Abstract
Lupus nephritis (LN) is a serious manifestation of SLE. Therapeutic studies in mouse LN models do not always predict outcomes of human therapeutic trials, raising concerns about the human relevance of these pre-clinical models. In this study we used an unbiased transcriptional network approach to define in molecular terms similarities and differences between three lupus models and human LN. Genome wide gene expression networks were generated using natural language processing and automated promoter analysis and compared across species via suboptimal graph matching.
The three murine models and human LN share both common and unique features. The 20 commonly shared network nodes reflect the key pathologic processes of immune cell infiltration/activation, endothelial cell activation/injury and tissue remodeling/fibrosis, with macrophage/dendritic cell activation as a dominant cross-species shared transcriptional pathway. The unique nodes reflect differences in numbers and types of infiltrating cells and degree of remodeling between the three mouse strains. To define mononuclear phagocyte derived pathways in human LN, gene sets activated in isolated NZB/W renal mononuclear cells were compared with human LN kidney profiles. A tissue compartment specific macrophage activation pattern was seen, with NFκB1 and PPARγ as major regulatory nodes in the tubulointerstitial and glomerular networks respectively.
Our study defines which pathologic processes in murine models of LN recapitulate the key transcriptional processes active in human LN and suggests that there are functional differences between mononuclear phagocytes infiltrating different renal microenvironments.
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune disorder characterized by loss of tolerance to nucleic acids and their binding proteins resulting in the production of autoantibodies that initiate inflammation or injury. Lupus nephritis (LN) is a major cause of end organ damage in SLE and is a risk factor for mortality (1, 2). Despite many advances in the diagnosis and management of LN, the incidence of end-stage renal disease secondary to SLE has not decreased in the last 20 years (3, 4). The complex and heterogeneous nature of SLE has represented a challenge for defining pathogenesis and developing effective therapeutics.
Murine models that spontaneously develop lupus (5, 6) have played a key role in our understanding of human disease (7) and have been used extensively for the identification of therapeutic targets. However there are fundamental differences in the genetic composition of mice and humans that extend to both the innate and the adaptive immune systems (8, 9); disappointingly, translating the therapeutic successes in animal models to successful clinical interventions for nephritis has been challenging. Therefore, there is a pressing need for a scientific approach that allows the exploration of the similarities and differences among species.
In order to define shared pathogenetic mechanisms in the development of LN in mice and humans and determine which mouse model most accurately reflects specific molecular pathways occurring in human disease, we compared gene expression profiles from microdissected human LN kidney biopsies and whole kidneys from three different SLE-prone murine models, NZB/W, NZM2410 and NZW/BXSB. The NZB/W female mouse (10) is characterized by hypercellular renal lesions and fibrinoid necrosis, similar to the lesions seen in Class IV human LN kidney specimens (11). NZM2410 mice are characterized by high levels of IL-4 and the production of autoantibodies of the IgG1 and IgE isotypes; they develop a rapidly progressive glomerulosclerosis with scant lymphocytic infiltrate in the kidneys (12). Males of the NZW/BXSB strain carry the Yaa (Y-linked autoimmune acceleration) locus that contains a reduplication of the Tlr7 gene (13). These mice develop an acute proliferative glomerulonephritis with severe tubulointerstitial inflammation (14). The three strains have different autoantibody profiles with the production of anti-dsDNA antibodies in the NZB/W mouse, anti-nucleosome antibodies in the NZM2410 mouse and anti-Sm/RNP and anti-phospholipid antibodies in the NZW/BXSB mouse. They also respond differently to therapeutic intervention (15); this underscores the effect of genetic heterogeneity not only on disease phenotype but also on responses to therapies. Analysis of comprehensive renal expression profiles with unbiased natural language processing tools (Genomatix Bibliosphere) and a suboptimal matching algorithm based approach (TALE) identified multiple shared key conserved regulatory network hubs (nodes) between the three mouse strains and the human LN samples. These pathway maps allow selection of the mouse model with the highest degree of similarity to the human disease or with activation of a pathway of interest, for further therapeutic interventions or mechanistic studies in the model system. We found that the sclerotic kidneys of the NZM2410 mice shared most renal transcriptional events with human LN kidneys, followed by the NZB/W and the NZW/BXSB.
We have previously shown that the onset of proteinuria in all three strains of mice is associated with the expansion and activation of a dominant population of resident renal mononuclear phagocytes that have a resting phenotype of CD11b+/CD11cint/F4/80hi/Gr1lo/MHChi and have variably been referred to as intrinsic renal macrophages or resident renal DCs due to their mixed function as both antigen presenting cells and phagocytic cells (16). At nephritis onset these cells markedly upregulate their expression of CD11b and they accumulate both in the interstitium and in the periglomerular space (14, 17, 18). In NZB/W mice we demonstrated a marked change in gene expression profile of isolated F4/80hi renal macrophages at proteinuria onset with the expression of pro-inflammatory, anti-inflammatory and tissue repair genes (17). We show here that this macrophage profile is shared by the human LN samples, but that there are significant differences in the gene expression profiles of the glomerular and interstitial compartments suggesting functional differences in this cell type in the two intrarenal microenvironments.
MATERIAL AND METHODS
NZB/W, NZM2410, NZW/BXSB SLE mouse strains
NZB/W F1 mice were purchased from Jackson Labs (Bar Harbor, ME), NZM2410 mice were purchased from Taconic (Hudson, NY) and NZW/BXSB F1 male mice were bred in our facility from the respective parents (Jackson Labs). All mice were housed in an SPF facility with 12 hr light/dark cycles and unlimited access to food and water. All animal experiments were approved by the IACUC Committee of the Feinstein Institute. Supplementary Table I provides details of the subgroups of mice analyzed from each mouse strain as described in previous publications (14, 18, 19). Briefly, the NZB/W F1 strain (Jackson Labs) included groups of the following ages: 16 weeks old (control group without any serum autoantibodies, immune complex deposition or proteinuria, N=8), 23 weeks old (pre-nephritic control group, with serum autoantibodies, minimal immune complex deposition, proteinuria <100mg/dl, N=11), 23 weeks old (with abundant immune complex deposition and new onset of proteinuria >300 mg/dL, N=6), 36 weeks old (with established proteinuria >300 mg/dL for >2 weeks, N=10) (20).
NZM2410 mice were 7 weeks old (control group without autoantibodies, renal immune complex deposition or proteinuria, N=5) and 22–30 weeks old (with proteinuria >300 mg/dL for 7–10 days, N=5) (18). Mice of this strain were harvested 7–10 days after proteinuria onset as most NZM2410 mice die within 14 days of proteinuria onset.
NZW/BXSB mice (14) were 8 weeks old (control group, without serum autoantibodies or proteinuria N=4), 17 weeks old (pre-nephritic control group, with serum autoantibodies, proteinuria ≤100mg/dl and histologic glomerular score ≤2, N=6) and 18–21 weeks old (with >7 days proteinuria >300 mg/dL and histologic glomerular score >2, N=6).
Human renal biopsy samples
Human renal biopsies were collected after informed consent was obtained, according to the guidelines of the respective local ethics committees. A total of 47 samples from the European Renal cDNA Bank (ERCB) (21) were processed and used for microarray analysis: pre-transplant healthy living donors (LD) (N=15) and LN patients (N=32). For real-time PCR, 11 LD and 9 LN samples were used from an independent cohort (of the ERCB). Demographic, clinical and histologic characteristics of these patients are provided in Supplementary Table I. There was no statistical difference in any parameters between the LN cohorts used in arrays and RT-PCR.
Murine and human RNA extraction, microarray preparation and processing
Kidneys were removed from perfused mice and immediately snap frozen. Tissues were homogenized in 3ml of Trizol and RNA was isolated according to manufacturers’ instructions. cDNA was generated from 5 μg total RNA and labeled with biotin using the Ovation Biotin system (NuGEN Technologies, Inc., San Carlos, CA). Biotin labeled cDNA was fragmented and hybridized to Affymetrix Mouse Genome 430 2.0 GeneChip arrays (Santa Clara, CA). After hybridization, GeneChip arrays were washed, stained, and scanned by GeneChip Scanner 3000 7G according to the Affymetrix Expression Analysis Technical Manual (http://media.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf). CD11bhi/CD11cint/F4/80hi mononuclear cells from perfused NZB/W mouse kidneys were isolated by fluorescence cell sorting as previously described (17) using antibodies to CD11b, CD11c and F4/80. RNA was extracted, amplified and analyzed using gene expression profiling as previously described (17). Normalized data are available in Gene Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under Accession Number [GEO: GSE27045].
Cortical tissue segments from human biopsy samples were manually microdissected into glomerular and tubulointerstitial compartments as previously described (22–24). Total RNA was isolated from the microdissected human tissues using the RNeasy mini kit (Qiagen), according to the manufacturer’s instructions. Gene expression profiling from microdissected human kidney biopsies was performed as previously described using the Human Genome U133A Affymetrix Genechip arrays (Santa Clara, CA) (22, 23).
Murine and human gene expression data processing and analysis
The mouse and human CEL files were processed using the GenePattern analysis pipeline (www.genepattern.com). CEL file normalization was performed with the Robust Multichip Average (RMA) method using the mouse and human Entrez-Gene custom CDF annotation from Brain Array version 10 http://brainarray.mbni.med.umich.edu/Brainarray/default.asp). The normalized files were log2 transformed and batch correction was performed for the NZB/W and NZW/BXSB murine data (25).
The poly-A RNA control kit was used in processing the mouse microarray data. The expression baseline was defined by calculating the gene expression median of each gene, and adding one standard deviation to the minimum value obtained. Of the 16539 mouse genes represented on the Affymetrix genechip, 13425, 13600 and 14252 were expressed above the defined expression baseline in NZB/W, NZM2410 and NZW/BXSB respectively. Of the 12029 human genes, 11285 and 11429 were expressed above the 27 Poly-A Affymetrix control expression baseline (negative controls) in the glomerular and tubulointerstitial compartments respectively and were used for further analyses. Mouse and human normalized data files are uploaded on Gene Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) and accessible under reference numbers [GEO: GSE32583 and GSE32591].
IgA nephropathy (IgAN) and hypertensive nephropathy (HT) gene expression profiles from ERCB cohorts were available to the investigators as part of an independent study (Wenjun Ju et al, manuscript under review) and were compared with the presented LN data. IgAN and HT data are available on GEO under the reference number GSE35488 and GSE37463 (the last one will be activated after acceptance of the manuscript).
