Summary
pH sensing is crucial for survival of most organisms, yet the molecular basis of such sensing is poorly understood. Here we present an atomic resolution structure of the periplasmic portion of the acid sensing chemoreceptor, TlpB, from the gastric pathogen Helicobacter pylori. The structure reveals a universal signaling fold, a PAS domain, with a molecule of urea bound with high affinity. Through biophysical, biochemical and in vivo mutagenesis studies, we show that urea and the urea binding site residues play critical roles in the ability of H. pylori to sense acid. Our signaling model predicts that protonation events at Asp114, affected by changes in pH, dictate the stability of TlpB through urea binding.
Introduction
All cells sense and respond to intra and extracellular pH changes, which influence an enormous number of important processes including metabolic reactions, ATP synthesis, cell motility, gene expression and apoptosis. The structure, stability and interactions of biological macromolecules are in many cases directly determined by charges on diverse titratable groups, emphasizing the necessity for pH homeostasis. Despite the importance of pH sensing for life, little is known about the underlying molecular mechanisms. pH sensing has been studied in diverse contexts including blood plasma pH homeostasis (Tresguerres et al., 2010), sperm activation (Pastor-Soler et al., 2003), and bacterial chemotaxis receptors (Kihara and Macnab, 1981). Although the players of many pH sensing signaling cascades, including in some cases the pH sensor itself, have been identified, the precise molecular mechanisms for detecting proton concentration and the subsequent signal relay have not been well described.
Candidate molecular moieties for pH sensing by a protein receptor include titratable side chains such as aspartate, glutamate and histidine that have pKa values near neutrality. The pKa for a sensor can be tuned over a wide range by the immediate environment of the protonatable residue. Protonation of critical residues is generally thought to lead to conformational changes that confer signaling states. Examples of bacterial pH sensors relevant to our studies include the histidine kinase HP165 (ArsS) of H. pylori, in which His94 has been identified as essential for pH sensitivity (Muller et al., 2009) and the chemotaxis receptors Tar and Tsr, where critical residues lie in the linker region between the transmembrane helices and the cytoplasmic domain (Umemura et al., 2002). In neither case are structural details available, nor have molecular mechanisms been proposed. Here we propose a molecular mechanism for pH sensing by a chemoreceptor of the acid-tolerant gastric pathogen H. pylori.
H. pylori is a Gram-negative bacterium that resides in the stomachs of over half the world’s population. Its gastric habitat contains a marked pH gradient from the highly acidic lumen, which can reach pH 2, to the more neutral environment adjacent to the epithelial lining, which is typically pH 7 (Sachs et al., 2003). Although they can survive in the lumen, H. pylori cells prefer more neutral environments, placing them in proximity to the tissue where they exert pathogenic effects (Schreiber et al., 2004). Chronic H. pylori infection can lead to gastric ulcers as well as gastric cancer, the second most common cause of terminal cancer worldwide (Marshall et al., 1988; Nomura et al., 1991; Parkin et al., 2005; Parsonnet et al., 1991; Shanks and El-Omar, 2009). Treatment of H. pylori infection involves a cocktail of medications that has unfortunately begun to fail in 20–30% of cases (Graham, 2009). The emergence of antibiotic resistant strains of H. pylori underscores the need to better understand processes important for H. pylori infection, such as chemotaxis (Foynes et al., 2000; Howitt et al., 2011; McGee et al., 2005; Terry et al., 2005), which could be targeted in the design of novel therapies to combat this prevalent pathogen.
H. pylori has evolved sophisticated strategies to survive within the harsh environment of the acidic stomach lumen and navigate to its preferred niche of the neutral gastric mucosa. In addition to surviving extreme acidity, H. pylori must be able to sense and move in response to chemicals in its environment, with pH being an important cue that determines its distribution (Schreiber et al., 2004). H. pylori chemotaxis mutants are impaired in colonization in both mouse and gerbil models, and have less intimate associations with the gastric epithelium (Foynes et al., 2000; Howitt et al., 2011; McGee et al., 2005; Terry et al., 2005; Williams et al., 2007).
Bacteria perceive and respond to chemical cues in their environment using chemoreceptors that transduce information about their ligand-bound state to direct the activity of the histidine kinase CheA. CheA activity modulates the phosphorylation state of the response regulator CheY, which directly interacts with the flagellar motor machinery to control swimming behavior. Based on genome sequence analysis, the H. pylori chemotaxis machinery resembles that of the well-studied model, E. coli, with a few notable variations including the absence of the methylation enzymes involved in receptor adaptation (Sweeney and Guillemin, 2011). H. pylori have four chemoreceptors, TlpA, B, C and D and several polar flagella that dictate smooth swimming behavior in the presence of an attractant and increased stopping behavior in the presence of a repellent (Lowenthal et al., 2009; Rader et al., 2011). Only a small number of chemotactic signals have been identified for H. pylori. The best characterized chemoreceptor is TlpB, which is required for chemorepulsive responses to acid, as well as the quorum sensing molecule auto-inducer-2 (AI-2) (Croxen et al., 2006; Rader et al., 2011).
Here we provide an atomic resolution structure of the periplasmic section of TlpB. The structure reveals a homodimer that contains Per-ARNT-Sim (PAS) domains, universal signaling modules. TlpB is the first bacterial chemoreceptor of known function shown by crystallography to contain an extracellular PAS domain. Strikingly, a molecule of urea is found within the canonical ligand binding site of the PAS domain, bound in a manner predicted to be sensitive to changes in pH. We report that urea is bound with extremely high affinity and specificity and is essential for the thermodynamic stability of the TlpB molecule. The urea binding site includes an aspartate group (Asp114), which we propose to be the key titratable residue responsible for pH sensing. Mutational and biophysical analyses of the urea binding site support a mechanism in which the urea cofactor, by binding in a pH-sensitive fashion, stabilizes the secondary structure of TlpB and signals a pH response.
