Abstract
Pathogen recognition receptors (PRRs) for fungi include dectin-1 and mannose receptor, and these mediate phagocytosis as well as production of cytokines, reactive oxygen species, and the lipid mediator leukotriene B4 (LTB4). The influence of G protein-coupled receptor (GPCR) ligands such as LTB4 on fungal PRR expression is unknown. Here we investigated the role of LTB4 signaling in dectin-1 expression and responsiveness in macrophages. Genetic and pharmacologic approaches showed that LTB4 production and signaling through its high-affinity GPCR BLT1 direct dectin-1-dependent binding, ingestion, and cytokine production both in vitro and in vivo. Impaired responses to fungal glucans correlated with lower dectin-1 expression in macrophages from LT- and BLT1-deficent mice than their WT counterparts. LTB4 increased the expression of the transcription factor responsible for dectin-1 expression, PU.1, and PU.1 siRNA abolished LTB4-enhanced dectin-1 expression. GM-CSF controls PU.1 expression, and this cytokine was decreased in LT-deficient macrophages. Addition of GM-CSF to LT-deficient cells restored expression of dectin-1 and PU.1 as well as dectin-1 responsiveness. In addition, LTB4 effects on dectin-1, PU.1 and cytokine production were blunted in GM-CSF−/− macrophages. Our results identify LTB4-BLT1 signaling as an unrecognized controller of dectin-1 transcription via GM-CSF and PU.1 that is required for fungi protective host responses.
INTRODUCTION
Protective innate immune responses require host recognition of microbes through the engagement of pattern-recognition receptors (PRRs), including Toll-like receptors (TLRs) and C-type lectins (1). PRRs recognize highly conserved microbial motifs known as pathogen-associated molecular patterns (PAMPs), which include carbohydrates, peptidoglycans, and lipopolysaccharides (2). Dectin-1, a C-type lectin, is the major receptor on macrophages for β-1,3-glucan, a polymer of glucose present in the fungal cell wall that stimulates phagocytosis and production of inflammatory cytokines (1). This receptor is predominantly expressed on cells of the monocyte/macrophage lineage, neutrophils, dendritic cells and a minor subpopulation of splenic T cells (1), and its expression can be enhanced by the cytokines IL-4 (3), IL-13 (4), IL-23 and GM-CSF (3), and decreased by corticosteroids and LPS (3). It is not presently known whether dectin-1 expression can be regulated by signals emanating from G protein-coupled receptors (GPCRs).
Dectin-1 is now recognized to be the main non-opsonic receptor involved in fungal binding and uptake (5). Signal transduction following dectin-1 ligation depends on its cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM), the phosphorylation of which by Src kinase leads to the recruitment of spleen tyrosine kinase (Syk) in phagocytes (6). Dectin-1 engagement also activates phospholipase A2 (PLA2) with subsequent production of eicosanoid lipid mediators including cyclooxygenase-derived prostanoids and 5-lipoxygenase (5-LO)-derived leukotrienes (LTs) such as LTB4 (7, 8). The latter, acting via its high-affinity GPCR BLT1, is best known as a leukocyte chemoattractant (9). It has been implicated in a variety of inflammatory disease states, such as atherosclerosis and ischemia-reperfusion injury (9). Importantly, LTB4 is also produced at sites of infection and participates in innate immune responses in vivo and in vitro (8). For example, we and others have previously shown that LTB4 enhances ingestion of IgG-opsonized targets (10) as well as unopsonized microbes including Leishmania amazonensis (11), Streptococcus pneumoniae (12), Candida albicans (8) and Histoplasma capsulatum (13). LTB4 can promote fungal ingestion in macrophages via both mannose and dectin-1 receptors (8). The role of specific 5-LO metabolites and receptors in modulating dectin-1-mediated responses is unknown. Here we demonstrate that LTB4 synthesis and signaling via BLT1 are necessary for optimal dectin-1 expression and responsiveness in macrophages in vivo and in vitro. This form of regulation involves LTB4-BLT1 control of GM-CSF production and subsequent expression of the dectin-1 transcription factor PU.1.
MATERIAL AND METHODS
Reagents
RPMI 1640, LTB4 and LTD4, 5-LO inhibitors (AA861 and zileuton), the dectin-1 antagonist laminarin prepared from Laminaria digitata, were from Enzo Life Sciences. The mannose receptor antagonist mannan prepared from Saccharomyces cerevisiae, Actinomycin D, Polymyxin B sulfate and pertussis toxin (PTX) were purchased from Sigma. The selective dectin-1 agonist curdlan from Alcaligenes faecalis and zymosan depleted of TLR agonists (by treatment with chloroform/methanol (14) were from Invivogen. U75302 (BLT1 antagonist) was from Cayman Chemicals. C5a and CXCL1 were from R&D. Compounds requiring reconstitution were dissolved in either ethanol or dimethyl sulfoxide (DMSO). Required dilutions of all compounds were prepared immediately before use, and equivalent quantities of vehicle were added to the appropriate controls.
Animals
8-week-old female 5-LO−/− mice (15) were bred in-house and strain-matched WT sv/129 mice were purchased from The Jackson Laboratory. GM-CSF−/− mice (16) were originally a gift from J. Whitsett (Children’s Hospital, Cincinnati OH) and were bred in-house. BLT1−/− mice (17) and strain-matched WT C57BL/6 mice were obtained from The Jackson Laboratory.
Ethics statement
Mice were treated according to NIH guidelines for the use of experimental animals, with the approval of the University of Michigan Committee for the Use and Care of Animals. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering by the attending veterinarian.
Cell isolation and culture
Elicited peritoneal macrophages were harvested from the peritoneal cavities of mice by lavage with PBS 4 days after the injection of 2 ml of 3% thioglycollate, as described previously (18). Resident murine alveolar macrophages were obtained by bronchoalveolar lavage (BAL) as described (18). Cells were cultured overnight in RPMI containing 10% fetal bovine serum and antibiotics and washed twice the next day with warm medium to remove nonadherent cells.
C. albicans culture
C. albicans strain CHN1 (a human pulmonary clinical isolate) was grown on Sabouraud dextrose agar plates and maintained at 4 °C. 72 h before the experiment, yeast were grown to stationary phase at 37 °C in Sabouraud dextrose broth (Difco; 1% neopeptone, 2% dextrose) with shaking. The cultures were washed in sterile nonpyrogenic PBS, counted with a hemocytometer, and diluted to 2 × 109 colony forming units (CFU)/ml in sterile nonpyrogenic PBS. C. albicans was killed through heating for 30 min at 56 °C and FITC-labeled as described (8).
In vitro binding assay
In vitro C. albicans binding assays were performed as previously described (19). In brief, overnight cultures of macrophages were cooled to 4°C and washed three times with pre-chilled serum-containing medium. FITCC. albicans was added to the macrophages at a ratio of 10 particles/cell for 1 h on ice and cells were washed three times to remove unbound FITC-yeast and then lysed with 3% Triton X-100. FITCC. albicans in lysates was quantified using a Spectramax Gemini EM fluorometer (Molecular Devices) at settings of 485 excitation/535 emission.
In vivo injection with curdlan
Curdlan (100 µg/kg) was reconstituted in PBS with 1% BSA and administered to the lungs of mice via oropharyngeal injection as described (20). BAL was performed by 3 successive instillations of 1 ml PBS with each, followed by gentle suction. BAL fluid from WT and 5-LO−/− mice was harvested after 24 h and levels of LTB4, cytokines, and chemokines were measured by ELISA or by antibody-based cytokine array. The pelleted cells were subjected to cytospin and cell counts and differentials for evaluation of neutrophil recruitment were determined by light microscopy.
Semi-quantitative cytokine array
WT and 5-LO−/− mice underwent intrapulmonary challenge with curdlan as described above and the BAL fluid was harvested 24 h later. Protein content was quantified by Bradford assay and 50 µg of protein were used for qualitative measurement of cytokine expression using the Mouse Cytokine Antibody Array, Panel A (ARY006), as recommended by the manufacturer (R&D Systems, Wiesbaden, Germany).
Measurement of LTB4
Levels of LTB4 in the BAL fluid obtained from WT mice 24 h after intrapulmonary challenge with curdlan were determined using EIA kits (Cayman Chemical Co.) as described (8).
Measurement of cytokine and chemokine levels
Levels of IL-12p40, IL-17A, GM-CSF, M-CSF, KC, IL-1β and TNF-α were determined by ELISA (R&D Duoset; R&D Systems) by the University of Michigan Cancer Center Cellular Immunology Core.
Flow cytometry
For flow cytometric analysis, cells were resuspended in PBS containing 2 mM EDTA and 0.5% FCS. Fc receptor–mediated and nonspecific antibody binding was blocked by addition of excess CD16/CD32 (BD Biosciences Pharmingen). For staining, macrophages were incubated with anti-dectin-1 conjugated to FITC (1:200, BD Bioscience Pharmigen) at 4°C in the dark for 15 min. Samples were stabilized with 1% paraformaldehyde and analyzed on the same day. A FACSCalibur flow cytometer (BD Biosciences) was used for flow cytometric characterization of cell populations, and data were analyzed with WinMDi and FlowJo Version 7.6.4 software (TreeStar).