Real-time PCR analysis of human samples
Reverse transcription (RT) and real-time PCR were performed as reported previously (22). Pre-developed TaqMan reagents (ABI Assays-on-demand) were used for all genes analyzed. The expression of each gene was normalized to the geometric mean of the housekeepers 18S rRNA, PGK1 and GAPDH (26). Those three reference genes were selected as described (27), as well as based on their low variance and their absence of regulation in LN compared to LD in arrays. The mRNA expression was analyzed by the ΔΔ-Ct method following the manufacturer instructions (User Bulletin #2, ABI Prism 7700 Sequence Detection System).
Cross-species comparison
To avoid ambiguity, the mouse genes were converted to the corresponding human orthologs using the NCBI homolog (Build 64) and Genomatix annotated ortholog databases. Differential gene expression was defined by a q-value <0.05 with a fold-change ≥1.2 for the up-regulated genes and ≤0.8 for the down-regulated genes.
Network and pathway analyses
Significantly regulated genes were analyzed by creating biological literature-based networks using Genomatix Bibliosphere software (www.genomatix.de). Canonical pathways were extracted using the Ingenuity Pathway Analysis Software (www.ingenuity.com).
TALE (Tool for Approximate LargE graph matching) analysis
TALE was employed to compare the resulting large transcriptional networks from humans and mice (see (28) for technical implementation). This algorithm allows comparison of large networks (often with 1000s of nodes and edges) and extraction of meaningful relationships between the query and database networks. TALE allows definition of the degree of mismatch tolerance in the resulting overlapping network. In addition to the query network and the database networks, TALE requires three fields of user inputs: 1. a similarity assignment for the genes in the networks; 2. the percentage of important nodes; and 3. the percentage of mismatch. The first parameter is required to group the nodes in the networks. For cross-species comparison, the orthologous information was used to group the human and mouse genes. The second parameter defines the set of seed genes in the query network to generate the overlapping network. In this setting the importance of a node (or a gene) is defined by the number of connections it has with the remaining genes in the human network. In the current study the top 10% nodes were used to build the core network elements. The third parameter defines the mismatch allowed while generating the neighborhood of the seed genes as well extending the network, in the current study a mismatch tolerance of 10% was set. The transcriptional networks (human and mice) generated from the literature-based Genomatix Bibliosphere software (gene/function word/gene (B3) filter) were used as input. The resulting mouse networks were populated into the database and the human network was used as the query network. The resulting TALE networks were visualized using Cytoscape version 2.7 (29).
Prior to the comparison of each murine m odel with human LN, a preliminary TALE analysis was performed within the NZB/W and NZW/BXSB models to define which group comparison (corresponding to different disease stages, see Table I) resembled most closely the human data sets for further analysis. The NZB/W analysis included the following comparisons: 36 weeks old with established nephritis compared to 16 weeks or 23 weeks pre-nephritic, 23 weeks old at onset of proteinuria compared to 16 weeks or 23 weeks old pre-nephritic. The 36 week old mice with established proteinuria compared to the 23 week old pre-nephritic mice shared the highest number of transcriptional network nodes with the human tubulointerstitium LN versus control; in the comparison with the human glomerular LN dataset, the 36 versus 16 week old mice had the highest overlap and were used for further analysis. Similarly, we compared the results for either 8 or 17 week prenephritic NZW/BXSB with 18–21 week nephritic NZW/BXSB mice and found that they were similar (Table I). We therefore used the larger 17 week pre-nephritic control group for the comparison analyses.
Table I.
Number of genes regulated between the groups compared in mouse models and human and overlapping with human datasets.
| Mouse model | Groups compared | Genes regulateda | Corresponding human orthologous genes | Overlap with 3159 genesa regulated in human glomerular LNb | Overlap with 2261 genesa regulated in human tubulointerstitial LNb | Genes regulated in murine and human LN in both renal compartmentsb |
|---|---|---|---|---|---|---|
| NZB/W | 23 wks Pre-N compared to 16 wks control | 234 | 203 | 58 | 56 | 29 |
| 23 wks N compared to 16 wks control | 919 | 826 | 315 | 242 | 177 | |
| 23 wks N compared to 23 wks Pre-N | 1382 | 1227 | 375 | 309 | 200 | |
| 36 wks N compared to 23 weeks Pre-N | 2642* | 2442 | 631 | 379 | 242 | |
| 36 wks N compared to 16 wks control | 3201 | 2972 | 674 | 385 | 244 | |
| NZM2410 | 30 wks N compared to 7wks control | 4015* | 3727 | 699 | 467 | 251 |
| NZW/BXSB | 17 wks Pre-N compared to 8 wks control | 319 | 302 | 95 | 79 | 48 |
| 18–21 wks N compared to 8 wks control | 1877 | 1757 | 568 | 384 | 246 | |
| 18–21 wks N compared to 17 wks Pre-N | 2900* | 2652 | 622 | 447 | 268 |
q-value<0.05 and fold-change ≥ 1.2 for the up-regulated genes and ≤ 0.8 for the down-regulated genes.
genes regulated in same directionality in regulation.
Groups with highest level of shared nodes in network analysis used for further studies. Legend: N: Nephritic (proteinuria > 300mg/dl, histologic nephritis score > 2), Pre-N: Pre-nephritic (serum autoantibodies, proteinuria ≤ 100mg/dl, histologic nephritis score ≤ 2), wks: weeks.
Immunohistochemistry
To localize macrophages and DCs in human lupus nephritis, archival sections from 10 renal biopsies with lupus nephritis class IV (ISN/RPS 2003 classification) and 5 pre-implantation biopsies were studied. Immunohistochemistry was performed as previously described (30). The monoclonal mouse anti-CD68 (Clone PG-M1; Dako, Germany, Hamburg), mouse anti-DC-SIGN (CD209, clone DCN46; BD Pharmingen, Heidelberg, Germany), and the polyclonal rabbit anti-S100 (Dako) were used.
Statistical analysis
For the array study, statistical unpaired analyses for each comparison between the relevant study groups were performed using Significance Analysis of Microarrays (SAM) method implemented in MultiExperiment Viewer (MeV) application (31, 32). Genes regulated between 2 groups with a q-value (False Discovery Rate) below 0.05 were considered significant and used for further transcriptional and pathway analyses. For the RT-PCR study, a student t-test was used; a p-value<0.05 was considered significant.
RESULTS
Renal LN cross-species functional analysis
Differentially regulated genes were identified from the kidneys of mice with LN compared with pre-diseased controls and from humans with LN compared to healthy kidneys. Human LN kidneys demonstrated 3159 glomerular and 2261 tubulointerstitial genes with mRNA expression significantly modified compared to living donor kidneys. Lupus mice were compared to their pre-nephritic counterparts, using the same significance criteria and fold-change used for the human study. 2642, 4015, and 2900 genes were differentially regulated in NZB/W mice (36 wks old with established proteinuria compared to 23 wks old pre-nephritic), NZM2410 mice (22–30 week nephritic compared to 7 week pre-nephritic controls), and NZW/BXSB mice (18–21 week nephritic compared to 17 week pre-nephritic), respectively. Real-time PCR validation of 120 genes confirmed > 85% of the genes identified in the microarray analysis (manuscript in preparation). Table I displays the number of genes significantly regulated in the murine disease groups available for analyses, as well as the number of regulated genes overlapping with the human data in each renal compartment. Murine data sets with highest degree of overlap in the network level comparison were used for further studies (See Materials and Methods for more details and Table I).
Gene expression profiles from whole mouse kidneys were compared with profiles from human microdissected kidneys. This study will compare the murine whole kidney expression data sets to the human tubulointerstitial compartment as the tubulointerstitial compartment constitutes more than 95% of total kidney mass, driving the majority of the signatures obtained from the mouse tissue. In addition, tubulointerstitial inflammatory lesions and fibrosis correlate with the decrease of renal function in LN and are markers of poor outcome (33–36).
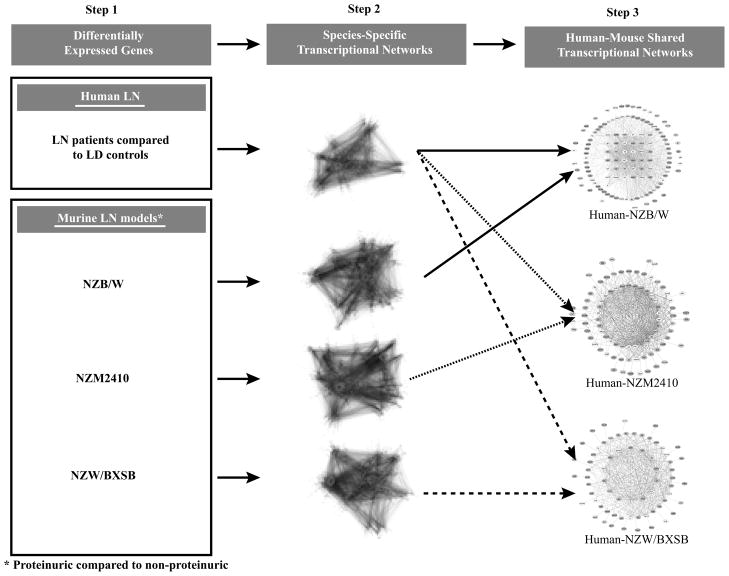
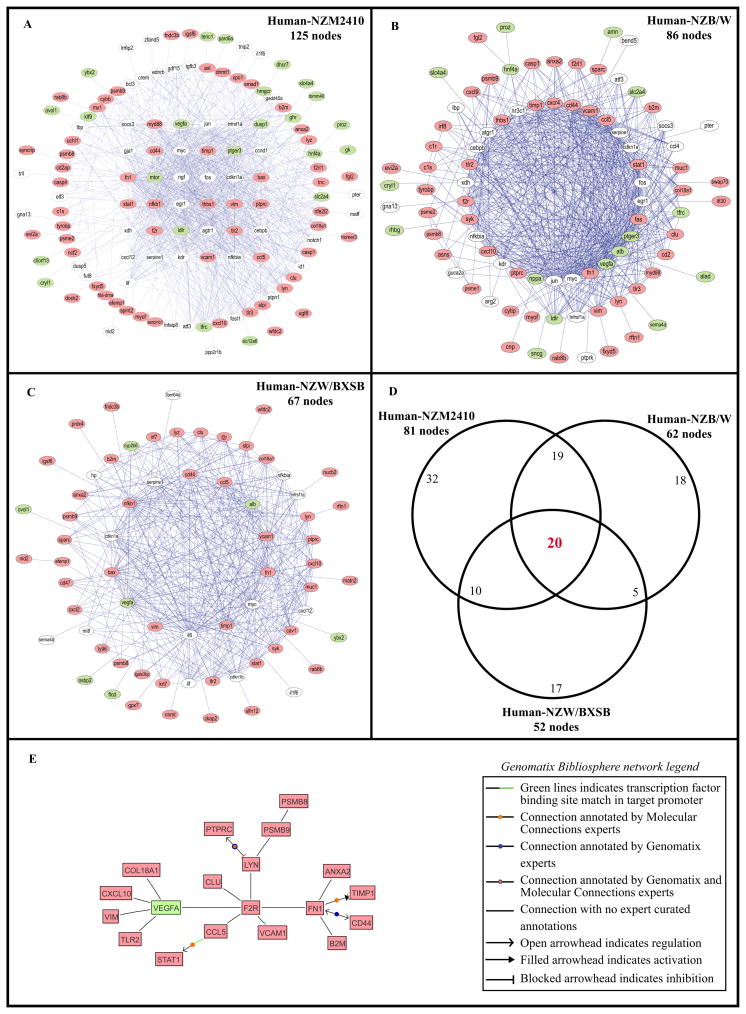
The strategy used for cross-species shared transcriptional network analysis is schematically depicted in Figure 1. A sequential knowledge extraction approach was applied first to generate a transcriptional network that integrates differential gene expression with public knowledge of gene interactions and automated promoter analysis to define transcriptional dependencies in each data set. Next, conservation of regulatory network elements was tested using a suboptimal graph matching approach (TALE) developed by Tian et. al. (28). TALE identifies core network elements shared between two data sets. TALE tolerates a pre-defined degree of mismatches in network substructure during consensus network generation, a critical feature shown to be robust for identifying the cross-species differences seen in many regulatory pathways between mouse and man. In the human–mouse comparison of LN, the NZM2410 glomerulosclerotic mouse model showed the highest number of cross-species conserved regulatory hubs (nodes) identified by TALE, followed by the NZB/W model and the NZW/BXSB model. The number of shared nodes was 125 (Figure 2A), 86 (Figure 2B) and 67 (Figure 2C), respectively. Of these, 81 for the NZM2410, 62 for the NZB/W and 52 for the NZW/BXSB were regulated in the same direction as in the human samples. The lists of nodes and their regulation in each species are provided in Supplementary Table II.
