A1 | 10q24.3 (5832) | 10 (100173488) | 26 (514759) | 1q54 (361755) | 19 (56454) | 6 (423976) | 12 (557186) | |
| 12 (100329417) | ||||||||
Numbers in parentheses indicate NCBI Entrez gene ID (GI). Records in bold text denote duplications compared with the human genome. Z, the sex Chr in birds (ZW system); cM, centiMorgans. Letter designations in mouse gene locations indicate chromosomal regions
*Zebrafish genes are named in accordance with nomenclature guidelines described at (http://www.zfin.org) and established by Mullins et al. [15].
We found evidence for several gene duplications. Table 3 lists all genes that show duplications, compared with genes in the human genome. This table provides a summary of existing information available within the NCBI gene entries, as well as recommended gene names based on our analyses and current nomenclature guidelines.
Table 3.
List of the Entrez Gene genes ID (GI), chromosome location, presence of introns, gene type and recommended gene name of all ALDH genes in this study that show evidence of gene duplication, compared with that in the human genome
| Gene (by homology) | Species | NCBI Gene ID | NCBI Gene name | Chromosome | Chromosomal location | Introns | Gene type | Recommended gene name | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref Seq ID | Range | |||||||||
| ALDH1A3 | Cow | 507093 | ALDH1A3 | 21 | NC_007319.4 | 4,261,104 | 4,301,275 | yes | Parent gene | ALDH1A3 |
| 534200 | LOC534200 | 28 | NC_007329.4 | 11,750,749 | 11,762,637 | yes | Pseudogene-- detritus | ALDH1A3P1 | ||
| ALDH2 | Zebrafish* | 393462 | aldh2a | 5 | NC_007116.4 | 71,734,127 | 71,754,941 | yes | Parent gene | aldh2.1* |
| 368239 | aldh2b | 5 | NC_007116.4 | 71,708,861 | 71,732,452 | yes | New gene | aldh2.2* | ||
| 100332355 | LOC100332355 | 5 | NC_007116.4 | 71,632,543 | 71,658,511 | yes | New gene | aldh2.3* | ||
| ALDH3A2 | Zebrafish* | 323653 | adh3a2 | 15 | NC_007126.4 | 21,001,391 | 21,009,951 | yes | Parent gene | aldh3a2.1* |
| 100000026 | LOC100000026 | 15 | NC_007126.4 | 20,970,670 | 20,976,922 | yes | New gene | aldh3a2.2* | ||
| 100329417 | LOC100329417 | 21 | NC_007132.4 | 40,585,351 | 40,617,892 | yes | New gene | aldh3a2.3* | ||
| 447920 | zgc:103715 | 21 | NC_007132.4 | 40,905,693 | 40,917,184 | yes | Pseudogene-- detritus | aldh3a2p1 | ||
| Zebra finch | 100230924 | LOC100230924 | 19 | NC_011483.1 | 8,354,898 | 8,361,968 | yes | Parent gene | ALDH3A2 | |
| 100226132 | LOC100226132 | 19 | NC_011483.1 | 8,364,080 | 8,368,708 | yes | New gene | ALDH3A3 | ||
| ALDH3B1 | Cow | 511469 | ALDH3B1 | 29 | NC_007330.4 | 47,708,146 | 47,722,523 | yes | Parent gene | ALDH3B1 |
| 508879 | LOC508879 | 29 | NC_007330.4 | 47,568,715 | 47,575,449 | yes | New gene | ALDH3B4 | ||
| Zebra finch | 100232483 | LOC100232483 | 5 | NC_011469.1 | 7,960,933 | 7,967,624 | yes | Parent gene | ALDH3B1 | |
| 100229547 | LOC100229547 | 5 | NC_011469.1 | 7,968,165 | 7,973,465 | yes | New gene | ALDH3B5 | ||
| ALDH3B2 | Rat | 688800 | ALDH3B2 | 1 | NC_005100.2 | 206,549,529 | 206,553,424 | yes | Parent gene | ALDH3B2 |
| 688778 | LOC688778 | 1 | NC_005100.2 | 206,500,430 | 206,510,746 | yes | New gene | ALDH3B3 | ||
| Mouse | 621603 | Aldh3b2 | 19 | NC_000085.5 | 3,972,328 | 3,981,665 | yes | Parent gene | Aldh3b2 | |
| 73458 | 1700055N04Rik | 19 | NC_000085.5 | 3,958,808 | 3,969,947 | yes | New gene | Aldh3b3 | ||
| ALDH5A1 | Zebrafish* | 565235 | aldh5a1 | 16 | NC_007127.4 | 35,584,243 | 35,592,745 | yes | Parent gene | aldh5a1.1* |
| 100330723 | LOC100330723 | 16 | NC_007127.4 | 35,723,717 | 35,735,263 | yes | New gene | aldh5a1.2* | ||
| ALDH7A1 | Macaque | 702749 | ALDH7A1 | 6 | NC_007863.1 | 122,937,640 | 122,989,782 | yes | Parent gene | ALDH7A1 |
| 716090 | LOC716090 | 14 | NC_007871.1 | 68,342,919 | 68,344,780 | no | Pseudogene-- RTevent | ALDH7A1P5 | ||
| ALDH9A1 | Zebrafish* | 100005587 | aldh9a1a | 8 | NC_007119.4 | 21,476,877 | 21,484,987 | yes | Parent gene | aldh9a1.1* |
| 399481 | aldh9a1b | 2 | NC_007113.4 | 4,838,438 | 4,863,128 | yes | New gene | aldh9a1.2* | ||
| 100006238 | LOC100006238 | 8 | NC_007119.4 | 21,464,110 | 21,473,710 | yes | New gene | aldh9a1.3* | ||
| ALDH18A1 | Zebrafish* | 557186 | aldh18a1 | 12 | NC_007123.4 | 29,670,615 | 29,686,508 | yes | Parent gene | aldh18a1.1* |
| 100332705 | LOC100332705 | 12 | NC_007123.4 | 29,643,982 | 29,661,436 | yes | New gene | aldh18a1.2* | ||
*Zebrafish genes are named in accordance with nomenclature guidelines described at (http://www.zfin.org) and established by Mullins et al. [15].