In vivo phagocytosis assay
WT and 5-LO−/− mice were subjected to intrapulmonary administration of 1 µg/ml zymosan as described above for curdlan, and 24 h later cells were harvested by BAL and subjected to cytospin and stained with Diff-Quick. The number of intracellular zymosan particles was determined microscopically. The phagocytic index was generated by counting the number of macrophages containing intracellular zymosan multiplied by the number of intracellular zymosan particles.
RNA isolation and semiquantitative real-time RT-PCR
RNA from cultured cells was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, and real-time RT-PCR was performed as previously described (18). Dectin-1 (Clec7A), dectin-2 (Clecsf10), GM-CSF (Gmcsf) or PU.1 (Spi1) mRNAs were normalized to β-actin or GAPDH, and the respective WT control was set at 100%. To determine the decay of Clec7A mRNA, WT and 5-LO−/− macrophages were treated with or without 2.5 µg/ml actinomycin D (Sigma-Aldrich), and the amount of mRNA was determined after harvesting at different time points. Clec7A mRNA was normalized to β-actin, and the respective WT control was set to 100%. Percentages were plotted against time, and decay curves were calculated.
Western blotting
2 × 106 macrophages were plated in 6-well tissue culture dishes and were incubated in the presence or absence of 100 nM LTB4 for 24 h and then lysed in buffer (50 mM Tris-HCl [pH 7.4], 25 mM KCl, 5 mM MgCl2, and 0.2% Nonidet P-40) supplemented with protease inhibitors (Roche Diagnostics). For immunoblot analysis, protein samples (30 µg) were mixed with loading buffer (50 mM Tris HCl (pH 6.8), 2% SDS, 100 mM DTT, 10% glycerol, and 0.1% bromphenol blue), boiled, applied to 10% SDS-polyacrylamide gels, and subjected to electrophoresis. Immunoblot analysis was performed as previously described (21), using primary antibodies against dectin-1 (1:1000, Biovision), Dectin-2 (1:500, Abcam), PU.1 and Sp.1 (both at 1:1000, Abcam) and GAPDH (1:10,000, Sigma). Densitometric analysis was as described (22); the intensity of of the protein band was divided by that of the GAPDH, and this ratio was then expressed relative to that of the untreated control, which was set at 100%. In all instances, density values of bands were corrected by subtraction of the background values.
RNA interference
RNA interference was performed according to a protocol provided by Dharmacon and as we have previously reported (18). WT and 5-LO−/− macrophages were transfected using DharmaFECT 1 reagent with 30 nM of nonspecific control or specific ON-TARGET SMARTpool Pu.1 siRNAs. After 48 h of transfection, macrophages were incubated with or without 100 nM LTB4 for 24 h and the cells harvested for mRNA or protein analysis.
Statistics
All experiments were performed at least 3 times unless otherwise specified, and data are presented as the mean ± SE of the values from all experiments. Within each experiment, triplicate values were used for each condition. Comparisons among three or more experimental groups were performed with ANOVA followed by the Bonferroni analysis. Differences were considered significant if p ≤ 0.05.
RESULTS
LTB4 regulates macrophage expression of dectin-1
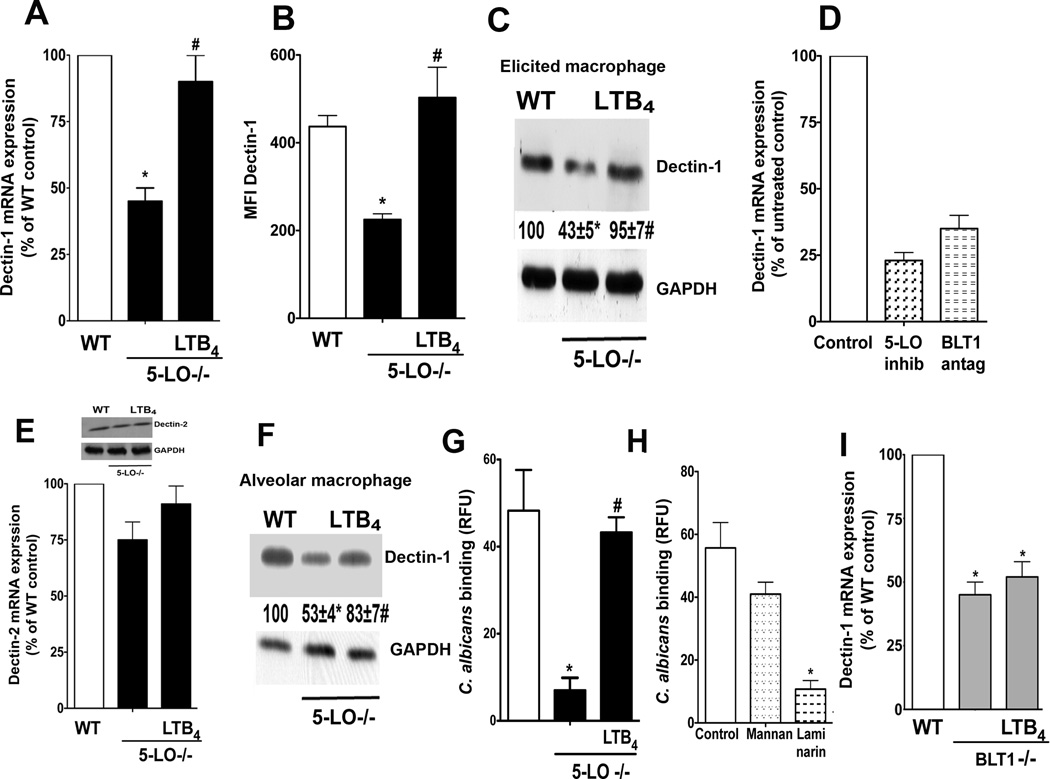
LTB4 can enhance PRR-mediated responses by regulating expression of MyD88 (18), and can also enhance expression of certain macrophage receptors, such as CD11b and CD11c (23). However, it is not known whether LTB4 can influence the expression of PRRs, including dectin-1. This possibility was first examined in elicited peritoneal macrophages from WT and 5-LO−/− mice, as this cell population is known to express high levels of dectin-1 (3). LT-deficient macrophages exhibited reduced baseline expression of dectin-1 mRNA, as determined by real-time RT-PCR (Fig. 1 A), and protein, as determined by both FACS (Fig. 1 B) and immunoblotting (Fig. 1 C). Reduced dectin-1 mRNA was confirmed in WT cells treated for 24 h with a 5-LO inhibitor (Fig. 1 D). 24 h treatment with 100 ng/ml LTB4 fully rescued dectin-1 mRNA (Fig. 1 A) and protein (Fig. 1 B and C) expression in 5-LO−/− macrophages back to the levels observed in WT cells. By contrast to its effects on dectin-1, neither endogenously produced nor exogenously added LTB4 modulated expression of dectin-2 mRNA or protein (Fig. 1 E and inset). Reduced expression of dectin-1 was also observed in alveolar macrophages (Fig. 1 F) and bone marrow-derived macrophages (not shown) from LT-deficient mice, and again, levels were significantly increased with 24 h treatment with LTB4. We next determined if the decreased dectin-1 expression correlated with lower macrophage binding of C. albicans. Alveolar macrophages from 5-LO−/− mice bound substantially less C. albicans than did WT macrophages, but LTB4 treatment for 24 h restored yeast binding to levels exhibited by WT cells (Fig. 1 G). The importance of dectin-1 in mediating yeast binding was confirmed by showing that treatment of WT cells with the dectin-1 receptor antagonist laminarin, but not the mannose receptor antagonist mannan, decreased C. albicans binding to levels approximating that observed in 5-LO−/− cells (Fig. 1 H). The specific role of BLT1 in controlling dectin-1 expression was confirmed by demonstrating reduced baseline dectin-1 mRNA in elicited macrophages from BLT1−/− mice (Fig. 1 I) and from WT mice treated overnight with a BLT1 antagonist. However, due to lack of BLT1, LTB4 was unable to restore deficient dectin-1 expression (Fig. 1 I), in contrast to 5-LO−/− cells. Together, these results show that LTB4 enhances basal expression of dectin-1 mRNA and protein in various macrophage populations.
Fig. 1. LTB4 is necessary for basal dectin-1 expression in macrophages.
