Abstract
Background. A recombinant canarypox vector expressing human immunodeficiency virus type 1 (HIV-1) Gag, Pro, and membrane-linked gp120 (vCP1521), combined with a bivalent gp120 protein boost (AIDSVAX B/E), provided modest protection against HIV-1 infection in a community-based population in Thailand (RV144 trial). No protection was observed in Thai injection drug users who received AIDSVAX B/E alone (Vax003 trial). We compared the neutralizing antibody response in these 2 trials.
Methods. Neutralization was assessed with tier 1 and tier 2 strains of virus in TZM-bl and A3R5 cells.
Results. Neutralization of several tier 1 viruses was detected in both RV144 and Vax003. Peak titers were higher in Vax003 and waned rapidly in both trials. The response in RV144 was targeted in part to V3 of gp120.vCP1521 priming plus 2 boosts with gp120 protein was superior to 2 gp120 protein inoculations alone, confirming a priming effect for vCP1521. Sporadic weak neutralization of tier 2 viruses was detected only in Vax003 and A3R5 cells.
Conclusion. The results suggest either that weak neutralizing antibody responses can be partially protective against HIV-1 in low-risk heterosexual populations or that the modest efficacy seen in RV144 was mediated by other immune responses, either alone or in combination with neutralizing antibodies.
It is widely believed that a neutralizing antibody (NAb) response of sufficient magnitude, breadth, and duration would be highly beneficial for human immunodeficiency virus type 1 (HIV-1) vaccines [1–3]. Indeed, Nabs protect against experimental challenge with simian human immunodeficiency virus (SHIV) in nonhuman primates [4–6], and they exert strong selective pressure on HIV-1 after infection in humans [7, 8]. Neutralization occurs when antibodies bind to functional envelope glycoprotein (Env) spikes on the virus surface to prevent entry into host cells [9–11]. Each Env spike consists of 3 surface gp120 molecules bound noncovalently to 3 transmembrane gp41 molecules [12, 13]. These glycoproteins exhibit an extraordinary degree of genetic and antigenic variability that poses major challenges for vaccine development [14, 15]. Moreover, the virus uses a number of mechanisms to evade NAbs [7, 12, 16]. As a result, a minor subset of circulating variants exhibit a highly neutralization-sensitive tier 1 phenotype and are often susceptible to vaccine-elicited NAbs, whereas most circulating strains exhibit a less sensitive tier 2 phenotype and have proven difficult to target with vaccines[1, 2, 15].
A recently completed HIV-1 vaccine efficacy trial in Thailand (RV144) showed that priming with a recombinant canarypox vector (vCP1521) and boosting with this vector plus bivalent gp120 protein (AIDSVAX B/E) can provide partial protection against the acquisition of HIV-1 infection in a community-based heterosexual population [17]. The same bivalent gp120 immunogen, when used alone and with an increased number of inoculations (Vax003 trial), showed no protection in a cohort of Thai injection drug users [18]. In addition, no overall protection was seen when a similar regimen of bivalent gp120 (AIDSVAX B/B) was used alone in a cohort of mostly men who have sex with men (MSM) in North America and the Netherlands (Vax004 trial) [19]. The gp120 protein component in all 3 clinical trials was designed to elicit NAbs [20, 21]. In Vax004, strong NAb responses were seen against a subset of tier 1 viruses, and sporadic weak responses were seen against tier 2 viruses [22]. Here we assessed the magnitude and breadth of NAb responses in RV144 and Vax003.
VOLUNTEERS, MATERIALS, AND METHODS
Clinical Trials
Trial designs and outcomes for RV144 and Vax003 were published previously [17, 18]. Participants in RV144 were administered vCP1521 (Sanofi Pasteur) at months 0, 1, 3, and 6, with coadministration of AIDSVAX B/E gp120 (Global Solutions for Infectious Diseases) at months 3 and 6. vCP1521 expressed the Gag and protease of MN (subtype B). It also expressed the gp120 of 92TH023 (CRF01_AE) linked to the transmembrane portion of subtype B gp41 that was devoid of the entire gp41 ectodomain. AIDSVAX B/E consisted of gp120 proteins from MN and the CRF01_AE strain, CM244 (A244), both produced in Chinese hamster ovary cells and administered at 600-μg doses (300 μg of each gp120) adsorbed onto alum. Participants in Vax003 received intramuscular injections of AIDSVAX B/E at months 0, 1, 6, 12, 18, 24, and 36 in the absence of vCP1521; antibody responses in Vax003 peaked after the fourth inoculation [18]. RV135 was a phase II trial of the same vaccines used in RV144 [23], where participants in the high-dose gp120 group studied here were immunized precisely as in RV144. All 3 clinical trials were conducted in accordance with the Declaration of Helsinki and local institutional review board requirements. Written informed consent was obtained from all subjects.
Serologic Specimens
Plasma in RV144 was obtained from 140 participants (112 vaccine recipients and 28 placebo recipients) at 2 weeks after the fourth (final) inoculation. Additional plasma was obtained from 30 vaccine recipients at 6, 12, 18, 24, 30, and 36 months after the final inoculation to examine the longevity of NAb responses. Serum from Vax003 was obtained from 90 vaccine recipients and 30 placebo recipients at baseline, 2 weeks after the second and fourth inoculations, and 6 months after the fourth inoculation. Serum from 45 vaccine recipients and 14 placebo recipients in RV135 was used to examine data obtained at a time point (2 weeks after the third inoculation) for which data were not available in RV144. Vaccine recipients were stratified by sex and selected randomly within each group. All participants were uninfected at the time of blood draw. Plasma and serum samples were stored at −80°C, thawed, and heat-inactivated at 56°C for 30 minutes prior to assay.