Figure 1. Analytical strategy of cross-species shared tubulointerstitial transcriptional networks using the Tool for Approximate LargE graph matching (TALE).
Individual transcriptional networks were generated using the literature-based Genomatix Bibliosphere software and were overlapped using TALE to define cross-species shared transcriptional networks.
Figure 2. Human-mouse shared tubulointerstitial transcriptional networks.
Networks sharing the most connections between human LN and (A) NZM2410 (nephritic vs. young control) (125 nodes), (B) NZB/W (36 wks with established nephritis vs. 23 wks pre-nephritic) (86 nodes), and (C) NZW/BXSB (nephritic vs. prenephritic) (67 nodes). (D) represents the overlap of the nodes between the three comparisons, using only the nodes regulated in the same direction in both species in each network (81/125 nodes in the NZM2410, 62/86 nodes in the NZB/W and 52/67 nodes in the NZW/BXSB). Legend: Each node represents a gene; each edge (blue line) represents a connection between two nodes. The nodes having more than 20 connections, between 2 and 19 connections and only one connection are displayed in the inner layer, the middle layer and the outside layer, respectively. In red, green and white are respectively the nodes up-regulated in both species, down-regulated in both species and discordantly regulated among species. (E) represents the transcriptional Genomatix Bibliosphere network from the 20 overlapping nodes of the TALE results.
20 nodes (19 induced and one repressed) were common to the three LN murine models and humans and regulated in the same direction (Figure 2D, 2E and Supplementary Table II). These nodes mainly reflect transcripts associated with cellular infiltration and activation and included genes involved in signal transduction (STAT1, LYN, ANXA2), cytokines/chemokines (CCL5, CXCL10), cellular reorganization/traffic (RAB8B, VIM), cell surface markers (PTPRC, TLR2, CD44) and antigen presentation/processing (B2M, and inducible immunoproteasome subunits PSMB8, PSMB9). Three nodes were related to extracellular matrix/GBM/fibrosis (FN1, COL18A1, TIMP1). An increase in lysosyme (LYZ) is most likely reflective of proximal tubular damage. Finally four nodes reflected endothelial cell activation (VCAM1), fibrinolysis (ANXA2), coagulation (F2R - thrombin receptor PAR1) and decreased angiogenesis (VEGF-A) (Figure 2A). A decrease in VEGF-A expression was confirmed at the protein level in the glomerular compartment of patients with lupus nephritis compared with control biopsies or biopsies from patients with vasculitis and a similar degree of kidney function (37). F2R (Thrombin receptor PAR1), FN1, LYN and VEGFA represented the main nodes of the transcriptional network built based on the literature knowledge, identifying these molecules as core elements in the shared regulatory network across models and species (Figure 2E).
Pathway analysis of the genes regulated in the same direction from each cross-species conserved network, further confirmed these findings, as the top pathways that were shared between the three models and human LN interstitium reflected innate and adaptive immune activation, immune cell infiltration and tissue remodeling (Table II and Supplementary Table III). In addition, upregulation of immunoglobulin genes was observed in all three models (particularly the NZB/W model) and in human LN, reflecting B cell and plasma cell infiltration. These genes did not appear in the TALE or ingenuity analyses due to species differences in the immunoglobulin repertoire.
Table II.
Top canonical pathways significantly regulated (p-value<0.05) from the genes shared and regulated in the same direction in human LN* tubulointerstitium and in each mouse model**, as assessed by IPA (Ingenuity Pathway Analysis).
| NZM2410 (467 genes) | NZB/W (379 genes) | NZW/BXSB (447 genes) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Canonical pathways (number of genes in the pathway) | Rank | p-value | Number of regulated genes in the pathway | Rank | p-value | Number of regulated genes in the pathway | Rank | p-value | Number of regulated genes in the pathway |
| Antigen Presentation Pathway (43) | 1 | 2.0E-09 | 12 | 1 | 1.2E-11 | 13 | 1 | 1.2E-09 | 12 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses (87) | 2 | 8.5E-08 | 15 | 5 | 6.8E-09 | 15 | 2 | 6.6E-09 | 16 |
| OX40 Signaling Pathway (90) | 3 | 2.6E-07 | 12 | 8 | 3.3E-08 | 12 | 3 | 1.7E-07 | 12 |
| Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells (81) | 4 | 6.2E-07 | 11 | 2 | 6.3E-10 | 13 | 4 | 4.2E-07 | 11 |
| T Helper Cell Differentiation (72) | 5 | 7.9E-07 | 13 | 13 | 7.1E-07 | 12 | 5 | 5.1E-07 | 13 |
| Allograft Rejection Signaling (91) | 6 | 1.9E-06 | 10 | 3 | 2.2E-09 | 12 | 6 | 1.4E-06 | 10 |
| Dendritic Cell Maturation (188) | 7 | 2.8E-06 | 19 | 4 | 5.6E-09 | 21 | 8 | 1.5E-06 | 19 |
| Type I Diabetes Mellitus Signaling (121) | 8 | 2.9E-06 | 16 | 9 | 2.4E-07 | 16 | 9 | 1.7E-06 | 16 |
| Cdc42 Signaling (174) | 9 | 3.4E-06 | 17 | 10 | 2.5E-07 | 17 | 12 | 8.8E-06 | 16 |
| Autoimmune Thyroid Disease Signaling (61) | 10 | 7.9E-06 | 9 | 6 | 1.1E-08 | 11 | 11 | 5.7E-06 | 9 |
| Graft-versus-Host Disease Signaling (50) | 11 | 1.6E-05 | 9 | 7 | 3.0E-08 | 11 | 13 | 1.2E-05 | 9 |
| Complement System (35) | 13 | 2.5E-05 | 8 | 15 | 6.2E-06 | 8 | 10 | 1.9E-06 | 9 |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation (147) | 16 | 3.7E-05 | 17 | 17 | 1.3E-05 | 16 | 7 | 1.4E-06 | 19 |
LN vs. LD;
Nephritic vs. Prenephritic.
TALE consensus networks generated from the comparison of the three mouse models and the glomerular human LN dataset are provided in Supplementary Figure 1. Similar to the results for the tubulointerstitial compartment, NZM2410 shared the most nodes with human LN and 24 nodes, representing a prominent macrophage/DC signature, were shared between all 3 models and LN.
Of the three models, the sclerotic kidneys of the NZM2410 mice shared most renal transcriptional events with human LN tubulointerstitium. Of the 125 shared nodes between NZM2410 and human LN kidneys and excluding the 20 common genes described above, 61 were regulated in the same direction (22 were downregulated and 39 upregulated) (Supplementary Table II). The downregulated nodes included many genes involved in cholesterol metabolism as well as genes involved in solute transport, consistent with loss of differentiated tubular cell function. The upregulated nodes reflected tissue injury processes (apoptosis, fibrosis, coagulation, tissue remodeling), expression of proteases, kinases and protease inhibitors and IFN induced genes (Supplementary Table II).
Each of the two other models shared unique nodes with human LN interstitium. The NZB/W mouse with diffuse proliferative glomerulonephritis shared 86 nodes with the human LN kidneys. Excluding the 20 shared genes described above, 42 of them were regulated in the same direction in both NZB/W and human tubulointerstitium (27 up and 15 down). 18 nodes were shared only by the NZB/W and human LN and included IFI30, CXCR4 and CCL9 (Supplementary Table II(D)). Many of the nodes unique to this human-NZB/W comparison reflected lymphocyte infiltration and activation, a feature of human LN not found in NZM2410.
The NZW/BXSB mouse with severe proliferative glomerulonephritis shared the least number of nodes with the human LN tubulointerstitium (67 nodes). Excluding the 20 genes common with the 2 other models as described above, 32 nodes were regulated in the same direction as in human disease (26 up and 6 down). 17 genes were shared exclusively by the NZW/BXSB and human LN (Supplementary Table II(D)). Unique nodes in this mouse included Muc1, Nid2 and Matn1 that are involved in extracellular matrix formation, the dendritic cell derived chemokine CXCL3, and the Type I interferon inducing transcription factor IRF7, likely reflecting the increase in TLR signaling that occurs both in this mouse and in human LN.