RT, reverse transcription
Table 4 lists additional information related to peptide sequences and calculated sequence identities. Additional genes (increase in gene number, compared with humans) show peptide divergence of as little as 0.4 per cent (zebrafish aldh2.2 and aldh2.3) and as much as 64.9 per cent (zebrafish aldh3a2.1 and aldh3a2.2). In most cases, gene duplications have similar sizes, are often nearby on the same chromosome (Chr) and show some degree of divergence (ie 70-95 per cent AA identity). Genes that have portions of the gene copied with no AA divergence include: cow ALDH1A3P1 (127 of 537 AAs), zebrafish aldh5a1.2 (404 of 514 AAs) and zebrafish aldh18a1.2 (782 of 782 AAs). Zebrafish aldh3a2.3 (169 of 514 AAs) represents a shortened copy which shows minor divergence (98.4 per cent identity). Ka/Ks ratios were calculated for all gene duplications. A value of < 1.0 indicates selective pressure to conserve the gene and suggests that it plays a functional role. All duplications were found to have a score of < 1.0, except macaque ALDH7A1P5 (to be discussed below).
Table 4.
Tabulation of all ALDH genes in this study that show evidence of gene duplication, compared with that in the human genome
| Species | Recommended gene name | RefSeq Protein ID | Protein length | Ka/Ks | Aligned sequences | % AA (unaligned) | % AA Identity (unaligned included) | % AA Identity (unaligned excluded) | Functional protein | Recommended protein name |
|---|---|---|---|---|---|---|---|---|---|---|
| Cow | ALDH1A3 | XP_583647.3 | 537 | 0.234 | - | - | - | - | Yes | ALDH1A3 |
| ALDH1A3P1 | XP_001789867.1 | 127 | 0.260 | (a)/(b) | 76.4 | 23.6 | 100 | No | Pseudogene | |
| Zebrafish | aldh2.1 | NP_956784.1 | 516 | 0.278 | - | - | - | - | Yes | Aldh2.1 |
| aldh2.2 | NP_998466.2 | 516 | 0.112 | (a)/(b) | 0 | 95.2 | 95.2 | Yes | Aldh2.2 | |
| aldh2.3 | XP_002662252.1 | 516 | 0.041 | (a)/(c) | 0 | 95.2 | 95.2 | Yes | Aldh2.3 | |
| (b)/(c) | 0 | 99.6 | 99.6 | - | - | |||||
| Zebrafish | aldh3a2.1 | NP_997814.1 | 488 | 0.175 | - | - | - | - | Yes | Aldh3a2.1 |
| aldh3a2.2 | XP_001335979.2 | 489 | 0.402 | (a)/(b) | 1.8 | 63.1 | 64.9 | Yes | Aldh3a2.1 | |
| aldh3a2.3 | XP_002666107.1 | 514 | 0.175 | (a)/(c) | 5.1 | 65.8 | 70.9 | Yes | Aldh3a2.3 | |
| aldh3a2p1 | NP_001004658.1 | 169 | 0.190 | (a)/(d) | 65.6 | 23.5 | 89.1 | No | Pseudogene | |
| (b)/(c) | 7.5 | 57.4 | 64.9 | - | - | |||||
| (b)/(d) | 66 | 18.9 | 84.9 | - | - | |||||
| (c)/(d) | 67.1 | 31.3 | 98.4 | - | - | |||||
| Zebra finch | ALDH3A2 | XP_002198810.1 | 510 | 0.396 | - | - | - | - | Yes | ALDH3A2 |
| ALDH3A3 | XP_002196134.1 | 526 | 0.625 | (a)/(b) | 5.6 | 84.1 | 89.7 | Yes | ALDH3A3 | |
| Cow | ALDH3B1 | NP_001068986.1 | 486 | 0.335 | - | - | - | - | Yes | ALDH3B1 |
| ALDH3B4 | XP_585724.2 | 486 | 0.550 | (a)/(b) | 4.5 | 80.9 | 85.4 | Yes | ALDH3B4 | |
| Zebra finch | ALDH3B1 | XP_002196917.1 | 450 | 0.308 | - | - | - | - | Yes | ALDH3B1 |
| ALDH3B5 | XP_002196928.1 | 341 | 0.434 | (a)/(b) | 39.9 | 53.2 | 93.1 | Yes | ALDH3B5 | |
| Rat | Aldh3b2 | XP_001068348.2 | 483 | 0.436 | - | - | - | - | Yes | ALDH3B2 |
| Aldh3b3 | XP_001068253.1 | 530 | 0.239 | (a)/(b) | 11 | 76.9 | 87.9 | Yes | ALDH3B3 | |
| Mouse | Aldh3b2 | NP_001170909.1 | 479 | 0.270 | - | - | - | - | Yes | ALDH3B2 |
| Aldh3b3 | XP_900106.1 | 479 | 0.229 | (a)/(b) | 0 | 86.4 | 86.4 | Yes | ALDH3B3 | |
| Zebrafish | aldh5a1.1 | NP_001103938.1 | 404 | < 0.001 | - | - | - | - | Yes | Aldh5a1.1 |
| aldh5a1.2 | XP_002664997.1 | 514 | 0.008 | (a)/(b) | 21.4 | 78.6 | 100 | Yes | Aldh5a1.2 | |
| Macaque | ALDH7A1 | XP_002804539.1 | 502 | 0.180 | - | - | - | - | Yes | ALDH7A1 |
| ALDH7A1P5 | XP_001111963.1 | 538 | 1.289 | (a)/(b) | 16.3 | 82.3 | 98.6 | No | Pseudogene | |
| Zebrafish | aldh9a1.1 | NP_958879.1 | 508 | 0.126 | - | - | - | - | Yes | Aldh9a1.1 |
| aldh9a1.2 | NP_958916.1 | 518 | 0.154 | (a)/(b) | 1.9 | 71.2 | 73.1 | Yes | Aldh9a1.2 | |
| aldh9a1.3 | NP_001119952.1 | 508 | 0.190 | (a)/(c) | 0 | 94.9 | 94.9 | Yes | Aldh9a1.3 | |
| (b)/(c) | 1.9 | 70.3 | 72.2 | - | - | |||||
| Zebrafish | aldh18a1.1 | NP_001077015.1 | 782 | 0.103 | - | - | - | - | Yes | Aldh18a1.1 |
| aldh18a1.2 | XP_002664020.1 | 782 | 0.826 | (a)/(b) | 0 | 100 | 100 | Yes | Aldh18a1.2 |