Results
The periplasmic portion of TlpB forms a dimer that contains urea binding PAS domains
The H. pylori transmembrane chemoreceptor TlpB belongs to the class of methyl-accepting chemotaxis (MCP) proteins that bacteria use to sense chemical cues in the environment and transduce signals to regulate their swimming behavior (Hazelbauer et al., 2008). Genetic analysis has demonstrated that it is responsible for sensing several environmental cues, including changes in pH and presence of autoinducer-2 (Croxen et al., 2006; Rader et al., 2011). The periplasmic sensing domain accounts for about one third of the total amino acids of TlpB (Figure 1A). We expressed a shortened, recombinant protein consisting of H. pylori strain SS1 periplasmic domain, residues 33–211 (referred to as TlpBpp throughout the paper) in E. coli. We purified the protein and found that it crystallized under multiple conditions. The crystal structure was solved using the SIRAS method and the atomic model ultimately refined at 1.38Å resolution (Table 1).
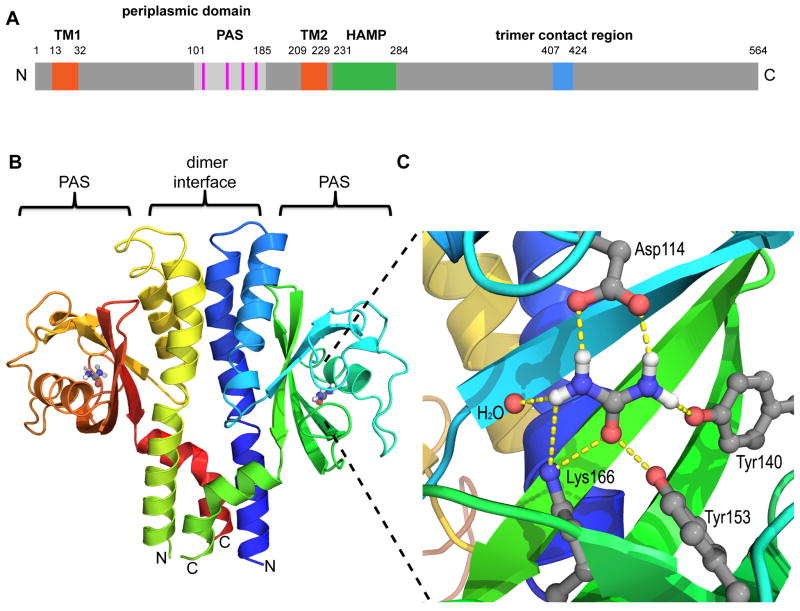
Figure 1.
TlpBpp forms a dimer that contains urea-binding PAS domains. (A) Schematic of estimated SS1 TlpB domains. TM (orange), trans-membrane region; PAS (light gray), per-ARNT-Sim domain; HAMP (green), Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases domain; trimer contact region (blue), chemoreceptor trimer of dimers contact region. The periplasmic domain is from amino acid ~32–209. (B) Ribbon diagram of TlpBpp homodimer and gray/blue/red/white urea molecules, with chain color gradation ranging from N-terminus (blue) to C-terminus (green) for the monomer on the right and N-terminus (green) to C-terminus (red) for the monomer on the left. For orientation, the bacterial inner membrane would be below the lower part of the protein model diagram. (C) Diagram of the TlpBpp urea binding site including hydrogen bonds (dashed lines) between urea (gray, white, blue and red) and the surrounding residues and water molecule. Oxygen atoms are shown as red spheres, nitrogen as blue and hydrogen as white. See also Table 1 and Figure S1.
Table 1.
Crystallographic data collection and refinement statistics.
| Table S1: Crystallographic Statistics | |||||
|---|---|---|---|---|---|
| Structure | Native | KI derivative | Hydroxyurea | Acetamide | Formamide |
| Data collection | |||||
| Cell dimensions (Å) | a = 79.6 b = 81.9 c = 94.5 |
a = 79.7 b = 82.5 c = 94.1 |
a = 79.6 b = 81.5 c = 94.6 |
a = 79.6 b = 82.0 c = 94.1 |
a = 79.6 b = 81.7 c = 94.5 |
| Total Reflections | 2151808 | 254955 | 653922 | 883763 | 596495 |
| Unique Reflections | 126318 | 33665 | 113775 | 107983 | 92912 |
| Resolutiona (Å) | 50–1.38 (1.4–1.38) | 50–2.12 (2.20–2.12) | 50–1.42 (1.45–1.42) | 50–1.40 (1.43–1.40) | 50–1.42 (1.45–1.42) |
| Rmergea,b | 0.071 (0.49) | 0.083 (0.436) | 0.062 (0.394) | 0.044 (0.517) | 0.052 (0.302) |
| I/σ(I)a | 30.4 (3.7) | 13.7 (2.8) | 18.4 (2.0) | 29.0 (2.5) | 23.3 (2.7) |
| Completenessa (%) | 99.4 (98.3) | 93.6 (92.5) | 97.6 (91.5) | 88.7 (89.3) | 79.6 (80.0) |
| Redundancya | 8.6 (6.6) | 3.3 (3.3) | 3.5 (2.6) | 4.0 (3.9) | 4.2 (2.8) |
| Refinement | |||||
| Space Group | P212121 | P212121 | P212121 | P212121 | |
| Resolution (Å) | 47.3–1.38 | 48.8–1.42 | 48.8–1.40 | 48.8–1.42 | |
| Rworkc | 12.3 | 13.3 | 13.8 | 13.9 | |
| Rfree | 14.6 | 15.7 | 16.2 | 16.5 | |
| Rfactor (all data) | 12.5 | 13.4 | 13.9 | 14.0 | |
| Number of moleculesd | 2 | 2 | 2 | 2 | |
| Number of protein atomsd | 2690 | 2664 | 2583 | 2535 | |
| Number of solvent atomsd | 575 | 473 | 453 | 428 | |
| Average B factors | |||||
| (Å2) | 18.3 | 17.2 | 17.1 | 16.7 | |
| Protein atoms | 15.4 | 15.3 | 15.3 | 15.0 | |
| Solvent atoms | 29.8 | 27.4 | 26.9 | 27.0 | |
| Rmsd bond lengths (Å) | 0.02 | 0.02 | 0.02 | 0.02 | |
| Rmsd bond angles (deg) | 1.5 | 1.6 | 1.5 | 1.5 | |
The values in parentheses indicate statistics for the highest resolution shell.