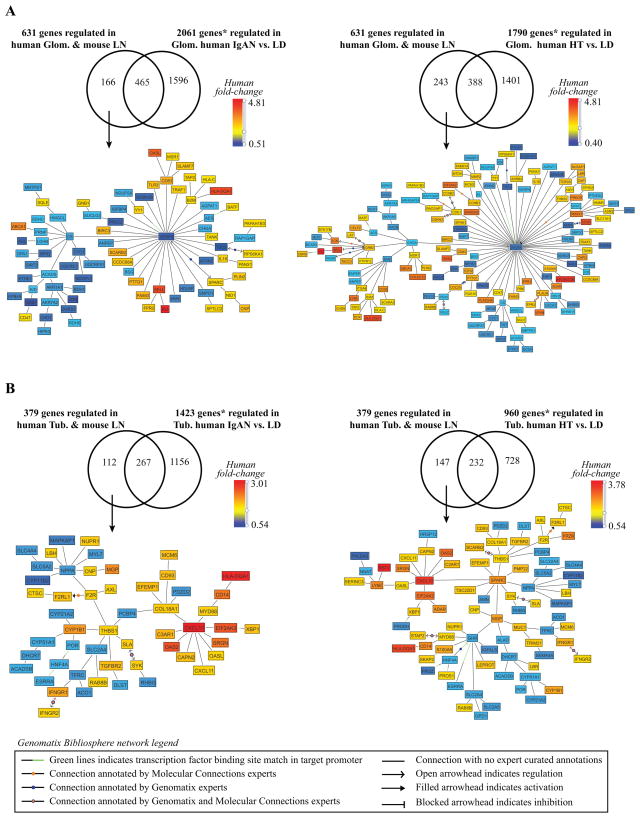
The gene expression pattern that we describe here in patients with LN might also be shared with other inflammatory renal diseases. To address this point, microarray data from ERCB cohorts available to the investigators as part of an independent study (manuscript under review) were compared with the LN data. Gene expression data from glomeruli and tubulointerstitial compartments of the inflammatory disease IgA nephropathy (IgAN) and the non-inflammatory disease hypertensive nephropathy (HT) were compared to the human/mouse overlapping gene expression profiles shown in Table I. For these comparisons we focused on NZB/W and NZM2410 mouse models, as they share most features with the human disease. As shown in Figure 3, a significant proportion of transcripts were found to be regulated in an LN specific manner (26–39% of the regulated genes in LN. As expected, more inflammatory genes were shared with the IgAN biopsies than the HT biopsies. Interestingly the LN glomeruli had more downregulation of VEGFA and of mitochondrial genes (citrate synthase node) than did the IgAN or HT biopsies (Figure 3A), suggesting a greater degree of oxidative stress in the LN biopsies. The LN interstitium had a more prominent Type I IFN signature (CXCL10 node) and more evidence of procoagulant activity (THBS1 node) than did the IgAN or HT biopsies (Figure 3B and data not shown)
Figure 3. Human-NZB/W mouse LN comparison with IGAN and HT.
A. Glomerular compartment. B. Tubulointerstitial compartment. The figures display the transcriptional networks (Genomatix Bibliosphere) obtained from the genes that were co-cited in PubMed abstracts in the same sentence linked to a function word (B2 filter) (145 of166 genes (A-left panel), 220 of 243 genes (A-right panel), 104 of 112 genes (B-left panel), 138 of 147 genes (Bright panel)). LN regulated transcripts were mapped into the transcriptional networks and included in the comparison * q-value<0.05 and fold-change ≥ 1.2 for the up-regulated genes and ≤ 0.8 for the down-regulated genes.
Renal LN macrophage functional analysis
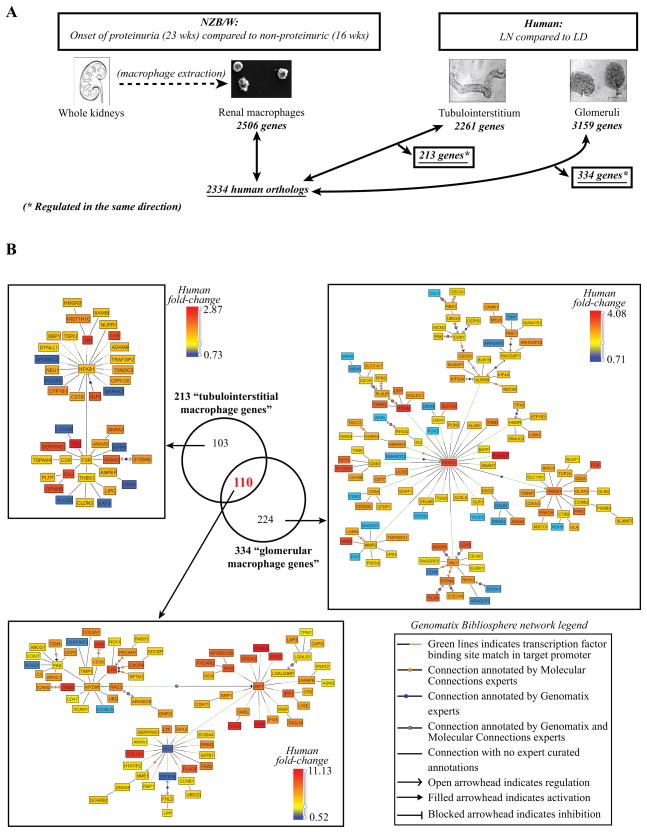
A key feature of the cross-species comparison in LN was the concordant regulation of macrophage/DC transcripts with a prominent antigen presentation and pattern recognition receptor signature shared between mouse and man (Table II and Supplementary Table III(A)). Mononuclear phagocytic cells have been extensively described as key players in both acute and chronic kidney diseases (38–40) but to our knowledge, no study has yet analyzed the macrophage transcriptome within the kidneys of LN patients. Purifying small subpopulations of cells from human LN kidney biopsies is technically challenging due to the limited amount available of kidney tissue from renal biopsy cores. We therefore applied a cross-species integration strategy of defining the transcriptome of the renal mononuclear phagocytes from nephritic mouse kidneys and using these data to define potential macrophage/DC derived genes from SLE biopsy samples. For an overview of the strategy employed see Figure 4.
Figure 4. Renal LN F4/80hi intrinsic functional analysis.
A. Analytical strategy of renal LN “macrophage” functional analysis (q-value <0.05, fold-change ≥1.2 for the up-regulated genes and ≤0.8 for the down-regulated genes). B. Overlap of the defined tubulointerstitial and glomerular “macrophage genes”. Transcriptional networks generated using the literature-based Genomatix Bibliosphere software from the 103 tubulointerstitial-restricted genes, the 224 glomerular-restricted genes and the 110 genes shared by both compartments and mouse F4/80hi intrinsic macrophages (q-value<0.05, fold-change ≥1.2 for the up-regulated genes and ≤0.8 for the down-regulated genes). Respectively, the pictures display the 42/103, 78/110 and 120/224 genes that were co-cited in PubMed abstracts in the same sentence linked to a function word (B2 filter).
We have previously shown in all three mouse models that activation of mononuclear phagocytes associated with upregulation of CD11b on CD11b/F4/80hi/CD11cint intrinsic renal “macrophages” (F4/80hi cells) is a cardinal feature of new onset proteinuria (14, 17, 18). Furthermore, in all three models F4/80hi cells are the dominant mononuclear phagocyte population both in pre-nephritic and nephritic kidneys (14, 17, 18). Although cells from the NZM2410 mouse kidneys, that share the most number of nodes with the human LN tubulointerstitium, might provide the most information, these mice have far fewer F4/80hi cells than the other two strains and die almost as soon as they become proteinuric (18). We therefore used the transcriptome from isolated F4/80hi cells from NZB/W kidneys as a probe for the human samples. Gene expression array data from these cells have previously been reported (17). A total of 2506 genes were differentially regulated in F4/80hi cells isolated from kidneys from nephritic mice compared to those from ≤16 week old NZB/W mice without any renal disease (q-value <0.05, fold-change ≥1.2 for the up-regulated genes and ≤0.8 for the down-regulated genes) (Figure 4A).
To analyze whether this murine renal F4/80hi intrinsic macrophage signature is present in human LN kidneys, we compared the regulated murine renal F4/80hi genes with the genes that were differentially expressed in the tubulointerstitium and glomeruli respectively of patients with LN compared to living donor controls. 213 murine macrophage genes were regulated in the same direction as in the human tubulointerstitium and 334 macrophage genes were concordantly regulated in human glomeruli (Figure 4B). Literature based networks were generated from these 213 “tubulointerstitial LN macrophage/DC genes” and 334 “glomerular LN macrophage/DC genes” for further interpretation.
A total of 110 macrophage/DC genes were expressed in both tubulointerstitium and glomeruli of human LN biopsies (Figure 4B) and were dominated by a Type I interferon signature with both MyD88 and IRF7 nodes (Figure 4B, lower panel). This signature included MDA-5 (IFIH1) and Viperin (RSAD2), genes involved in nucleic acid sensing and the interferon pathway. Fourteen of the “macrophage” genes found in both human renal compartments had a binding-site for the transcription factor IRF7 in their promoter (as assessed by Genomatix Inc, Table III), suggesting a role for TLR activation in tissue injury (30). Other concordantly induced genes included the src family kinases Hck and Lyn, the chemokine receptor CXCR4, complement factor C3, and Factor B, an important amplifier of complement activation through the alternative complement pathway. Myc transcription factor consensus binding sites were found to be significantly enriched in the promoter regions of macrophage genes and concordant with this observation the transcriptional repressor Myc was found to be downregulated in the isolated murine F4/80hi cells.
Table III.
Transcription factor analysis from the defined “tubulointerstitial and glomerular macrophage genes”, as assessed by Genomatix Bibliosphere software.
| 25 genes from the 224 “glomerular-restricted macrophage genes” having a binding site for PPARG (V$PERO) in their promoter | HMOX1 | CCR2 | SOAT1 | ID2 | ALAS1 | ITGAM | ACSL5 | |
| HMMR | ENO2 | VAV1 | BATF | PLIN2 | CD40 | AURKB | ||
| MMP2 | MAP4K4 | CD8A | OLR1 | SMAD7 | CFLAR | PLA2G7 | ||
| PCK2 | TGM2 | SMARCD3 | LMO4 | |||||
|
| ||||||||
| From the 110 “shared tubulointerstitial and glomerular macrophage genes”: | 14 genes having a binding site for IRF7 (V$IRFF) in their promoter | MGP | MYC | DOCK2 | IFI35 | LGALS3BP | ISG20 | OASL |
| MYD88 | IGFBP6 | IFIT1 | IFIH1 | RSAD2 | IFI44 | |||
| NRP1 | ||||||||
|
| ||||||||
| 27 genes having a binding site for MYC (V$EBOX) in their promoter | NCK1 | S100A4 | FAS | SERPING1 | CD44 | LGALS1 | ANXA1 | |
| DKK3 | VCAM1 | LTF | TIMP1 | CXCR4 | PLAC8 | CD38 | ||
| LYN | ZBTB16 | COL1A2 | RRM2 | NME1 | CCNB1 | HCK | ||
| MYD88 | SATB1 | IRF7 | RBP1 | LGALS3BP | HTATIP2 | |||
|
| ||||||||
| 24 genes from the 103 “tubulointerstitial- restricted macrophage genes” having a binding site for NFKB1 (V$NFKB) in their promoter | S100A6 | NPPA | C1R | ANXA2 | APOBEC2 | XBP1 | DPP4 | |
| THBS1 | SERPINE2 | TRAF3IP2 | TXNDC5 | GPR126 | PDGFB | TNC | ||
| MAP4K2 | VIM | TSPO | ADAM9 | ANXA5 | DYNLL1 | CLU | ||
| L1CAM | CD9 | ANXA3 | ||||||
224 unique “macrophage” genes were expressed in the glomerular profile from human LN biopsies. In this compartment PPARγ, MMP2, and the CD11b alpha chain ITGAM were key nodes in the transcriptional network (Figure 4B). PPARγ binding sites were found to be enriched in promoter regions of 25 glomerular macrophage genes, further supporting a role of this transcriptional activator for macrophage transcript regulation in LN (Table III). Multiple phagocytic receptors were upregulated in the glomerular profile (Trem2, MERTK and TGM2), as well as tissue repair genes (MMP2, Heparanase, the anti-oxidant HMOX1 and SMAD7) and proinflammatory genes (Trem1, FPR1, CD40). Induction of multiple elements of the proteasome resulted in ‘protein ubiquitination’ being scored as the top ranking pathway (Supplementary Table III(B)) found in the shared transcriptome induced in both murine LN macrophages and glomeruli from human lupus nephritis.