Included are protein lengths (in number of amino acids [AAs]), Ka/Ks values, RefSeq protein IDs and recommended protein names. '% AA identity' denotes the absolute number of identical AAs relative to the absolute number of AA locations. '% AA unaligned' indicates the percentage of AAs that are represented by either a gap in the alignment of either sequence or an overhang if one sequence is longer than the other. '% AA identity (unaligned excluded)' indicates the percentage of AA locations that are identical when unaligned AAs are excluded from the total number of AA locations. For example, a 127-AA fragment of a 537-AA protein, which is identical except for the truncation, would have 127/537 = 23.6 per cent identity, of which 410/537 = 76.4 per cent is represented by unaligned residues (AAs in the longer sequence that have no correlation with the shorter sequence) but, excluding those residues, 127/127 = 100 per cent paired AAs are identical. The final column indicates which sequences are being compared for percentage identity, percentage gaps and percentage identity (excluding gaps)
ALDH1
ALDH1A1 is present in all species except zebrafish, confirming earlier studies [23]. In cow, there are two distinct records for ALDH1A3: the gene found on Chr 21 is full length (537 AAs) and represents the putatively functional parent gene (ALDH1A3), whereas the second is a detritus pseudogene on Chr 28 which appears to the product of a partial gene-duplication event (ALDH1A3P1). The shorter genomic sequence would translate a peptide sharing 100 per cent sequence identity to only the 127 carboxy-terminal AAs of the full-length parent protein. Several gene duplications appear to have been conserved in rodents. One such gene is ALDH1A7, found in rats and mice. In both cases, the ALDH1A7 gene is present on the same chromosome and in close proximity to ALDH1A1. Mouse ALDH1A7 shares 92 per cent AA identity with mouse ALDH1A1, and studies have confirmed that the gene encodes inducible tissue-specific mRNA [24]. ALDH1B1 is present in mammals but missing from birds and fish. ALDH1L1 is missing from both bird species (zebra finch and chicken) but present in other species examined and thus may represent a deletion in the avian lineage.
ALDH2
ALDH2 appears to be one of many genes duplicated in zebrafish. It has been suggested that an entire genome duplication event may have occurred after the divergence of teleosts and mammals;[25] this may explain the increased ALDH gene number in zebrafish. A second gene-duplication event appears to have occurred, giving rise to three zebrafish aldh2 gene records (aldh2.1-3). The aldh2.1 gene is believed to be the parent, based on homology with orthologous ALDH2 protein sequences. Both aldh2.2 and aldh2.3 potentially encode full-length peptides. Aldh2.2 is 95.2 per cent similar to Aldh2.1 and aldh2.3 may represent a more evolutionarily recent duplication of aldh2.2, as evidenced by 99.6 per cent AA identity between the Aldh2.2 and Aldh2.3 proteins.
ALDH3
The ALDH3 genes show the most variation in gene number of any ALDH family among the organisms studied. ALDH3A1 is missing from birds and fish but is present in every mammalian genome analysed in this study. The zebra finch has a duplicate ALDH3A2 (ALDH3A3) entry which encodes a full-length peptide that shares 84.1 per cent identity with the parent protein. Four ALDH3A2 homologues were identified within the zebrafish genome. The aldh3a2.1 is considered the parent gene. The aldh3a2.2 and aldh3a2.3 full-length gene products, respectively, share 64.9 per cent and 70.9 per cent sequence identity with that of Aldh3a2.1 and 64.9 per cent identity with each other. Zebrafish aldh3a2p1 represents a partial gene duplication; the resulting 169-AA peptide would most likely undergo proteolytic degradation if translated.
ALDH3B1 is duplicated in cow and zebra finch, as well as in zebrafish, on the proviso that D. rerio aldh3d1 is also considered an ALDH3B1 homologue. Zebrafish Aldh3d1 shares 44 per cent AA identity with Aldh3b1 and is listed in NCBI HomoloGene as a homologue of ALDH3B1 (HomoloGene, data not shown) [12]. Zebra finch ALDH3B5 encodes a 341-AA peptide that shares 100 per cent sequence identity with the 228 amino-terminal AAs of the parent gene's protein. Cow and zebra finch ALDH3B4 and ALDH3B5 proteins share 80.9 per cent and 53.2 per cent sequence identity with their respective parent genes, and 39.7 per cent with one another, indicating that none of the genes is an orthologue. Zebra finch ALDH3B5 is shorter than ALDH3B1 (341 versus 450 AAs) and, without this sequence gap, they share 93.1 per cent AA identity; it is unknown whether this smaller gene product is functional.