(A) WT and 5-LO−/− macrophages were treated ± 100 nM LTB4 for 24 h and dectin-1 mRNA was determined by real-time RT-PCR. (B) WT and 5-LO−/− elicited peritoneal macrophages were probed with anti-dectin-1 antibody and the cells subjected to FACS analysis as described in Materials and Methods. Mean fluorescence intensity (MFI) is expressed as the mean ± SEM from 3 individual experiments. (C) Elicited macrophages were incubated ± LTB4 for 24 h and the expression of dectin-1 and GAPDH determined by immunoblot analysis. Numbers under lanes indicate the relative density of dectin-1, determined from densitometric analysis and expressed as the mean ± SEM from 3 individual experiments, with the values of the WT control group set as 100%. (D) WT macrophages were pretreated with the 5-LO inhibitor AA-861 (10 µM) or the BLT1 antagonist U7532 (1 µM) for 24 h and dectin-1 mRNA levels were determined by real-time RT-PCR. (E) WT and 5-LO−/− macrophages were incubated ± LTB4 for 24 h and the expression of dectin-2 was determined by real-time RT- PCR. Inset: Dectin-2 protein abundance in WT and 5-LO−/− macrophages determined by immunoblotting. (F) Alveolar macrophages were incubated ± LTB4 for 24 h and the expression of dectin-1 and GAPDH determined by immunoblot analysis. Data are expressed and analyzed as in (C). G) Alveolar macrophages from WT and 5-LO−/− mice were incubated ± 100 nM LTB4 for 24 h and the binding capacity for 10:1 heat-killed FITCC. albicans was determined as described in the Materials and Methods. (H) Alveolar macrophages from WT mice were pretreated with mannan or laminarin (both at 100 µg/ml) for 30 min before the addition of 10:1 heat-killed FITCC. albicans and yeast binding capacity was determined as described in (G). (I) WT and BLT1−/− elicited peritoneal macrophages were incubated ± LTB4 for 24 h and dectin-1 mRNA was determined by real-time RT-PCR. In all circumstances, data represent the mean ± SEM from 3 individual experiments, each performed in triplicate. *p < 0.05 versus WT control or untreated control; # p < 0.001 versus untreated 5-LO−/− macrophages by ANOVA.
LTB4 regulates macrophage responses via dectin-1 in vitro and in vivo
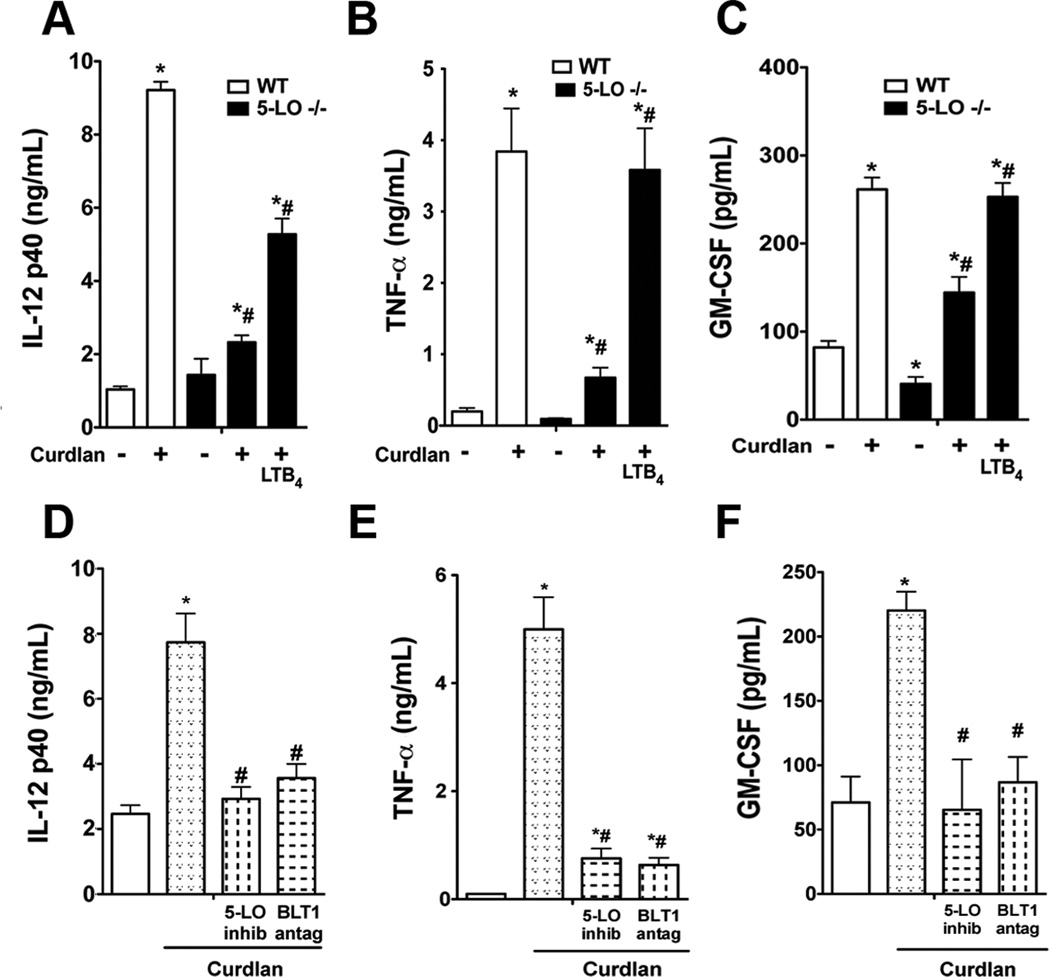
Since LTB4 and BLT1 signaling control dectin-1 expression, we reasoned that LTB4 would also control host responses to dectin-1 engagement. This was tested both in vitro and in vivo. In the in vitro experiments, we employed the dectin-1-selective agonist curdlan or zymosan which is able to ligate dectin-1 but whose TLR ligands were removed by treatment with choloform/methanol (14). Initially, we performed dose-response experiments in which elicited peritoneal macrophages from WT and 5-LO−/− mice were stimulated with concentrations of curdlan or treated zymosan. TNF-α production increased in dose-dependent fashion, and peaked at a dose of 100 µg/mL for both agonists. In all circumstances, 5-LO−/− cells exhibited lower responsiveness to both agonists than WT macrophages (Suppl. Fig. 1 A and B). To determine if LTB4 is the 5-LO product involved in dectin-1 responsiveness, 5-LO−/− cells were pretreated in the presence or absence of LTB4 for 24 h and stimulated with curdlan for another 24 h. Curdlan stimulation induced IL-12p40, TNF-α and GM-CSF in WT macrophages, as expected, but the response to curdlan in 5-LO −/− cells was significantly reduced (Fig. 2 A–C). Pharmacologic inhibition of 5-LO by 24 h pretreatment with AA-861 likewise resulted in attenuation of curdlan-induced cytokine generation in WT macrophages (Fig. 2 D–F). The specific role of endogenous LTB4 in regulating responses to curdlan was evidenced by the facts that overnight LTB4 pretreatment restored the ability of curdlan to induce IL-12p40, GM-CSF and TNF-α levels in 5-LO −/− macrophages (Fig. 2 A–C), and that overnight pretreatment with the selective BLT1 antagonist U7532 prevented curdlan-induced cytokine generation to the same extent as did pharmacologic inhibition of 5-LO (Fig. 2 D–F). Since curdlan preparations could be contaminated with endotoxins, we pretreated WT macrophages with the LPS inhibitor polymyxin B sulfate (10 µg/ml) prior to curdlan stimulation. Polymyxin B did not alter curdlan-induced TNF-α production (data not shown), which excludes a possible role for contaminating endotoxin in our curdlan preparations. Also, the primary role of dectin-1 in mediating curdlan effects was demonstrated by showing that the dectin-1 selective antagonist laminarin, which blocks dectin-1, but not complement receptor 3 and mannose receptor (19, 24), impaired curdlan-induced TNF-α secretion by ~70% (Suppl. Fig. 2).
Fig. 2. LTB4 is necessary for dectin-1 responses in macrophages.
(A–C) WT and 5-LO−/− macrophages were pretreated ± LTB4 for 24 h, then incubated with the dectin-1 selective agonist curdlan (100 µg/ml) for another 24 h prior to determination of IL-12 p40 (A), TNF-α (B), and GM-CSF (C) levels by ELISA. (D–F) WT macrophages were incubated with the 5-LO inhibitor AA-861 (10 µM) or the BLT1 antagonist U7532 (1 µM) for 24 h, followed by curdlan stimulation for 24 h and IL-12 p40 (D), TNF-α (E), and GM-CSF (F) levels determined by ELISA. Data represent the mean ± SEM from 3 individual experiments, each performed in triplicate. *p < 0.05 versus WT control or untreated control; #p < 0.01 versus untreated 5-LO−/− macrophages or untreated WT and stimulated with curdlan by ANOVA.