Viruses
Tier 1 Env-pseudotyped viruses expressed the entire gp160 of MN.3 (subtype B), SF162.LS (subtype B), Bal.26 (subtype B), MW965.26 (subtype C), TH023.6 (CRF01-AE), and NP03.13 (CRF01-AE). TH023.6 and NP01.13 gp160 genes were cloned after adaptation of the parental viruses in A3R5 cells and H9 cells, respectively. TH023.6 contains 4 amino acid substitutions in gp120 and 1 in gp41 as compared to the strain used in vCP1521. Additional CRF01-AE Env pseudotyped viruses expressed gp160 genes of circulating strains from Thailand (Table 1). Seven infectious molecular clones were used that expressed the entire ectodomain of circulating CRF01-AE viruses from Thailand and that carried a Renilla luciferase (Luc) reporter gene (Env.IMC.LucR viruses) [24] (Table 1). Env-pseudotyped viruses were prepared by cotransfecting 293T/17 cells (American Type Culture Collection [ATCC], Manassas, VA) with an Env-expressing plasmid plus an Env-defective backbone plasmid (pSG3Δenv) as described elsewhere [25]. Env.IMC.LucR viruses were also prepared by transfection in 293T/17 cells but without a backbone plasmid.
Table 1.
Circulating CRF01-AE Human Immunodeficiency Virus Type 1 (HIV-1) Envs Used to Measure Neutralization
| Namea | Accession No. | Infection Year | Infection Statusb | Source/Methodc | Assayd | Tier (TZM-bl) | Tier (A3R5)e |
|---|---|---|---|---|---|---|---|
| C1080.c03 | JN944660 | 1999 | Chronic | Plasma/SGA | TZM-bl/A3R5 | 2 | 1 |
| C3347.c11 | AF259954 | 1999 | Chronic | Plasma/SGA | TZM-bl/A3R5 | 2 | 1 |
| C2101.c01 | JN966661 | 1999 | Chronic | Plasma/SGA | TZM-bl | 2 | NA |
| CM246.c1 | JN944663 | 1990 | Chronic | ccPBMC | TZM-bl | 2 | NA |
| 427299.c12 | JN944655 | 2006 | T/F | Plasma/SGA | TZM-bl/A3R5 | 2 | 1 |
| 816763.c02 | JN944659 | 2006 | T/F | Plasma/SGA | TZM-bl | 2 | NA |
| 703357.c02 | JN944658 | 2005 | T/F | Plasma/SGA | TZM-bl | 2 | NA |
| 620345.c10 | JN944656 | 2005 | T/F | Plasma/SGA | TZM-bl | 2 | NA |
| 356272.c02 | JN944654 | 2005 | T/F | Plasma/SGA | TZM-bl | 3 | NA |
| CM244.ec1 | AY713425 | 1990 | Chronic | ccPBMC | A3R5 | 2 | 2 |
| R2184.c04 | JN944665 | 2001 | Chronic | Plasma/SGA | A3R5 | 2 | 2 |
| CM235-2 | JN944662 | 1990 | Chronic | ccPBMC | A3R5 | 2 | 2 |
| 644039.c01b | JN944656 | 2006 | T/F | Plasma/SGA | A3R5 | 2 | 2 |
| M066.07 | JN944664 | 1996 | Chronic | ccPBMC/SGA | TZM-bl | 2 | NA |
a All viruses are from Thailand and exhibit an R5 biological phenotype.
b Envs are from viruses present during chronic infection, or they are transmitted/founder (T/F) Envs.
c Envs were derived by single genome amplification (SGA) of viral RNA from plasma or by polymerase chain reaction amplification of DNA from patient peripheral blood mononuclear cells (PBMCs) that were cocultured with PBMCs from a healthy HIV-1–negative donor (ccPBMC).
d Unless otherwise specified, Envs were used either as Env-pseudotyped viruses in the TZM-bl assay or as Env.IMC.LucR viruses in the A3R5 assay. Note that for some Envs, both types of virus were used (TZM-bl/A3R5 assay). Env.IMC.LucR viruses were made using an NL3-4 backbone, except for 427299.c12 and 644039.c01b, which were made using a backbone derived from CRF01_AE strainCM235. Also, the CM244.ec1Env.IMC.LucR virus was made using a backbone derived from CRF01-AE strain ETH2220.
e Values were not applicable (NA) for some cells because the corresponding Env.IMC.LucR constructs were not available for virus characterization.
Neutralization Assays
Neutralization was measured in 96-well culture plates by using Tat-regulated Luc reporter gene expression to quantify reductions in virus infection in either TZM-bl or A3R5 cells. Assays in TZM-bl cells were performed with Env-pseudotyped viruses in a validated format as described elsewhere [25, 26]. TZM-bl is a genetically engineered HeLa cell line (also known as JC53-BL) that expresses CD4, CCR5, and CXCR4 [27] and carries Tat-regulated reporter genes for firefly Luc and Escherichia coli β-galactosidase under regulatory control of an HIV-1 long-terminal-repeat sequence [28]. A3R5 (A3.01/CCR5) is a derivative of the A3.01 human lymphoblastoid cell line that naturally expresses CD4 and CXCR4 [29] and was engineered to express CCR5 (R. J. McLinden et al, unpublished data). The A3R5 assay was performed with Env.IMC.LucR viruses as described elsewhere (R. J. McLinden et al., unpublished data). Serum and plasma samples were assayed at 3-fold dilutions ranging from 1:20 to 1:43 740. In some cases, the samples were diluted with an equal volume of phosphate-buffered saline (PBS), pH 7.4, and incubated with peptide (50 μg/mL) for 1 hour at 37°C prior to assay. Neutralization titers are the sample dilution at which relative luminescence units (RLU) were reduced by 50% as compared to RLU in virus control wells after subtraction of background RLU in cell control wells. Some samples were tested at a 1:10 dilution in triplicate against tier 2 viruses in TZM-bl cells. Neutralization was calculated as the percentage reduction in RLU in wells containing postimmunization sample relative to RLU in wells containing the corresponding preimmune sample. All samples were blinded until the end of the study.
Isolation of Monoclonal Antibodies by Flow Cytometry
Antigen-specific sorting of memory B cells was performed as described elsewhere [30], with the following modifications. Group M consensus gp140ConS Env was labeled with Pacific Blue and Alexa Fluor 647, using fluorochrome labeling kits (Invitrogen, Carlsbad, CA). Thawed peripheral blood mononuclear cells (PBMCs) were stained as described previously [30], and memory B cells stained with gp140ConS in both colors were sorted as single cells. Immunoglobulin genes were recovered as described elsewhere [31, 32]. Gene analysis was performed as previously described [33], and isolated immunoglobulin V(D)J gene pairs were assembled by polymerase chain reaction into linear full-length immunoglobulin heavy- and light-chain gene expression cassettes and expressed in 293T cells (ATCC) and purified for use in the neutralization assay [33].