Rmerge = Σ|I − <I>|/Σ<I>, where I is the observed intensity and <I> is the average intensity from multiple observations of symmetry-related reflections.
R = Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure amplitudes, respectively.
Per asymmetric unit.
The asymmetric unit of the crystal contains a symmetric homodimer, the core of which forms a four helix bundle as expected for a bacterial chemoreceptor (Hazelbauer et al., 2008). For each monomer within the homodimer, residues 40–203 are well ordered and are included in the final model (Figures 1B and S1A). Electron density is discernable but weak for additional residues at either end of the fragment, consistent with partial disorder, or fraying, of the helical termini of the cloned fragment.
The structure of the periplasmic domain of the TlpB monomer, in contrast to those of the S. typhimurium and E. coli aspartate chemoreceptors Tar and the E. coli serine receptor Tsr (Bowie et al., 1995; Kim et al., 1999; Milburn et al., 1991), consists of a PAS domain inserted into the second helix of the bundle (residues ~101–185, Figure 1B). The TlpBpp structure resembles that of two unpublished structures deposited in the Protein Data Bank (PDB) of the periplasmic domains of two putative MCPs of unknown function from Vibrio cholerae and Vibrio parahaemolyticus, both of which contain PAS domains bound to small molecules (Figure S1B, C and D). Interestingly, the putative MCP in Vibrio parahaemolyticus likely binds pyruvate and may be a pyruvate chemoreceptor. The TlpBpp PAS domain also nicely overlays with the PAS domain from photoactive yellow protein (PYP) (Figure S1E). For in-depth analysis and discussion of the structural overlays of TlpBpp with the two receptors and PYP, see Supplementary Experimental Discussion S1 and Supplementary Procedure S3.
Clearly evident in the final electron density maps of TlpBpp is a small, trigonal planar molecule bound within the PAS domain, which could be modeled as either urea or bicarbonate (Figure 1C). The ligand was positively identified as urea through an in vitro assay in which urea was released from denatured TlpBpp and detected by a colorimetric reaction (Figure S2A and B). This assay is specific to urea as other urea-like molecules do not react (Figure S2A). Using this assay, urea was determined to be present in roughly 1:1 molar stoichiometry to TlpBpp while control proteins (BSA and lysozyme) showed no detectable urea bound (Figure S2B). Urea was not added to any of the crystallization or protein purification buffers, so we presume it was scavenged from the medium during cell growth and/or lysis, indicating that TlpBpp binds urea with extremely high affinity. Attempts to prepare apo-TlpBpp have so far resulted in protein precipitation, suggesting that urea plays an important role in the stability of the protein fold. We were able to exchange the urea bound to TlpB with formamide via dialysis, which allowed us to measure a relative binding affinity of urea to TlpBpp. We dialyzed the formamide-bound TlpBpp against increasing concentrations of urea and determined the fraction of urea-bound TlpBpp. From these values we calculated a relative urea binding curve for TlpBpp with an apparent dissociation constant of 700 ± 300 nM (Figure S2C and Supplemental Experimental Procedure S4).
Urea binding in TlpBpp is coordinated by hydrogen bonds to four residues: Asp114, Tyr140, Tyr153 and Lys166 (Figure 1C). In this configuration, Asp114 must accept two hydrogen bonds from the amide nitrogens of the urea ligand and must therefore be negatively charged. Analysis of the total ligation shell allows all proton positions to be inferred and also suggests that Lys166 must be positively charged, leading to a urea ligation sphere with net charge of zero.
In solution, aspartate has a side chain pKa of about 4. We reasoned that at the range of pHs H. pylori experiences in the stomach (pH ~2–7), Asp114 in the urea binding pocket could switch between protonated and unprotonated forms. Protonation of Asp114 should in turn cause steric interference with the amide nitrogens of bound urea, perturb the binding of urea and alter the stability and structural state of TlpB. In this model, urea would function as an acid-sensing cofactor by altering the structural state of the protein in a pH dependent manner. We tested this model using thermal denaturation, mutation, and in vivo chemotaxis studies.
Urea binds with specificity and confers thermal stability to TlpBpp in a pH dependent manner
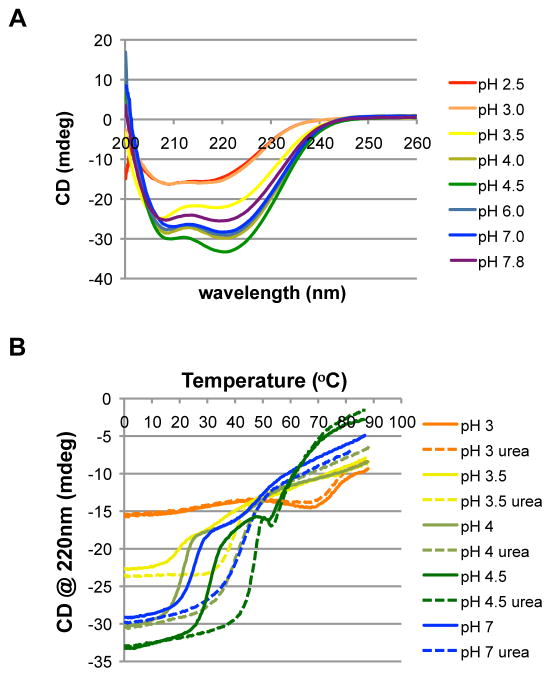
If urea were to function as a specific cofactor in the receptor response, then its binding should have a discernible effect on the structure or stability of the protein. To explore the thermal stabilization of TlpBpp in response to urea binding, we used circular dichroism (CD) spectroscopy to quantify secondary structure. To assess the effect of changes in pH on the secondary structure, CD spectra of TlpBpp were recorded from 260-200nm in buffers ranging from pH 2.5 to 7.8 at ~ 2°C (Figure 2A). The pronounced minima observed in the spectra near 220nm indicate predominately α-helical structure, consistent with the crystal structure, with the largest α-helical signal being observed at pH 4.5. This suggests that TlpBpp is most folded at pH 4.5, less folded at more neutral pHs and mostly unfolded at pH less than 3.0. Interestingly, pH 4.5 is near the middle of the pH range experienced in the stomach environment (Sachs et al., 2003) and close to the unperturbed pKa (4.0) of the Asp114 side chain. Consistent with our model, at pH less than 4, protonation of Asp114 would perturb the binding of urea in the PAS domain, in turn altering the thermodynamic stability of the periplasmic region of TlpB and thus leading to a change in the overall structure of the receptor.
