Abstract
The mammalian rod photoreceptor phosphodiesterase (PDE6) holoenzyme is isolated in both a membrane-associated and a soluble form. Membrane binding is a consequence of prenylation of PDE6 catalytic subunits, whereas soluble PDE6 is purified with a 17-kDa prenyl-binding protein (PDEδ) tightly bound. This protein, here termed PrBP/δ, has been hypothesized to reduce activation of PDE6 by transducin, thereby desensitizing the photoresponse. To test the potential role of PrBP/δ in regulating phototransduction, we examined the abundance, localization, and potential binding partners of PrBP/δ in retina and in purified rod outer segment (ROS) suspensions whose physiological and biochemical properties are well characterized. The amphibian homologue of PrBP/δ was cloned and sequenced and found to have 82% amino acid sequence identity with mammalian PrBP/δ. In contrast to bovine ROS, all of the PDE6 in purified frog ROS is membrane-associated. However, addition of recombinant frog PrBP/δ can solubilize PDE6 and prevent its activation by transducin. PrBP/δ also binds other prenylated photoreceptor proteins in vitro, including opsin kinase (GRK1/GRK7) and rab8. Quantitative immunoblot analysis of the PrBP/δ content of purified ROS reveals insufficient amounts of PrBP/δ (<0.1 PrBP/δ per PDE6) to serve as a subunit of PDE6 in either mammalian or amphibian photoreceptors. The immunolocalization of PrBP/δ in frog and bovine retina shows greatest PrBP/δ immunolabeling outside the photoreceptor cell layer. Within photoreceptors, only the inner segments of frog double cones are strongly labeled, whereas bovine photoreceptors reveal more PrBP/δ labeling near the junction of the inner and outer segments (connecting cilium) of photoreceptors. Together, these results rule out PrBP/δ as a PDE6 subunit and implicate PrBP/δ in the transport and membrane targeting of prenylated proteins (including PDE6) from their site of synthesis in the inner segment to their final destination in the outer segment of rods and cones.
Sensory transduction of light stimuli begins in the rod and cone photoreceptor cells of the vertebrate retina. The extensive membrane system in the outer segment is where proteins involved in visual excitation are located. Activation of the visual receptor, opsin, by light causes it to catalyze the exchange of GDP for GTP on the α-subunit of the heterotrimeric G-protein, transducin (Tα).1 Upon dissociation of Tα-GTP from Tβγ, the effector enzyme, photoreceptor phosphodiesterase (PDE6) is activated by displacement of its inhibitory γ-subunit (Pγ) by Tα-GTP. As activated PDE6 lowers cytoplasmic cGMP levels, cGMP-gated ion channels in the plasma membrane are closed, causing membrane hyperpolarization (for reviews, see Refs. 1 and 2).
Rod photoreceptor PDE6 is a catalytic heterodimer of very similar α and β catalytic subunits (Pαβ) to which two identical inhibitory γ-subunits (Pγ) are tightly bound. Each catalytic subunit has a catalytic domain responsible for cGMP hydrolysis and two regulatory GAF domains, one of which binds cGMP with high affinity. Pγ interacts allosterically with both the catalytic and regulatory domains. Each catalytic subunit is post-translationally modified at its C terminus with an isoprenyl group (either farnesyl or geranylgeranyl) that is responsible for its high affinity association with outer segment disk membrane (for review, see Ref. 3).
A significant fraction of the total rod PDE6 (and all cone PDE6) extracted from bovine retina is found in a soluble form. What distinguishes this soluble PDE6 from membrane-associated PDE6 is the binding of a 17-kDa protein that was originally named the “δ-subunit” of PDE6 (4–6). It should be noted that soluble PDE6 is derived from a retinal extract containing broken rod and cone outer segments, and therefore the cellular origin of the 17-kDa protein binding to PDE6 is uncertain. The primary site of interaction of this 17-kDa protein (referred to in this report as prenyl-binding protein/δ (PrBP/δ)) with PDE6 is at the prenylated, carboxymethylated C terminus of the catalytic subunits (6, 7). Based on peptide competition experiments, PrBP/δ binding to PDE6 requires not only the prenyl moiety but also the modified, carboxymethylated C-terminal amino acids of rod PDE6 α or β (7). In contrast, the rod and cone opsin kinases (GRK1 and GRK7) can be prevented from binding to PrBP/δ by preincubation with prenyl compounds altogether lacking the C-terminal peptide sequence (8). This suggests that the affinity and/or specificity of PrBP/δ for individual prenylated proteins may depend not only on prenyl binding, but also on interactions with amino acids in the C-terminal domain of the prenylated protein.
The crystal structure of PrBP/δ-subunit (9) is structurally similar to a known prenyl-binding protein, RhoGDI (10). Comparison of the two structures reveals that practically the entire sequence of PrBP/δ is an immunoglobulin-like domain that forms a highly hydrophobic prenyl binding pocket. The outer surface of PrBP/δ also forms a complex with Arl2 (9), but this interaction shows no structural homology to the binding site of Cdc42 on the surface of RhoGDI (11). Hence, this family of prenyl-binding proteins (which also includes the homologous unc119/RG4 protein (10, 12)) consists of a highly conserved prenyl binding pocket, along with protein-specific interaction sites that confer unique functions for each prenyl-binding protein.
Unlike most phototransduction proteins, PrBP/δ is also expressed outside the retina in numerous other tissues that are not known to express photoreceptor PDE6 (6, 13–15). Furthermore, PrBP/δ interacts with proteins unrelated to known phototransduction proteins, such as Rab13 (14), the retinitis pigmentosa GTPase regulator (16), and the Arf-like proteins Arl2 and Arl3 mentioned above (9, 17). This highly conserved protein (99% amino acid identity within mammals) has homologues in lower vertebrates and invertebrates, including eyeless animals (e.g. Caenorhabditis elegans, 65% amino acid identity) (6, 13, 18, 19). This suggests that PrBP/δ may have different functions and/or biological activities depending upon the tissue in which it is expressed.
Previous biochemical studies in retinal photoreceptors have shown that exogenous bovine PrBP/δ can bind to rod PDE6 with high affinity and cause release of PDE6 from the disk membrane (6). Interaction of bovine PrBP/δ with rod PDE6 also alters cGMP binding to the regulatory GAF domains, converting a non-exchangeable cGMP binding site to one that readily exchanges bound and free nucleotide (20). The solubilization of PDE6 by PrBP/δ is accompanied by a decrease in its activation by transducin (21). These in vitro results are all consistent with the hypothesis that PrBP/δ allosterically regulates PDE6, perhaps serving as a desensitization mechanism during light adaptation (21).
To critically examine the role of PrBP/δ in phototransduction, we have studied the binding properties, regulation, abundance, and subcellular localization of PrBP/δ in both bovine and frog photoreceptors. Having cloned, sequenced, and expressed the frog homologue of PrBP/δ, we confirm that it binds to PDE6 in vitro, releases it from the disk membrane, and effectively uncouples transducin activation of PDE6, similarly to its bovine counterpart. However, purified frog ROS containing a full complement of soluble and membrane proteins (22) lacks sufficient PrBP/δ to bind and solubilize more than a few percent of the PDE6 that is present in ROS. Furthermore, ~90% of total PrBP/δ immunoreactivity in frog or bovine retinal samples does not co-purify with intact ROS. We conclude that PrBP/δ is not a genuine subunit of the PDE6 holoenzyme, and is unlikely to regulate PDE6 during phototransduction.
Immunocytochemical examination of the localization of PrBP/δ reveals that most PrBP/δ is found outside the photoreceptor cells of the retina. Of the PrBP/δ immunoreactivity seen in the photoreceptor layer, careful examination of its subcellular distribution refines previous observations (6, 8) and localizes most of the PrBP/δ to either the inner segment (frog) or to the junction between the inner and outer segment (bovine) of photoreceptor cells. Furthermore, immunoelectron microscopy places PrBP/δ distal to the connecting cilium, associated with axonemes extending into bovine rod and cone outer segments. Together, these results indicate that PrBP/δ is most likely functioning in the intracellular trafficking of newly synthesized prenylated proteins from the inner to the outer segment of retinal photoreceptors.
EXPERIMENTAL PROCEDURES
Materials
Frogs (Rana catesbeiana) were obtained from Niles Biologicals Co. or Charles D. Sullivan Co. and maintained on a 12-h dark-light cycle for 2 weeks before use in experiments (23). Frozen bovine retinas were obtained from W. L. Lawson Inc., and fresh bovine eyes were kindly provided by Lemay & Sons (Goffstown, NH). Sucrose- and Ficoll-purified bovine ROS from freshly isolated retinas (24) were a kind gift of Dr. Paul Schnetkamp (University of Calgary). [3H]cGMP was purchased from PerkinElmer Life Sciences. Supplies for immunoblotting were purchased from Schleicher & Schuell, Pierce, or Bio-Rad. Chemicals were obtained from Sigma. The bovine recombinant GST-PrBP/δ fusion protein was a kind gift of Drs. Terry Cook and Joe Beavo (University of Washington).
Antibodies
Rabbit polyclonal antiserum to the PrBP/δ protein was raised in rabbits by immunization with the full-length recombinant bovine protein and was affinity-purified on PrBP/δ-Sepharose (FL antibody). Other rabbit polyclonal antibodies used in this study and raised in our laboratory include a PDE6 anti-peptide antibody directed to the GAFb domain that is conserved in all PDE6 catalytic subunit sequences (NC) and an affinity-purified antibody to the C terminus of the Pγ-subunit of PDE6 (CT-9710) (23). Commercial antibodies to other proteins included the following: rod transducin α-subunit (K-20, Santa Cruz Biotechnology), rhodopsin (1D4, Chemicon) (25), opsin kinase (G-8, Affinity Bioreagents) (26), and Rab8 protein (Rab8, BD Biosciences). Other antibodies to phototransduction proteins were generously provided by the following individuals: rod and cone PDE6 catalytic subunits (ROS1, Dr. R. L. Hurwitz (27)), cone arrestin (7G6, P. R. MacLeish) (28), rod arrestin (SC-128, W. C. Smith), and cone opsin (COS-1, A. Szel) (29). Horseradish peroxidase-conjugated secondary antibodies for Western blotting were from Pierce, whereas fluorochrome-conjugated or biotinylated secondary antibodies (Alexa-350, Alexa-488, or fluorescein isothiocyanate) and the tyramide amplification kit for immunohistochemistry were from Molecular Probes.