ALDH3B2 is present as a single distinct gene in human, chimpanzee and macaque, whereas two copies occur in mouse and rat. ALDH3B2 is absent from common marmoset, cow, zebra finch, chicken and zebrafish. Mouse and rat ALDH3B3 share 86.4 per cent and 76.9 per cent AA identity, respectively, with the corresponding parent ALDH3B2 proteins and 83.4 per cent identity with each other. The two ALDH3B3 genes are found on corresponding syntenic chromosomes within their respective genomes. Presently, the protein product of Entrez Gene ID 688778 (R. norvegicus) is annotated as 'ALDH3B1 (predicted)'. Based on a phylogenetic clustering of ALDH3B1 and ALDH3B2 protein sequences (Figure 1), however, we believe it is better to name this protein ALDH3B3; this shows that both mouse and rat ALDH3B3 proteins are in the ALDH3B2 clade and are more similar to each other than to rodent or human ALDH3B2 proteins. The alignment used for phylogenetic clustering can be seen in Supplementary Table S1 (Table 5).
Figure 1.
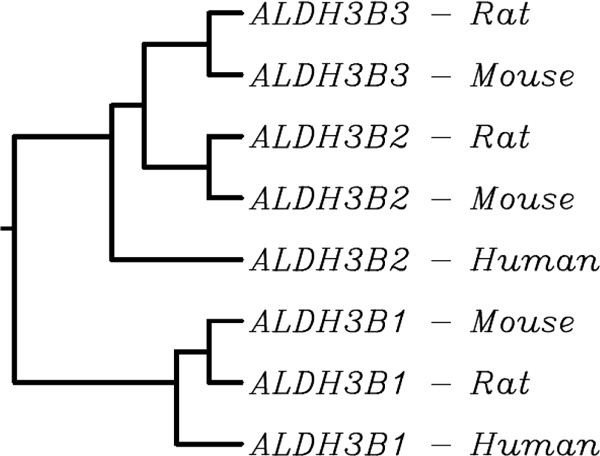
Neighbour-joining dendrogram (with branch lengths representing relative protein sequence similarity) of ALDH3B sequences in human, rat and mouse, indicating the likely homology and identity of the genes assigned 'Aldh3b3' in rat and mouse.
Table S1.
Known copy number variations in humans
| Variation ID | ALDH | Type | Gain/loss | Site | Sample size (variant/controls) | Chr |
|---|---|---|---|---|---|---|
| 26310 | 16A1 | InDel | Gain | Intron | 1/1 | 19q13.33 |
| 26311 | 16A1 | InDel | Gain | Intron | 1/1 | 19q13.33 |
| 26312 | 16A1 | InDel | Gain | Intron | 1/1 | 19q13.33 |
| 26313 | 16A1 | InDel | Loss | Intron | 1/1 | 19q13.33 |
| 109892 | 1A1 | InDel | Gain | Intron | 1/1 | 9q21.13 |
| 102109 | 1A2 | CNV | Loss | Intron | 1/1 | 15q22.1 |
| 25534 | 1A2 | InDel | Loss | Intron | 1/1 | 15q22.1 |
| 40101 | 1A2 | InDel | Loss | Intron | 1/1 | 15q22.1 |
| 41386 | 1A2 | InDel | Loss | Intron | 1/1 | 15q22.1 |
| 45349 | 1A2 | InDel | Loss | Intron | 1/1 | 15q22.1 |
| 45350 | 1A2 | InDel | Loss | Intron | 1/1 | 15q22.1 |
| 102186 | 1A3 | CNV | Loss | Intron | 1/1 | 15q26.3 |
| 11819 | 1A3 | InDel | Loss | Intron | 1/36 | 15q26.3 |
| 25599 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 25600 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 25601 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 40124 | 1A3 | InDel | Loss | Intron | 1/2 | 15q26.3 |
| 42429 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 42898 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 45395 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 61482 | 1A3 | InDel | Loss | Intron | 1/1 | 15q26.3 |
| 68446 | 1L1 | InDel | Loss | Intron | 1/39 | 3q21.2 |
| 106822 | 1L2 | CNV | Gain | Intron | 1/1 | 12q23.3 |
| 42760 | 3A2 | InDel | Loss | Intron | 1/1 | 17p11.2 |
| 24787 | 3B2 | InDel | Loss | Intron | 1/1 | 11q13.2 |
| 44926 | 3B2 | InDel | Loss | Intron | 1/1 | 11q13.2 |
| 81276 | 5A1 | InDel | Gain | Intron | 1/90 | 6p22.2 |
| 93550 | 5A1 | CNV | Loss | Intron | 2/90 | 6p22.2 |
| 99466 | 5A1 | CNV | Loss | Intron | 1/1 | 6p22.2 |
| 33982 | 7A1 | InDel | Gain | Intron | 1/1 | 5q23.2 |
| 97538 | 9A1 | InDel | Gain | Intron | 1/1 | 1q24.1 |
| 23991 | 9A1 | InDel | Gain | Intron | 1/1 | 1q24.1 |