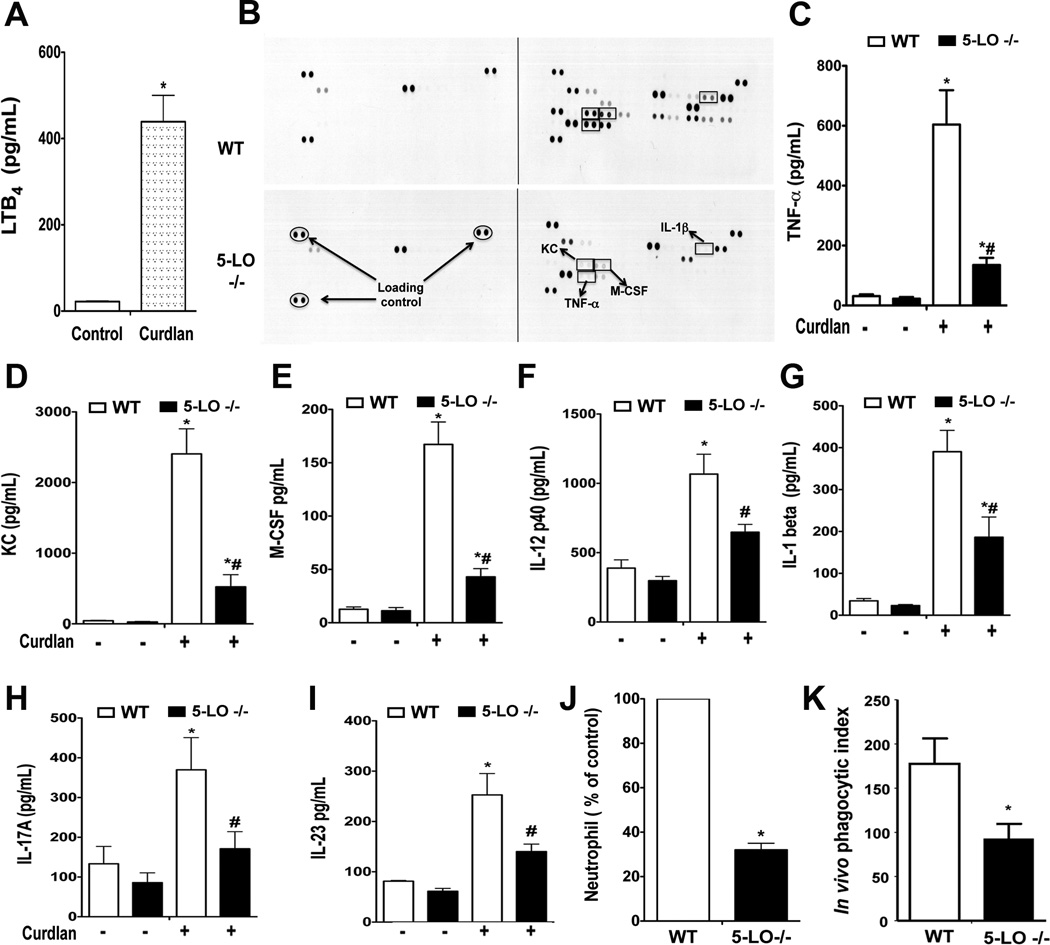
The dectin-1-dependent production of proinflammatory mediators in the lung in vivo was determined by oropharyngeal injection of curdlan in WT and 5-LO−/− mice. Twenty-four h after curdlan injection, high levels of LTB4 were measured in BAL fluid of WT mice (Fig. 3 A), verifying that generation of this lipid mediator is a component of the host response to dectin-1 ligation. Next we determined the pattern of cytokine/chemokine secretion in the BAL fluid of WT and 5-LO−/− mice using an antibody-based array. 5-LO−/− mice were globally less responsive to curdlan than were WT mice (Fig. 3 B). This finding was confirmed by ELISA determination of individual mediators in BAL fluid. Levels of TNF-α, KC, and M-CSF were decreased by at least 70% in fluid from 5-LO−/− mice, whereas levels of IL-12p40, IL-1β, IL-17A, and IL-23 were decreased by ~ 30–50% (Fig. 3 C–I). The recruitment of neutrophils to the lung of 5-LO−/− mice in response to curdlan challenge was also lower than in WT animals (Fig. 3 J). Following intrapulmonary challenge with zymosan, LT-deficient mice also manifested significantly lower in vivo ingestion of the yeast particles by macrophages (Fig. 3 K), but not by neutrophils (data not shown). These findings indicate that LTB4 produced in response to dectin-1 engagement amplifies macrophage phagocytosis, cytokine secretion, and neutrophil recruitment.
Fig. 3. LT biosynthesis is required for optimal pulmonary anti-fungal responses.
(A) WT mice were oropharyngeally administered 100 µg/ml curdlan, and 24 h later BAL fluid was harvested and LTB4 was measured by EIA. (B) BAL fluid was harvested from WT and 5-LO−/− mice 24 h after intrapulmonary administration of curdlan and subjected to cytokine/chemokine antibody array as described in Materials and Methods. Adjacent dots represent cytokine/chemokine expression from two independent experiments. Equal protein loading is demonstrated by loading controls, as indicated by the oval labels. Identities of mediators highlighted by the rectangles are as labeled in the bottom panel. Experiment representative of two independent experiments. (C–I) BAL fluid from WT and 5-LO−/− mice challenged with curdlan as in (B) was harvested and levels of TNF-α (C), KC (D), M-CSF (E), IL-12p40 (F), IL-1β (G), IL-17A (H), and IL-23 (I) were determined by ELISA. (J) BAL fluid from curdlan-challenged WT and 5-LO−/− mice was subjected to cytospin, stained with Diff-Quick and the neutrophil counts were determined microscopically; values are expressed as a percentage of the neutrophil count found in WT mice, which was 60 ± 4/mouse when at least two hundreds cells were counted. (K) WT and 5-LO−/− mice were challenged with zymosan (10 µg/ml) via the oropharyngeal route and 24 h later BAL cells were subjected to cytospin and the number of intracellular zymosan particles determined microscopically from cells stained with Diff-Quick; phagocytic index was determined as described in Materials and Methods. Data represent the means ± SEM from at least three independent experiments with 6 mice per genotype. *p < 0.05 versus WT control; #p < 0.01 versus untreated 5-LO−/− macrophages by ANOVA.
LTB4 enhances dectin-1 expression by a transcriptional mechanism involving PU.1
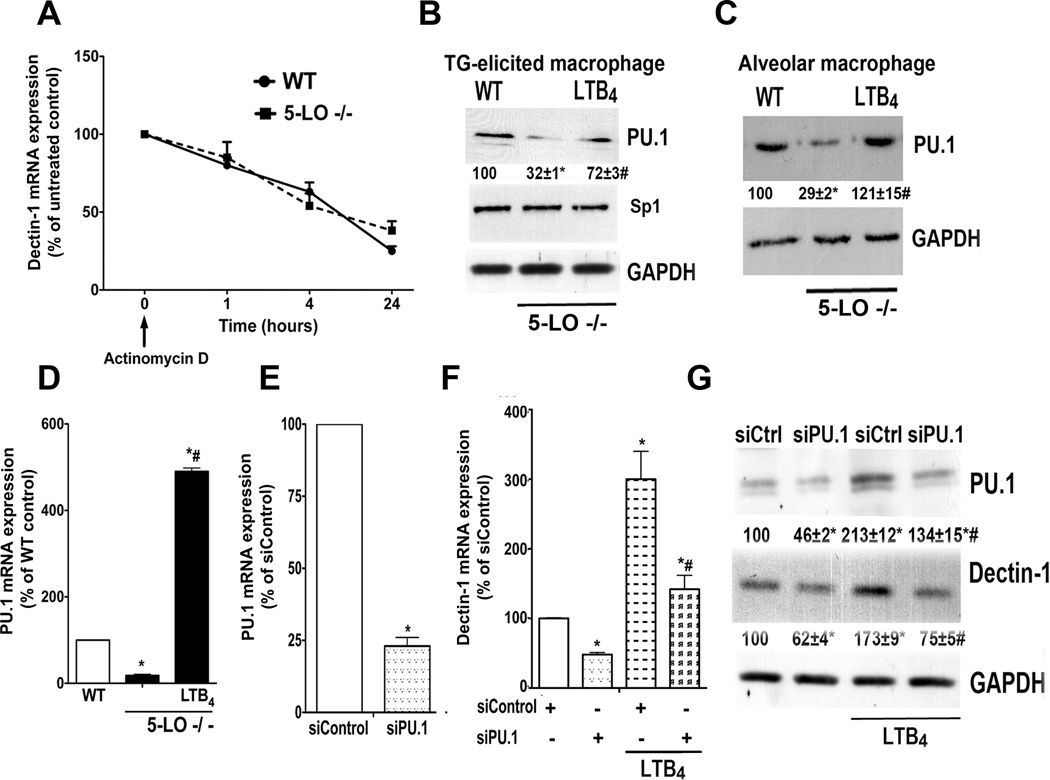
Since LTB4 can modulate the mRNA turnover rate of SOCS-1 in macrophages (18), we considered the possibility that it may increase dectin-1 mRNA expression by enhancing message stability. This was examined by comparing its decay in WT and 5-LO−/− macrophages. At various time points following addition of actinomycin D to block the formation of new transcripts, cells were processed for real-time RT-PCR analysis. No difference in mRNA stability was observed between 5-LO−/− and WT macrophages (Fig. 4 A), which suggests instead that reduced dectin-1 mRNA in 5-LO−/− cells reflects a transcriptional defect.
Fig. 4. PU.1 mediates LTB4-enhanced dectin-1 expression in macrophages.