Cloning and Expression of Frog PrBP/δ
A frog (Rana pipiens) retinal cDNA library was prepared as described elsewhere (30). Frog PrBP/δ was cloned by screening the cDNA library with the bovine PrBP/δ nucleic acid sequence. The resulting clone, fpded1, contained a 2.7-kb cDNA insert in pBluescript SK. The 450-bp coding region of PrBP/δ was PCR-amplified with primers 5′-GGATCCATGTCTAGTAATGAGCGA and 5′-GAATTCTCAAACATAAAACAGCCTG, containing BamH1 and EcoR1 restriction sites. This PCR product was inserted into the TOPO cloning vector (Invitrogen) and then transferred into the pGEX-KG expression plasmid (Amersham Biosciences). Sequencing of both strands confirmed that the coding region contained the authentic nucleotide sequence. The GenBank™ accession number for the frog PrBP/δ sequence is AY044177.
The pGEX-KG/PrBP/δ construct was transformed into the Escherichia coli strain BL21(DE3) for protein expression. Frog PrBP/δ expression was induced by addition of 1.0 mm isopropyl-β-d-thiogalactopyranoside to log phase cultures. Growth continued for 1 h at 37 °C prior to extraction of the recombinant fusion protein by sonication and centrifugation. GST-PrBP/δ was purified on a glutathione-agarose column. Following thrombin cleavage of the fusion protein from PrBP/δ and subsequent removal of thrombin with the Thrombin Cleavage Capture Kit (Novagen), PrBP/δ was separated from GST by glutathione-agarose chromatography, and the flow-through fraction was concentrated by ultrafiltration (Millipore BioMax 5K MWCO filter). Recombinant protein concentration was determined by a colorimetric protein assay (31), which agreed with spectrophotometric estimates. Purity of the protein was >95%, as judged by SDS-PAGE. Bovine GST-PrBP/δ was expressed and purified in an identical manner.
Preparation of Retinal Extracts and Purified ROS
To prepare retinal extracts, frog or bovine retinas were placed in extraction buffer (10 mm Tris, pH 8.0, 150 mm NaCl, 1% Triton X-100, 2 mm dithiothreitol, and mammalian protease inhibitor mixture (Sigma)) and homogenized with a motor-driven pestle in a glass mortar. Detergent-solubilized proteins were separated from particulate matter by centrifugation for 5 min at 100,000 × g in an Airfuge. The concentrations of protein (31), rhodopsin (32), and PDE6 (23) were determined. Under these conditions, >90% of the visual pigment was solubilized, indicating extraction of most intrinsic membrane proteins. For immunoblotting, the retinal homogenate was added to SDS-PAGE gel sample buffer. For Fig. 2B, freshly isolated retinas were immediately frozen in liquid nitrogen, pulverized with a liquid nitrogen-cooled ceramic mortar and pestle, and directly transferred to boiling gel sample buffer to minimize sample degradation.
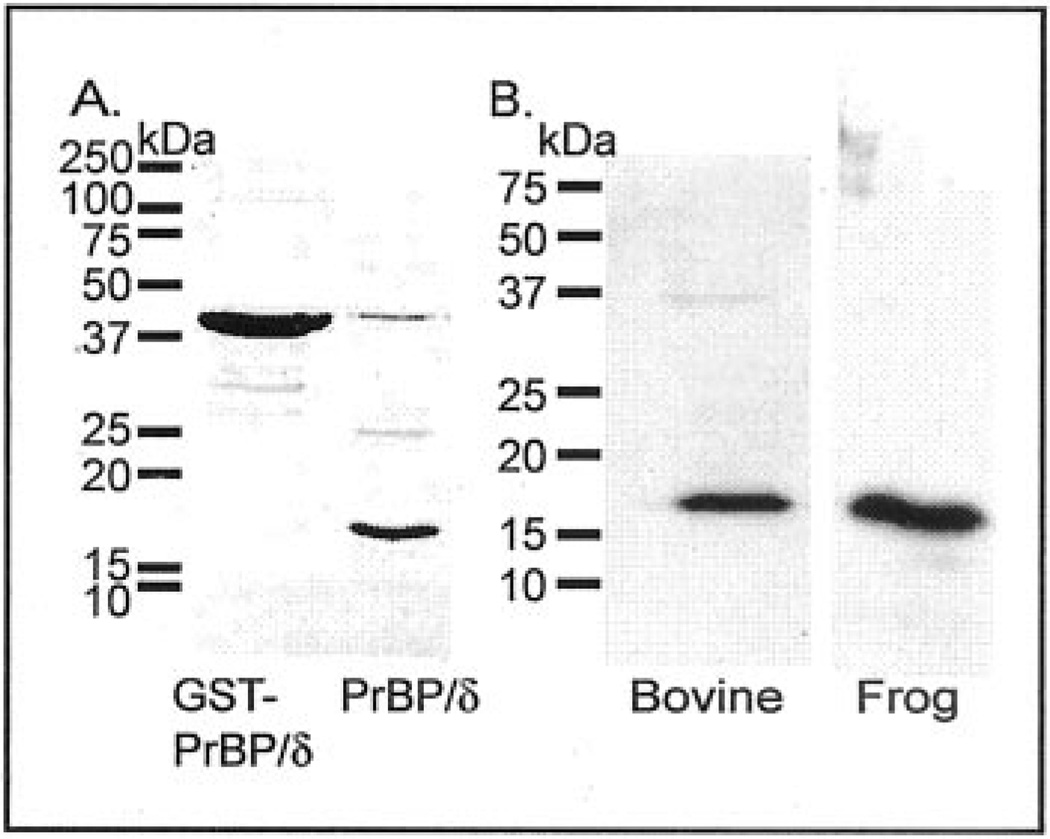
Fig. 2. Recombinant expression of frog PrBP/δ and antibody specificity of the PrBP/δ antibody (FL).
A, bacterially expressed frog GST-PrBP/δ fusion protein was purified by glutathione agarose affinity chromatography and then treated with thrombin to cleave the fusion partner from PrBP/δ. Samples were analyzed by SDS-PAGE and Coomassie staining. Lane 1: 20 µg of frog GST-PrBP/δ. Lane 2: 20 µg of PrBP/δ. B, the specificity of the PrBP/δ antibody (FL) was tested by immunoblot analysis of bovine and frog retinal extracts (11 µg of protein) run on SDS-PAGE, transferred to polyvinylidene difluoride membranes, and the membranes probed with 1:1000 dilution of the FL antibody and 1:3000 dilution of secondary antibody.
Osmotically intact frog ROSs were purified on a discontinuous Percoll gradient exactly as described previously (23). Purified ROSs were then homogenized in a pseudo-intracellular medium containing 77 mm KCl, 35 mm NaCl, 2.0 mm MgCl2, 1.0 mm CaCl2, 1 mm EGTA, 10 mm HEPES (pH 7.5), 2 mm dithiothreitol, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 µm leupeptin, and 1 µm pepstatin.
Bovine ROSs were purified on a discontinuous sucrose gradient as previously described (33). Bovine ROSs were homogenized as described for frog ROSs above, except that complete disruption of ROS structure required trituration through a 26-gauge syringe needle.
Subcellular fractionation of ROS homogenates, where carried out, was accomplished by centrifugation at 110,000 × g in an Airfuge. Greater than 95% of the rhodopsin was recovered in the ROS membrane pellet under these conditions. Purification of PDE6 followed established procedures in our laboratory (23, 34).
PDE6 cGMP Binding and Activity Assays
The PDE6 concentration of frog ROS homogenates was measured by the known ability of PDE6 to bind 2.0 mol of [3H]cGMP per mole of holoenzyme under defined assay conditions (PDE6 being the sole high affinity cGMP-binding protein in ROS (23, 35)). In brief, PDE6-containing samples were incubated for 10 min at room temperature in the presence of 1 µm [3H]cGMP, 200 µm zaprinast, and a 2-fold molar excess of Pγ. Samples were filtered onto pre-wet nitrocellulose membranes (Millipore HAWP25) and immediately washed three times with 1 ml of ice-cold intracellular medium (23).
The rate of transducin-activated PDE6 hydrolysis of cGMP was measured by a coupled-enzyme phosphate release assay (23). GTPγS (equal in concentration to the amount of rhodopsin) was added to the PDE6 sample for 1 min prior to the addition of 10 mm cGMP. For each experiment, rate measurements were obtained at a minimum of three individual time points, during which <30% of substrate was consumed. Measurements of the activity were made in the following buffer: 100 mm Tris (pH 7.5), 10 mm MgCl2, 0.5 mm EDTA, 2 mm dithiothreitol, and 0.5 mg/ml bovine serum albumin. This PDE6 activity assay was also used to estimate the bovine PDE6 concentration under assay conditions where the enzyme was fully activated by trypsin-induced proteolysis of its inhibitory Pγ-subunits (20, 36).
PrBP/δ Solubilization Assay
Recombinant PrBP/δ or GST-PrBP/δ was added to ROS homogenates at various molar ratios relative to the PDE6 concentration. After overnight incubation, ROS membranes were pelleted, and the solubilized PDE6 in the supernatant was quantitated. In some cases, pull-down assays were then performed using either glutathione-agarose beads (to pull down GST-PrBP/δ) or ROS1-Sulfolink beads (to pull down PDE6-binding proteins). The ROS1 antibody was covalently coupled to Sulfolink beads (Pierce) following the manufacturer’s recommended procedure.
Pull-down Assays of PrBP/δ-interacting Proteins
To identify potential binding partners for PrBP/δ, recombinant frog or bovine PrBP/δ was coupled to cyanogen bromide-activated Sepharose 4B (Sigma) following the manufacturer’s protocol. Detergent-solubilized ROS homogenates (containing 10 mm CHAPS) were incubated with PrBP/δ-Sepharose beads for 12 h, then pelleted, and washed three times with TMN buffer (10 mm Tris, pH 7.5, 1 mm MgCl2, 300 mm NaCl, 1 mm dithiothreitol). Proteins bound to the resin were eluted with gel sample buffer and analyzed by SDS-PAGE and immunoblotting. Unbound proteins in the supernatant were trichloroacetic acid-precipitated before loading onto the gel. Control samples were pretreated with a large excess of recombinant PrBP/δ prior to mixing the beads with the ROS homogenate to test for nonspecific binding of proteins to the resins.
Immunoblot Analysis
Samples for immunoblotting were prepared in Laemmli gel sample buffer. The proteins were separated on SDS-PAGE and then transferred to Immobilon-PSQ (Millipore) membranes. The blots were blocked with 1% bovine serum albumin in TTBS (20 mm Tris, pH 7.6, 137 mm NaCl, 0.5% Tween 20) prior to incubation for 1 h with the primary antibody. Binding of the primary antibody was detected with the appropriate goat anti-host secondary antibody coupled to horseradish peroxidase (1:5000 dilution, 1-h incubation), followed by chemiluminescent detection (Pierce SuperSignal West Pico Chemiluminescent Substrate). For quantitative analysis of the PrBP/δ content of total retinal homogenates and ROSs, known concentrations of recombinant frog or bovine PrBP/δ were run on the same gel, and the density of immunoreactive bands from Western blots was analyzed with Quantiscan (Biosoft).