| 11004 | 9A1 | InDel | Loss | Intron | 15/50 | 1q24.1 |
| 35661 | 16A1 | CNV | Gain | Part | 1/1 | 19q13.33 |
| 114045 | 1A3 | CNV | Gain | Part | 1/30 | 15q26.3 |
| 72379 | 1A3 | CNV | Loss | Part | 1/39 | 15q26.3 |
| 4352 | 1L1 | CNV | 2G 1L | Part | 3/95 | 3q21.2 |
| 59786 | 1L1 | Inv | Inversion | Part | 1/1 | 3q21.2 |
| 68445 | 1L1 | CNV | Loss | Part | 1/39 | 3q21.2 |
| 107014 | 1L2 | CNV | Loss | Part | 1/1 | 12q23.3 |
| 88379 | 3A2 | CNV | Loss | Part | 1/90 | 17p11.2 |
| 88381 | 3A2 | CNV | Loss | Part | 1/90 | 17p11.2 |
| 3140 | 3A2 | CNV | Loss | Part | 4/270 | 17p11.2 |
| 65982 | 3B2 | CNV | Gain | Part | 2/450 | 11q13.2 |
| 85827 | 3B2 | CNV | Loss | Part | 2/90 | 11q13.2 |
| 53128 | 3B2 | CNV | Loss | Part | 2/1064 | 11q13.2 |
| 3055 | 6A1 | CNV | Gain | Part | 1/270 | 14q24.3 |
| 66668 | 6A1 | CNV | Loss | Part | 2/450 | 14q24.3 |
| 6793 | 9A1 | CNV | Loss | Part | 2/50 | 1q24.1 |
| 3856 | 3B1 | CNV | Gain/loss | Whole | 3/270 | 11q13.2 |
| 113072 | 3B1 | CNV | Gain | Whole | 1/30 | 11q13.2 |
| 30558 | 3B1 | CNV | Gain | Whole | 1/1 | 11q13.2 |
| 5275 | 3B2 | CNV | Gain | Whole | 1/272 | 11q13.1-11q13.2 |
| 5111 | 16A1 | CNV | Loss | Whole | 25/95 | 19q13.33 |
| 32261 | 16A1 | CNV | Loss | Whole | 18/30 | 19q13.32-19q13.33 |
| 5110 | 16A1 | CNV | Loss | Whole | 4/95 | 19q13.33 |
| 2201 | 1A3 | CNV | Loss | Whole | 3/269 | 15q26.3 |
| 47939 | 1B1 | CNV | Loss | Whole | 6/2906 | 9p13.1 |
| 30022 | 3A1 | CNV | Loss | Whole | 2/485 | 17p11.2 |
| 53160 | 3B1 | CNV | Loss | Whole | 2/1064 | 11q13.2 |
| 2931 | 3B1 | CNV | Loss | Whole | 8/270 | 11q13.2 |
| 29913 | 3B1 | CNV | Loss | Whole | 1/485 | 11q13.2 |
| 29914 | 3B1 | CNV | Loss | Whole | 1/485 | 11q13.2 |
| 47969 | 5A1 | CNV | Loss | Whole | 9/2906 | 6p22.2 |
Included are the variation ID from the Database of Genomic Variants, ALDH family member, type (CNV - copy number variation with changes > 1 kb; InDel - insertions and deletions with changes 100-999 bp; inv --inversions with changes that invert the nucleotide sequence), whether the change was a loss or gain, site (intron -- change only affects an intronic region; part -- change affects one or more exons; whole -- change affects the entire gene), sample size and chromosomal location
ALDH4
ALDH4A1 is missing from chimpanzee and common marmoset but is present in all others. Previously, rat ALDH4A1 had been conspicuously absent from the major databanks but it was recently added. During a BLAST search of the rat genome using various individual exon segments from mouse Aldh4a1, significant hits for Aldh4a1 in the rat genome were identified on Chr 5q36 and it was determined to be a part of the fusion gene LRRP Ba1-651 [26]. Figure 2 shows an assembled structure of this fusion gene with the Aldh4a1 exons highlighted in red. Although it appears that these exons are transcribed and contain the conserved ALDH catalytic domain, it is not clear whether the gene product retains aldehyde dehydrogenase activity.
Figure 2.
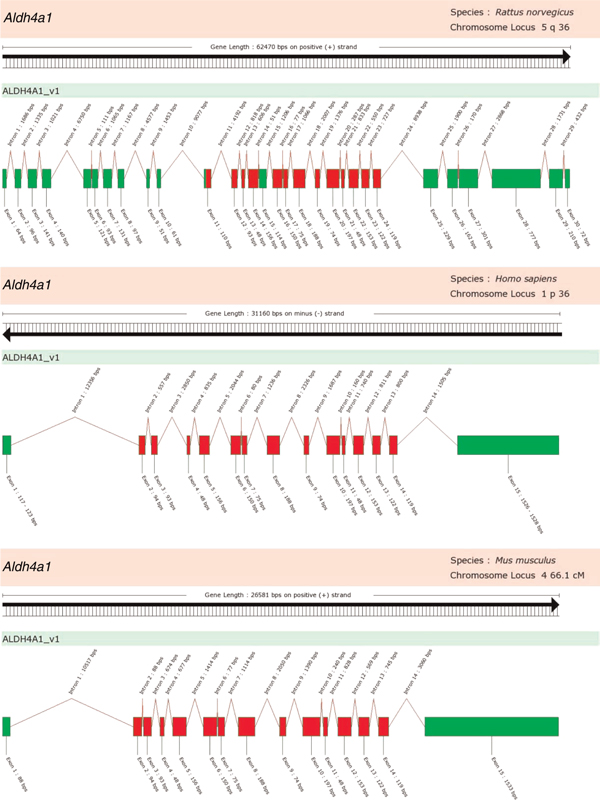
Comparison of ALDH4A1 from human and rat. Rat Aldh4a1 is part of the larger fusion gene LRRP Ba1-651 [26]. The exons representing the Aldh4a1 portion of this gene with homology to mouse and human are highlighted.