(A) Dectin-1 mRNA decay in WT and 5-LO−/− macrophages harvested after treatment with actinomycin D (2.5 µg/ml). Data are from 3 experiments in triplicate; values are relative to untreated macrophages from both genotypes. (B) WT and 5-LO−/− elicited peritoneal macrophages or (C) resident alveolar macrophages were treated ± LTB4 for 24 h and the expression of PU.1, Sp1 and GAPDH were determined by immunoblotting. Numbers under lanes indicate relative density of PU.1 from three independent experiments. (D) PU.1 mRNA expression was determined by real-time RT-PCR in WT and 5-LO−/− elicited macrophages incubated ± LTB4 for 24 h. (E) WT macrophages were treated for 48 h in the presence of scrambled siRNA (siControl) or PU.1 siRNA, and the expression of PU.1 mRNA was determined by real-time RT-PCR and expressed relative to values in siControl cells. (F) Dectin-1 mRNA and (G) protein expression in WT macrophages treated with PU.1 and control siRNA, as determined by real-time RT-PCR and immunoblotting, respectively; mRNA levels are expressed relative to those in siControl-treated WT cells. Immunoblot is representative of 3 independent experiments. Numbers under lanes indicate relative density of dectin-1 or PU.1 from three independent experiments. Data represent the mean ± SEM from 3 individual experiments, each performed in triplicate. *p < 0.05 versus WT or WT siControl; #p < 0.05 versus 5-LO−/− control or LTB4-stimulated siControl macrophages by ANOVA
Transcription factors for dectin-1 include PU.1 (25), Sp1 (25) and PPAR-γ (4). We examined the effects of LT deficiency and exogenous LTB4 on levels of these transcription factors. Elicited peritoneal (Fig. 4 B) and resident alveolar (Fig. 4 C) macrophages from 5-LO−/− mice both exhibited less PU.1 than did cells from WT mice. Overnight treatment of 5-LO−/− cells with LTB4 largely restored PU.1 protein expression to WT levels in both populations of macrophages (Fig. 4 B and C) and drove PU.1 mRNA in elicited peritoneal macrophages to levels that far exceeded WT (Fig. 4 D). By contrast, neither Sp1 protein (Fig. 4 B) nor mRNA (data not shown) levels were reduced compared to WT cells. Likewise, no reduction in PPAR-γ expression was observed in 5-LO−/− macrophages (data not shown). To investigate the importance of PU.1 for BLT1-mediated dectin-1 expression, we utilized siRNA to knock down this transcription factor in elicited WT macrophages. We achieved approximately 75% knockdown of PU.1 mRNA (Fig. 4 E) and ~ 55% knockdown of protein (Fig. 4 G), when compared to control siRNA. PU.1 silencing decreased dectin-1 mRNA (Fig. 4 F) and protein (Fig. 4 G) expression by approximately 55%. In addition, PU.1 siRNA abolished LTB4 enhancement of dectin-1 expression (Fig. 4 F–G). These data show that LTB4 enhancement of dectin-1 involves upregulated expression of its transcription factor, PU.1.
BLT1/Gαi plays a non-redundant role in enhancing dectin-1 and PU.1 expression
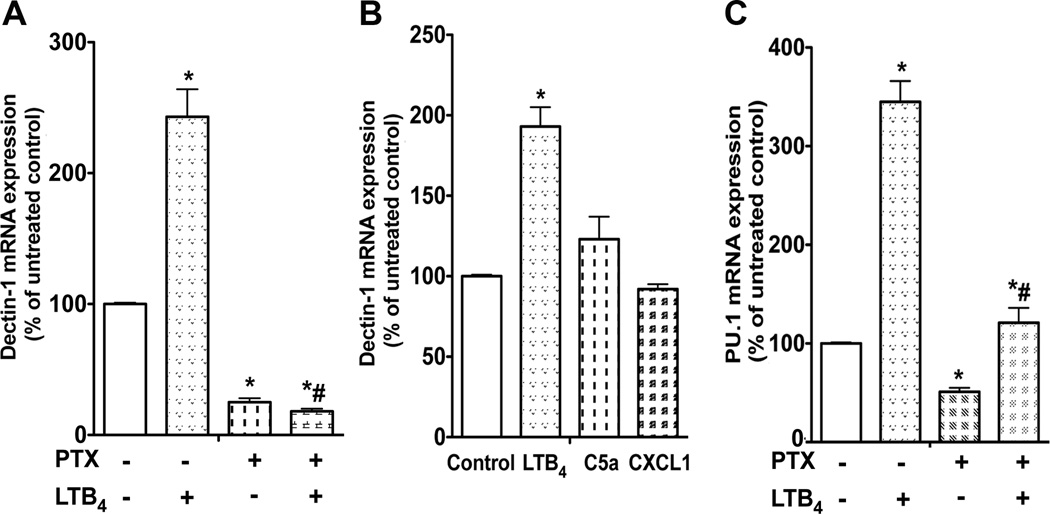
Although BLT1 can couple to both Gαi and Gαq in macrophages (26), numerous activation responses in macrophages preferentially involve Gαi signaling (18). To determine the importance of Gαi in LTB4/BLT1 control of dectin-1 expression, we tested the ability of pretreatment with the Gαi inhibitor pertussis toxin (PTX) to interfere with basal and LTB4-enhanced dectin-1 mRNA. PTX treatment for 24 h decreased basal dectin-1 expression and also prevented the enhancement in dectin-1 expression elicited by LTB4 (Fig. 5 A), suggesting that constitutive Gαi signaling is required for dectin-1 expression and that LTB4/BLT1 signaling requires Gαi. Since other Gαi-coupled receptors besides BLT1 are expressed and promote activation responses in macrophages, we tested whether other selected ligands could also enhance dectin-1 expression. Neither C5a nor CXCL1 was capable of increasing dectin-1 mRNA expression (Fig. 5 B), suggesting a non-redundant role for BLT1/Gαi signaling in controlling the expression of this PRR. The importance of Gαi signaling in controlling PU.1 expression was also studied. PTX treatment of elicited macrophages revealed that Gαi signaling is necessary for baseline PU.1 expression and for LTB4/BLT1 enhancement of PU.1 expression (Fig. 5 C), as it was for dectin-1 expression (Fig. 5 A).
Fig. 5. Gαi signaling is required for LTB4/BLT1-induced dectin-1 and PU.1 expression in macrophages.
(A) WT elicited macrophages were pretreated with PTX (600 ng/ml) for 24 h and incubated with or without LTB4 for another 24 h, after which RNA was isolated for dectin-1 mRNA determination by real-time RT-PCR. (B) WT macrophages were treated for 24 h with LTB4 (100 nM), C5a (50 ng/ml) or CXCL1 (20 ng/ml) and dectin-1 mRNA was determined by real-time RT-PCR. (C) WT macrophages were pretreated with PTX for 24 h and incubated with or without LTB4 for another 24 h, after which RNA was isolated for PU.1 mRNA determination by real-time RT-PCR. Data represent the mean ± SEM from 3 individual experiments, each performed in triplicate, and are expressed relative to untreated macrophages. *p < 0.05 versus WT control or untreated control; # p < 0.001 versus macrophages incubated with LTB4 only by ANOVA.
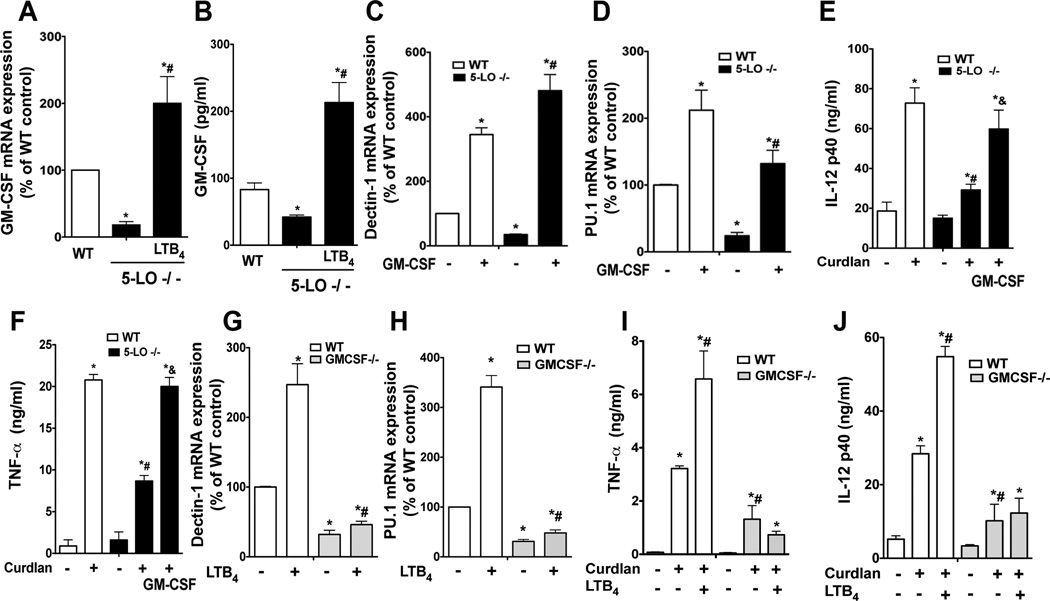
GM-CSF is a critical mediator of LTB4-enhanced PU.1 and dectin-1 expression
GM-CSF upregulates PU.1 (27) and dectin-1 (3) expression in macrophages. However, it is unknown if endogenously produced GM-CSF is also required for dectin-1 expression. It is also unknown if GM-CSF participates in the LTB4 amplification of dectin-1 and PU.1. To evaluate this possibility, we initially determined the levels of GM-CSF in 5-LO−/− and WT elicited peritoneal macrophage cultures. Expression of both mRNA (Fig. 6 A) and protein (Fig. 6 B) was decreased in LT-deficient cells when compared to WT macrophages. Overnight treatment of 5-LO−/− cells with LTB4 restored GM-CSF mRNA and protein levels beyond the WT range (Fig. 6 A and B). To determine whether the lower GM-CSF expression in 5-LO−/− cells was responsible for their decreased PU.1 and dectin-1 expression, we pretreated LT-deficient cells with GM-CSF for 24 h and determined dectin-1 and PU.1 mRNA expression. GM-CSF treatment (10 ng/ml) significantly augmented baseline expression of both dectin-1 (Fig. 6 C) and PU.1 (Fig. 6 D) in WT cells and also overcame their deficient expression in 5-LO−/− macrophages. As expected, the restored dectin-1 expression achieved by GM-CSF treatment also rescued responsiveness to curdlan in LT-deficient cells, as shown by the production of IL-12p40 (Fig. 6 E) and TNF-α (Fig. 6 F).