Immunohistochemistry and Immunoelectron Microscopy
Light-adapted bovine eyes were hemisected, and the lens and vitreous were removed. The posterior eyecup was everted and fixed in freshly made 4% paraformaldehyde in PBS, pH 7.4, for 1–2 h at room temperature. Fixative was removed by three changes of PBS. Dissected areas of central retina with or without attached eyecup were cryoprotected in 30% sucrose/PBS, oriented, and snap-frozen in optimal cutting temperature compound (OCT, VWR International). Blocks were held at −80 °C until sectioned. Frog eyes from dark- and light-adapted animals were treated in essentially the same manner: following hemisectioning and removal of the lens, eyecups were fixed, rinsed, and cryoprotected as described. The entire eyecup was then oriented and snap-frozen in OCT.
Frozen sections (10 µm) were mounted on SuperFrost Plus slides (VWR International) and processed for standard indirect immunofluorescence. Briefly, sections were incubated in primary antibody, either for 2–4 h at room temperature or overnight at 4 °C. All antibodies were diluted in PBS containing 5% normal goat serum and 0.5% Triton X-100. Following primary antibody incubation, slides were washed in PBS three times for 5 min, then incubated for 1 h at room temperature in the appropriate secondary antibody, and finally washed as above. (For Fig. 7C, additional amplification of the PrBP/δ primary antibody was obtained using a biotin-labeled secondary antibody followed by a streptavidin-Alexa 488 conjugate (Molecular Probes). For Fig. 8J, tyramide amplification (Molecular Probes) was used for enhancing the detection of the PrBP/δ primary antibody.) Sections were mounted in ProLong Gold (Molecular Probes), with 4′,6-diamidino-2-phenylindole included (as a nuclear stain) in some cases. Slides were viewed by phase-contrast and epifluorescence microscopy on a Nikon Optiphot 2, and images were photographed with a SPOT-RT digital camera (Diagnostic Instruments). All antibodies were tested at several concentrations to optimize specific binding. Tissue autofluorescence and background staining were minimal in our experiments. Secondary reagents were found to be species-specific, and there was no overlap of the fluorescent signals from two different fluorophores. The specificity of PrBP/δ staining by the FL antibody was confirmed by preincubating the primary antibody with equimolar recombinant PrBP/δ; no detectible fluorescence above background was observed (data not shown).
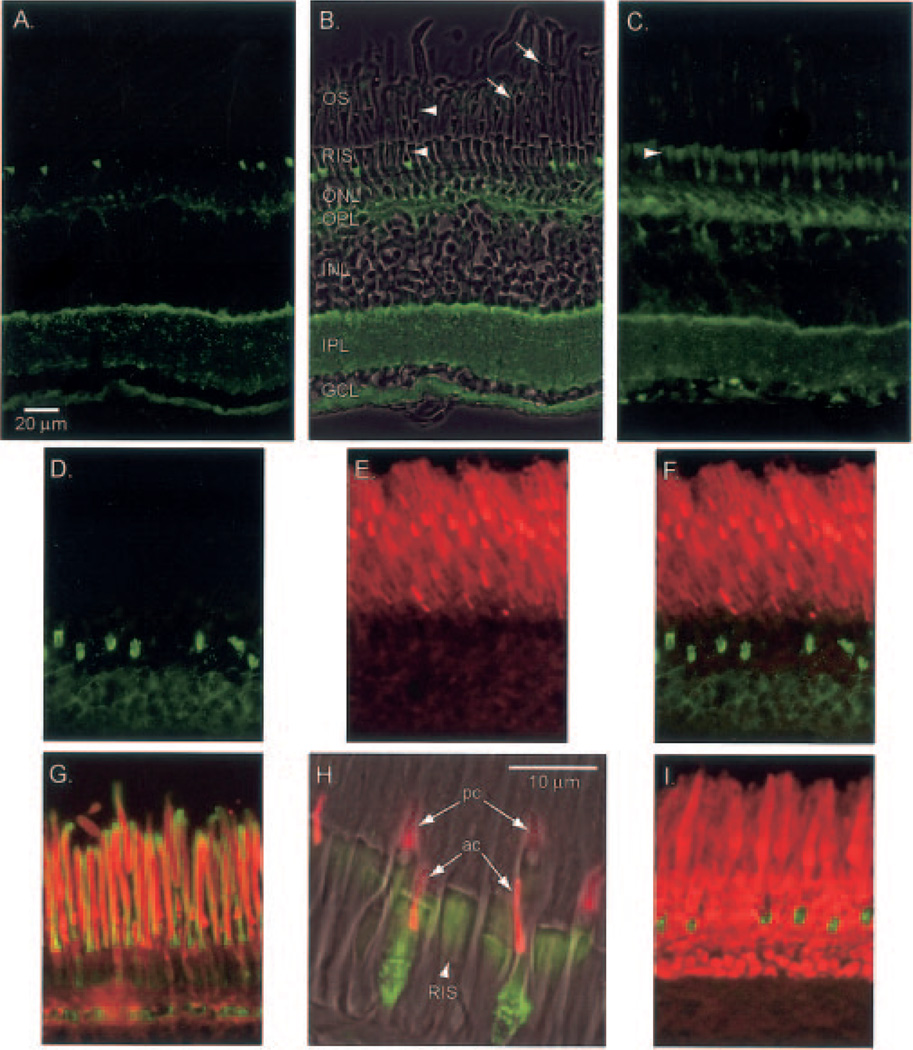
Fig. 7. Immunohistochemical localization of PrBP/δ in frog retina.
Frog retina was isolated, fixed, and sectioned as described under “Experimental Procedures” and incubated with primary antibodies to the indicated proteins: A, PrBP/δ antibody (FL, green). B, same as A with phase contrast superimposed and retinal layers identified: OS, outer segment; RIS, rod inner segment; ONL, outer nuclear layer, OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Stain appears greatest in certain inner segments and stratum 1 of the IPL. Arrowheads indicate cone outer segments, while arrows indicate rod outer segment. C, FL antibody signal amplified with biotin-streptavidin detection reagents permits labeling of all rod inner segments (arrowhead). D–F, double labeling with FL (green) and ROS1 (a PDE6 catalytic subunit antibody, red) stainings do not co-localize. G, superimposition of PDE6 (ROS1, red) and Pγ-subunit (CT-9710, green) show co-localization in the outer segments. H, superimposition of phase contrast image with FL (green) and red/green cone opsin (COS-1, red). Arrows point to two double cones: pc, principal cone; ac, accessory cone. Arrowhead points to rod inner segment (RIS). I, double staining of FL (green) and rod arrestin (SCT-128, red).
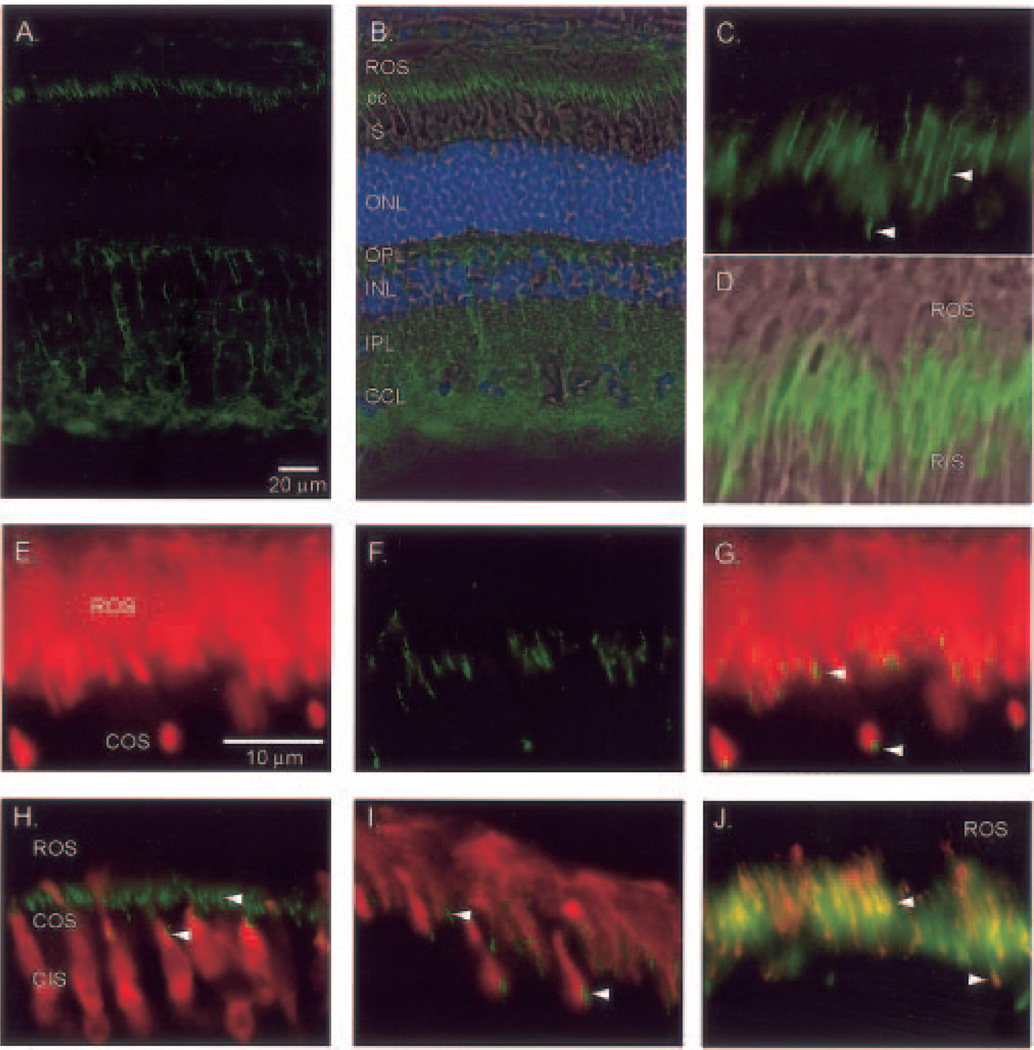
Fig. 8. Immunohistochemical localization of PrBP/δ in bovine retina.