Figure 2.
Urea thermally stabilizes TlpB in a pH dependent manner. (A) Circular dichroism spectra of TlpBpp with endogenous urea at multiple pHs reveals α-helical nature (spectra taken at 2ºC). TlpB concentration was 1 μM. (B) CD thermal melts of TlpBpp at multiple pHs. Dashed lines refer to samples with 5mM exogenously added urea. TlpB concentration for each curve was 1 μM. See also Figure S2.
To determine if urea binding affects the thermal stability of TlpBpp, we used a CD thermal denaturation assay. Although TlpBpp co-purifies with bound urea, we tested whether additional, exogenous urea would increase the melting temperature of TlpBpp. The α-helical content of TlpBpp was monitored by CD at 220nm for 1μM samples and slowly heated from 1.2°C to 86°C. In the absence of added urea, the CD measurements were consistent with two structural transitions, a fully reversible transition with an apparent Tm of about 20°C and a second, irreversible transition occurring at about 50°C. As the urea concentration increased over the range of 0 to 5000μM, the Tm of only the first transition increased (Figure S2D), and the overall curve shape resembled a single structural transition. Urea is therefore important for the thermal stability of TlpBpp. A control experiment using full length TlpD protein, a soluble H. pylori chemoreceptor, showed no thermal stabilization upon addition of 5000μM urea (data not shown).
The finding that the reversible transition at 20°C is stabilized by addition of urea is consistent with commonly observed protein-ligand systems that obey a simple equilibrium model (Bowie and Sauer, 1989; Celej et al., 2005a). According to this model, increasing temperature favors the unfolded, ligand free form of TlpBpp (U), thus reducing the concentration of folded TlpBpp (F) that is competent to bind urea (L) (Equation 1). However, specific ligand binding invariably stabilizes a protein toward thermal denaturation. As added urea shifts the binding equilibrium to favor bound TlpBpp (FL) relative to F, the U → F equilibrium in turn reduces U at any given temperature. Equation 1 can accommodate the pH dependence of urea binding by assuming that Kd depends on pH.
| Equation 1 |
We demonstrated that the stabilizing effect of urea is expected even for the case that TlpB affinity for urea is very high using computational simulations of a reversible denaturation process as described (Celej et al., 2005b). The free energy of unfolding of TlpB was calculated according to equation (14) of Rees & Robertson (Rees and Robertson, 2001). Figure S2 illustrates the simulated unfolding for TlpB ([TlpB] = 2 μM) with a melting transition for ligand free protein at an assumed Tm of 300K, for assumed binding constants ranging from “weak” Kd = 1 mM (Figure S2E), “moderate” Kd = 1 μM (S2F) to “tight” Kd = 1 nM (S2G). In all cases, as [urea] increases well above Kd, the apparent Tm increases as expected. Interestingly Figure S2G (Kd = 10−9 M) shows a bimodal transition, because at 1 μM total urea, the protein (2 μM) is half saturated with ligand. Unbound protein melts at the assumed, unperturbed Tm of 300 K while the bound species is stabilized by roughly 20 K. The results of these calculations are in accord with our experimental observations and a measured urea binding dissociation constant of ~700 nM (relative to formamide). See Supplemental Experimental Procedures for a more detailed discussion of Equation 1, together with supporting numerical simulations.
The experimentally derived CD melt data of Supplemental Figure S2D shows a strikingly similar bimodal transition to the calculated curve in Figure S2G. The upper transition in the experimental data, however, is not sensitive to the concentration of urea as seen in the simulation. We conclude that the higher-temperature transition, which is not reversible, represents a urea-independent unfolding transition of TlpBpp. One possible interpretation of the experimental data is that the PAS domain reversibly unfolds and that its stability depends on the presence of urea, whereas the four helix bundle formed by the dimer interaction unfolds at high temperatures, independently of the PAS domain.
To determine if urea stabilizes TlpBpp in a pH-sensitive manner, we performed thermal melts at five different pH values (pH 3–7) with and without 5mM urea added (Figure 2B). For all pH values except pH 3, the addition of 5mM urea stabilized TlpBpp by about 20°C and reduced the number of structural transitions from two to one. At pH 3, the CD spectra of TlpBpp showed no significant stabilization by urea, suggesting a denatured structure. The absence of a stabilizing effect of urea at pH 3 is consistent with a model wherein low pH environments lead to protonation of residue Asp114, resulting in a decreased affinity of TlpBpp for urea and structural destabilization of the protein.
As discussed above, exogenously added urea stabilizes the fold of TlpBpp against thermal denaturation. For urea to function as a cofactor in chemotactic signaling, its effects on TlpBpp stability should be dynamic and reversible. To test this, we asked whether the addition of urea could re-fold partially melted TlpBpp. The CD signal at 220nm was used to quantify the secondary structure of TlpBpp in a pH 4 buffer at 28°C, a point after the initial melting transition, but prior to loss of all α-helical signal (Figure 2B). Addition of 1mM urea to a partially denatured TlpBpp solution at 28°C restored the α-helical CD signal to greater than 95% of the maximum seen at 1.2°C (Figure S2H). When a second aliquot of urea was added for a final concentration of 2mM, the signal was restored to almost 100% of the maximum. The sample was then cooled to 1.2°C, urea was added to a final concentration of 5mM, and a second thermal melt from 1.2°C 86°C was performed (Figure S2I). The sample so treated behaved identically to a sample that had not been subjected to prior thermal cycling. Urea is thus capable of assisting the reversible re-folding of partially denatured TlpBpp.