Epitope Mapping of Vaccine-Induced Monoclonal Antibodies
Enzyme-linked immunosorbant assays (ELISAs) were performed as described elsewhere [34], with the following modifications. ELISA plates (384 wells; Corning Life Sciences, Lowell, MA) were coated with either purified HIV-1gp120 (A244, MN, TH023) or 15-mer overlapping peptides spanning MNgp120 or TH023gp120 and blocked with assay diluent (PBS containing 4% [w/v] whey protein/15% normal goat serum/0.5% Tween 20/0.05% sodium azide) for 1 hour at room temperature. A total of 10 µL/well of serial 3-fold dilutions of purified antibodies, starting at 100 µg/mL, were incubated for 2 hours at room temperature. Plates were developed as previously described [34].
Statistical Methods
The analysis focused on 2 types of readouts: the NAb titer for individual isolates and the area under the curve of the magnitude-breadth plot (AUC-MB). Box plots are used to graphically display distributions of log10 NAb titers for individual isolates. NAb responses to an individual isolate were summarized by the percentage of subjects who had a positive response, defined as a 50% inhibitory dose (ID50) of >1:20 (ie, positive response rate). An overall false-positive response rate was calculated as the percentage of positive response rate, combining data for vaccine recipients at baseline and placebo recipients at all time points and averaging across isolates. The magnitude of NAb responses to an individual isolate was summarized by the geometric mean titer (GMT) and its 95% confidence interval. The Wilcoxon rank sum test was used to test for a difference in magnitude of NAb titers between 2 independent groups. The Fisher exact test was used to test for a difference in positive response rate between 2 independent groups.
The magnitude (NAb titer) and breadth (number of isolates neutralized) of an individual plasma sample assayed against a panel of isolates were characterized by magnitude-breadth (M-B) curves [35]. The x-axis of a M-B curve is the threshold of neutralization that is considered positive, and the y-axis is the fraction of isolates neutralized. The AUC-MB was calculated as the average of the log10 NAb titer over the panel of isolates. The Wilcoxon rank sum test, stratified by sex, with weight inversely proportional to stratum size, was used to test for a difference in AUC-MB distribution between 2 independent groups. The paired-data-stratified Wilcoxon signed rank test was used to test for a difference in AUC-MB distribution between 2 time points.
All P values are 2-sided. Multiple hypotheses performed were adjusted for by controlling the false discovery rate (FDR) [36] at the 0.05 level.
RESULTS
Properties of the CRF01-AE Viruses Used in NAb Assays
Envs from 14 CRF01-AE viruses from Thailand were used as Env-pseudotyped viruses, Env.IMC.LucR viruses, or both. The Envs represented a spectrum of genetic diversity within CRF01-AE and composed a mixture of transmitted/founder (T/F) and chronic viruses (Table 1). Most exhibited a tier 2 neutralization phenotype in TZM-bl cells, except for 356272.c02, which was tier 3. Among the viruses that were evaluated in A3R5 cells, 4 were tier 2 and 3 were tier 1. Tier phenotypes were determined with a panel of 11 CRF01-AE plasmas from chronically infected individuals in Bangkok, Thailand (S. Tovanabutra, unpublished data). Of note, 3 Env.IMC.LucR viruses (C3347.c11, C1080.c03, 427299.c12) were tier 1 in A3R5 cells but were tier 2 in TZM-bl cells, where the corresponding Env-pseudotyped viruses also exhibited a tier 2 phenotype.