103 “macrophage” genes were shared with the genes differentially regulated in the LN tubulointerstitium and included molecules from the coagulation and fibrinolytic systems: F2R, Serpine 2 (PAI-1), TNC, THBS1, ANXA5 (Figure 4B). Transcription factor binding analysis indicated a significant enrichment of NFκB binding sites in promoter regions of the shared regulated transcripts (Table III).
In sum, the comparative analysis of murine derived transcriptome of renal macrophages with human LN signatures allowed us to define transcriptional networks operating ubiquitously in LN including IRF7 dependent transcripts, as well as those operating in distinct microenvironments. Signatures of glomerular resident macrophages showed a specific enrichment for PPARγ dependent regulation, whereas tubulo-interstitial macrophages showed a predominance of NFκB dependent transcripts.
Much of the data from the macrophages of NZB/W mice has previously been validated using real-time RT-PCR (17) and real-time RT-PCR data for whole kidneys from each of the three mouse strains will be reported separately (manuscript in preparation). Human microarray data were validated by processing TaqMan real-time PCR on tubulointerstitial samples from an independent cohort for several of the specific genes of interest described in the above paragraphs (Supplementary Table I and Table IV). Since glomerular material is very limited, we focused validation on the most relevant gene (ITGAM) in these samples (Table IV). All genes evaluated by RT-PCR were significantly regulated with high fold-change in LN compared to healthy pre-transplant living donors (fold-change between 1.48 and 22.21, p-value < 0.05), except CD14, CXCR4 and GPNMB for which there was a trend towards upregulation as observed in the arrays. TaqMan real-time PCR was also performed on a subset of 9 LN from the 32 used on arrays and 6 LDs. All genes were significantly up-regulated in this technical replicate (data not shown).
Table IV.
TaqMan real-time PCR of selected genes of interest as validation of microarrays data. The comparison LN vs. LD represents the Fold-change.
| Compartment | Gene | Microarrays | Real-time PCR | ||
|---|---|---|---|---|---|
| Fold-change | q-value | Fold-change | p-value | ||
| Tubulointerstitium | CCR1 | 1.29 | 0.004 | 2.18 | 0.047 |
| CD14 | 1.81 | 0.001 | 1.48 | 0.332 | |
| CCL5 | 1.81 | 0.001 | 12.81 | 0.049 | |
| CTSS | 1.86 | 0.000 | 10.57 | 0.001 | |
| CXCL10 | 3.01 | 0.000 | 16.20 | 0.003 | |
| STAT1 | 3.62 | 0.000 | 9.28 | 0.000 | |
| CXCR4 | 1.86 | 0.001 | 1.95 | 0.237 | |
| IRF7 | 1.66 | 0.000 | 6.99 | 0.002 | |
| HCK | 1.37 | 0.000 | 3.35 | 0.003 | |
| LYN | 1.63 | 0.000 | 2.54 | 0.009 | |
| CFB | 2.74 | 0.000 | 5.22 | 0.000 | |
| IFI44 | 10.16 | 0.000 | 22.21 | <0.0001 | |
| GPNMB | 2.10 | 0.000 | 3.36 | 0.058 | |
| Glomeruli | ITGAM | 3.21 | 0.000 | 3.82 | 0.009 |
Immunohistochemical staining of kidneys
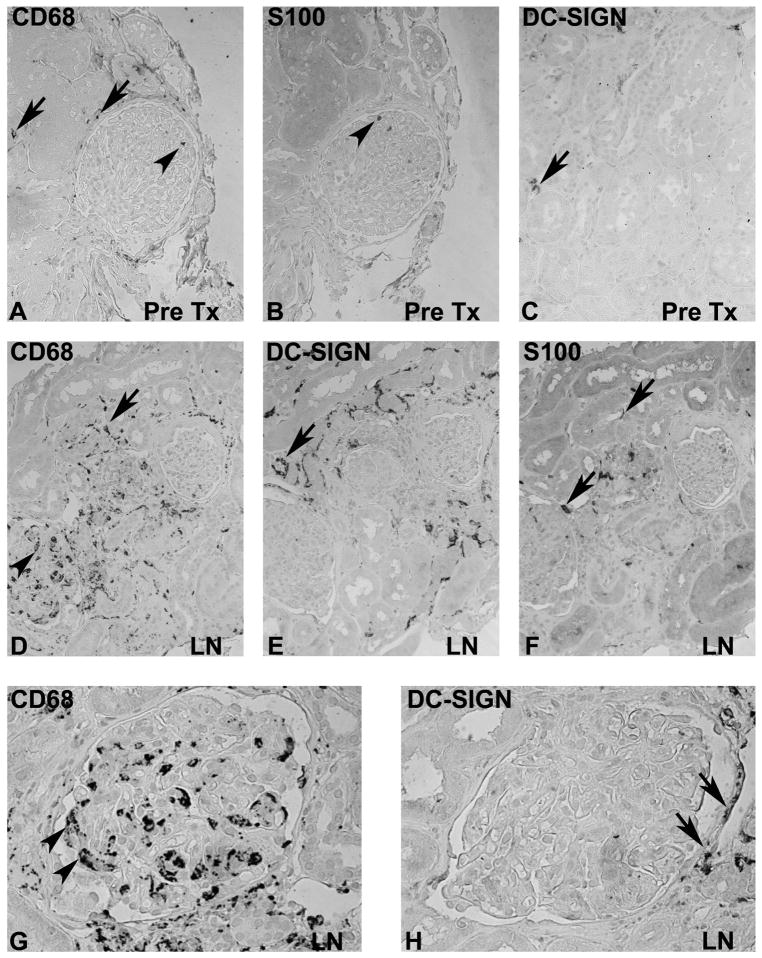
To illustrate differences between mononuclear cells in microcompartments of the human kidney a series of 10 biopsies with lupus nephritis were compared with 5 pretransplant controls (Figure 5). Immunohistochemistry for DC-SIGN (CD209), the myeloid DC marker S100 and the macrophage scavenger receptor CD68 was performed in human LN (Figure 5). As predicted by the transcriptional analysis, SLE patients had an increase in cells staining with all three markers. CD68 positive cells were found both within the glomeruli and in the interstitium, whereas DC-SIGN positive cells were restricted to the interstitium. S100 positive cells were loosely scattered in both glomeruli (circulating cells within glomerular capillaries) and the interstitium. These findings confirm our hypothesis generated from the transcriptional data that distinct renal mononuclear phagocytes infiltrate the different intrarenal compartments in human LN.
Figure 5. Localization of macrophage/DC markers in human lupus nephritis.
Immunohistochemistry for CD68 (A, D, G), S100 (B, E) and DC-SIGN (C, F, H) was performed on consecutive sections from a pre-transplant biopsy (A–C), and biopsies with lupus nephritis class IV (ISN/RPS 2003 classification, original magnifications X200 in A–F, X400 in G and H). Scattered CD68 and lower numbers of S100 positive cells were found in the tubulointerstitium (arrow) and occasionally in glomerular capillaries (arrowhead) in pretransplant biopsies (A, B). A low number of DC-SIGN positive cells were present in the tubulointerstitium (C, arrow). In contrast prominent numbers of CD68 and DC-SIGN positive cells where present in biopsies from patients with lupus nephritis (D–G). Consecutive sections demonstrate a prominent number of CD68 positive cells in glomerular capillaries (arrowheads in D and G), but also a prominent accumulation of CD68 positive cells in the tubulointerstitium (arrows). DC-SIGN positive cells were restricted to the tubulointerstitium (arrow in H), but no DC-SIGN was expressed on glomerular CD68 positive cells.
In sum, our data suggest that there are different functional features of various mononuclear phagocytic populations that infiltrate different renal microenvironments in LN.
DISCUSSION
While much of the focus in the classification of human lupus has been on the glomerulus, there is an increasing understanding that renal outcomes correlate best with the degree of tubulointerstitial inflammation and damage (33–35). After the deposition of immune complexes in the glomeruli, periglomerular macrophages may be attracted by locally produced chemokines or may be activated by inflammatory mediators to proliferate in situ. In addition, because the effluent blood flow from the glomerulus provides the sole blood supply for peritubular capillaries, glomerular hypertension and hypertrophy compromise peritubular blood flow resulting in hypoxia, tubular activation and tubular epithelial cell death. This induces resident macrophage activation and peritubular inflammation and is associated with tissue remodeling, fibrosis (33) and finally, irreversible renal damage. Release of damaged tissue, cytokines, and other inflammatory mediators amplify the inflammatory process. These effector inflammatory processes are likely to be shared between individuals and may be addressed therapeutically.
Our study therefore had two major goals. The first was to identify common expression profiles that reflect renal events shared between the three murine models and human LN. Defining such common pathways between mouse models and human diseases will most likely result in therapeutic targets with broad specificity. Our second goal was to define which molecular pathways found in the three disparate murine models with proliferative (NZB/W and NZW/BXSB) or sclerotic (NZM2410) glomerulonephritis are shared with human LN. This will facilitate experimental studies in murine LN models by allowing the a priori selection of the model system most closely mimicking the human LN situation for the specific pathway under interrogation. Our study showed that the sclerotic kidneys of the NZM2410 mice shared most renal transcriptional events with human LN kidneys but also identified unique features of each of the murine models that were shared with human LN, stressing the need to have multiple model systems with matching molecular data available for further experimental validation of human LN pathways.