Fig. 6. GM-CSF mediates the enhancement by LTB4 of PU.1 and dectin-1 in macrophages.
(A) GM-CSF mRNA expression was determined by real-time RT-PCR in WT and 5-LO−/− macrophages incubated ± LTB4 for 24 h. (B) GM-CSF protein was determined by ELISA in WT and 5-LO−/− macrophages incubated ± LTB4 for 24 h. (C and D) WT and 5-LO−/− macrophages were treated with GM-CSF (10 ng/ml) for 24 h and the expression of dectin-1 (C) or PU.1 (D) mRNA was determined. (E and F) WT and 5-LO−/− macrophages were pretreated with GM-CSF for 24 h and then stimulated with curdlan (100 µg/ml) for another 24 h, after which the supernatant levels of IL-12p40 (E) or TNF-α (F) were determined by ELISA. (G and H) WT and GM-CSF−/− macrophages were treated ± LTB4 for 24 h and the expression of dectin-1 (G) and PU.1 (H) mRNA determined by real-time RT-PCR. (I and J) WT and GM-CSF−/− macrophages were pretreated ± LTB4 for 24 hours followed by curdlan for another 24 h and the supernatant harvested to determine levels of TNF-α (I) and IL-12p40 (J) by ELISA. Data represent the mean ± SEM from 3 individual experiments, each performed in triplicate. *p < 0.05 versus WT Control; #p < 0.05 versus 5-LO−/− alone or LTB4-stimulated macrophages; & p < 0.05 versus 5-LO−/− macrophages incubated with curdlan only by ANOVA.
To determine if endogenous GM-CSF is required for PU.1 and dectin-1 expression, we measured the expression of these mRNAs in elicited macrophages from GM-CSF-deficient and WT mice by real-time RT-PCR. As expected (28) PU.1 (Fig. 6 G) expression was lower in GM-CSF−/− than WT macrophages. Accordingly, dectin-1 (Fig. 6 H) expression was also markedly lower in GM-CSF−/− than WT cells. Importantly, overnight treatment with LTB4 was unable to enhance either PU.1 (Fig. 6 G) or dectin-1 (Fig. 6 H) expression in GM-CSF deficient macrophages, as it was in WT cells. As predicted on the basis of decreased dectin-1 expression, GM-CSF−/− macrophages exhibited lower cytokine generation in response to curdlan than did WT macrophages (Fig. 6 I and J). Moreover, overnight LTB4 treatment was unable to potentiate curdlan-induced cytokine production in GM-CSF-deficient cells as it was in WT cells (Fig. 6 I and J). These results indicate that LTB4/BLT1 regulation of dectin-1 expression and responsiveness in macrophages depends on an autocrine loop involving GM-CSF potentiation of PU.1.
DISCUSSION
We provide evidence here that the GPCR BLT1 is a central determinant of dectin-1 expression and of host responses to fungi recognized by this PRR, and perhaps others that recognize the β-glucan moiety. Our findings also reveal a previously unrecognized interplay between the cytokine GM-CSF and LTB4 that mediates this effect. More specifically, we have shown that: (1) homeostatic LTB4 production is required for dectin-1 responsiveness in vivo and in vitro; (2) LTB4/BLT1/Gαi signaling is necessary for basal dectin-1 expression; (3) LTB4 enhances the expression of the transcription factor PU.1, which in turn controls dectin-1 expression; and (4) GM-CSF is a key mediator of LTB4-induced PU.1 and dectin-1 expression in macrophages. A scheme illustrating the relevant events is depicted in Fig. 7. Since LTB4 production is a component of the host response to fungal infections (8) and since dectin-1 is a major recognition receptor for numerous fungi, including species of Candida, Aspergillus, Histoplasma, Cryptococcus, Coccidioides, and Pneumocystis carinii (29), our findings suggest a potentially broad role for LTB4 in anti-fungal defense. Indeed, a protective role for endogenous LTB4 has been described in a murine model of pulmonary histoplasmosis (30). Moreover, dectin-1 may also participate in recognition of other microbes, including mycobacteria (31), and it is interesting to note that potential roles for LTB4 have been identified in defense against mycobacterial infections in both humans (32) and mouse models (33).
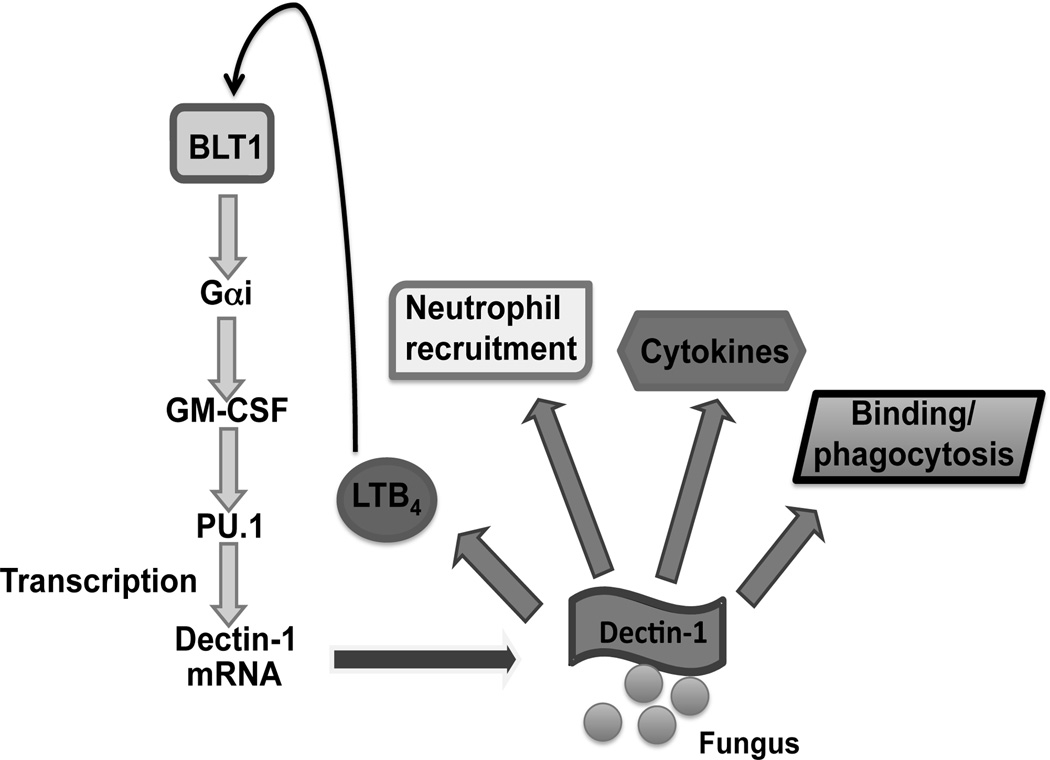
Fig. 7. Proposed model of LTB4/BLT1 regulation of GM-CSF and PU.1 expression and enhancement of dectin-1 expression and responsiveness.
Basal or dectin-1-activated LTB4/BLT1 signaling elicits Gαi activation that results in enhanced expression of GM-CSF. This cytokine potentiates expression of PU.1, which carries out transcription of dectin-1, augmenting dectin-1 protein expression and responsiveness, as evidenced by binding, phagocytosis, cytokine secretion, and neutrophil recruitment in response to challenge with C. albicans, zymosan, or the fungal glucan curdlan. LTB4 generation in response to dectin-1 ligation represents an autocrine self-amplifying loop.
We employed both genetic and pharmacologic approaches to interrupt either LTB4 synthesis or signaling via its GPCR, BLT1. Together, these establish that basal elaboration of LTB4 and ligation of BLT1 was necessary for optimal dectin-1-dependent responses, including in vitro macrophage binding of C. albicans and cytokine production in response to curdlan, as well as in vivo phagocytosis, cytokine generation, and neutrophil recruitment. Indeed, the concentration of LTB4 elaborated constitutively by elicited peritoneal macrophages (100 pg/ml, equivalent to 0.5 nM) (18) substantially exceeds that necessary to amplify macrophage antimicrobial functions via BLT1 (0.01 nM) (26).