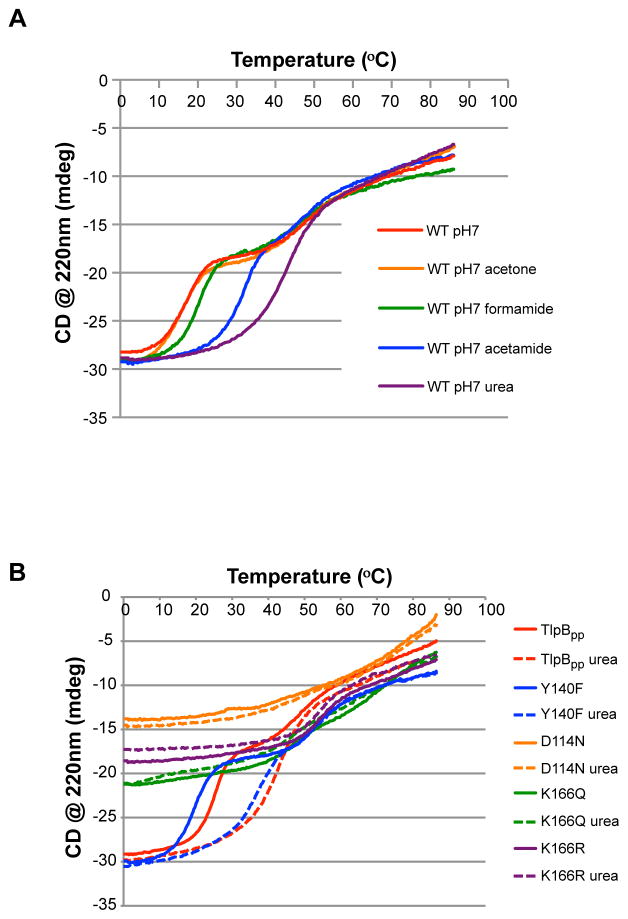
Next we explored the specificity of the stabilizing effect of urea by asking whether related small molecules could function similarly. We performed thermal denaturation assays on TlpBpp in the presence of 5mM acetone, formamide and acetamide (Figure 3A). The addition of acetone resulted in no change in the melting temperature of TlpBpp, while formamide and acetamide increased the melting temperature to a lesser extent than urea. These results demonstrate that urea specifically stabilizes TlpBpp, supporting the hypothesis that urea is the endogenous ligand of TlpB. In addition, we obtained crystal structures of TlpBpp with acetamide, formamide and hydroxyurea bound (Figure S3 and Table 1). Although these molecules do replace co-purified urea in the binding site when added at high concentrations, the electron density maps are consistent with some positional disorder compared to urea. To improve the fit with the X-ray data, the ligands were each modeled as a superposition of two copies, at 50% occupancy each. These compounds are unable to form as many specific hydrogen bonds as urea, explaining their decreased affinities.
Figure 3.
Urea stabilizes TlpBpp specifically, and mutations in the urea binding pocket of TlpBpp abrogate the structure of TlpBpp and urea’s thermal stability effect. (A) CD thermal melts of TlpBpp with urea-like compounds (acetone, formamide and acetamide) added or urea added exogenously (all chemicals added at 5mM). TlpB concentration for each curve was 1 μM. (B) CD thermal melts of TlpBpp wild type (TlpBpp), Y140F, K166Q, K166R and D114N with urea (dashed lines) or without 5mM urea (solid lines). All samples were at pH 7. See also Figure S3.
These studies demonstrate that TlpBpp binds urea specifically and that this binding reversibly stabilizes the fold of TlpBpp in a pH sensitive manner.
Engineered mutations disrupt the binding and thermal stabilization of TlpB by urea
We next verified that the side chains identified from the crystal structure as urea ligands are in fact required for urea binding and thermal stability of TlpBpp. Four mutant TlpBpp proteins were prepared with conservative substitutions (Y140F, K166Q, D114N and K166R), selected to support the three-dimensional structure of the protein but to modify the charge or hydrogen bonding capabilities within the binding site. The mole percent of urea that co-purified with the protein was determined in each case. Wild type TlpBpp was found to be approximately 72% saturated with urea, while TlpBpp Y140F contained only 11% bound urea (Figure S2B). Urea was not detectable in samples of the K166Q, D114N and K166R mutants. These latter three point mutations thus substantially decrease the affinity of TlpBpp for urea.
We also studied the effect of added urea on the thermal stability of these four TlpB mutants. The Y140F mutant, which contained a significant amount of urea when purified, behaved similarly to wild type TlpBpp. The CD signal was consistent with mostly α-helical structure and the addition of 5mM urea increased the melting temperature by approximately 20°C (Figure 3B). Note that in Y140F the substitution is not only very conservative in size and shape, but also does not affect the charge balance of the active site, consistent with the observation that the mutation did not significantly destabilize the protein. CD spectra of TlpBpp K166Q and D114N, however, in which the charge balance of the urea binding site is perturbed, indicated that these protein were significantly unstructured and they were not thermally stabilized by urea. TlpBpp K166R does not have a change in charge balance, however steric hindrance of a larger side chain likely contributed to the disrupted secondary structure and a decrease in urea stabilization. Strikingly, the secondary structure of TlpBpp D114N was the most disrupted, with the lowest amount of α-helical structure, and no thermally stability conferred by urea. The instability of these mutants supports the model that urea binding plays a critical role in the structural integrity of TlpBpp, with the D114N mutation, although extremely conservative from a structural point of view, being the most deleterious.