Transcriptional analyses of human and mouse kidneys with lupus nephritis and proteinuria using the Tool for Approximate LargE graph matching (TALE) allowed us to extend our analysis beyond a simple gene-gene comparison by overlapping transcriptional networks from the two different species and then defining shared functional relationships. This analysis showed that macrophages/DCs are key players in the development of LN disease. The small amount of tissue obtained from kidney biopsies is a limitation for studying the role of isolated cell populations in the pathogenesis of human LN disease. The strategy of analyzing gene expression profiles from infiltrating macrophage populations obtained from mouse kidneys allowed us to attribute mRNA signature to those cells in the human samples even when such information might not be derived from the human tissue homogenate initially. These studies suggest differences in functional activities between mononuclear phagocytes in different renal sub-compartments.
The shared transcriptional profile
Using TALE analysis, we identified 20 common transcriptional network nodes that were shared by all three LN murine models and the tubulointerstitium of human LN and regulated in the same direction. These nodes reflect key processes in chronic renal injury in SLE nephritis, namely immune cell infiltration and activation, macrophage and dendritic cell activation, endothelial cell activation and damage and tissue remodeling/fibrosis, processes that were confirmed by pathway analysis.
Of the three models, the sclerotic kidneys of the NZM2410 mice shared most renal transcriptional events with human LN kidneys. Histologically, the kidneys of NZM2410 mice model have a limited degree of inflammation and more glomerulosclerosis than those of the other two strains (12). The highest similarity of this model to human LN could be because patients may have, in contrast to the murine models, accumulated significant chronic end-organ damage before they are referred to a nephrologist and undergo renal biopsy. In addition, most patients are already partially treated by the time biopsies are performed, thus repressing the severity of acute inflammation compared to the untreated mouse models. Thus the NZM2410 mouse may be an appropriate mouse model for testing add-on therapies directed at chronic progression of LN.
Most of the transcriptional network nodes unique to the NZB/W mouse with diffuse proliferative glomerulonephritis reflected lymphocyte infiltration and activation, a feature found in many SLE biopsies; this feature of nephritis is not found in NZM2410. In particular the expression of CXCR4 was elevated; this chemokine receptor is expressed on plasma cells in mice and is overexpressed on multiple leukocyte subsets in active lupus patients; its ligand CXCL12 is overexpressed in human LN kidneys (41, 42). Tubulointerstitial inflammation with lymphoid aggregates that may include germinal centers and abundant plasma cells is commonly found in human LN and is associated with worse renal outcome (43, 44). Indeed, a prominent immunoglobulin signature was found both in mouse kidneys and in the human tubulointerstitium. Compared with NZM2410 there was less expression of nodes that included proteases, pro-apoptotic molecules and extracellular matrix. Thus this model appears to have more inflammation and less tissue remodeling than NZM2410.
The NZW/BXSB mouse model shared the least number of nodes with the human LN kidneys and surprisingly, given that both NZB/W and NZW/BXSB have severe proliferative disease, overlapped less with the NZB/W mouse than it did with NZM2410. This may reflect, as discussed above, the partially treated nature of the human disease. Unique nodes in this mouse were associated with extracellular matrix formation and myeloid cell activation, consistent with the large numbers of infiltrating myeloid cells in this model (14).
Several nodes were discordantly regulated in the murine models and human samples. These included nodes indicating a response to ER stress or hypoxia (SERPINE1, EGR1, ATF3, MYC, CDKN1A, Fos and Jun), and genes that downregulate acute inflammatory cytokines (SOCS3, ATF3, LIF, NFKBIA). These findings suggest a greater degree of hypoxia in the mouse models than in human disease, perhaps reflecting the aggressive nature of the untreated murine disease.
In summary, the three regulatory networks representing the transcriptional events shared between the human LN tubulointerstitium and kidneys of three different LN mouse models showed significant overlaps, but also characteristics specific to each model, each of which is shared with the human disease. These findings underscore the heterogeneity of lupus nephritis and the pitfalls of extrapolating from a single mouse model to human disease. Our study illustrates the necessity of studying different mouse models, as specific questions about human disease might best be answered using a particular model. Importantly, the transcriptional profile highlighted a large number of genes associated with processes involved in chronic renal injury and disease progression that may be resistant to systemic immune suppression. Furthermore, comparison of the LN profiles with those of two other human renal diseases showed both similarities and differences. In particular, the differences between LN and IgA nephropathy reflected more prominent tissue hypoxia and tissue remodeling in the LN biopsies, features that might require a different type of therapeutic intervention in order to prevent a poor longterm outcome seen in LN. Our findings stress the need for including management strategies in SLE nephritis directed at slowing the progression of the renal impairment that may continue even after systemic inflammation has resolved.
This approach has a number of intrinsic limitations. First, TALE has been designed to provide the similarities between large networks (from two species here). Indeed, it aligns networks to define the conserved network. By definition, “approximate large graph matching” is useful because it has a defined mismatch tolerance and thereby can build a consensus signature; however, this feature is not optimized to define differences between networks. Second, as our transcriptional network generation integrates differential gene expression with automated promoter analysis and natural language processing using PubMed abstracts, it is biased towards the current literature, thereby enriching for known biological interactions between transcripts. Third, as most of the patients were at least partially treated at the time of biopsy, leading to treatment induced alterations of the expression profiles they may not display the natural disease history observed in the murine models, masking important aspects of disease pathogenesis.
Despite these constraints, our study has identified a prominent macrophage/DC activation signature as a key feature of all three lupus models and of human LN. Indeed, our study confirms a previous report (44) of a prominent myelomonocytic signature in the glomeruli of most LN patients, with expression of a set of genes similar to that identified here. Our findings therefore suggest not only that further study of these cell types, their origins and the mechanisms by which they become activated in the kidneys may yield new therapeutic strategies, but that mouse models such as NZB/W are suitable for performing these studies.
The macrophage profile
Pathway analysis of the shared nodes identified mononuclear phagocyte activation as a key player in LN. We therefore investigated the contribution of these cells to the total mRNA expression profile by comparing the human compartment specific gene expression profiles with the profile of the dominant F4/80hi intrinsic macrophage population isolated from control and nephritic NZB/W kidneys. This strategy allowed us to detect transcripts of the macrophage signature in the heterogenous human renal tissue.
Resident renal mononuclear phagocytes have variably been called resident or intrinsic renal macrophages or resident renal dendritic cells (16). In mice there are at least 5 subpopulations of these cells in the kidneys ((17) and Sahu and Davidson, in preparation) but the major population that forms a network throughout the interstitium expresses high levels of F4/80, CD11b, intermediate levels of CD11c and low/intermediate levels of Ly6C and they are positive for MHC Class II but express low levels of costimulatory molecules (17). The generation of CX3CR1-GFP labeled mice has allowed visualization of analogous CX3CR1 positive cells in a network surrounding glomerular tubules (45). These cells are capable of phagocytosis and constantly retract and extend dendritic processes into the interstitium (46, 47). Their role is probably a sentinel one under physiologic circumstances, but they can contribute to renal injury once activated. We have recently shown that F4/80hi resident renal cells increase in number in the periglomerular area and/or throughout the interstitium in all three lupus models during the course of the disease; they become activated with disease onset (17, 18, 19) and revert to their physiologic state upon induction of remission. The expression profile of these cells reveals a mixed functional signature with both M1 and M2 like characteristics that likely reflects exposure of SLE kidney resident macrophages to multiple stimuli including immune complexes, multiple cytokines, TLR signals, fibrinogen, dead and dying cells, hypoxia and other danger signals. This aberrant activation profile is associated with chronic and progressive renal injury (17).
In humans, only limited phenotypic analyses of macrophages and DC have been performed (30, 48, 49), mostly using immunohistochemistry. Human tubulointerstitial CD68 positive mononuclear phagocytes have a mixed phenotype and include CX3CR1 positive cells and cells variably positive for the C type lectins DC-SIGN and BDCA-1. As we show here, in SLE biopsies CD68 positive cells infiltrate both the glomerular and the tubulointerstitial compartment but interestingly the tubulointerstitial cells tend to be positive for DC-SIGN whereas cells that accumulate in and around glomeruli are negative for DC-SIGN (reviewed Nössner et al, Nephron exp. Nephrology submitted), suggesting potential functional differences between the two compartments. A previous study of gene expression of microdissected glomeruli form SLE patients revealed that nearly all lupus biopsies have a myelomonocytic expression signature (44) that includes genes that we have also detected in F4/80hi cells from nephritic kidneys. Functional studies in humans are however still lacking.
Our strategy allows us to separately query the glomerular/periglomerular and tubulointerstitial compartments of human LN biopsy samples for genes that are regulated in activated renal mononuclear phagocytes from nephritic NZB/W mice. This strategy allows us to extract gene expression patterns that reflect differences in expression rather than changes in cell number. The genes shared by murine activated F4/80hi macrophages and both human intrarenal compartments are dominated by a prominent Type I interferon signature, activation of the alternative complement pathway through Factor B, and upregulation of the src family kinases Hck and Lyn that are important effectors of integrin mediated macrophage adhesion and Fc receptor mediated phagocytosis (47, 50–52). The chemokine receptor CXCR4 that is expressed by multiple leukocyte subsets in SLE kidneys (41, 42) is also highly upregulated. CXCR4 blockade has been reported to reduce renal leukocyte infiltration in murine lupus nephritis (41, 53). Transcription factor analysis revealed a large number of genes regulated by the transcriptional repressor Myc that is itself downregulated in the murine cells. Downregulation of Myc is associated with monocyte to macrophage differentiation although the functional target genes are not well defined (54, 55).
Unique profiles of the “macrophage signature” in the glomerular and interstitial compartments also suggest functional differences. The 224 unique genes shared between the NZB/W macrophage signature and human LN glomeruli are dominated by a PPARγ transcriptional profile. PPARγ is expressed by alternatively activated macrophages and suppresses the production of inflammatory cytokines while inducing production of IL-10 (56, 57); it also mediates phagocytosis of apoptotic material (58). PPARγ agonists decrease extracellular matrix accumulation and prevent renal macrophage accumulation in response to injury. PPARγ deficient macrophages also fail to acquire an anti-inflammatory phenotype upon engulfment of apoptotic cells, suggesting a role of PPARγ in immune clearance. Several recent studies have shown remarkably beneficial effects of PPARγ agonists in murine models of SLE nephritis, suggesting that these drugs could be appropriate therapies for preventing chronic renal damage in SLE (59, 60); our study suggests that these drugs may enhance a protective function of glomerular macrophages in humans as well. Other phagocytic receptors were also highly upregulated in the glomerular profile suggesting that the glomerular macrophages are actively involved in phagocytosis as are the mouse F4/80hi cells (17). Pathway analysis revealed a significant enrichment of protein ubiquitination transcript regulation in glomeruli. We have previously shown that F4/80hi cells from proteinuric NZB/W mice accumulate large numbers of autophagocytic vesicles (17). An increase in both protein ubiquitination and autophagy pathways, together with the increase in cell surface molecules involved in phagocytosis, suggest an important role for these cells in intracellular proteolysis, presumably of oxidatively modified and ubiquitinated proteins. The glomerular “macrophage” profile also uniquely expressed a tissue repair signature, suggesting a role for these cells in tissue remodeling. Together, these data suggest that the glomerular mononuclear phagocyte is an actively phagocytic cell with potentially protective functions but that it may also contribute to aberrant tissue remodeling.