In addition to modulating the expression of dectin-1, LTB4 could also potentiate the signals derived from this and other fungal PRRs. Dectin-1 signaling is mediated mainly by Syk, which is responsible for optimal TNF-α secretion, and Raf/MAPK, which is responsible for IL-12 production (6). The fact that TNF-α production in the lungs of 5-LO−/− mice was more impaired relative to WT mice than was IL-12p40 production suggests that LTB4 may exert an additional potentiating effect directed at activation of Syk. Such a potentiating effect by LTB4/BLT1 on Syk activation has been previously noted in the context of macrophage phagocytosis via the Fcγ receptor (21), which, like dectin-1, also signals via an ITAM-Syk mechanism (6). In any case, our findings demonstrate that LTB4 is essential in order for fungal engagement of dectin-1 to elicit Th1- and Th17-type responses that participate in anti-fungal immunity (34). These effects on dectin-1 are not the only mechanism by which LTB4 can potentiate PRR pathways. We have recently reported that by enhancing expression of the adaptor protein MyD88, BLT1 signaling increases MyD88-dependent NFkB activation that is an integral component of the host responses to various TLR and cytokine receptors (18). This effect on MyD88 expression would be expected to have broad implications for enhancing innate immunity, and it is possible that other PRRs or their downstream partners might also be targets for modulation by LTB4. Although LTB4 has also been reported to enhance the expression of certain leukocyte cell surface receptors, including CD11b and CD11c in human monocytes and IL-2 receptor beta in human lymphocytes (23), we are not aware of any previous reports indicating its capacity to specifically regulate expression of a PRR. Among the transcription factors that controls dectin-1 expression, only PU.1 expression was downregulated in 5-LO−/− macrophages, and LTB4 was capable of enhancing its expression. PU.1 is an ets-family transcription factor that regulates myeloid lineage development (35). PU.1 gene disruption abolishes macrophage and B lymphocyte production and delays neutrophil and T lymphocyte production (36). PU.1 also participates in the transcriptional control of various genes involved in macrophage activation, such as TLR4 (37), CD14 (38), mannose receptor (39), CLEC5A (40) and FcRI-III (41). Our finding that LTB4 controls PU.1 expression represents a means by which this lipid mediator might similarly promote the transcription of other PRRs and functionally related receptors. This will be the subject of future studies.
Gαi signaling and GM-CSF production elicited by LTB4/BLT1 were critical for its ability to enhance PU.1 expression and subsequent dectin-1 expression. Since a variety of Gαi-coupled receptors are present in macrophages, one could speculate that other Gαi-coupled ligands should exert similar effects as LTB4 on dectin-1 and PU.1 expression. Surprisingly, neither C5a nor CXCL1 enhanced dectin-1 expression, which supports the findings from BLT1−/− cells – in which expression of other Gαi-coupled receptors are intact – that LTB4/BLT1/Gαi signaling controls dectin-1 transcription in a non-redundant manner. The reasons for this non-redundant role are unknown, but could reflect unique signaling programs or efficiency of BLT1, or specific spatially-defined molecular interactions with dectin-1.
In addition to its ability to induce PU.1 expression, LTB4 could also potentiate its transcriptional activation. For instance, PU.1 activation is known to be controlled by PKC-δ-mediated phosphorylation (42), and LTB4 activates PKC-δ in macrophages to enhance phagocytosis (43). The possible role of PKC-δ in this axis remains to be clarified.
The regulation of dectin-1 expression is not extensively studied, but it is known that GM-CSF (3) is among the cytokines that can enhance its expression. GM-CSF exhibits a wide range of effects in macrophages, promoting maturation (44), differentiation (44), cytokine secretion (45), and phagocytosis of opsonized (41) and nonopsonized targets (46). Generation of the GM-CSF-deficient mouse was instrumental in elucidating the role of this cytokine in host defense (44). These mice exhibit reduced pulmonary clearance of various microbial pathogens, including group B Streptococcus (47), Pneumocystis carinii (48), Mycobacterium tuberculosis (49), Leishmania major (50) and Cryptococcus neoformans (51). Since both GM-CSF and LTB4 play pivotal roles in host defense, it is possible that cross-talk between these two molecules contributes to their capacities to enhance macrophage function. Indeed, GM-CSF protein and mRNA levels were lower in 5-LO−/− macrophages than in WT cells and LTB4 challenge enhanced GM-CSF production. The molecular mechanisms by which LTB4/BLT1 controls GM-CSF mRNA expression await future investigation. However, that lower GM-CSF production is indeed responsible for lower dectin-1 and PU.1 expression was evidenced by the fact that addition of this cytokine to LT-deficient cells restored dectin-1 and PU.1 production as well as curdlan responsiveness. Although we have previously reported that GM-CSF enhances macrophage LT generation (52), its ability to enhance dectin-1 expression in 5-LO−/− macrophages indicate that this effect is independent of LTB4 synthesis.
Our findings reveal a novel form of regulation in which BLT1 – a GPCR ligated at sites of infection – modulates transcription of the important fungal PRR dectin-1 via a GM-CSF/PU.1 cascade. Cross-talk between BLT1 signaling and PRRs would be anticipated to participate in shaping nascent innate immune responses to infections. However, this network is likely disabled in states of immunosuppression characterized by deficient LTB4 synthesis, such as malnutrition (9), infection with human immunodeficiency virus (9), cigarrete smoking (9), and bone marrow transplantation (9). These data provide important insights and new opportunities to modulate innate immune and inflammatory responses to pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Peters-Golden laboratory and Ana Paula Moreira for their thoughtful input.
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and National Institutes of Health HL-058897 (MP-G) and HL-103777-01 (CHS).
Abbreviations
- 5-LO
5-lipoxygenase
- AM
alveolar macrophage
- PTX
pertussis toxin
- BLT1
leukotriene B4 receptor 1
- cysLT1
cysteinyl leukotriene receptor 1
Footnotes
The authors have no financial conflict of interest.
REFERENCES
- 1.Reid DM, Gow NA, Brown GD. Pattern recognition: recent insights from Dectin-1. Curr Opin Immunol. 2009;21:30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. International reviews of immunology. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 3.Willment JA, Lin HH, Reid DM, Taylor PR, Williams DL, Wong SY, Gordon S, Brown GD. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J Immunol. 2003;171:4569–4573. doi: 10.4049/jimmunol.171.9.4569. [DOI] [PubMed] [Google Scholar]
- 4.Gales A, Conduche A, Bernad J, Lefevre L, Olagnier D, Beraud M, Martin-Blondel G, Linas MD, Auwerx J, Coste A, Pipy B. PPARgamma controls dectin-1 expression required for host antifungal defense against Candida albicans. PLoS Pathog. 2010;6:e1000714. doi: 10.1371/journal.ppat.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Balestrieri B, Hsu VW, Gilbert H, Leslie CC, Han WK, Bonventre JV, Arm JP. Group V secretory phospholipase A2 translocates to the phagosome after zymosan stimulation of mouse peritoneal macrophages and regulates phagocytosis. J Biol Chem. 2006;281:6691–6698. doi: 10.1074/jbc.M508314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morato-Marques M, Campos MR, Kane S, Rangel AP, Lewis C, Ballinger MN, Kim SH, Peters-Golden M, Jancar S, Serezani CH. Leukotrienes target F-actin/cofilin-1 to enhance alveolar macrophage anti-fungal activity. J Biol Chem. 2011 doi: 10.1074/jbc.M111.235309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters-Golden M, Henderson WR., Jr Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 10.Canetti C, Aronoff DM, Choe M, Flamand N, Wettlaufer S, Toews GB, Chen GH, Peters-Golden M. Differential regulation by leukotrienes and calcium of Fc gamma receptor-induced phagocytosis and Syk activation in dendritic cells versus macrophages. J Leukoc Biol. 2006;79:1234–1241. doi: 10.1189/jlb.0705374. [DOI] [PubMed] [Google Scholar]
- 11.Serezani CH, Perrela JH, . Russo M, Peters-Golden M, Jancar S. Leukotrienes are essential for the control of Leishmania amazonensis infection and contribute to strain variation in susceptibility. J Immunol. 2006;177:3201–3208. doi: 10.4049/jimmunol.177.5.3201. [DOI] [PubMed] [Google Scholar]
- 12.Mancuso P, Lewis C, Serezani CH, Goel D, Peters-Golden M. Intrapulmonary administration of leukotriene B4 enhances pulmonary host defense against pneumococcal pneumonia. Infect Immun. 2010;78:2264–2271. doi: 10.1128/IAI.01323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolete R, Secatto A, Pereira PA, Soares EG, Faccioli LH. Leukotriene B4-loaded microspheres as a new approach to enhance antimicrobial responses in Histoplasma capsulatum-infected mice. Int J Antimicrob Agents. 2009;34:365–369. doi: 10.1016/j.ijantimicag.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda Y, Adachi Y, Ishii T, Miura N, Tamura H, Ohno N. Dissociation of Toll-like receptor 2-mediated innate immune response to Zymosan by organic solvent-treatment without loss of Dectin-1 reactivityx. Biol Pharm Bull. 2008;31:13–18. doi: 10.1248/bpb.31.13. [DOI] [PubMed] [Google Scholar]