ALDH5 and beyond
ALDH5A1 is missing in marmoset and duplicated in zebrafish. The zebrafish duplication, aldh5a1.2, encodes a slightly truncated peptide (404 versus 514 AAs) which shares 100 per cent AA identity with the first 426 AAs and resides on the same Chr as aldh5a1.1.
ALDH7A1 is duplicated in the macaque. The ALDH7A1P5 duplication is located on Chr 14 and contains the complete ALDH7A1 coding sequence; however, the sequence lacks any intronic regions, suggesting a reverse transcriptase-mediated duplication event. Furthermore, this gene has a Ka/Ks score of 1.289, indicating a lack of selective pressure to conserve this gene. This provides further evidence that ALDH7A1P5 does not code for a functional protein.
In zebrafish, aldh9a1 has three additional copies. The parent gene aldh9a1.1 and aldh9a1.3 reside on Chr 8; aldh9a1.2 is found on Chr 2. Both aldh9a1.2 and aldh9a1.3 encode putative full-length proteins which respectively share 71.2 per cent and 94.9 per cent AA identity with Aldh9a1.1 and 70.3 per cent sequence identity with each other. Zebrafish also contains a duplication of aldh18a1. The aldh18a1.2 is found on the same chromosome and encodes a protein that is 100 per cent identical with that of the parent gene.
The naming of zebrafish genes required further genomic analyses in order to determine whether duplications originated from the ray-finned lineage whole-genome duplication event. Many of the duplicated genes reside within close proximity on the same chromosome, suggesting that they are segmental duplications that resulted from misguided recombination processes during meiosis and not a product of the whole genome duplication that took place within the ray-fin lineage [27]. These include the aldh2, aldh5a1 and aldh18a1 paralogues, which are located in close proximity on Chr 5, 16 and 12, respectively. It also includes aldh3a2.1 and aldh3a2.2, located on Chr 15, as well as aldh9a1.1 and aldh9a1.3, found on Chr 8. The gene architecture surrounding aldh3a2.3 on Chr 21 does not support a duplicated chromosome, in that the region lacks other duplicated genes from Chr 15. Furthermore, studies looking at zebrafish gene duplications found that a high frequency of genes found on Chr 21 are duplicated on Chr 5 and none were identified on Chr 15, suggesting that Chr 5, rather than Chr 15, is the paralogous chromosome [28,29]. A similar situation was identified with respect to aldh9a1.2 on Chr 2. Uridine-cytidine kinase-2 homologues (uck2a and uck2b) are found upstream of both aldh9a1.1 and aldh9a1.2, supporting a tandem gene-duplication event; however, other genes in close proximity to this duplication do not show any homology between chromosomes 2 and 8.
Alternatively spliced transcriptional variants and CNVs of human ALDH genes
In addition to the increase in ALDH identification through genomic sequencing, other sources of complexity in the ALDH superfamily are being studied. Transcript sequencing has revealed that many ALDH genes encode multiple mRNA splice variants (for a review of human ALDH splice variants, see Black et al. [30]). Besides splice variants, CNVs have been reported for human ALDH genes. By querying the Database of Genomic Variants, 35 CNVs, 28 InDels and one inversion have been detected in the ALDH family, although these records are usually representative of one or several individuals (Supplementary Table S1 (Table 5)). Of these 64 events, 33 were InDels entirely within intronic regions and may be silent. Others are likely to cause loss of function of the enzyme involved, including loss of the whole gene (11 events; occurred in ALDHs 1A3, 1B1, 3A1, 3B1, 5A1 and 16A1) or duplication, loss or inversion of exons within the coding sequence (16 events; occurred in ALDHs 1A3, 1L1, 1L2, 3A2, 3B2, 6A1 and 9A1). Finally, in a few cases, a region containing the entire gene and surrounding region was duplicated (four events; occurred in ALDH3B1 and ALDH3B2).
Discussion
The ALDH superfamily shows considerable diversity among vertebrate genomes, with species in the current study showing between 14 and 25 putatively protein-encoding genes. Many of the gene duplications discussed here probably encode functional proteins. There are also a number of duplication events that give rise to non-functional pseudogenes. Names were assigned to the 'new genes' and 'pseudogenes' (Table 3) according to the ALDH nomenclature system established in 1999 [14]. The species-specific nomenclature system was used for zebrafish genes [15]. Pseudogenes were also named according to the standardised protocol [20].
In the cow genome, ALDH1A3P1 resembles the product of a partial gene duplication event. The coding region would translate a peptide sharing 100 per cent sequence identity to the 127 carboxy-terminal AAs of the full-length parent gene. Such a high degree of sequence identity is suggestive of a relatively recent evolutionary duplication. Even if the truncated gene encodes the 127-AA peptide; however, it lacks many highly conserved residues required for ALDH activity. Thus, the truncated peptide would probably be targeted for rapid degradation. As such, this gene represents a nonfunctional pseudogene and has been named accordingly.
ALDH1B1 is present in mammals but missing from birds and fish. The high degree of AA sequence conservation between ALDH2 and ALDH1B1 suggests that the latter may be the product of a gene duplication event that occurred some time after the avian-land animal split around 310 MYA. Future analyses should consider other species, including amphibians and reptiles, in order to verify and more accurately pinpoint this evolutionary event.