Light-adapted bovine retinas were fixed and sectioned as described under “Experimental Procedures,” and then sections were incubated with the following primary antibodies. Arrowheads in some panels indicate the connecting cilium (cc) region. A, PrBP/δ (FL, green) alone. B, same as A with superimposition of phase contrast image and 4′,6-diamidino-2-phenylindole (blue) staining, with layers identified as in Fig. 7. C, FL staining of connecting cilium region of rods (upper arrowhead) and cones (lower arrowhead). D, superimposition of C with phase contrast image. E–G, double labeling of PDE6 antibody (ROS1, red) and FL (green); PrBP/δ-staining is greatest at the base of the outer segment labeled by ROS1. H, cone arrestin (7G6, red) stains cones (COS and CIS) but not rods, whereas FL staining (green) in cones is at the junction of COS and CIS. I, GRK1/7 (G8, red) stains rod and cone outer segments, whereas FL staining (green) is observed at the base of outer segments. J, RP1 (anti-RP1, red) stains the connecting cilium region and overlaps with tyramide-amplified FL staining (green).
For immunoelectron microscopy, after cryoprotection, retinas were frozen and thawed three times (to enhance reagent penetration) and mounted in 4% agarose. Radial Vibratome sections (100 µm) were immunostained as above, except that Triton X-100 (0.1%) was added only to the preincubation medium. (Complete elimination of Triton X-100 gave no staining.) Sections were incubated with secondary antibody conjugated to horse-radish peroxidase, developed using diaminobenzidene and hydrogen peroxide, and intensified using the gold-substitution silver intensification method (37). The sections were then treated with osmium tetroxide (2%; 60 min), stained with 2% uranyl acetate in 70% ethanol, dehydrated in graded ethanol series (70–100%), cleared in propylene oxide, and embedded in Epon 812. Ultrathin sections were mounted on Formvar-coated slot grids and stained with uranyl acetate prior to viewing.
RESULTS AND DISCUSSION
The Frog 17-kDa Prenyl-binding Protein (PrBP/δ) Is Highly Conserved
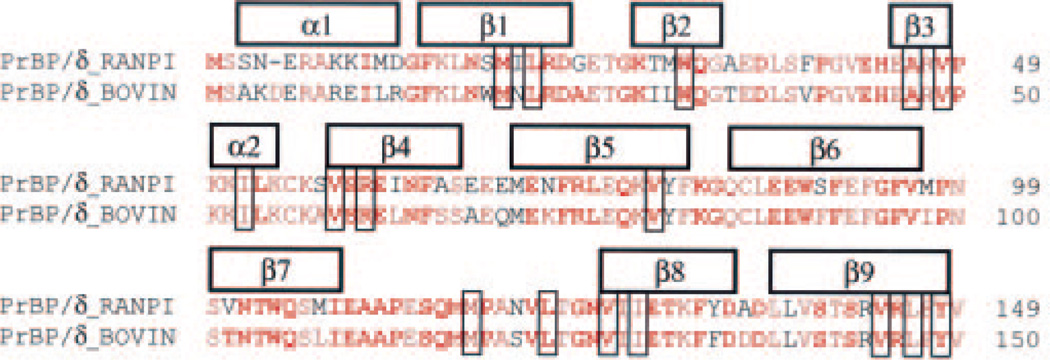
To determine the sequence of the frog homologue of the mammalian PrBP/δ protein, an R. pipiens retinal cDNA library was screened using the bovine nucleotide sequence as a probe. Clones containing the complete open reading frame were identified and sequenced. The deduced amino acid sequence of frog PrBP/δ (Fig. 1) is 82% identical to known mammalian sequences. When five mammalian, one chicken, and two fish PrBP/δ sequences are aligned with the frog sequence, 78% of the positions are identical for at least eight of the nine vertebrate sequences (Fig. 1, red letters). Orthologs of the vertebrate PrBP/δ protein have been identified in invertebrates as well (38, 39). Inclusion of six invertebrate PrBP/δ sequences in the multiple sequence alignment shows that the frog PrBP/δ sequence also has >61% identity with each of the invertebrate sequences. Indeed, at 45% of the total sites, the frog sequence is identical to at least 14 of the 15 sequences compiled for the sequence analysis (Fig. 1, red boldface letters). Of the 16 residues representing likely contact sites within the prenyl binding pocket (based on structural homology of PrBP/δ to RhoGDI (9)), 12 are identical in 15 of 16 PrBP/δ sequences, and the remaining 4 are identical among the vertebrate sequences analyzed here (Fig. 1, blue shaded sites). Based on the highly conserved amino acid sequence and structural homology modeling, it is evident that practically the entire sequence of frog PrBP/δ folds into a functional prenyl group-binding pocket. Sequence and structural analysis of PrBP/δ with the related proteins unc119/HRG4 (12, 40) and RhoGDI (9, 11) support the hypothesis that PrBP/δ is the basic prototype for a family of proteins containing a hydrophobic pocket that binds prenyl groups.
Fig. 1. The frog homolog of mammalian PrBP/δ is highly conserved.
The amino acid sequence of R. pipiens PrBP/δ was aligned and compared with eight other vertebrate sequences: bovine (shown), human, dog, rat, mouse, chick, fugu (partial sequence from genomic data base), and zebrafish (from genomic data base). Residues in red are identical in at least eight out of nine vertebrate species. Multiple sequence alignment was also performed, including six invertebrate, full-length PrBP/δ orthologous sequences: C. elegans, C. briggsae, C. intestinalis, D. melanogaster, A. gambiae, and Halocynthia roretzi. Residues in boldface red are identical in at least 14 out of 15 species. Secondary structure (above sequence) is based on the PrBP/δ crystal structure, and sites that are structurally equivalent to the prenyl interacting sites in the crystal structure of RhoGDI are boxed (9).
Recombinant Frog PrBP/δ Quantitatively Releases PDE6 from ROS Membranes
Expression of frog PrBP/δ as a GST fusion protein in E. coli yielded ~4 mg of affinity-purified GST-PrBP/δ per liter of culture. Following cleavage of the fusion protein, a 17-kDa protein that migrated identically to native frog PrBP/δ on SDS-PAGE was recovered (Fig. 2A). A polyclonal antibody (FL) raised against the full-length bovine recombinant PrBP/δ protein recognized a single 17-kDa band on immunoblots of both frog and bovine retinal extracts (Fig. 2B), demonstrating the specificity of the FL antibody for PrBP/δ.
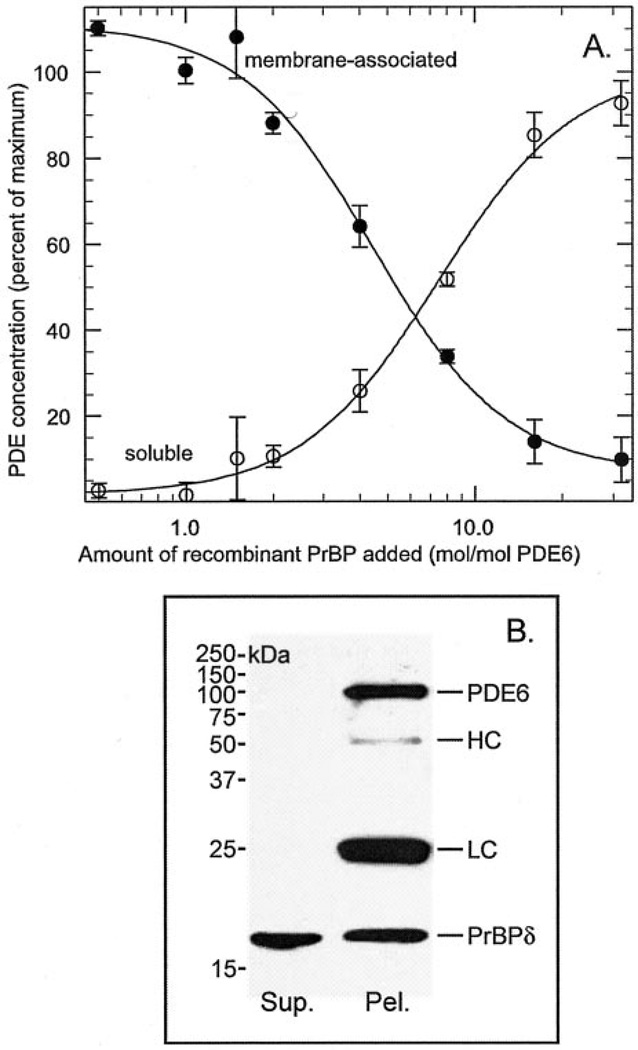
To test the functional activity of the expressed frog PrBP/δ, we examined its ability to solubilize PDE6 from ROS membranes. Frog ROS membranes were prepared from Percoll-purified ROS, and the soluble proteins were removed by centrifugation (see “Experimental Procedures”). In contrast to bovine ROSs where ~25% of the total PDE6 is recovered as a soluble protein (7), Percoll-purified frog ROSs have negligible amounts (1.0 ± 0.2%, n = 5) of PDE6 in the soluble fraction. When increasing amounts of recombinant frog PrBP/δ were added to ROS membranes, PDE6 was released from the membrane in a dose-dependent manner (Fig. 3A). The IC50 for PrBP/δ solubilization of PDE6 from frog ROS membranes (128 ± 11 nm) represents a substantial excess of PrBP/δ relative to the PDE6 concentration. Interestingly, the time course of PDE6 solubilization was very slow: only 30% of the PDE6 was released after a 5-h incubation, and 17 h were needed to observe complete solubilization. This slow process was not due to PrBP/δ binding to ROS membranes, because immunoblot analysis showed no increase in PrBP/δ immunoreactivity in ROS membranes following incubation of PrBP/δ with ROS homogenates (data not shown).
Fig. 3. Recombinant frog PrBP/δ solubilizes PDE6 from ROS membranes and forms a stable complex in vitro.
A, purified frog ROS homogenates (30 nm PDE) were incubated with increasing amounts of recombinant frog PrBP/δ for ≥17 h at 4 °C. Each sample was centrifuged to separate membrane-associated (●) from soluble (○) PDE6. The PDE6 concentration was determined by quantifying cGMP binding (see “Experimental Procedures”) and normalized to the PDE6 concentration in the absence of added PrBP/δ (<5% soluble). Data are mean ± S.E. for six separate experiments. B, frog ROS homogenates were incubated overnight at 4 °C with an 8-fold molar excess of frog PrBP/δ relative to PDE6. Solubilized PDE6 was separated from ROS membranes by centrifugation. ROS1-Sulfolink was then incubated with the supernatant fraction for 2 h at 22 °C. Precipitated proteins were recovered by brief centrifugation of the antibody beads. The pellet and supernatant fractions were mixed with SDS-PAGE gel sample buffer, electrophoresed, and transferred to membranes. The immunoblot was probed with a mixture of PrBP/δ (FL) and PDE6 (NC) antibodies. HC and LC refer to IgG heavy and light chains, which cross-reacted with the secondary reagent.