Urea binding mutants have reduced chemotactic responses to acid
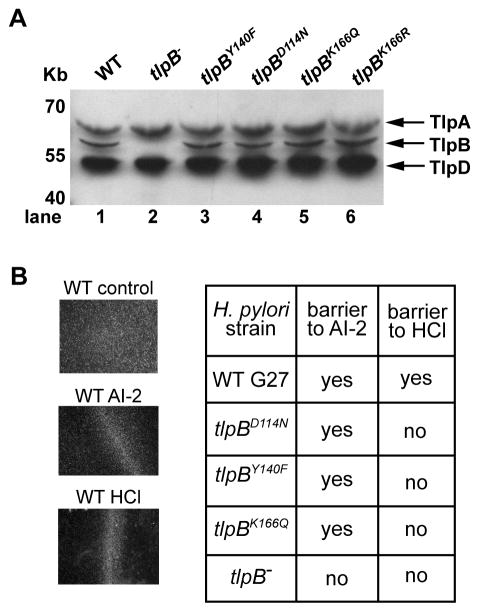
To test the model of urea acting as a cofactor in TlpB for acid sensing, we engineered H. pylori strains in which the wild type tlpB gene was replaced with each of the four urea-binding TlpB point mutations (Y140F, D114N, K166Q, and K166R). We confirmed that the mutant TlpB proteins were expressed at similar levels to wild type TlpB by western blot analysis (Figure 4A). This indicates that in the context of the full-length TlpB protein, the mutant periplasmic domains are folded and persist in the cell. We previously reported that TlpB is responsible for detecting the quorum sensing molecule AI-2 (Rader et al., 2011). As a further demonstration that the mutant proteins were expressed and retained some function, we found that strains tlpBY140F, tlpBK166Q and tlpBD114N in H. pylori retained responsiveness to AI-2, as assayed in a wet mount preparation in which motile barriers of responsive cells form upon addition of a repellent chemical (Figure 4B, Movie S1 and Figure S4A). The three strains failed to form barriers upon addition of HCl, indicating that they were defective in pH chemotaxis. The formation of a pH gradient in this assay was confirmed using the pH sensitive dye bromophenol blue (Figure S4B). The tlpBK166R mutant had an altered swimming behavior with an increased frequency of stopping and did not produce reliable barriers in response to AI-2 or acid. However, the mutant protein tlpBK166R was expressed at normal levels in H. pylori (Figure 4A) and the strain did respond to acid in the video chemotaxis assay, indicating TlpB was present and functional (see next section).
Figure 4.
H. pylori strains containing TlpB with urea binding point mutations show normal protein expression levels and remain responsive to AI-2. (A) Western blot of H. pylori G27 wild type (WT), tlpB knock out (tlpB−) or the four point mutants (tlpBY140F, tlpBD114N, tlpBK166Q, tlpBK166R) using an antibody to the common, cytoplasmic region among all four chemoreceptors. (B) Results for the chemotaxis barrier assay. Images on the left show that wild type (WT) H. pylori respond to AI-2 and HCl by forming a barrier. The chart on the right shows responses of wild type, the tlpB− strain and three of the TlpB urea binding mutant strains to AI-2 and HCl in the barrier assay. Images of AI-2 response to the five strains can be found in Figure S4A. See also Movie S1 and Figure S4.
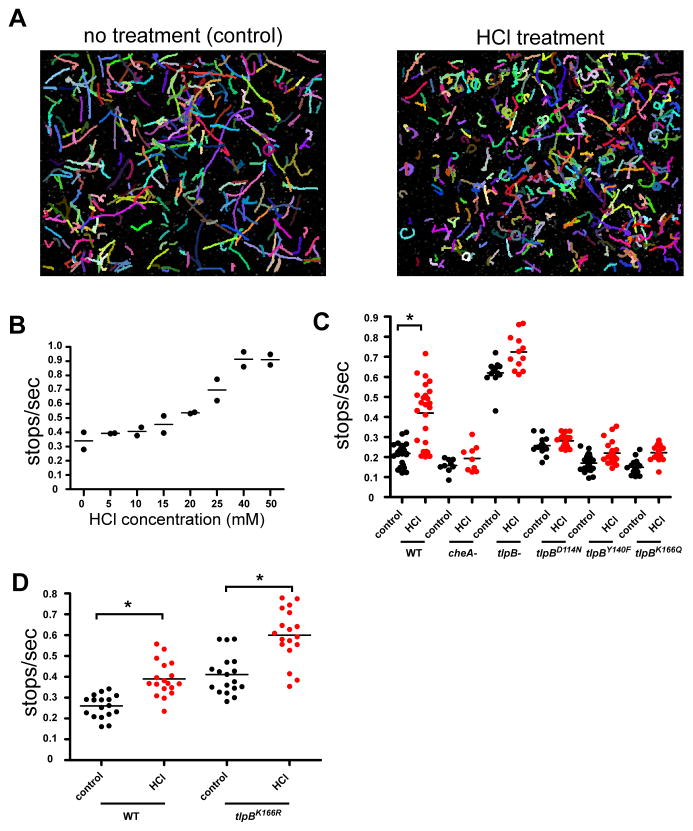
To further investigate how mutations in the urea-binding site of TlpB affected the ability of the protein to sense acid, we used a video chemotaxis assay from which we could quantify features of the cells’ swimming behavior. In the presence of chemical repellants, individual H. pylori stop more frequently and change direction. We incubated wild type and mutant bacteria with either no treatment (control) or HCl for approximately three minutes and then recorded 10 second videos. Using custom particle tracking software, the trajectories of individual H. pylori cells were determined and the stop rates were calculated (Figure 5). For each video, 40–80 tracks were followed and an average number of stops per second was determined (the averaged tracks for each video corresponds to one dot in Figure 5B–D). To identify a concentration of HCl that elicited a response without causing detrimental effects, we performed an HCl titration curve. With exposure to 25mM HCl, corresponding to a pH of approximately 4.5, we observed a marked increase in stop rate but no significant increase in the number of non-motile H. pylori (Figure 5B); this concentration was used in our subsequent video chemotaxis assays. We next compared the movement of wild type H. pylori to tlpBD114N, tlpBY140F and tlpBK166Q strains when exposed to control treatment or 25mM HCl (Figure 5C). Wild type cells responded to the acid treatment with roughly a two-fold increase in stops per second, whereas tlpBY140F, tlpBK166Q and tlpBD114N had no response to HCl. Control H. pylori strains entirely lacking TlpB (tlpB−) or the chemotrasnduction protein CheA (cheA−) had no response to HCl treatment. The overall increase in stops seen with the tlpB− strain and decrease in stops seen with the cheA− strain had been observed previously and are most likely due to increased or decreased activation of the chemotaxis pathway, respectively (Croxen et al., 2006; Rader et al., 2011). Collectively, these data suggest that residues of the TlpB PAS pocket bind urea and confer pH sensitivity.