The unique tubulointerstitial-restricted “macrophage” signature was characterized by expression of genes involved in the coagulation and fibrinolytic systems. These genes might therefore be useful as markers for the presence of tubulointerstitial infiltrates and possibly a risk for fibrosis in LN. Another gene highly expressed in the interstitium is GPNMB (osteoactivin) that is upregulated during macrophage differentiation from monocytes and appears to function as a negative regulator of inflammation (61). Upregulation of osteoactivin has been observed in monocytes obtained from uremic patients or in normal monocytes cultured in uremic serum (61) as well as in CD11b+ cells within inflammatory myocardial infiltrates in a model of adverse myocardial remodeling (62). In addition this compartment was characterized by a striking NFκB transcriptional profile that was absent in the glomerular “macrophage” signature. These findings in sum suggest that although mononuclear phagocytes in different renal locations share some functional characteristics, the different microenvironments are associated with unique gene expression profiles that may profoundly influence cell function.
Several genes that were highly upregulated in F4/80hi cells from NZB/W nephritic kidneys (17) were not regulated in human LN samples. Some of this discrepancy may have been due to the dilution effects of whole tissue vs. purified cells, for example IL-10 expression was not regulated in whole mouse kidneys although it was highly expressed in isolated F4/80hi cells from proteinuric mice. Nevertheless, these differences are a reminder that the animal models do not necessarily reflect the human processes in their entirety. Our studies allow us to determine which pathways best reflect the human disease and can be further investigated in the appropriate mouse models.
An obvious limitation is that a “specific transcriptional macrophage signature” cannot be definitively attributed in the context of whole organ sampling since some of the mRNAs expressed by isolated F4/80hi cells may also be expressed either in the same or opposite direction by other resident cells. For example, while Myc expression is downregulated in isolated F4/80hi cells, reflecting their maturation status, it is upregulated in whole mouse kidneys. Nevertheless our study has generated hypotheses that can now be tested in the relevant murine models. In particular, by enhancing or antagonizing specific functions of these cells it may be possible to identify which of their functions are protective and which are pathogenic. This could lead to new strategies that harness their therapeutic potential.
In summary our study identified the renal transcriptional features common in human LN and three LN murine models; this strategy will help the scientific community to select the appropriate model to study pathways or pathogenic processes of interest. These common characteristics can further form the basis for new therapeutic strategies. Conversely, characteristics unique to each of the three murine models might be exploited to subset patients for further studies based on their renal molecular profiles. Our study highlights the contribution of renal mononuclear phagocytic cells in both mouse and man, with both phenotypic similarities and differences in gene expression between the glomerular and tubulointerstitial compartments. We can now explore the functional and therapeutic implications of these findings, confirmed in humans, by further experimentation in our mouse models. This bidirectional flow of information should allow us not only to discover new therapeutic opportunities, but also to identify genes that are biomarkers for clinical disease stage or outcome.
Supplementary Material
Acknowledgments
We thank the University of Michigan Microarray Core Facility (Cancer Center) for human DNA chip hybridizations and Greg Khitrov for mouse DNA chip hybridizations.
We thank Dr Nicolas Kozakowski (Department of Surgical Pathology, University of Vienna, Austria) for providing archival biopsies from lupus nephritis.
We thank also Jignesh M. Patel for their tool development (TALE: Tool for Approximate LargE graph matching).
We thank Stefanie Gaiser (Division of Nephrology, University Hospital, Zurich, Switzerland) for her help with TaqMan real-time PCR.
We are grateful to all the members of the European Renal cDNA Bank-Kroener-Fresenius biopsy bank at the time of the study: C.D. Cohen, M. Fischereder, H. Schmid, P.J. Nelson, M. Kretzler, D. Schloendorff, Muenich; J.D. Sraer, P. Ronco, Paris; M.P. Rastaldi, G. D’Amico, Milano; F. Mampaso, Madrid; P. Doran, H.R. Brady, Dublin; D. Moenks, Goettingen; P. Mertens, J. Floege, Aachen; N. Braun, T. Risler, Tuebingen; L. Gesualdo, F.P. Schena, Bari; J. Gerth, G. Wolf, Jena; R. Oberbauer, D. Kerjaschki, Vienna; B. Banas, B.K. Kraemer, Regensburg; W. Samtleben, Muenich; H. Peters, H.H. Neumayer, Berlin; K. Ivens, B. Grabensee, Duesseldorf; R.P. Wuethrich, Zuerich; V. Tesar, Prague.
Abbreviations used
- anti-Sm/RNP
anti-Sm antigen/ribonucleoprotein
- DC
dendritic cell
- DC-SIGN
dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- IACUC
institute animal care and use committee
- LN
lupus nephritis
- NZB/W
New Zealand Black/New Zealand White (mouse hybrid)
- NZM
New Zealand mixed; NZW/BXSB
- SLE
systemic lupus erythematosus
- SPF
specific pathogen free
- TALE
Tool for Approximate LargE graph matching
Footnotes
This study was supported by the National Institute of Health: R01 DK085241 to A.D. and by R01 DK079912 and P30 DK081943 to M.K. and by the Alliance for Lupus Research to A.D. and M.K.; C.C.B. was a supported by the research fellowship FLB1245 from the National Kidney Foundation. R.B. was supported by a Kirkland Scholar Award (Rheuminations). S.S. is supported by a Grant of the University of Zurich, a Clinical Evidence Council grant from Baxter Healthcare Corporation, and a grant by the Swiss National Science Foundation (SNF 32003B_129710).
References
- 1.Doria A, Iaccarino L, Ghirardello A, Zampieri S, Arienti S, Sarzi-Puttini P, Atzeni F, Piccoli A, Todesco S. Long-term prognosis and causes of death in systemic lupus erythematosus. Am J Med. 2006;119:700–706. doi: 10.1016/j.amjmed.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 2.Cook RJ, Gladman DD, Pericak D, Urowitz MB. Prediction of short term mortality in systemic lupus erythematosus with time dependent measures of disease activity. J Rheumatol. 2000;27:1892–1895. [PubMed] [Google Scholar]
- 3.Ward MM. Changes in the incidence of end-stage renal disease due to lupus nephritis, 1982–1995. Arch Intern Med. 2000;160:3136–3140. doi: 10.1001/archinte.160.20.3136. [DOI] [PubMed] [Google Scholar]
- 4.Ward MM. Changes in the incidence of endstage renal disease due to lupus nephritis in the United States, 1996–2004. J Rheumatol. 2009;36:63–67. doi: 10.3899/jrheum.080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grande JP. Experimental models of lupus nephritis. Contrib Nephrol. 2011;169:183–197. doi: 10.1159/000319241. [DOI] [PubMed] [Google Scholar]
- 6.Davidson A, Aranow C. Lupus nephritis: lessons from murine models. Nat Rev Rheumatol. 2010;6:13–20. doi: 10.1038/nrrheum.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry T, Mohan C. Systemic lupus erythematosus--recent clues from congenic strains. Arch Immunol Ther Exp (Warsz) 2005;53:207–212. [PubMed] [Google Scholar]
- 8.Lu Y, Rosenfeld R, Nau GJ, Bar-Joseph Z. Cross species expression analysis of innate immune response. J Comput Biol. 2010;17:253–268. doi: 10.1089/cmb.2009.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 10.Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148:1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helyer BJ, Howie JB. Renal disease associated with positive lupus erythematosus tests in a cross-bred strain of mice. Nature. 1963;197:197. doi: 10.1038/197197a0. [DOI] [PubMed] [Google Scholar]
- 12.Singh RR, Saxena V, Zang S, Li L, Finkelman FD, Witte DP, Jacob CO. Differential contribution of IL-4 and STAT6 vs STAT4 to the development of lupus nephritis. J Immunol. 2003;170:4818–4825. doi: 10.4049/jimmunol.170.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen N, Fu Q, Deng Y, Qian X, Zhao J, Kaufman KM, Wu YL, Yu CY, Tang Y, Chen JY, Yang W, Wong M, Kawasaki A, Tsuchiya N, Sumida T, Kawaguchi Y, Howe HS, Mok MY, Bang SY, Liu FL, Chang DM, Takasaki Y, Hashimoto H, Harley JB, Guthridge JM, Grossman JM, Cantor RM, Song YW, Bae SC, Chen S, Hahn BH, Lau YL, Tsao BP. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn P, Ramanujam M, Bethunaickan R, Huang W, Tao H, Madaio MP, Factor SM, Davidson A. Prevention of murine antiphospholipid syndrome by BAFF blockade. Arthritis Rheum. 2008;58:2824–2834. doi: 10.1002/art.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanujam M, Davidson A. Targeting of the immune system in systemic lupus erythematosus. Expert Rev Mol Med. 2008;10:e2. doi: 10.1017/S1462399408000562. [DOI] [PubMed] [Google Scholar]
- 16.Ferenbach D, Hughes J. Macrophages and dendritic cells: what is the difference? Kidney Int. 2008;74:5–7. doi: 10.1038/ki.2008.189. [DOI] [PubMed] [Google Scholar]
- 17.Bethunaickan R, Berthier CC, Ramanujam M, Sahu R, Zhang W, Sun Y, Bottinger EP, Ivashkiv L, Kretzler M, Davidson A. A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J Immunol. 2011;186:4994–5003. doi: 10.4049/jimmunol.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanujam M, Bethunaickan R, Huang W, Tao H, Madaio MP, Davidson A. Selective blockade of BAFF for the prevention and treatment of systemic lupus erythematosus nephritis in NZM2410 mice. Arthritis Rheum. 2010;62:1457–1468. doi: 10.1002/art.27368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiffer L, Bethunaickan R, Ramanujam M, Huang W, Schiffer M, Tao H, Madaio MP, Bottinger EP, Davidson A. Activated renal macrophages are markers of disease onset and disease remission in lupus nephritis. J Immunol. 2008;180:1938–1947. doi: 10.4049/jimmunol.180.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiffer L, Sinha J, Wang X, Huang W, von Gersdorff G, Schiffer M, Madaio MP, Davidson A. Short term administration of costimulatory blockade and cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice by a mechanism downstream of renal immune complex deposition. J Immunol. 2003;171:489–497. doi: 10.4049/jimmunol.171.1.489. [DOI] [PubMed] [Google Scholar]