Neutralization in TZM-bl Cells
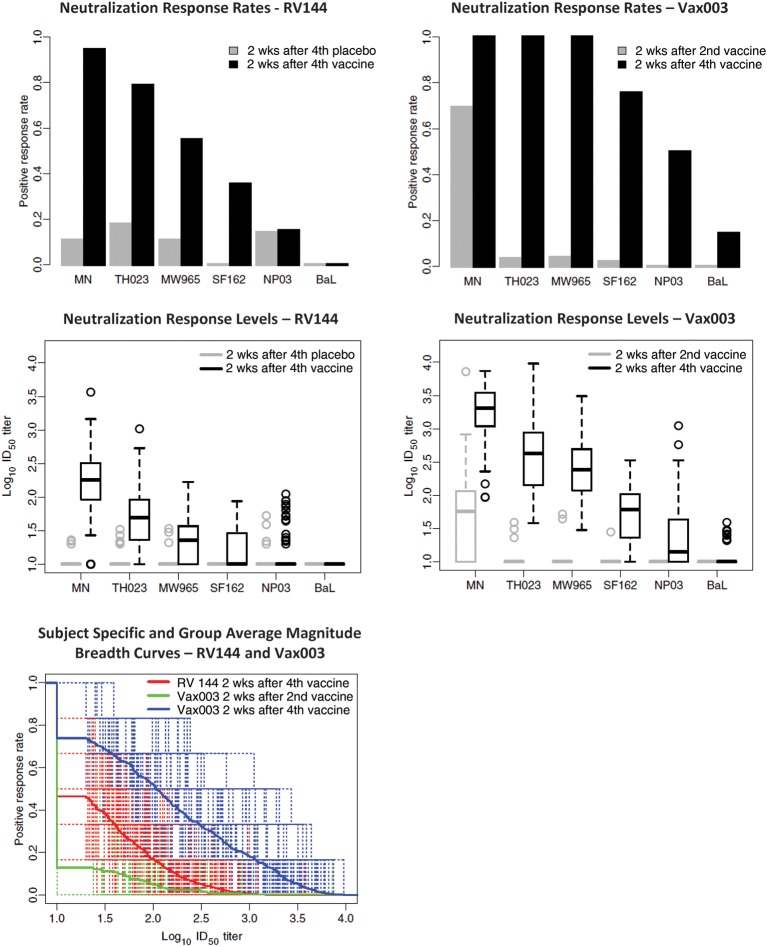
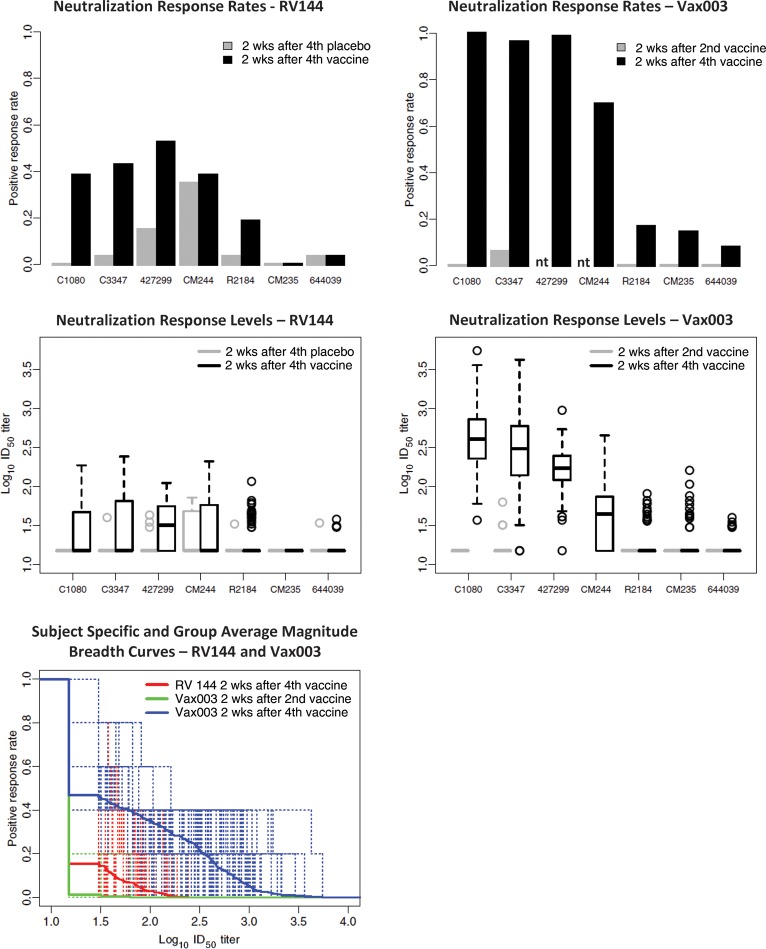
NAbs were first assessed in a validated TZM-bl assay that has gained wide acceptance in the field [25, 26]. Across the 6 tier 1 viruses evaluated, the overall false-positive response rate was 8.9% for RV144 and 0% for Vax003. Vaccines in RV144 and Vax003 generated NAbs against multiple tier 1 viruses at peak immunity (2 weeks after the final inoculation in RV144; 2 weeks after the fourth inoculation in Vax003) (Figure 1). Responses were most frequent and strongest against MN.3 and TH023.6, followed by MW965.26, SF162.LS, NP03.13, and Bal.26 for both trials. Overall positive response rates were higher in Vax003, although a majority of vaccine recipients in both trials had NAbs against MN.3 and TH023.6. GMTs of NAbs were 2–10 times higher in Vax003 for all viruses except Bal.26, which was neutralized poorly in both trials. A comparison of the AUC-MB plots showed a significantly stronger NAb response in Vax003 (P < .001 after FDR adjustment). No neutralization of tier 2 viruses was detected in either trial when the TZM-bl assay was used (data not shown). Additionally, no neutralization was detected in RV144 when a PBMC assay was used (C1080.c03 and R2184.c04 as Env.IMC.LucR viruses; data not shown).
Figure 1.
Neutralizing antibody (NAb) responses as measured in TZM-bl cells. NAbs were assessed against a panel of 6 tier 1 reference strains of Env-pseudotyped viruses in the TZM-bl assay. Samples for RV144 included plasma from 112 vaccine recipients and 20 placebo recipients obtained at 2 weeks after the fourth inoculation (visit 8). Samples for Vax003 included serum from 90 vaccine recipients and 30 placebo recipients obtained at 2 weeks after the second (visit 5) and fourth (visit 9) inoculation. All results for placebo recipients in Vax003 were negative (not shown). Top, Positive response rates (frequency of positive results at ≥1:20 plasma dilution) against each of 6 tier 1 reference viruses (listed on the x-axis). Middle, Box plots of NAb titers against each virus. For the box plots, 25% of values lie below the box, 25% lie above the box, and 50% lie below the horizontal line (the median) inside the box. Vertical lines above the box extend to a distance 50% greater than the height of the box; points beyond this are unusually high values (outliers). Bottom, Magnitude-breadth (M-B) curves of 50% inhibitory dose (ID50) NAb titers against all 6 viruses (x-axis, neutralization titers; y-axis, fraction of viruses neutralized). Dashed lines represent subject-specific responses. Solid lines represent group averages.
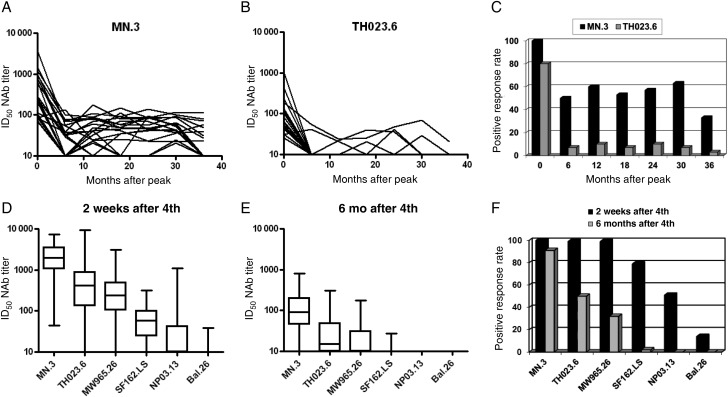
Longevity of the response was examined in RV144 using MN.3 and TH023.6 and plasma samples collected every 6 months for 3 years after the final boosting. Longevity was examined in Vax003 using 6 tier 1 viruses and serum collected 6 months after the fourth inoculation. Responses in both trials waned considerably after 6 months (Figure 2). After an initial decline, low titers of NAbs against MN.3 were detected for at least 3 years in 32% of subjects in RV144 (Figure 2), where a higher peak response was associated with a greater probability of persistence (P = .01, by the Wilcoxon rank sum test). Positive neutralization of TH023.6 after 3 years in RV144 was less frequent (<4%).
Figure 2.