Figure 5.
TlpB mutants in the urea binding site have reduced or no response to acid while a reengineered urea-mimetic mutant remains responsive to acid. (A) Video chemotaxis assay tracks of individual wild type H. pylori shown for 10sec videos with no treatment (control) or HCl treatment. The bacteria showed more linear motion in the control compared to more curving and pausing in the presence of HCl. (B) Results of the chemotaxis video assay for a range of different HCl concentrations using wild type G27 H. pylori. Each dot represents a single video with the stops/sec of each track averaged. (C) Results of the chemotaxis video assay performed with wild type H. pylori (WT), cheA knock out (cheA−), tlpB knock out (tlpB−), tlpBD114N, tlpBY140F and tlpBK166Q with and without HCl treatment. The HCl treatment results are shown as red dots, the control treatments as black dots. (D) Results of the chemotaxis video assay performed with wild type H. pylori (WT) and tlpBK166R. An asterisk in C or D indicates that the pair is statistically significantly different using ANOVA with Tukey’s pairwise comparisons (Alpha = 0.01).
Re-engineering the urea binding pocket of TlpB to create a new urea-independent mechanism of acid sensing
If, according to our model, urea occupies a pocket of the TlpB PAS domain and confers stability to the protein in a pH dependent manner, we reasoned that it should be possible to engineer the PAS domain pocket to contain a pH sensitive residue that would mimic urea. We replaced Lys166 with Arg, whose side chain is substantially longer than that of Lys and contains a terminal positively charged guanidinium moiety, which is similar in shape and hydrogen bonding capability to urea. Arg could thus act as a urea mimetic by extending into the urea pocket. This substitution is predicted to maintain the overall charge neutrality of the binding site, with Asp114 in the anionic form and may thus form an internal salt bridge with the carboxylate of Asp114 (Figure 1C), which would prevent urea binding. As predicted, urea did not copurify with TlpB K166R and the mutant protein was not thermally stabilized by urea (Figure 3B). However, when introduced into the H. pylori genome, the tlpBK166R gene conferred responsiveness to HCl treatment in the video chemotaxis assay (Figure 5D). The acid-responsiveness of the tlpBK166R strain is consistent with the introduced arginine residue contributing to a urea-mimetic acid sensing PAS domain.
Discussion
Before the Nobel-winning work of Marshall and Warren (Marshall and Warren, 1984), the human stomach was presumed to be a sterile environment. We now know that the bacterium, H. pylori, inhabits this organ where it can cause a spectrum of ailments. To navigate and persist within its chosen habitat, H. pylori must be able to sense and respond to its harsh acidic environment. A better understanding of H. pylori’s adaptations to life in low pH will propel the design of more effective antibiotic therapies to treat H. pylori infections and prevent infection-associated diseases.
By sequence analysis, H. pylori TlpB is organized like a typical member of the MCP superfamily of transmembrane receptors (Wuichet et al., 2007) with two transmembrane helices per subunit (tm1, tm2) bracketing an extracellular sensing domain. The extracellular sensing domain is responsible for detecting ligands directly or indirectly via interactions with periplasmic binding proteins. Continuing from tm2, the C-terminal portion of the MCP is cytoplasmic and mostly helical. It contains a HAMP domain, followed by a helical domain and a segment that binds to the CheA/CheW histidine autokinase complex (domains identified through ExPASY Prosite and NCBI Conserved Domain Database) (Marchler-Bauer et al., 2011; Sigrist et al., 2010). Phosphorylation of CheA in response to an extracellular signal in turn controls downstream components that modulate activity of the flagellar motor. MCPs dimerize at the membrane, forming a four helix bundle with ligand binding domains in the periplasm, while trimers of dimers assemble with CheA and CheW to form a high-performance signaling array (Briegel et al., 2009) located in the cytoplasm.
The S. typhimurium and E. coli aspartate (Tar) and E. coli serine (Tsr) chemoreceptors have long served as models for bacterial MCPs (Bowie et al., 1995; Kim et al., 1999; Milburn et al., 1991). Their periplasmic sensing domains consist of a four-helix bundle that dimerizes to form an intermolecular four-helix bundle interaction, with the ligand binding sites at the dimer interface. Recent evidence now suggests that this architecture is not the most common and that MCPs will more often incorporate PAS domains, a universal family of signaling modules (60–120 amino acids) found in all kingdoms of life (Glekas et al., 2010; Henry and Crosson, 2010; Taylor and Zhulin, 1999). In their roles as sensors, PAS domains specifically recognize chemically active or light-sensitive cofactors, relatively inert small molecules and/or mediate protein-protein interactions.
Until recently, PAS domains were thought to be present only in cytoplasmic proteins (Taylor and Zhulin, 1999) but have now been discovered in extracellular segments of bacterial receptors of the two-component histidine kinase family, such as the CitA receptor of Klebsiella pneumoniae (Reinelt et al., 2003) and the LuxQ quorum-sensing receptor from Vibrio harveii (Neiditch et al., 2006). Glekas et al. (Glekas et al., 2010) used a variety of techniques to convincingly argue that the periplasmic portion of the B. subtilis asparagine chemoreceptor McpB should contain two tandem PAS domains, one of which specifically binds asparagine. Furthermore, careful sequence analysis suggests that extracellular PAS domains have been systematically underpredicted (Chang et al., 2010). This report directly establishes the presence of an extracellular PAS domain (residues 99–185 of TlpB) in a chemotaxis receptor of known function.