- 15.Chen XS, Sheller JR, Johnson EN, Funk CD. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372:179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- 16.Wilkins HJ, Crane MM, Copeland K, Williams WV. Hypereosinophilic syndrome: an update. Am J Hematol. 2005;80:148–157. doi: 10.1002/ajh.20423. [DOI] [PubMed] [Google Scholar]
- 17.Tager AM, Bromley SK, Medoff BD, Islam SA, Bercury SD, Friedrich EB, Carafone AD, Gerszten RE, Luster AD. Leukotriene B4 receptor BLT1 mediates early effector T cell recruitment. Nat Immunol. 2003;4:982–990. doi: 10.1038/ni970. [DOI] [PubMed] [Google Scholar]
- 18.Serezani CH, Lewis C, Jancar S, Peters-Golden M. Leukotriene B4 amplifies NF-kappaB activation in mouse macrophages by reducing SOCS1 inhibition of MyD88 expression. J Clin Invest. 2011;121:671–682. doi: 10.1172/JCI43302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, Wong SY, Gordon S. Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rand TG, Sun M, Gilyan A, Downey J, Miller JD. Dectin-1 and inflammation-associated gene transcription and expression in mouse lungs by a toxic (1,3)-beta-D glucan. Arch Toxicol. 2010;84:205–220. doi: 10.1007/s00204-009-0481-4. [DOI] [PubMed] [Google Scholar]
- 21.Canetti C, Hu B, Curtis JL, Peters-Golden M. Syk activation is a leukotriene B4-regulated event involved in macrophage phagocytosis of IgG-coated targets but not apoptotic cells. Blood. 2003;102:1877–1883. doi: 10.1182/blood-2003-02-0534. [DOI] [PubMed] [Google Scholar]
- 22.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 23.Flamand N, Mancuso P, Serezani CH, Brock. TG. Leukotrienes: mediators that have been typecast as villains. Cell Mol Life Sci. 2007;64:2657–2670. doi: 10.1007/s00018-007-7228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Wang SH, Liao CP, Shao S, Lasbury ME, Durant PJ, Lee CH. Downregulation of PU.1 leads to decreased expression of Dectin-1 in alveolar macrophages during Pneumocystis pneumonia. Infect Immun. 2010;78:1058–1065. doi: 10.1128/IAI.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peres CM, Aronoff DM, Serezani CH, Flamand N, Faccioli LH, Peters-Golden M. Specific leukotriene receptors couple to distinct G proteins to effect stimulation of alveolar macrophage host defense functions. J Immunol. 2007;179:5454–5461. doi: 10.4049/jimmunol.179.8.5454. [DOI] [PubMed] [Google Scholar]
- 27.Bonfield TL, Raychaudhuri B, Malur A, Abraham S, Trapnell BC, Kavuru MS, Thomassen MJ. PU.1 regulation of human alveolar macrophage differentiation requires granulocyte-macrophage colony-stimulating factor. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1132–L1136. doi: 10.1152/ajplung.00216.2003. [DOI] [PubMed] [Google Scholar]
- 28.Shibata Y, Berclaz PY, Chroneos ZC, Yoshida M, Whitsett JA, Trapnell BC. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity. 2001;15:557–567. doi: 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 29.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nature reviews. Immunology. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 30.Medeiros AI, Sa-Nunes A, Soares EG, Peres CM, Silva CL, Faccioli LH. Blockade of endogenous leukotrienes exacerbates pulmonary histoplasmosis. Infect Immun. 2004;72:1637–1644. doi: 10.1128/IAI.72.3.1637-1644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schorey JS, Lawrence C. The pattern recognition receptor Dectin-1: from fungi to mycobacteria. Current drug targets. 2008;9:123–129. doi: 10.2174/138945008783502430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keefe DM, Schubert MM, Elting LS, Sonis ST, Epstein JB, Raber-Durlacher JE, Migliorati CA, McGuire DB, Hutchins RD, Peterson DE. Updated clinical practice guidelines for the prevention and treatment of mucositis. Cancer. 2007;109:820–831. doi: 10.1002/cncr.22484. [DOI] [PubMed] [Google Scholar]
- 33.Peres CM, de Paula L, Medeiros AI, Sorgi CA, Soares EG, Carlos D, Peters-Golden M, Silva CL, Faccioli LH. Inhibition of leukotriene biosynthesis abrogates the host control of Mycobacterium tuberculosis. Microbes Infect. 2007;9:483–489. doi: 10.1016/j.micinf.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Netea MG, Marodi L. Innate immune mechanisms for recognition and uptake of Candida species. Trends Immunol. 2010;31:346–353. doi: 10.1016/j.it.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Oikawa T, Yamada T, Kihara-Negishi F, Yamamoto H, Kondoh N, Hitomi Y, Hashimoto Y. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 1999;6:599–608. doi: 10.1038/sj.cdd.4400534. [DOI] [PubMed] [Google Scholar]
- 36.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 37.Rehli M, Poltorak A, Schwarzfischer L, Krause SW, Andreesen R, Beutler B. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J Biol Chem. 2000;275:9773–9781. doi: 10.1074/jbc.275.13.9773. [DOI] [PubMed] [Google Scholar]
- 38.Berclaz PY, Carey B, Fillipi MD, Wernke-Dollries K, Geraci N, Cush S, Richardson T, Kitzmiller J, O'Connor M, Hermoyian C, Korfhagen T, Whitsett JA, Trapnell BC. GM-CSF regulates a PU.1-dependent transcriptional program determining the pulmonary response to LPS. Am J Respir Cell Mol Biol. 2007;36:114–121. doi: 10.1165/rcmb.2006-0174OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichbaum Q, Heney D, Raveh D, Chung M, Davidson M, Epstein J, Ezekowitz RA. Murine macrophage mannose receptor promoter is regulated by the transcription factors PU.1 and SP1. Blood. 1997;90:4135–4143. [PubMed] [Google Scholar]
- 40.Batliner J, Mancarelli MM, Jenal M, Reddy VA, Fey MF, Torbett BE, Tschan MP. CLEC5A (MDL-1) is a novel PU.1 transcriptional target during myeloid differentiation. Mol Immunol. 2011;48:714–719. doi: 10.1016/j.molimm.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berclaz PY, Shibata Y, Whitsett JA, Trapnell BC. GM-CSF, via PU.1, regulates alveolar macrophage Fcgamma R-mediated phagocytosis and the IL-18/IFN-gamma -mediated molecular connection between innate and adaptive immunity in the lung. Blood. 2002;100:4193–4200. doi: 10.1182/blood-2002-04-1102. [DOI] [PubMed] [Google Scholar]
- 42.Hamdorf M, Berger A, Schule S, Reinhardt J, Flory E. PKCdelta-induced PU.1 phosphorylation promotes hematopoietic stem cell differentiation to dendritic cells. Stem cells. 2011;29:297–306. doi: 10.1002/stem.564. [DOI] [PubMed] [Google Scholar]
- 43.Campos MR, Serezani CH, Peters-Golden M, Jancar S. Differential kinase requirement for enhancement of Fc gammaR-mediated phagocytosis in alveolar macrophages by leukotriene B4 vs. D4. Mol Immunol. 2009;46:1204–1211. doi: 10.1016/j.molimm.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Trapnell BC, Whitsett JA. Gm-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annual review of physiology. 2002;64:775–802. doi: 10.1146/annurev.physiol.64.090601.113847. [DOI] [PubMed] [Google Scholar]
- 45.Page AV, Liles WC. Granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, and other immunomodulatory therapies for the treatment of infectious diseases in solid organ transplant recipients. Current opinion in organ transplantation. 2008;13:575–580. doi: 10.1097/MOT.0b013e3283186b80. [DOI] [PubMed] [Google Scholar]
- 46.Richardson MD, Brownlie CE, Shankland GS. Enhanced phagocytosis and intracellular killing of Candida albicans by GM-CSF-activated human neutrophils. J Med Vet Mycol. 1992;30:433–441. [PubMed] [Google Scholar]
- 47.LeVine AM, Reed JA, Kurak KE, Cianciolo E, Whitsett JA. GM-CSF-deficient mice are susceptible to pulmonary group B streptococcal infection. J Clin Invest. 1999;103:563–569. doi: 10.1172/JCI5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paine R, 3rd, . Preston AM, Wilcoxen S, Jin H, Siu BB, Morris SB, Reed JA, Ross G, Whitsett JA, Beck JM. Granulocyte-macrophage colony-stimulating factor in the innate immune response to Pneumocystis carinii pneumonia in mice. J Immunol. 2000;164:2602–2609. doi: 10.4049/jimmunol.164.5.2602. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Juarrero M, Hattle JM, Izzo A, Junqueira-Kipnis AP, Shim TS, Trapnell BC, Cooper AM, Orme IM. Disruption of granulocyte macrophage-colony stimulating factor production in the lungs severely affects the ability of mice to control Mycobacterium tuberculosis infection. J Leukoc Biol. 2005;77:914–922. doi: 10.1189/jlb.1204723. [DOI] [PubMed] [Google Scholar]
- 50.Solbach W, Greil J, Rollinghoff M. Anti-infectious responses in Leishmania major-infected BALB/c mice injected with recombinant granulocyte-macrophage colony-stimulating factor. Annales de l'Institut Pasteur. Immunology. 1987;138:759–762. doi: 10.1016/s0769-2625(87)80033-8. [DOI] [PubMed] [Google Scholar]
- 51.Chen GH, Olszewski MA, McDonald RA, Wells JC, Paine R, 3rd, Huffnagle GB, Toews GB. Role of granulocyte macrophage colony-stimulating factor in host defense against pulmonary Cryptococcus neoformans infection during murine allergic bronchopulmonary mycosis. Am J Pathol. 2007;170:1028–1040. doi: 10.2353/ajpath.2007.060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brock TG, McNish RW, Coffey MJ, Ojo TC, Phare SM, Peters-Golden M. Effects of granulocyte-macrophage colony-stimulating factor on eicosanoid production by mononuclear phagocytes. J Immunol. 1996;156:2522–2527. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.