Longevity of the neutralizing antibody (NAb) response in RV144 and Vax003. A and B, Titers of NAbs against MN.3 and TH023.6 at 2 weeks and at 6, 12, 18, 24, 30, and 36 months after final boosting in RV144. C, Positive response rates (frequency of positive results at ≥1:20 plasma dilution) against MN.3 and TH023.6 at each time point shown in A and B. D and E, Box plots of ID50 NAb titers against 6 tier 1 viruses at 2 weeks and 6 months after the fourth inoculation. F, Positive response rates against the 6 tier 1 viruses at 2 weeks and 6 months after the fourth inoculation. Six-month longevity was the longest period available in the Vax003 trial design. Abbreviation: ID50, 50% inhibitory dose.
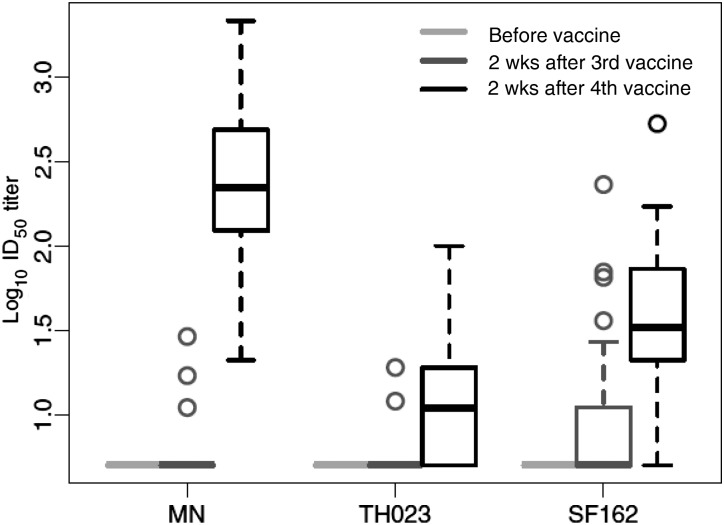
We asked whether a single inoculation with gp120 protein was sufficient to induce detectable NAbs in RV144. Because samples from this time point were not available in RV144, plasmas from participants in the high-dose group of RV135 were used. A weak NAb response was detected in a small subset of subjects after 1 protein inoculation (Figure 3). Two protein inoculations induced a much stronger response that resembled the peak response in RV144. Thus, a single protein inoculation was relatively ineffective, compared with 2 inoculations. Vector priming was apparent because the peak response in RV144 was significantly stronger than the response seen after 2 gp120 protein inoculations in Vax003 (Figure 1; P < .001 after FDR adjustment).
Figure 3.
Kinetics of the neutralizing antibody (NAb) response in RV135. Serum NAbs for 45 vaccine recipients and 14 placebo recipients were assessed at baseline and at 2 weeks after the third and fourth inoculations (after the first and second protein inoculation) in the TZM-bl assay. All results for placebo recipients were negative (data not shown). Abbreviation: ID50, 50% inhibitory dose.
Neutralization in A3R5 Cells
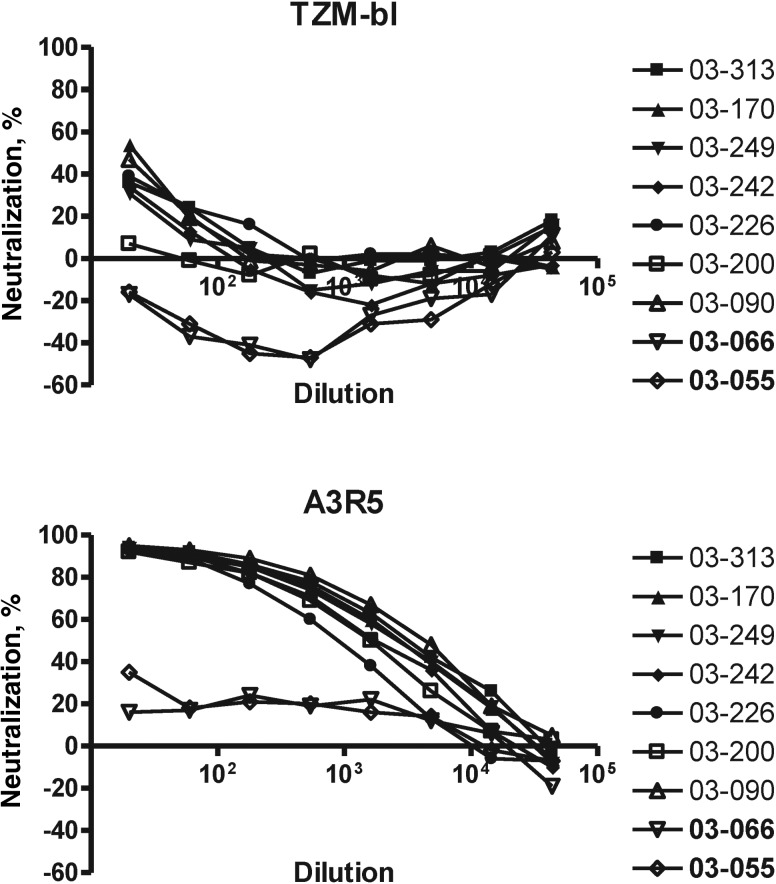
The lack of detection of NAbs against tier 2 viruses in the TZM-bl assay by using samples from RV144 and Vax003 prompted an investigation of alternative cell lines that express lower levels of CD4 and CCR5 for possible increased assay sensitivity [37]. One such cell line, A3R5, was chosen that is substantially more sensitive than TZM-bl for detecting NAbs in HIV-1-positive sera (R. J. McLinden et al., unpublished data). In a preliminary experiment that used an identical stock of C1080.c03 Env.IMC.LucR virus in both assays, sera from vaccine recipients in Vax003 scored strongly positive in A3R5 cells and were weak or negative in TZM-bl-cells (Figure 4). Subsequently, A3R5 cells were used to assess NAbs at peak immunity in RV144 and Vax003. Across the 7 viruses used for RV144 and Vax003, the overall false-positive response rate was 6.3% for RV144 and 0.6% for Vax003. The overall response after 4 inoculations in Vax003 was substantially stronger than the peak response in RV144 (Figure 5). Positive neutralization in RV144 was seen against 3 tier 1 viruses: C1080, C3347, and 427299. Responses against these 3 viruses were substantially stronger in Vax003, where occasional weak neutralization was also detected against 4 tier 2 viruses (CM244, R2184, CM235, and 644039). The stronger response in Vax003 was highly significant when AUC-MB curves were compared (P < .001 after FDR adjustment). As in the TZM-bl assay, the peak response in RV144 was significantly stronger than the response after 2 gp120 protein inoculations in Vax003 (P < .001 after FDR adjustment for comparing AUC-MB), suggesting a priming effect for vCP1521 in RV144.