To determine whether the PrBP/δ remained associated with PDE6 following solubilization of the enzyme from ROS membranes, we used the ROS1 antibody (41) coupled to Sulfolink beads to immunoprecipitate the soluble PDE6. As shown in Fig. 3B, quantitative immunoprecipitation of PDE6 by the ROS1 antibody also co-precipitated PrBP/δ. In a similar experiment, we solubilized PDE6 with GST-PrBP/δ, and then used glutathione-agarose beads to pull down the GST-PrBP/δ. We found that all of the PDE6 co-precipitated with PrBP/δ (data not shown), confirming that PrBP/δ forms a stable complex with PDE6. These results preclude each molecule of PrBP/δ releasing multiple PDE6 molecules from the membrane and then dissociating from the enzyme.
PrBP/δ Is Able to Disrupt Transducin Activation of PDE6
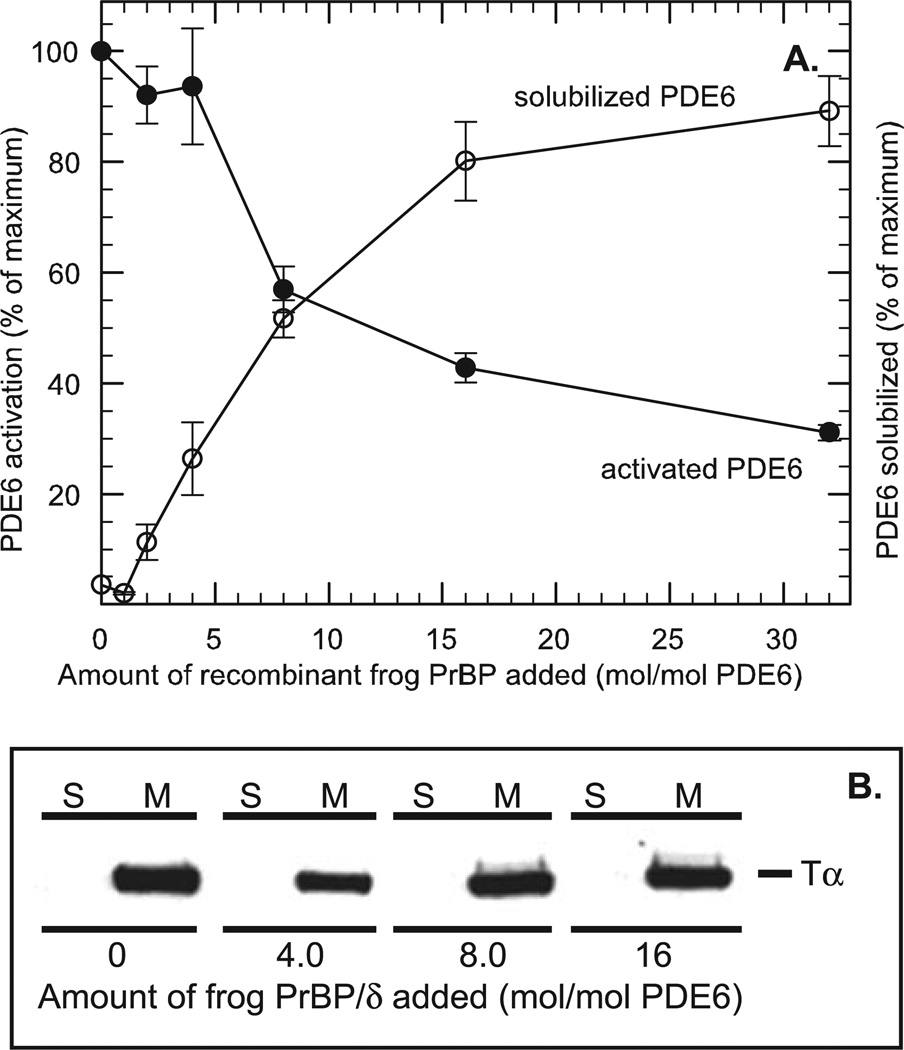
A proposed role of PrBP/δ in mammalian phototransduction is to decrease the ability of transducin to activate PDE6, thereby desensitizing the response to light stimulation (21). To test whether this mechanism is relevant to PDE6 activation in frog ROS, we first treated ROS homogenates with PrBP/δ, and then assayed the maximum extent of transducin activation of PDE6 in light-exposed ROS homogenates that had been incubated with GTPγS (to irreversibly activate transducin). As seen in Fig. 4A, increasing amounts of PrBP/δ cause a progressive reduction in PDE6 activation by transducin in light-exposed ROS. Under conditions where 90% of the PDE6 had been released from ROS membranes, a 70% reduction in PDE6 activation by transducin was seen. This behavior is generally consistent with a previous report with bovine ROSs (21). The IC50 values for disruption of transducin activation and for solubilization of membrane-associated PDE6 were also observed to be similar (Fig. 4A). The simplest explanation is that the reduced ability of transducin to activate PDE6 is a direct consequence of PrBP/δ removing PDE6, but not transducin, from ROS membranes, consistent with Cook et al. (21). (We verified that the Tα-subunit of transducin remains fully membrane-associated at all of the PrBP/δ concentrations tested (Fig. 4B).) This is consistent with earlier studies with purified, reconstituted ROS proteins showing that efficient activation of PDE6 by transducin requires membrane attachment of both proteins on the ROS disk membrane (42–45).
Fig. 4. PrBP/δ blocks transducin activation of PDE6 in vitro without releasing Tα*-GTPγS from ROS membranes.
A, nucleotide-depleted, light-exposed frog ROSs (40 nm PDE6) were incubated overnight at 4 °C with the indicated amount of PrBP/δ. To one portion (●), 13 µm GTPγS was added to determine the maximum extent to which PDE6 could be activated by transducin. Another portion (○) was centrifuged to determine the extent to which non-activated PDE6 was solubilized (as in Fig. 3). B, immunoblot analysis of ROS homogenates treated with the indicated amounts of PrBP/δ prior to centrifugation. Supernatants (S) and membrane pellets (M) were analyzed on immunoblots using a transducin (Tα) antibody.
PrBP/δ Associates with Other Prenylated Photoreceptor Proteins in Vitro
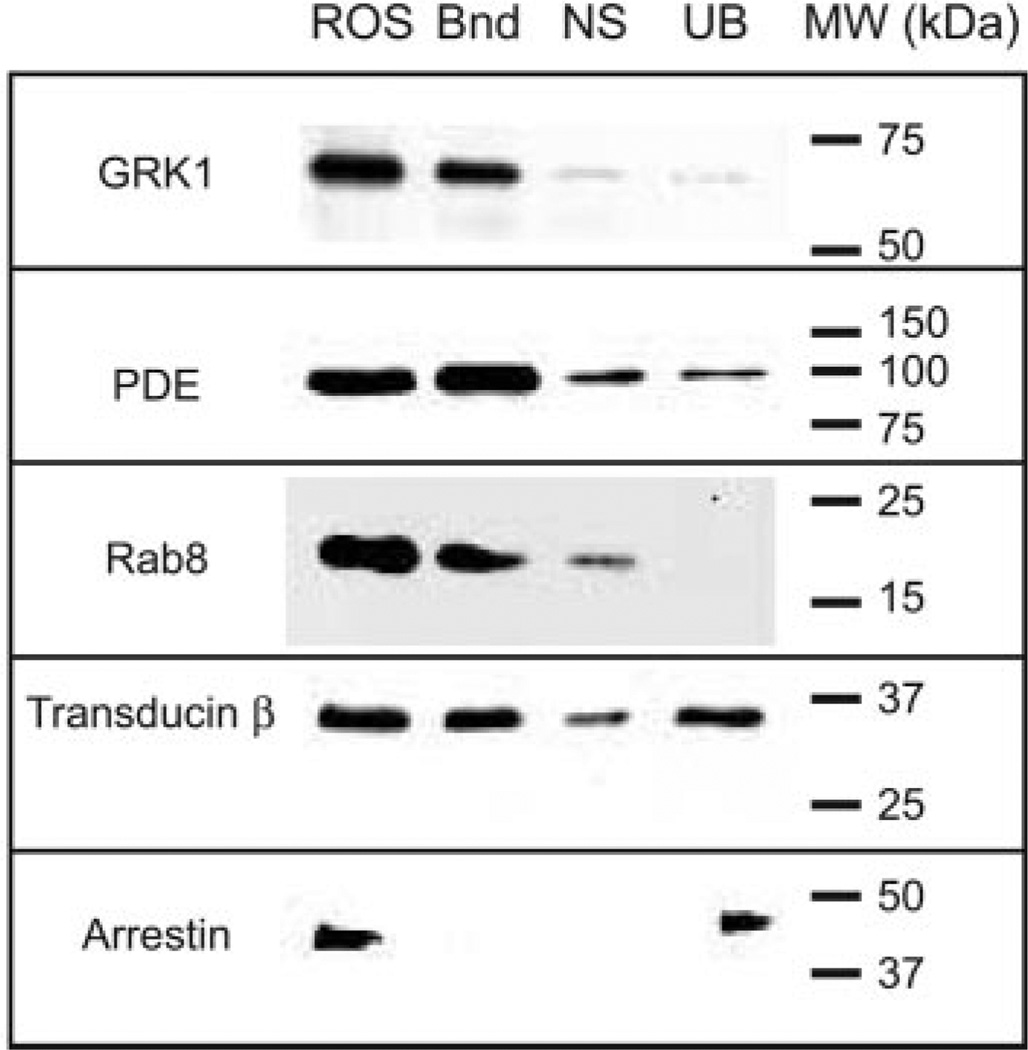
In addition to PDE6, a number of other photoreceptor proteins contain post-translational modifications that result in prenylation and carboxymethylation. These include the transducin γ-subunit (farnesylated), opsin kinases (GRK1, farnesylated; GRK7, geranylgeranylated), and rab8 (farnesylated). To test the ability of PrBP/δ to interact with photoreceptor proteins other than PDE6, we prepared PrBP/δ covalently coupled to Sepharose beads (PrBP/δ-Sepharose) and then incubated the immobilized PrBP/δ with detergent-solubilized bovine ROS homogenates. (Bovine ROSs were used for these experiments because of the greater availability of antibodies that recognize bovine, but not frog photoreceptor proteins.) As a control for the specificity of the interaction with the PrBP/δ-Sepharose, samples were preincubated with an excess of recombinant PrBP/δ. As seen in Fig. 5, PrBP/δ is able to form stable, specific complexes with GRK1, PDE6, Rab8, and the transducin β-subunit. In all of these cases, addition of excess PrBP/δ blocked most or all of the specific interaction with these proteins (Fig. 5, “NS”). Although the transducin β-subunit lacks a prenyl modification itself, it tightly associates with the farnesylated transducin γ-subunit, thus explaining its ability to be pulled down by PrBP/δ. In contrast, arrestin, an abundant soluble photoreceptor protein, was not pulled down by PrBP/δ-Sepharose. These results demonstrate that exogenous PrBP/δ is capable of forming stable interactions with several photoreceptor proteins other than PDE6, all of which contain C-terminal farnesyl or geranylgeranyl groups, or, in the case of transducin β-subunit, are tightly associated with a prenylated protein.