- 21.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Grone HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M European Renal cDNA Bank (ERCB) Consortium. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 22.Cohen CD, Frach K, Schlondorff D, Kretzler M. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61:133–140. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 23.Lindenmeyer MT, Kretzler M, Boucherot A, Berra S, Yasuda Y, Henger A, Eichinger F, Gaiser S, Schmid H, Rastaldi MP, Schrier RW, Schlöndorff D, Cohen CD. Interstitial vascular rarefaction and reduced VEGF-A expression in human diabetic nephropathy. J Am Soc Nephrol. 2007;18:1765–1776. doi: 10.1681/ASN.2006121304. [DOI] [PubMed] [Google Scholar]
- 24.Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, Boucherot A, Neusser MA, Cohen CD, Carter-Su C, Argetsinger LS, Rastaldi MP, Brosius FC, Kretzler M. Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes. 2009;58:469–477. doi: 10.2337/db08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative real-time PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;2:113–120. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abruzzo LV, Barron LL, Anderson K, Newman RJ, Wierda WG, O’Brien S, Ferrajoli A, Luthra M, Talwalkar S, Luthra R, Jones D, Keating MJ, Coombes KR. doi: 10.2353/jmoldx.2007.070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian YPJ. TALE: A Tool for Approximate Large Graph Matching. ICDE, IEEE 24th International Conference on Data Engineering; 2008. pp. 963–972. [Google Scholar]
- 29.Clin MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segerer S, Heller F, Lindenmeyer MT, Schmid H, Cohen CD, Draganovici D, Mandelbaum J, Nelson PJ, Grone HJ, Grone EF, Figel AM, Nössner E, Schlöndorff D. Compartment specific expression of dendritic cell markers in human glomerulonephritis. Kidney Int. 2008;74:37–46. doi: 10.1038/ki.2008.99. [DOI] [PubMed] [Google Scholar]
- 31.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 32.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 33.Park MH, D’Agati V, Appel GB, Pirani CL. Tubulointerstitial disease in lupus nephritis: relationship to immune deposits, interstitial inflammation, glomerular changes, renal function, and prognosis. Nephron. 1986;44:309–319. doi: 10.1159/000184012. [DOI] [PubMed] [Google Scholar]
- 34.Fujii K, Kobayashi Y. Quantitative analysis of interstitial alterations in lupus nephritis. Virchows. Arch A Pathol Anat Histopathol. 1988;414:45–52. doi: 10.1007/BF00749737. [DOI] [PubMed] [Google Scholar]
- 35.Hill GS, Delahousse M, Nochy D, Thervet E, Vrtovsnik F, Remy P, Glotz D, Bariety J. Outcome of relapse in lupus nephritis: roles of reversal of renal fibrosis and response of inflammation to therapy. Kidney Int. 2002;61:2176–2186. doi: 10.1046/j.1523-1755.2002.00357.x. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh C, Chang A, Brandt D, Guttikonda R, Utset TO, Clark MR. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care Res (Hoboken) 2011;63:865–874. doi: 10.1002/acr.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thacker SG, Berthier CC, Mattinzoli D, Rastaldi MP, Kretzler M, Kaplan MJ. The detrimental effects of IFN-alpha on vasculogenesis in lupus are mediated by repression of IL-1 pathways: potential role in atherogenesis and renal vascular rarefaction. J Immunol. 2010;185:4457–4469. doi: 10.4049/jimmunol.1001782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duffield JS. Macrophages and immunologic inflammation of the kidney. Semin Nephrol. 2010;30:234–254. doi: 10.1016/j.semnephrol.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duffield JS. Macrophages in kidney repair and regeneration. J Am Soc Nephrol. 2011;22:199–201. doi: 10.1681/ASN.2010121301. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Huen S, Nishio H, Nishio S, Lee HK, Choi BS, Ruhrberg C, Cantley LG. Distinct macrophage phenotypes contribute to kidney injury and repair. J Am Soc Nephrol. 2011;22:317–326. doi: 10.1681/ASN.2009060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang A, Fairhurst AM, Tus K, Subramanian S, Liu Y, Lin F, Igarashi P, Zhou XJ, Batteux F, Wong D, Wakeland EK, Mohan C. CXCR4/CXCL12 hyperexpression plays a pivotal role in the pathogenesis of lupus. J Immunol. 2009;182:4448–4458. doi: 10.4049/jimmunol.0801920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang A, Guilpain P, Chong BF, Chouzenoux S, Guillevin L, Du Y, Zhou XJ, Lin F, Fairhurst AM, Boudreaux C, Roux C, Wakeland EK, Davis LS, Batteux F, Mohan C. Dysregulated expression of CXCR4/CXCL12 in subsets of patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62:3436–3446. doi: 10.1002/art.27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang A, Henderson SG, Brandt D, Liu N, Guttikonda R, Hsieh C, Kaverina N, Utset TO, Meehan SM, Quigg RJ, Meffre E, Clark MR. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol. 2011;186:1849–1860. doi: 10.4049/jimmunol.1001983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson KS, Huang JF, Zhu J, D’Agati V, Liu X, Miller N, Erlander MG, Jackson MR, Winchester RJ. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soos TJ, Sims TN, Barisoni L, Lin K, Littman DR, Dustin ML, Nelson PJ. CX3CR1+ interstitial dendritic cells form a contiguous network throughout the entire kidney. Kidney Int. 2006;70:591–596. doi: 10.1038/sj.ki.5001567. [DOI] [PubMed] [Google Scholar]
- 46.Kruger T, Benke D, Eitner F, Lang A, Wirtz M, Hamilton-Williams EE, Engel D, Giese B, Muller-Newen G, Floege J, Kurts C. Identification and functional characterization of dendritic cells in the healthy murine kidney and in experimental glomerulonephritis. J Am Soc Nephrol. 2004;15:613–621. doi: 10.1097/01.asn.0000114553.36258.91. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki T, Kono H, Hirose N, Okada M, Yamamoto T, Yamamoto K, Honda Z. Differential involvement of Src family kinases in Fc gamma receptor-mediated phagocytosis. J Immunol. 2000;165:473–482. doi: 10.4049/jimmunol.165.1.473. [DOI] [PubMed] [Google Scholar]
- 48.Lindenmeyer M, Noessner E, Nelson PJ, Segerer S. Dendritic cells in experimental renal inflammation – Part I. Nephron Exp Nephrol. 2011;119:e83–90. doi: 10.1159/000332029. [DOI] [PubMed] [Google Scholar]
- 49.Noessner E, Lindenmeyer M, Nelson PJ, Segerer S. Dendritic cells in human renal inflammation – Part II. Nephron Exp Nephrol. 2011;119:e91–98. doi: 10.1159/000332032. [DOI] [PubMed] [Google Scholar]
- 50.Xiao W, Hong H, Kawakami Y, Lowell CA, Kawakami T. Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J Clin Invest. 2008;118:924–934. doi: 10.1172/JCI34013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Totani L, Piccoli A, Manarini S, Federico L, Pecce R, Martelli N, Cerletti C, Piccardoni P, Lowell CA, Smyth SS, Berton G, Evangelista V. Src-family kinases mediate an outside-in signal necessary for beta2 integrins to achieve full activation and sustain firm adhesion of polymorphonuclear leucocytes tethered on E-selectin. Biochem J. 2006;396:89–98. doi: 10.1042/BJ20051924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitzer-Attas CJ, Lowry M, Crowley MT, Finn AJ, Meng F, DeFranco AL, Lowell CA. Fcgamma receptor-mediated phagocytosis in macrophages lacking the Src family tyrosine kinases Hck, Fgr, and Lyn. J Exp Med. 2000;191:669–682. doi: 10.1084/jem.191.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chong BF, Mohan C. Targeting the CXCR4/CXCL12 axis in systemic lupus erythematosus. Expert Opin Ther Targets. 2009;13:1147–1153. doi: 10.1517/14728220903196761. [DOI] [PubMed] [Google Scholar]
- 54.Amanullah A, Liebermann DA, Hoffman B. Deregulated c-Myc prematurely recruits both Type I and II CD95/Fas apoptotic pathways associated with terminal myeloid differentiation. Oncogene. 2002;21:1600–1610. doi: 10.1038/sj.onc.1205231. [DOI] [PubMed] [Google Scholar]
- 55.Van Dang C, McMahon SB. Emerging Concepts in the Analysis of Transcriptional Targets of the MYC Oncoprotein: Are the Targets Targetable? Genes Cancer. 2010;1:560–567. doi: 10.1177/1947601910379011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rigamonti E, Chinetti-Gbaguidi G, Staels B. Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler Thromb Vasc Biol. 2008;28:1050–1059. doi: 10.1161/ATVBAHA.107.158998. [DOI] [PubMed] [Google Scholar]
- 57.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 58.Majai G, Sarang Z, Csomos K, Zahuczky G, Fesus L. PPARgamma-dependent regulation of human macrophages in phagocytosis of apoptotic cells. Eur J Immunol. 2007;37:1343–1354. doi: 10.1002/eji.200636398. [DOI] [PubMed] [Google Scholar]
- 59.Aprahamian T, Bonegio RG, Richez C, Yasuda K, Chiang LK, Sato K, Walsh K, Rifkin IR. The peroxisome proliferator-activated receptor gamma agonist rosiglitazone ameliorates murine lupus by induction of adiponectin. J Immunol. 2009;182:340–346. doi: 10.4049/jimmunol.182.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao W, Thacker SG, Hodgin JB, Zhang H, Wang JH, Park JL, Randolph A, Somers EC, Pennathur S, Kretzler M, Brosius FC, 3rd, Kaplan MJ. The peroxisome proliferator-activated receptor gamma agonist pioglitazone improves cardiometabolic risk and renal inflammation in murine lupus. J Immunol. 2009;183:2729–2740. doi: 10.4049/jimmunol.0804341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pahl MV, Vaziri ND, Yuan J, Adler SG. Upregulation of monocyte/macrophage HGFIN (Gpnmb/Osteoactivin) expression in end-stage renal disease. Clin J Am Soc Nephrol. 2010;5:56–61. doi: 10.2215/CJN.03390509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Psarras S, Mavroidis M, Sanoudou D, Davos CH, Xanthou G, Varela AE, Panoutsakopoulou V, Capetanaki Y. Regulation of adverse remodelling by osteopontin in a genetic heart failure model. Eur Heart J. 2011 doi: 10.1093/eurheartj/ehr119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.