Figure 4.
Heightened sensitivity of A3R5 cells for detecting neutralizing antibody (NAbs). Serum from 7 vaccine recipients and 2 placebo recipients in Vax003 (2 weeks after the fourth inoculation) were assayed for neutralizing activity against C1080.c03 Env.IMC.LucR in TZM-bl and A3R5 cells by using an identical stock of virus in both assays. Placebo recipients are labeled 03–066 and 03–055.
Figure 5.
Neutralizing antibody (NAb) responses as measured in A3R5 cells. NAbs were assessed against a panel of tier 1 and tier 2 reference strains of Env.IMC.LucR viruses in the A3R5 assay. Plasma (RV144) and serum (Vax003) samples are the same as those described in Figure 1. Top, Positive response rates (frequency of positive results at ≥1:20 plasma dilution) against each virus (listed on the x-axis). Because of limited sample volumes, 2 viruses (427299 and CM244) were not assayed at visit 5 for Vax003 (nt, not tested). Middle, Box plots of NAb titers against each virus (7 viruses for RV144 and 5 viruses for Vax003). For the box plots, 25% of values lie below the box, 25% lie above the box, and 50% lie below the horizontal line (the median) inside the box. Vertical lines above the box extend to a distance 50% greater than the height of the box; points beyond this are unusually high values (outliers). Bottom, Magnitude-breadth (M-B) curves of NAbs against the 5 viruses that were common to the assays performed for RV144 and Vax003 (C1080, C3347, R2184, CM235, and 644039). Neutralization titer is shown on the x-axis; the fraction of viruses neutralized is shown on the y-axis. Dashed lines represent subject-specific responses. Solid lines represent group averages. Abbreviation: ID50, 50% inhibitory dose.
Epitope Mapping of Vaccine-Elicited Nabs
From the circulating memory B cells of 2 subjects in RV135, the monoclonal Abs (mAbs) CH21, CH22 (subject T141485), and CH23 (subject T143859) were isolated that exhibited neutralizing activity in TZM-bl cells. The 3 mAbs exhibited variable neutralizing activity against a subset of tier 1 viruses but had no detectable neutralizing activity against tier 2 viruses (Table 2). CH21 and CH22 preferentially neutralized subtype B tier 1 viruses, whereas CH23 showed a strong preference for TH023.6 (CRF01_AE). CH21 and CH22 bound MNgp120 with EC50s of 2.1 nM and 0.4 nM, respectively, whereas CH23 bound to A244gp120 and 92TH023gp120 (IC50 values of 0.4 nM and 0.3 nM, respectively). CH22 and CH23 mapped to V3 linear peptides RKRIHIGPGRAFYTT and NTRTSINIGPGQVFY, respectively. CH21 failed to bind any linear peptides spanning gp120 and was therefore considered to recognize a conformational epitope yet to be defined. The 3 mAbs used diverse V-gene segments (IGVH 1 and 3 families) (Table 3). Mutation frequencies of the heavy and light chains ranged from 1.5% to 4.5% and from 1% to 2.2%, respectively. The length of the heavy and light complementarity-determining region 3 (CDR3) was 11–16 amino acids and 10–11 amino acids, respectively. All 3 antibodies used a λ light chain.
Table 2.
Functional Characteristics of Vaccine-Induced Neutralizing Monoclonal Antibodies CH21, CH22, and CH23
| IC50 NAb Titer,a μg/mL |
|||||
|---|---|---|---|---|---|
| Virus | Clade | Tier | CH21 | CH22 | CH23 |
| MN.3 | B | 1 | 5.5 | <0.02 | >50 |
| SF162.LS | B | 1 | 25.7 | >50 | >50 |
| Bal.26 | B | 1 | >50 | 0.35 | >50 |
| W61D-TCLA.71 | B | 1 | 0.31 | <0.02 | 40.4 |
| 92RW020.2 | A | 1 | >50 | >50 | >50 |
| TV1.21 | C | 1 | >50 | >50 | >50 |
| TH023.6 | AE | 1 | >50 | >50 | <0.02 |
| NP03.13 | AE | 1 | >50 | >50 | >50 |
| CM244.ec1 | AE | 2 | >50 | >50 | >50 |
| C3347.c11 | AE | 2 | >50 | >50 | >50 |
| Q23.17 | A | 1 | >50 | >50 | >50 |
| Q842.d12 | A | 2 | >50 | >50 | >50 |
| Q461.e2 | A | 2 | >50 | >50 | >50 |
| Q769.d22 | A | 2 | >50 | >50 | >50 |
| 6535.3 | B | 2 | >50 | >50 | >50 |
| QH0692.42 | B | 2 | >50 | >50 | >50 |
| WEAU-d15.410.787 | B | 2 | >50 | >50 | >50 |
| BB1006-11.C3.1601 | B | 2 | >50 | >50 | >50 |
| Du422.1 | C | 2 | >50 | >50 | >50 |
| ZM197M.PB7 | C | 2 | >50 | >50 | >50 |
| Ce2010_F5 | C | 2 | >50 | >50 | >50 |
| 704809221_1B3 | C | 2 | >50 | >50 | >50 |
aPositive values are shown in bold.
Abbreviation: IC50, 50% inhibitory concentration.
Table 3.
Characteristics of the V(D)J Rearrangements of Vaccine-Induced Neutralizing Monoclonal Antibodies (NAbs)
| V Heavy Chain |
V Light Chain |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NAb | V | D | J | Mutation Frequency, % | HCDR3a Length, aa | Isotype | V | J | Mutation Frequency, % | LCDR3b Length, aa |
| CH21 | 3-11*01 | 2-8*02 | 5*02 | 1.5 | 16 | G1 | 3-21*02 | 3*02 | 1.0 | 11 |
| CH22 | 1-f*01 | 3-16*01,02 | 3*02 | 3.7 | 11 | G1 | 2-14*04 | 3*02 | 1.0 | 11 |
| CH23 | 3-66*01 | 3-OR15*3 | 1*01 | 4.5 | 11 | G1 | 6-57*01 | 3*02 | 2.2 | 10 |
Abbreviations: HCDR3, heavy chain complementarity region 3; LCDR3, light chain complementarity region 3.