Analysis of the aldh2 gene duplications in zebrafish indicates that these represent protein-coding genes and not pseudogenes. As mentioned above, translation of either gene would result in a full-length peptide. The aldh2.2 gene would encode a product 95.2 per cent identical to that of the parent gene aldh2.1. At 95.2 per cent AA identity, aldh2.2 represents a new gene. The aldh2.3 homologue may represent a more evolutionarily recent duplication of aldh2.2, as evidenced by the ~99.6 per cent sequence identity noted. Therefore, aldh2.3 is likely to be a gene-duplication event of aldh2.2. All three protein products include the conserved ALDH motifs and residues required for enzyme activity.
The ALDH3 family showed the greatest variability among species. ALDH3A1 facilitates cell cycle regulation and scavenging of reactive oxygen species, and acts as a corneal crystallin by filtering UV irradiation in the eye. ALDH3A1 is missing from birds and fish but is present in every mammalian genome analysed in this study, suggesting that the gene evolved some time after 310 MYA. ALDH3A1 is conserved among mammals and shows no apparent duplications. In some species, such as rabbit, it appears that ALDH1A1 is expressed as a corneal crystallin instead of ALDH3A1 [31]. Interestingly, zebrafish is the only species in this study that apparently lacks both ALDH3A1 and ALDH1A1. Studies have suggested that zebrafish use scinla (cytosolic gelsolin) as a corneal crystallin instead [32-34].
Zebra finch ALDH3A3 encodes a full-length peptide that shares 84.1 per cent similarity with the ALDH3A2 parent gene. Zebrafish has three aldh3a2 duplications, which include two full-length genes (aldh3a2.2 and aldh3a2.3) and a significantly truncated partial duplication (aldh3a2p1). The degree of sequence identity that Aldh3a2.2 and Aldh3a2.3 share with the parent peptide (64.9 per cent and 70.9 per cent, respectively) suggests that they diverged sufficiently long ago to be considered new ALDH3A family members. They also share 64.9 per cent identity with each other and less than 60 per cent identity with zebra finch ALDH3A3, suggesting that all three genes are paralogues rather than orthologues. Zebra finch ALDH3A5 should also be considered a new functional ALDH family member. In addition, the zebrafish pseudogene aldh3a2p1, if translated, would share the highest degree of sequence identity with aldh3a2.3. Thus, the pseudogene most likely reflects a more recent partial duplication of this gene.
ALDH3B1 is duplicated in both cow and zebra finch. The cow ALDH3B4-encoded protein would be full length and share 85.4 per cent identity to ALDH3B1, suggesting that it is a new ALDH3B family member. Zebra finch ALDH3B5 shares an extremely high degree of homology with the amino-terminus of ALDH3B1. However, it lacks ~150 AAs that comprise the carboxy-terminus needed for enzyme oligomerisation. The truncated protein would still contain the conserved motifs required for ALDH activity. Until more experimental evidence becomes available, the ALDH3B5 gene should be considered as putatively functional.
The mouse and rat Aldh3b3 genes appear to represent new orthologous ALDH family members; the genes reside in syntenic chromosomal regions and share a high degree (83.4 per cent) of sequence identity with one another. The two proteins are more divergent than the rodent ALDH3B2 orthologues, which share 89.9 per cent sequence identity.
Aldh5a1 is another duplicated ALDH gene within the zebrafish genome. The duplication aldh5a1.2 resides on the same chromosome as the aldh5a1.1 parent gene, and the two share 100 per cent sequence identity. Aldh5a1.2 encodes a peptide containing an additional 22 amino-terminal and 88 carboxy-terminal residues. It also shares greater sequence identity with the human ALDH5A1 orthologue than Aldh5a1.1 (65.5 per cent versus 51.4 per cent). This suggests that aldh5a1.2 might actually be the parent gene and aldh5a1.1 a slightly truncated version formed as the result of gene duplication.
As mentioned above, the macaque ALDH7A1P5 genomic sequence lacks intronic regions, suggesting that a reverse transcriptase-mediated event gave rise to this pseudogene (ie having no adjacent promoter or other regulatory sequences). Four additional ALDH7A1 pseudogenes have been identified on chromosomes 5q14 (ALDH7A1P1), 2q31 (ALDH7A1P2), 7q36 (ALDH7A1P3) and 10q21 (ALDH7A1P4) [19]. Macaque ALDH7A1P5 is located on Chr 14, which is not syntenic with human Chr 11 and does not share common origins with any of the human pseudogenes. Therefore, the event that gave rise to ALDH7A1P5 must have taken place within the last 25 million years.
Three full-length ALDH9A1 homologues were identified in zebrafish. The Aldh9a1.2 peptide shares 71.2 per cent and 70.3 per cent identity with Aldh9a1.1 and Aldh9a1.3, respectively. Aldh9a1.3 is 94.9 per cent identical to the parent Aldh9a1.1 peptide, suggesting that this duplication was a relatively recent event when compared with the duplication that gave rise to Aldh9a1.2. Hence, aldh9a1.1, aldh9a1.2 and aldh9a1.3 represent three distinct protein-coding ALDH9 family members. The zebrafish genome also contains two copies of aldh18a1, which are found in very close proximity on Chr 12. Both genes are considered protein coding and would give rise to peptides of the same length which share 100 per cent sequence identity, suggesting a relatively recent duplication event.
ALDH gene-naming conventions dictate that (i) ALDH superfamily members sharing more than ~40 per cent AA identity belong to the same family (eg ALDH1A, ALDH1B, etc.), and (ii) ALDH family members that share greater than 60 per cent AA identity belong to the same subfamily (eg ALDH1A1, ALDH1A2, etc). This provides a convenient and systematic naming system for an entire superfamily. Interestingly, this does not always indicate homology properly; these rules in the cytochrome P450 (CYP) gene superfamily are known to break down when one includes evolutionarily distantly related animals [27]. For example, whereas zebrafish Aldh3d1 and Aldh3b1 share only 50 per cent AA identity, HomoloGene evidence and alignments suggest that aldh3d1 is probably a duplication of aldh3b1 (data not shown). Although aldh3d1 has diverged considerably, it is likely to be more closely related to aldh3b1 than the naming convention would suggest.