Fig. 5. PrBP/δ binds to several prenylated photoreceptor proteins in bovine ROS homogenates.
Detergent-solubilized bovine ROS (3.5 pmol of PDE6) were incubated with PrBP/δ-Sepharose (2 mg of PrBP/δ per milliliter of resin), and proteins bound to the resin (Bnd) were separated from unbound proteins (UB) by centrifugation. As a control for nonspecific binding to the resin (NS), identical samples were incubated with a large excess of PrBP/δ. Equivalent amounts of the starting sample (ROS) were also loaded. After SDS-PAGE and transfer to polyvinylidene difluoride, blots were probed with primary antibodies to GRK1, PDE6, Rab8, transducin β-subunit, and arrestin.
Quantitative Analysis of PrBP/δ Reveals Low Abundance in Retina and in ROSs
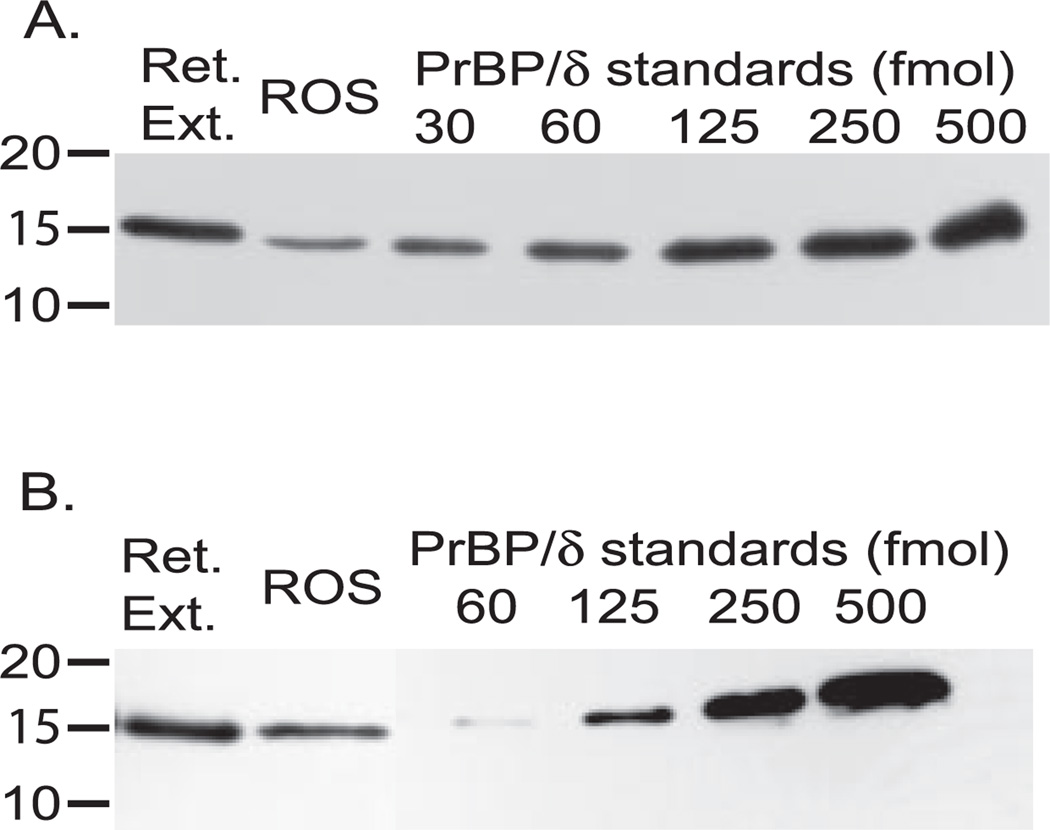
In order for PrBP/δ to serve as a regulatory protein for PDE6 during phototransduction, there must be enough PrBP/δ present to bind a significant fraction of the PDE6. Previous studies have failed to quantify the ratio of PrBP/δ relative to PDE6. To examine this, we prepared retinal extracts and purified ROS samples of known rhodopsin and PDE concentration, along with a series of PrBP/δ recombinant protein standards, and performed quantitative immunoblot analysis. Fig. 6A shows that a frog retinal extract containing 1.0 pmol of PDE6 has sub-stoichiometric amounts of PrBP/δ (0.4 mol of PrBP/δ per mol of PDE6; Table I) relative to PDE6. The amount of frog PrBP/δ relative to PDE6 is ~10-fold lower in samples of osmotically intact, purified frog ROS (Fig. 6A and Table I). (Note that Percoll-purified frog ROSs retain the full complement of soluble proteins (22), and hence loss of PrBP/δ from the outer segment cannot explain this result.) Of the PrBP/δ that is found in frog ROSs, >80% is recovered as a soluble protein following centrifugation of ROS homogenates in an isotonic buffer. Under these conditions, ~99% of the PDE6 is membrane-associated, indicating that only minute amounts of PrBP/δ are bound to PDE6 in frog ROSs.
Fig. 6. Quantitating PrBP/δ content in retinal extracts and in ROS.
A, detergent-treated frog retinal extracts (1.0 pmol of PDE6) and Percoll-purified frog ROS (1.0 pmol of PDE6) were examined for their PrBP/δ content by reference to the indicated amounts of purified, recombinant frog PrBP/δ. B, detergent-treated bovine retinal extracts (0.25 pmol of PDE6) and sucrose-purified ROS (0.5 pmol of PDE6) were analyzed as in panel A.
Table I. The content of PrBP/δ in retina and purified ROSs.
The amounts of PrBP/δ in retinal extracts, purified ROSs, and purified soluble rod PDE6 were determined by quantitative immunoblot analyses of samples where the concentration of PDE6 was independently determined and the PrBP/δ standards fell within the range of the unknown samples. Each value represents the mean ± S.E. for at least three different experiments.
| Sample | PrBP/δ content | |
|---|---|---|
| Bovine | Frog | |
| mol per mol PDE6 | ||
| Retinal extract | 0.6 ± 0.1 | 0.4 ± 0.1 |
| Purified ROS | 0.09 ± 0.01 | 0.03 ± 0.01 |
| Purified soluble PDE6 | 1.0 ± 0.1 | NAa |
NA, not applicable.
Qualitatively similar results are obtained for bovine retinal extracts or bovine ROSs (Fig. 6B and Table I). Overall, the total content of PrBP/δ is slightly higher for bovine compared with amphibian retinal extracts (0.6 PrBP/δ per PDE6). In bovine ROS, there is <0.1 PrBP/δ per PDE6 (Table I). Similar values for PrBP/δ relative to PDE6 were obtained for bovine ROSs purified from freshly isolated or frozen retinas, and for ROSs purified on a Ficoll gradient (24).
We also examined the stoichiometry of PrBP/δ bound to purified, soluble bovine rod PDE6 isolated from the bovine retinal extracts. Using quantitative immunoblot analysis (Table I), we estimated that 1.0 ± 0.1 PrBP/δ is bound to the catalytic dimer of bovine rod PDE6. We also estimated the Pγ content of our PDE6 samples using quantitative immunoblot analysis (1.8 ± 0.2 Pγ per PDE6; n = 4) to confirm that the PDE6 concentration loaded on the gel was accurate. This leads to the interesting conclusion that PrBP/δ binds only one of the two prenyl groups attached to the PDE6 catalytic αβ dimer. Although PrBP/δ can bind to PDE6 stoichiometrically, the low abundance of PrBP/δ in photoreceptors (Table I) precludes a significant amount of PDE6 from being bound to PrBP/δ in intact ROSs. We think it most likely that most soluble rod PDE6 (containing bound PrBP/δ) isolated from bovine retinal extracts results from binding of PrBP/δ derived from other retinal cells during homogenization of the retina.
Immunohistochemical Localization of PrBP/δ in Frog Retina
Based on our finding that most of the PrBP/δ present in frog retina is not recovered in purified, sealed ROSs, we performed indirect immunofluorescent labeling of PrBP/δ in frog retinal cryosections. The FL antibody to PrBP/δ was used in conjunction with a panel of well characterized antibodies to other retinal proteins to localize PrBP/δ within the retina. Fig. 7 (A and B) shows that in frog retina the majority of PrBP/δ is located in the inner plexiform layer (IPL, particularly stratum 1), in the optic nerve fiber layer beneath the ganglion cell layer, and, to a lesser extent, in the outer plexiform layer (OPL). Within the photoreceptor layer, we observe intense staining in inner segments of a subpopulation of photoreceptor cells (Fig. 7, A and B). In an effort to detect low levels of PrBP/δ in the photoreceptor layer, we amplified the signal by using a biotin-conjugated secondary antibody, which was then visualized with streptavidin conjugated to the Alexa 488 fluorophore. This method amplified PrBP/δ staining in the rod inner segments (Fig. 7C, arrowhead). Faint labeling of the ROS was also occasionally observed, but usually occurred in sections where background staining was greater. These immunohistochemical results are fully consistent with the quantitative immunoblot analysis (Table I) showing that <10% of the total PrBP/δ immunoreactivity in frog retina co-purifies with ROSs.
To better define the subcellular localization of PrBP/δ within the photoreceptor layer of the frog retina, we performed co-localization studies with several antibodies specific for rod and/or cone phototransduction proteins. Although PrBP/δ is widely regarded in the literature as a subunit of PDE6, the rod/cone PDE6 catalytic subunit antibody (ROS1) shows that PDE6 fails to co-localize with PrBP/δ (Fig. 7, D–F). In contrast, an antibody to the bona fide Pγ-subunit does indeed show co-localization with the PDE6 catalytic subunits in rods and cones under identical immunostaining conditions (Fig. 7G). We next identified the cell type that stained intensely with the FL antibody in the inner segment layer using the COS-1 antibody that is specific for medium/long wavelength-sensitive cone opsins in many vertebrate species (46). This antibody preferentially labels the outer segment of the large single cone and the double cone (consisting of primary and accessory members) of Xenopus retina (47). As seen in Fig. 7H, the bright, punctuate staining for PrBP/δ in the inner segment is almost always situated just below a COS-1-immunoreactive outer segment that lacks an oil droplet in its inner segment. This identifies the PrBP/δ-reactive photoreceptor as the accessory member of the double cone. Note that other COS-1-stained outer segments are joined to inner segments containing oil droplets and represent either the principal member of the double cone or single large cones that predominate in frog retina. Faint PrBP/δ immunoreactivity was also seen in the principal (“red”) rod inner segments (Fig. 7H). Finally, an antibody specific for rod arrestin (SCT-128) did not stain the PrBP/δ-positive cone inner segments at all, confirming that PrBP/δ in frog photoreceptors is primarily located in accessory cone inner segments. We conclude that the preponderance of PrBP/δ in frog retina is in the synaptic regions of all classes of retinal neurons. Within the photoreceptor cells themselves, strong staining for PrBP/δ is detected only in the inner segment of the accessory cone of the double cone pair, with lesser staining of rod inner segments, and very little immunoreactivity in photoreceptor outer segments.