Additional epitope mapping was performed with 6 plasma samples from RV144 that contained high NAb titers against MN.3 and with another 6 samples that contained high NAb titers against TH023.6. Four peptides that frequently bound in PepStar microarray array experiments (unpublished data) were synthesized by JPT Peptide Technologies (Germany) and added to 1:2-diluted plasma at a concentration of 50 μg/mL for 1 hour prior to assay. MN V3 peptide (NYNKRKRIHIGPGRAFYTTKNIKGT) blocked an average of 65% (range, 48%–100%) of neutralizing activity against MN.3. TH023 V3 peptide (SNNTRTSINIGPGQVFYRTGDIIGD) blocked 100% of neutralizing activity against TH023.6 in all 6 samples. Peptides from the C1, V2, and C5 regions of these 2 viruses had little or no effect on neutralization.
DISCUSSION
We assessed the NAb response in 2 HIV-1 vaccine efficacy trials in which either partial protection (RV144) [17] or no protection (Vax003) [18] was seen. By all measures, the peak NAb response in RV144 was substantially weaker than the peak response in Vax003. Only tier 1 viruses were neutralized in RV144, as mediated in part by Abs against the V3 loop of gp120. Weak neutralization of tier 2 viruses was occasionally seen in Vax003, but only in the more sensitive A3R5 assay. Peak responses in both trials waned considerably after 6 months, possibly reflecting a rate of decay for other antibody responses as well and suggesting that regular boosting will be required to maintain adequate titers.
After initial waning, low titers of NAbs persisted in a subset of RV144 vaccine recipients for at least 3 years. This observation, together with the isolation of neutralizing mAbs CH21, CH22, and CH23 from circulating memory B cells, suggest that the RV144 vaccine is able to induce, at least in a subset of vaccine recipients, long-term memory responses through both memory B cells and long-lived plasma cells. An earlier study of recombinant canarypox prime/gp120 protein boosting showed that low titers of NAbs can persist for at least 4–5 years and that recall of peak titers is possible by boosting again with gp120 protein [38].
A priming effect was seen for vCP1521 in that the peak NAb response after final boosting in RV144 (2 gp120 inoculations) was stronger than the response seen after 2 gp120 protein inoculations alone in Vax003 (Figure 3). Priming was relatively weak as compared to that for another canarypox vector, vCP1452, which required only a single protein boost for strong NAb response [39, 40]. RV144 (vCP1521) and the trials that tested vCP1452 used the same schedule of 2 vector inoculations followed by 2 boosts with vector plus gp120 protein. Moreover, they used the same protein dose and adjuvant for bivalent gp120. Multiple reasons might explain the weaker priming in RV144, including a lower dose of vector, different Envs in the vectors and as protein boosts, and the presence of vaccinia virus E3L and K3L genes in vCP1452.
Several HIV-1 vaccine concepts have been tested for efficacy in human trials. One concept tested a vectored Gag-Pol-Nef immunogen that, despite robust virus-specific CD8+ T-cell responses, failed to protect in a predominantly MSM risk group [41, 42]. The Vax003 and Vax004 trials tested bivalent gp120 proteins that elicited high titers of Env binding antibodies but failed to protect when administered to injection drug users and MSM, respectively [18, 19]. In Vax004, robust NAbs responses were seen against a subset of tier 1 viruses, with occasional weak neutralization seen against tier 2 viruses [22]. The response in Vax004 was much stronger than the response in RV144 and was similar to the response in Vax003, with the exception that occasional neutralization of tier 2 viruses in the more stringent TZM-bl assay was only seen in Vax004 [22].
RV144 is the first efficacy trial of a vector prime/gp120 protein boost vaccine for HIV-1. It is also the only trial to date in a heterosexual risk group and in which some level of protection was seen. That the NAb response in RV144 was weaker than the responses in Vax003 and Vax004 raises the question of whether NAbs might be sufficient but not necessary for prevention of HIV-1 infection. Our studies did not address other potentially protective antibody activities, such as Fc receptor–mediated effector functions [43–45] and antibodies that impede virus transmission across intact mucosa [46–48]. One Fc receptor–mediated function, antibody-dependent cell mediated cytotoxicity, was detected in a majority of vaccine recipients in RV135 [49]. It is also possible that NAbs were present in RV144 that were below the level of detection in our assays (eg, ID50 titers of <1:20). Low levels of NAbs might have a greater impact on heterosexual transmission in RV144 than in the higher-risk groups that participated in Vax003 and Vax004. Lower-risk behavior has been associated with lower multiplicity of transmitted/founder viruses [50] and therefore might be more easily countered by antibodies. The efficacy seen in RV144 might be improved by eliciting stronger NAb responses, particularly against tier 2 viruses, and/or by maintaining peak antibody responses for longer periods. Detailed studies of NAbs and other potential antiviral antibody activities in future clinical trials may provide new insights into the requirements for vaccine-mediated protection against HIV-1.
Notes
Acknowledgments. We thank Hagen von Briesen, for assistance in preparing large assay stocks of virus, and Viseth Ngauy, for critical review of the manuscript. We also thank Lautaro G. Perez, Amanda Martelli, Xing Yuan, Wenhong Feng, Corinne Dary, Francesca Suman, Joseph Jackson, Kara John, Alicia Gaitan, and Miroslawa Bilska, for excellent technical assistance. TZM-bl cells were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. We gratefully acknowledge the Thai volunteers who participated in RV144, RV135, and Vax003. We also acknowledge the generosity of the Ministry of Public Health, Thailand, and the Thai AIDS Vaccine Evaluation Group in providing the RV144 clinical trial materials through the Henry M. Jackson Foundation for the Advancement of Military Science.
Disclaimer. The opinions herein are those of the authors and should not be construed as official or representing the views of the US Department of Health and Human Services, the National Institute for Allergy and Infectious Diseases (NIAID), the Centers for Disease Control and Prevention, the Department of Defense, or the Department of the Army.