Many of these proteins have been defined based on genomic or dbEST data and have not been studied extensively. Many records remain in databases that are listed as 'protein-coding' but which instead may represent pseudogenes of various types. Furthermore, although the genes here do not have internal stop codons, without functional analysis, it is difficult to determine whether the genes might have other inactivating mutations or if they experience selective pressure. Although automated prediction and naming of ALDH proteins from completely sequenced genomes have achieved a great deal of information in a short amount of time, the alignment, curation and naming of these genes remains an important task. The fact that no new human ALDH genes have been identified over the past six years and that most other vertebrates seem to have settled close to this number suggests that identification of ALDH superfamily members in vertebrates is nearing completion. Determining the function and biological importance of each family member still requires additional work, however. As more information becomes available, the web database resource at (http://www.aldh.org) (the aldehyde dehydrogenase gene superfamily resource center)[35] will be updated to reflect our current understanding of this diverse and essential gene superfamily.
Figure S1.

Alignment of ALDH3B2 genes in human, rat and mouse created by ClustalW. Dashes (-) represent sequence gaps, asterisks (*) represent identical amino acids (AAs), colons (:) represent very similar AAs, periods (.) represent less similar AAs, whereas spaces () represent dissimilar AAs.
Acknowledgements
We would like to thank our colleagues for critically reviewing this manuscript. This work was supported, in part, by the following NIH grants: R01EY17963 (V.V.), R21AA017754 (V.V.), F31AA018248 (C.B.) and P30 ES06096 (D.W.N.). In addition, travel was supported by USPHS NIH grant R13-AA019612 to present this work at the 15th International Meeting on Enzymology and Molecular Biology of Carbonyl Metabolism in Lexington, KY, USA.
References
- Sophos NA, Pappa A, Ziegler TL, Vasilou V. Aldehyde dehydrogenase gene superfamily: The 2000 update. Chem Biol Interact. 2001;130-132:323–337. doi: 10.1016/s0009-2797(00)00275-1. [DOI] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasilou V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425255.4.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129:1–19. doi: 10.1016/S0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Sobreira TJ, Marletaz F, Simoes-Costa M, Schechtman D. et al. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc Natl Acad Sci USA. 2011;108:226–231. doi: 10.1073/pnas.1011223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey T, Cantore M, Weston PA, Carpenter JF. et al. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–4392. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- Estey T, Piatigorsky J, Lassen N, Vasilou V. ALDH3A1: A corneal crystallin with diverse functions. Exp Eye Res. 2007;84:3–12. doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Brocker C, Donald E. et al. Aldehyde dehydrogenase 1B1: Molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Orlicky DJ, Matsumoto A, Singh S. et al. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405:173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ. et al. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: The 2002 update. Chem Biol Interact. 2003;143-144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P. et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barratt T, Benson DA, Bolton E. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: Human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–434. [PubMed] [Google Scholar]
- Mullins M. Genetic nomenclature guide. Zebrafish. Trends Genet. 1995;11:31–32. [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Liberles DA. Evaluation of methods for determination of a reconstructed history of gene sequence evolution. Mol Biol Evol. 2001;18:2040–2047. doi: 10.1093/oxfordjournals.molbev.a003745. [DOI] [PubMed] [Google Scholar]
- Zhang J, Feuk L, Duggan GE, Khaja R. et al. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL. et al. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Goodman M. The genomic record of Humankind's evolutionary roots. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittlik S, Domingues S, Meyer A, Begemann G. Expression of zebrafish aldh1a3 (raldh3) and absence of aldh1a1 in teleosts. Gene Expr Patterns. 2008;8:141–147. doi: 10.1016/j.gep.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008;101:51–64. doi: 10.1093/toxsci/kfm280. [DOI] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P. et al. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizzano M, Sbarbati A. Is rat LRRP Ba1-651 a Delta-1-pyrroline-5-carboxylate dehydrogenase activated by changes in the concentration of sweet molecules? Med Hypotheses. 2007;68:864–867. doi: 10.1016/j.mehy.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A. et al. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P. et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WJ, Stagos D, Marchitti SA, Nebert DW. et al. Human aldehyde dehydrogenase genes: Alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genomics. 2009;19:893–902. doi: 10.1097/FPC.0b013e3283329023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Cantore M, Jester JV. et al. Corneal aldehyde dehydrogenases: Multiple functions and novel nuclear localization. Brain Res Bull. 2010;81:211–218. doi: 10.1016/j.brainresbull.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YS. et al. Evidence for gelsolin as a corneal crystallin in zebrafish. J Biol Chem. 2000;275:24645–24652. doi: 10.1074/jbc.M001159200. [DOI] [PubMed] [Google Scholar]
- Jia S, Omelchenko M, Garland D, Vasiliou V. et al. Duplicated gelsolin family genes in zebrafish: A novel scinderin-like gene (scinla) encodes the major corneal crystallin. FASEB J. 2007;21:3318–3328. doi: 10.1096/fj.07-8172com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling TM, Clark JI. The transparent lens and cornea in the mouse and zebra fish eye. Semin Cell Dev Biol. 2008;19:94–99. doi: 10.1016/j.semcdb.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black W, Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum Genomics. 2009;4:136–142. doi: 10.1186/1479-7364-4-2-136. [DOI] [PMC free article] [PubMed] [Google Scholar]