Re-examination of the Immunolocalization of PrBP/δ in Bovine Retina
The low level of PrBP/δ staining in the photoreceptors of frog retina (Fig. 7) contrasts with previous, conflicting reports localizing PrBP/δ either to ROSs (6) or to outer and inner segments of both rods and cones (8). Because these differences might represent real species differences or, alternatively, differences in the light history or methods of staining retina for immunohistochemistry, we re-examined the localization of PrBP/δ in bovine retina using the FL antibody along with other well characterized antibodies that recognize bovine photoreceptor proteins.
As seen in panels A and B of Fig. 8, the majority of PrBP/δ in light-adapted bovine retina is found, not in the photoreceptor cells, but in the synaptic termini of the OPL, IPL, and beneath the ganglion cell layer. The nuclear layers have significantly less PrBP/δ staining (Fig. 8B). In these regards, PrBP/δ distribution in the bovine retina is quite similar to what is seen in frog retina. However, bovine retina exhibits a unique labeling pattern for PrBP/δ at the junction between the inner and outer segments of rod and cone photoreceptors (Fig. 8B, connecting cilium region (cc)). It shows a predominantly filamentous staining pattern for PrBP/δ that extends part way into the outer segments, a pattern consistent with PrBP/δ being present in the highest concentrations at the ciliary axoneme of both rod and cone photoreceptors (Fig. 8, C and D).
To further refine the localization of bovine PrBP/δ within photoreceptor cells, we first looked for co-localization of PrBP/δ with PDE6. Fig. 8E shows strong labeling of both rod and cone outer segments with the ROS1 antibody, consistent with previous work showing that this antibody recognizes both rod and cone PDE6 (6, 41). The PrBP/δ staining extends from the base of the outer segment part way into the outer segment of both rods and cones (Fig. 8, F and G). The merged image (Fig. 8G) shows PrBP/δ alone (green) and both PrBP/δ and PDE6 (yellow) signals, indicating that some PrBP/δ is found at the juncture of the inner and outer segments where there is little PDE6. The widely spaced PrBP/δ immunoreactivity (lower arrowhead) in Fig. 8H is shown to co-localize with a cone-specific arrestin antibody (red), beneath the major ribbon of PrBP/δ labeling (upper arrowhead) present in rods. Because opsin kinases have been shown to interact with PrBP/δ in pull-down assays (Fig. 5) and by yeast two-hybrid screens (8), we also labeled retinal sections with an antibody recognizing bovine rod (GRK1) and cone (GRK7) opsin kinase. GRK1/7 distribution, like PDE6 staining, is confined to the outer segments of both rods and cones in light-adapted retina, whereas PrBP/δ can be clearly seen at the base of the outer segments, suggesting the stained structures are the connecting cilia of rods and cones (Fig. 8I).
Finally, to further define PrBP/δ staining relative to the connecting cilium region, we defined the position of PrBP/δ relative to the RP1 protein, known to be localized to the axonemes immediately distal to the connecting cilium (48, 49). To enhance detection of PrBP/δ in this co-localization experiment, we used a tyramide amplification kit (see “Experimental Procedures”), which causes the normally filamentous PrBP/δ labeling to appear more diffuse (but did not alter overall localization of PrBP/δ in the retina). Fig. 8J shows substantial overlap in RP1 and PrBP/δ labeling (yellow) with additional PrBP/δ labeling (green) below and some RP1 labeling (red) above the region of co-localization. This places PrBP/δ in the immediate vicinity of the connecting cilium of rods and cones. Furthermore, semi-thin sections (0.5 µm) of bovine retina at lower magnification confirm that greater staining of PrBP/δ is detected at the junction of the inner and outer segments of rods and cones (Fig. 9A). To determine the localization of PrBP/δ at the ultrastructural level, we performed immunoelectron microscopy. PrBP/δ labeling was greatest at the base of rod and cone outer segments, just above the basal bodies and connecting cilia (Fig. 9, B and C). The PrBP/δ labeling appears strongly associated with ciliary axonemal microtubules. In both rods and cones, PrBP/δ staining extends part way into the outer segment before the labeling trails off.
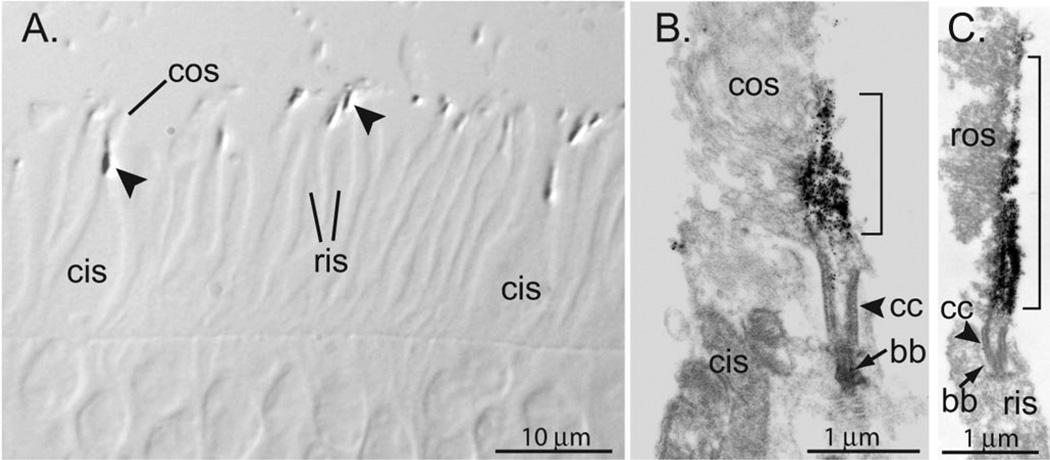
Fig. 9. Immunoelectron microscopy localizes PrBP/δ to the ciliary axonemes of bovine rods and cones.
A, semithin sections from bovine retina were stained for PrBP/δ (see “Experimental Procedures”). Immunoreactivity concentrated in the connecting cilium region (arrowheads) that joins cone inner segments (cis) and outer segments (cos) or rod inner segments (ris) and outer segments. B and C, ultrathin sections stained for PrBP/δ. Labeling is present in both rod (B) and cone (C) axonemal microtubules. Stain is greatest above the basal body (bb) and connecting cilium (cc), where it extends up along the axonemes (brackets).
This primary localization of PrBP/δ within bovine photoreceptors to the connecting cilium region (Figs. 8 and 9) differs from earlier reports of its predominant location in the rod outer segment (6) or in rod and cone outer segments (8). Several factors might account for these differing results, including different primary antibodies used, technical issues in detecting ciliary axonemal proteins (e.g. RPGR (50)) that may be sensitive to subtle variations in specimen preparation, and epitope masking that may result from PrBP/δ-interacting proteins binding to this small protein and preventing primary antibody binding.
The species difference observed in the immunohistochemical localization of PrBP/δ, i.e. in the connecting cilium and axonemes of bovine, but not amphibian, photoreceptors, is not understood. We cannot presently exclude the possibility that our experimental conditions for immunohistochemistry failed to detect PrBP/δ in the connecting cilium region and axonemes of frog photoreceptors. (Use of higher antibody concentrations, addition of increasing amounts of detergent, and attempts to immunolabeled purified ROS or isolated axonemal preparations did not show additional PrBP/δ labeling of photoreceptors (data not shown).) Another possibility is that PrBP/δ interacts with different proteins in frog photoreceptors compared with bovine photoreceptors and that epitope masking greatly hampers visualization of PrBP/δ by immunohistochemistry of frog retinal sections. Preliminary results using serial sectioning and immunoblot analysis (51) confirm that a 17-kDa, PrBP/δ-immunoreactive protein can be detected in the inner segment region of rods in frog retina.2 Although the total content of PrBP/δ in frog and bovine retinas is not very different (Table I), it is also possible that the function of PrBP/δ differs in the two species. For example, amphibian retinas contain distinct classes of photoreceptors not found in mammalian retinas (e.g. double cones) and frog photoreceptors are larger in diameter than their mammalian counterparts. On the other hand, the very high sequence and structural similarities of all vertebrate PrBP/δ proteins suggests that this prenyl-binding protein may serve identical functions in all vertebrate photoreceptors.
Summary
The ability of the frog homologue of PrBP/δ to completely solubilize membrane-attached frog PDE6 and to disrupt the ability of transducin to activate PDE6 demonstrates that the high sequence identity observed between the amphibian and mammalian PrBP/δ proteins reflects very similar functional properties of this prenyl-binding protein throughout tetrapod evolution. Although PrBP/δ can bind stoichiometrically to PDE6 with high affinity in vitro, the low concentration of PrBP/δ (<0.1 PrBP/δ per PDE6) in vertebrate outer segments precludes a role for this protein in the phototransduction pathway. Because PrBP/δ is not a bona fide subunit of PDE6, it should be identified based on its primary function, namely as a prenyl-binding protein.
The primary location of PrBP/δ in photoreceptor cells at the junction between the inner segment and the proximal region of the outer segment (defined as the connecting cilium) argues for a function for PrBP/δ related to transport of material through this connecting cilium. Specific binding of PrBP/δ with proteins that are located at or near the connecting cilium of photoreceptor cells has been reported, including the retinitis pigmentosa GTPase regulator (16, 52), rab8 (Ref. 53 and our Fig. 5), and Arl3 (9, 17, 54). Other proteins known to be involved in protein trafficking have also been shown to interact with PrBP/δ (although their presence in the connecting cilium has not been determined), especially ras-related GTPases (9, 10, 14). Hence, we propose that PrBP/δ most likely serves a role in protein transport through the connecting cilium of rods and cones. We envision two possible processes in which PrBP/δ may function: 1) transport of prenylated phototransduction proteins (e.g. transducin γ-subunit, PDE6 α-, β-, and α′-subunits, GRK1, GRK7) from their site of biosynthesis in the inner segment to the outer segment disk membrane; 2) light-driven protein translocation of prenylated proteins (e.g. transducin (51, 55)) between outer and inner segments. PrBP/δ and a structurally related protein, RG4/unc119 (18, 56), are both found in the synaptic layers of vertebrate retinas and may play a role in transport of synaptic vesicle proteins (57). The fact that defects in protein transport at the connecting cilium and at the synaptic vesicle have been correlated with photoreceptor degenerative diseases (e.g. retinitis pigmentosa, Usher’s syndrome, and Leber congenital amaurosis) emphasizes the importance of better understanding the functional roles of PrBP/δ and related prenyl-binding proteins in targeting proteins to specific photoreceptor membranes.