Financial support. This study was supported by the Bill & Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery, by the Center for HIV/AIDS Vaccine Immunology (grant AI678501 from the Division of Acquired Immunodeficiency Syndrome, NIAID, NIH), and by an interagency agreement (Y1-AI-2642-12) between the US Army Medical Research and Materiel Command and the NIAID. This work was also supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the US Department of Defense.
Potential conflicts of interest. P. W. B., D. F., F. S., and C. L. are former employees of VaxGen, and P. G. and S. G. S. received consulting fees from VaxGen in the past. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Ann Rev Immunol. 2010;28:413–44. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 2.Hoxie JA. Toward an antibody-based HIV-1 vaccine. Ann Rev Med. 2010;61:135–52. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- 3.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 4.Shibata R, Igarashi T, Haigwood N, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–10. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 5.Mascola JR, Stiegler G, VanCott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 6.Hessell AJ, Poignard P, Hunter M, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–5. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape. Nature. 2003;422:307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 8.Moore PL, Ranchobe N, Lambson BE, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Lipchina I, Cocklin S, Chaiken I, Sodroski J. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J Virol. 2006;80:11404–8. doi: 10.1128/JVI.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooks ET, Moore PL, Richman D, et al. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Human Antibodies. 2005;14:101–13. [PMC free article] [PubMed] [Google Scholar]
- 11.Moore PL, Crooks ET, Porter L, et al. The nature of non-functional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515–28. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 13.White TA, Bartesaghi A, Borgnia MJ, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaschen B, Taylor J, Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–60. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 15.Seaman MS, Janes H, Hawkins N, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessments of neutralizing antibodies. J Virol. 2010;84:1439–52. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong PD, Doyle ML, Casper DJ, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 17.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 18.Pitisuttithum P, Gilbert PB, Gurwith M, et al. Randomized, placebo-controlled efficacy trial of a bivalent rgp120 HIV-1 vaccine among injecting drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 19.Flynn NM, Forthal DN, Harro CD, et al. Placebo-controlled phase 3 trial of recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 20.Berman PW. Development of bivalent rgp120 vaccines to prevent HIV type 1 infection. AIDS Res Hum Retroviruses. 1998;14:S277–89. [PubMed] [Google Scholar]
- 21.Francis DP, Gregory T, McElrath MJ, et al. Advancing AIDSVAX to phase 3. Safety, immunogenicity, and plans for phase 3. AIDS Res Hum Retroviruses. 1998;14:S325–31. [PubMed] [Google Scholar]
- 22.Gilbert P, Wang M, Wrin T, et al. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J Infect Dis. 2010;202:595–05. doi: 10.1086/654816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitayaphan S, Pitisuttithum P, Karnasuta C, et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190:702–6. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 24.Edmonds TG, Ding H, Yuan X, et al. Molecular clones of HIV-1 expressing a Renilla luciferase reporter: new approaches for assessing immune responses elicited by HIV vaccine immunogens. Virol. 2010;408:1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Gao F, Mascola JR, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd CA, Greene KM, Yu X, et al. Development and implementation of an international proficiency testing program for a neutralizing antibody assay for HIV-1 in TZM-bl cells. J Immunol Methods. 2012;375:57–67. doi: 10.1016/j.jim.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophage tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei X, Decker JM, Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folks T, Benn S, Rabson A, et al. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci U S A. 1985;82:4539–43. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray ES, Moody MA, et al. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J Virol. 2011;85:7719–29. doi: 10.1128/JVI.00563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao HX, Levesque MC, Nagel A, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–9. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrammert J, Smith K, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moody MA, Zhang R, et al. H3N2 Influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonsignori M, Hwang KK, Xi C, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Gilbert PB, Montefiori DC, Self SG. Simultaneous evaluation of the magnitude and breadth of a left- and right-censored multivariate response, with application to HIV vaccine development. Stat Biopharm Res. 2009;1:81–91. doi: 10.1198/sbr.2009.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 37.Choudhry V, Zhang M-Y, Harris I, et al. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem Biophys Res Comm. 2006;348:1107–15. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans TG, Frey S, Israel H, et al. Long term memory B-cell responses in recipients of candidate human immunodeficiency virus type 1 vaccines. Vaccine. 2004;22:2626–30. doi: 10.1016/j.vaccine.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Russell ND, Graham BS, Keefer M, et al. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J Acquir Immune Defic Syndr. 2007;44:203–12. doi: 10.1097/01.qai.0000248356.48501.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleghorn F, Pape JW, Schechter M, et al. Lessons from a multisite international trial in the Caribbean and South America of an HIV-1 Canarypox vaccine (ALVAC-HIV vCP1452) with or without boosting with MN rgp120. JAIDS. 2007;46:222–30. doi: 10.1097/QAI.0b013e318149297d. [DOI] [PubMed] [Google Scholar]
- 41.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case–cohort analysis. Lancet. 2008;372:1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baum LL, Cassutt KJ, Knigge K, et al. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J Immunol. 1996;157:2168–73. [PubMed] [Google Scholar]
- 44.Forthal DN, Landucci G, Daar ES. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J Virol. 2001;75:6953–61. doi: 10.1128/JVI.75.15.6953-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hessell AJ, Hangartner L, Hunter M, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–5. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 46.Lai SK, Hida K, Shukair S, et al. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervicovaginal mucus. J Virol. 2009;83:11196–200. doi: 10.1128/JVI.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bomsel M, Heyman M, Hocini H, et al. Intracellular neutralization of HIV transcytosis across tight epithelial barriers by anti-HIV envelope protein dIgA or IgM. Immunity. 1998;9:277–87. doi: 10.1016/s1074-7613(00)80610-x. [DOI] [PubMed] [Google Scholar]
- 48.Huang YT, Wright A, Gao X, Kulick L, Yan H, Lamm ME. Intraepithelial cell neutralization of HIV-1 replication by IgA. J Immunol. 2005;174:4828–35. doi: 10.4049/jimmunol.174.8.4828. [DOI] [PubMed] [Google Scholar]
- 49.Karnasuta C, Parisa RM, Cox JH, et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX® B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine. 2005;23:2522–29. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Bar KJ, Wang S, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]