Acknowledgments
We thank Drs. Joe Beavo, Rich Hurwitz, Peter MacLeish, Eric Pierce, Clay Smith, and Agosten Szel, who kindly provided their antibodies, and Dr. Dave Townsen for use of his cryostat.
Footnotes
This work was supported by National Institutes of Health Grants EY05798 (to R. H. C.), EY08123 (to W. B.), and EY11105 (to N. V.) and is Scientific Contribution #2250 from the New Hampshire Agricultural Experiment Station. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY044177.
The abbreviations used are: Tα, transducin; PDE6, phosphodiesterase; PrBP, prenyl-binding protein; GST, glutathione S-transferase; PBS, phosphate-buffered saline; GTPγS, guanosine 5′-3-O-(thio)-triphosphate; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; IPL, inner plexiform layer; OPL, outer plexiform layer; ROS, rod outer segment.
M. Sokolov, S. Hosier, V. Y. Arshavsky, and R. H. Cote, unpublished data.
REFERENCES
- 1.Arshavsky VY, Lamb TD, Pugh EN., Jr Annu. Rev. Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 2.Stavenga DG, DeGrip WJ, Pugh EN., Jr . Molecular Mechanisms in Visual Transduction. Amsterdam: Elsevier Science; 2002. [Google Scholar]
- 3.Cote RH. In: Handbook of Cell Signaling. Bradshaw RA, Dennis EA, editors. San Diego, CA: Academic Press; 2003. pp. 453–458. [Google Scholar]
- 4.Gillespie PG, Beavo JA. J. Biol. Chem. 1988;263:8133–8141. [PubMed] [Google Scholar]
- 5.Gillespie PG, Prusti RK, Apel ED, Beavo JA. J. Biol. Chem. 1989;264:12187–12193. [PubMed] [Google Scholar]
- 6.Florio SK, Prusti RK, Beavo JA. J. Biol. Chem. 1996;271:1–12. doi: 10.1074/jbc.271.39.24036. [DOI] [PubMed] [Google Scholar]
- 7.Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. Biochemistry. 2000;39:13516–13523. doi: 10.1021/bi001070l. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HB, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. J. Biol. Chem. 2004;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 9.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nancy V, Callebaut I, El Marjou A, de Gunzburg J. J. Biol. Chem. 2002;277:15076–15084. doi: 10.1074/jbc.M109983200. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman GR, Nassar N, Cerione RA. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 12.Higashide T, Murakami A, McLaren MJ, Inana G. J. Biol. Chem. 1996;271:1797–1804. doi: 10.1074/jbc.271.3.1797. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz B, Migliaccio C, Lichtner P, Meyer C, Strom TM, D’Urso M, Becker J, Ciccodicola A, Meitinger T. Eur. J. Hum. Genet. 1998;6:283–290. doi: 10.1038/sj.ejhg.5200215. [DOI] [PubMed] [Google Scholar]
- 14.Marzesco AM, Galli T, Louvard D, Zahraoui A. J. Biol. Chem. 1998;273:22340–22345. doi: 10.1074/jbc.273.35.22340. [DOI] [PubMed] [Google Scholar]
- 15.Wang WQ, Zhang Q, Acland GM, Mellersh C, Ostrander EA, Ray K, Aguirre GD. Gene (Amst.) 1999;236:325–332. doi: 10.1016/s0378-1119(99)00246-2. [DOI] [PubMed] [Google Scholar]
- 16.Linari M, Ueffing M, Manson F, Wright A, Meitinger T, Becker J. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linari M, Hanzal-Bayer M, Becker J. FEBS Lett. 1999;458:55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Florio SK, Pettenati MJ, Rao PN, Beavo JA, Baehr W. Genomics. 1998;49:76–82. doi: 10.1006/geno.1998.5210. [DOI] [PubMed] [Google Scholar]
- 19.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 20.Mou H, Grazio HJ, Cook TA, Beavo JA, Cote RH. J. Biol. Chem. 1999;274:18813–18820. doi: 10.1074/jbc.274.26.18813. [DOI] [PubMed] [Google Scholar]
- 21.Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. J. Biol. Chem. 2001;276:5248–5255. doi: 10.1074/jbc.M004690200. [DOI] [PubMed] [Google Scholar]
- 22.Hamm HE, Bownds MD. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- 23.Cote RH. Methods Enzymol. 2000;315:646–672. doi: 10.1016/s0076-6879(00)15873-2. [DOI] [PubMed] [Google Scholar]
- 24.Schnetkamp PPM. Methods Enzymol. 1981;81:110–116. doi: 10.1016/s0076-6879(82)81019-7. [DOI] [PubMed] [Google Scholar]
- 25.Molday RS, MacKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Huang J, Khani SC, Palczewski K. J. Biol. Chem. 1998;273:5124–5131. doi: 10.1074/jbc.273.9.5124. [DOI] [PubMed] [Google Scholar]
- 27.Hurwitz RL, Beavo JA. Adv. Cyclic Nucleotide Protein Phosphorylation Res. 1984;17:239–248. [PubMed] [Google Scholar]
- 28.Zhang H, Cuenca N, Ivanova T, Church-Kopish J, Frederick JM, MacLeish PR, Baehr W. Invest. Ophthalmol. Vis. Sci. 2003;44:2858–2867. doi: 10.1167/iovs.03-0072. [DOI] [PubMed] [Google Scholar]
- 29.Rohlich P, Szel A, Papermaster DS. J. Comp. Neurol. 1989;290:105–117. doi: 10.1002/cne.902900107. [DOI] [PubMed] [Google Scholar]
- 30.Pittler SJ, Fliesler SJ, Baehr W. FEBS Lett. 1992;313:103–108. doi: 10.1016/0014-5793(92)81422-i. [DOI] [PubMed] [Google Scholar]
- 31.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 32.Bownds D, Gordon-Walker A, Gaide Huguenin AC, Robinson W. J. Gen. Physiol. 1971;58:225–237. doi: 10.1085/jgp.58.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDowell JH. Methods Neurosci. 1993;15:123–130. [Google Scholar]
- 34.Pentia DC, Hosier S, Collupy RA, Valeriani BA, Cote RH. Methods Mol. Biol. 2005 doi: 10.1385/1-59259-839-0:125. in press. [DOI] [PubMed] [Google Scholar]
- 35.Norton AW, D’Amours MR, Grazio HJ, Hebert TL, Cote RH. J. Biol. Chem. 2000;275:38611–38619. doi: 10.1074/jbc.M004606200. [DOI] [PubMed] [Google Scholar]
- 36.Mou H, Cote RH. J. Biol. Chem. 2001;276:27527–27534. doi: 10.1074/jbc.M103316200. [DOI] [PubMed] [Google Scholar]
- 37.Johnson MA, Vardi N. Vis. Neurosci. 1998;15:743–753. doi: 10.1017/s0952523898154135. [DOI] [PubMed] [Google Scholar]
- 38.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 39.Li N, Baehr W. FEBS Lett. 1998;440:454–457. doi: 10.1016/s0014-5793(98)01501-4. [DOI] [PubMed] [Google Scholar]
- 40.Maduro MF, Gordon M, Jacobs R, Pilgrim DB. J. Neurogenet. 2000;13:191–212. doi: 10.3109/01677060009084494. [DOI] [PubMed] [Google Scholar]
- 41.Hurwitz RL, Bunt Milam AH, Chang ML, Beavo J. J. Biol. Chem. 1985;260:568–573. [PubMed] [Google Scholar]
- 42.Tyminski PN, O’Brien DF. Biochemistry. 1984;23:3986–3993. doi: 10.1021/bi00312a028. [DOI] [PubMed] [Google Scholar]
- 43.Clerc A, Bennett N. J. Biol. Chem. 1992;267:6620–6627. [PubMed] [Google Scholar]
- 44.Malinski JA, Wensel TG. Biochemistry. 1992;31:9502–9512. doi: 10.1021/bi00154a024. [DOI] [PubMed] [Google Scholar]
- 45.Melia TJ, Malinski JA, He F, Wensel TG. J. Biol. Chem. 2000;275:3535–3542. doi: 10.1074/jbc.275.5.3535. [DOI] [PubMed] [Google Scholar]
- 46.Rohlich P, Szel A. Curr. Eye Res. 1993;12:935–944. doi: 10.3109/02713689309020400. [DOI] [PubMed] [Google Scholar]
- 47.Rohlich P, Szel A. Microsc. Res. Tech. 2000;50:327–337. doi: 10.1002/1097-0029(20000901)50:5<327::aid-jemt2>3.3.co;2-g. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Zhou J, Daiger SP, Farber DB, Heckenlively JR, Smith JE, Sullivan LS, Zuo J, Milam AH, Pierce EA. Invest. Ophthalmol. Vis. Sci. 2002;43:22–32. [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Zuo J, Pierce EA. J. Neurosci. 2004;24:6427–6436. doi: 10.1523/JNEUROSCI.1335-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong DH, Pawlyk B, Sokolov M, Strissel KJ, Yang J, Tulloch B, Wright AF, Arshavsky VY, Li T. Invest. Ophthalmol. Vis. Sci. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 51.Sokolov M, Lyubarsky AL, Strissel KJ, Savchenko AB, Govardovskii VI, Pugh EN, Arshavsky VY. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 52.Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deretic D, Huber LA, Ransom N, Mancini M, Simons K, Papermaster DS. J. Cell Sci. 1995;108:215–224. doi: 10.1242/jcs.108.1.215. [DOI] [PubMed] [Google Scholar]
- 54.Grayson C, Bartolini F, Chapple JP, Willison KR, Bhamidipati A, Lewis SA, Luthert PJ, Hardcastle AJ, Cowan NJ, Cheetham ME. Hum. Mol. Genet. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Huang W, Zhang H, Zhu X, Craft CM, Baehr W, Chen CK. Mol. Vis. 2003;9:231–237. [PubMed] [Google Scholar]
- 56.Higashide T, McLaren MJ, Inana G. Invest. Ophthalmol. Vis. Sci. 1998;39:690–698. [PubMed] [Google Scholar]
- 57.Kobayashi A, Kubota S, Mori N, McLaren MJ, Inana G. FEBS Lett. 2003;534:26–32. doi: 10.1016/s0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]