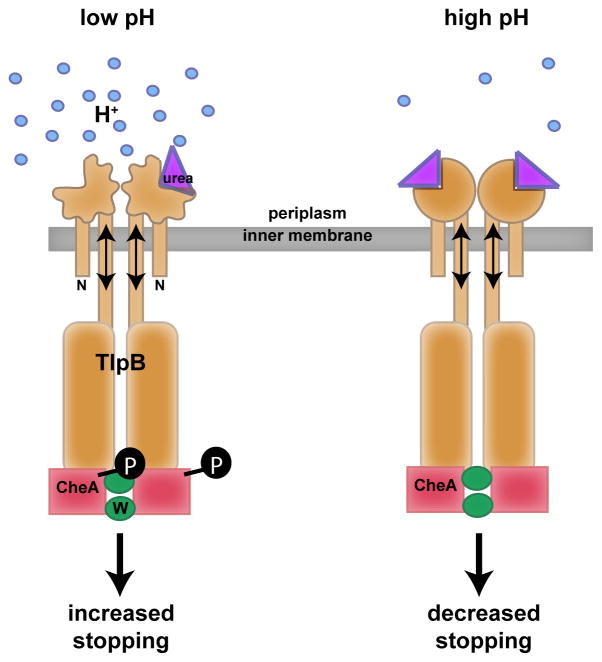
Model for pH sensing by TlpB
We presented evidence that a molecule of urea serves as a cofactor for detection of changes in pH by the PAS domain in TlpB. At pH near neutrality, Asp114 is negatively charged and accepts two hydrogen bonds from the amide nitrogen atoms of urea, which is thus tightly bound and stabilizes the fold of the PAS domain. However, at low pH, we predict that protonation of Asp114 disrupts or weakens urea binding, leading to a structural transition within the PAS domain (Figure 6). We demonstrated that urea stabilizes TlpBpp against thermal denaturation in a pH sensitive fashion and showed that mutagenesis of three urea ligands (Asp114, Lys166 and Tyr140) disrupt this interaction. Our model is informed by in vitro experiments on the periplasmic domain of TlpB and mutational analysis of the full length protein in vivo. We observe some difference between these two systems. For example, whereas TlpBpp D114N is very unstable in solution, the full length protein is present at normal levels in the cell. Also, although TlpBpp Y140F is only partially impaired for urea binding, the mutant protein is completely unable to mediate a chemotaxis response to acid in vivo. Additionally, one might expect increased basal levels of stopping in mutant H. pylori containing TlpB proteins with impaired urea binding, mimick the protonated state, which we do not observe. Predicting basal stopping frequency, however, is difficult because it is dictated by the sum of all chemoreceptor activities and is sensitive to the levels of TlpB, as seen by the high basal levels of the tlpB deficient cells (Figure 5C and Rader 2011).
Figure 6.
Model for how TlpB senses acid. In low pH conditions (shown on the left), TlpB’s periplasmic domain is in a “relaxed” or expanded state due to decreased hydrogen bonding to urea and consequent lowered urea binding affinity. However, in high or more neutral pH conditions, TlpB’s periplasmic domain is in a “tense” or condensed state with increased urea binding affinity. The state of the periplasmic domain is relayed through the transmembrane region which affects CheA’s phosphorylation state, ultimately affecting the flagellar motor and dictating stopping behavior. TlpB dimer (orange), CheA (red), CheW (green), urea (purple), protons (blue), phosphate (black).
Our model predicts that the signaling state of TlpB should be sensitive to changes in structure or charge associated with key urea ligands. Indeed, the conservative point mutation D114N, which closely mimics the protonated state of Asp114 by substitution of NH2 for O−, is remarkably deleterious to both the activity and structural integrity of TlpBpp. Even in the presence of 5mM urea, CD spectra indicate that TlpBpp D114N loses a large fraction of the wild-type helical signal, suggesting that it is not properly folded. Likewise, the substitution K166Q eliminates the positive charge and strongly destabilizes the fold of TlpBpp. In contrast, the conservative substitution Y140F, which substitutes H for OH and substantially lowers the affinity for urea, behaves similarly to TlpBpp with respect to thermal denaturation. The Y140F substitution does not change the charge balance in the ligand binding site and presumably for that reason is less disruptive of the structure. However, urea affinity may be reduced to the point that under the conditions of the in vivo assays, the binding site of Y140F is not fully occupied by urea and is thus unable to signal a response to acid treatment.
Key side chains that interact with the urea ligand can thus exert profound influence on the stability and structure of the PAS domain, and link the structural transition to the state of ligation. In this process, charge neutrality of the binding site appears to be an important determinant for structural stability. We attempted to capture different signaling states by crystallizing TlpBpp at different pHs, however all structures were the same (with urea bound), most likely due to crystal packing constraints (data not shown).
Our biophysical studies were conducted with a soluble fragment of TlpB. Since it can be argued that these measurements are not representative of transitions that take place in the intact receptor, we showed that when these same mutations are introduced into the intact tlpB gene, they interfere with or abrogate the chemotactic response of live bacteria to acidification of the medium. Furthermore, we demonstrated that urea is dispensable as a cofactor by replacement of Lys166 with arginine, the terminal guanidinium moiety of which resembles the shape of the urea cofactor but maintains the positive charge associated with lysine. We showed that although TlpBpp K166R does not bind urea, bacteria expressing tlpBK166R are capable of mounting a chemotaxis response to acid in vivo.
Mechanisms of signal transduction
What is the structural signal generated by the PAS domain and how is this signal transmitted through the membrane? Extensive studies of transmembrane receptors of the chemotaxis and histidine kinase families have led to the generally accepted view that signal transmission across the membrane consists of a piston-like motion of transmembrane helix 2 (tm2) relative to tm1, with amplitude 1–2Å (Chervitz and Falke, 1996; Cheung and Hendrickson, 2009; Falke and Erbse, 2009; Moore and Hendrickson, 2009; Ottemann et al., 1999). Within the cytoplasm, the HAMP domain evidently constitutes a switch region that translates the piston-like motion into a different type of transition within the distal portions of the receptor (see recent summary by Watts et al. and references therein (Watts et al., 2011)).
In TlpB, the PAS domain is inserted into the second helix of the bundle such that the C-terminal strand of the central β-sheet is directly connected to a helix (Figure 1B). Presuming this helix is continuous with tm2 in the intact receptor; motions of the β-strand could thus induce a piston-like displacement of tm2 in either direction. The packing of the four-helix bundle that makes up the dimer interface of TlpB differs from that observed in Tsr and Tar. Within the E. coli Tar chemoreceptor, transmembrane helix tm1 interacts more closely with tm1′ (Pakula and Simon, 1992) whereas within TlpB, the two helices that interact most closely correspond to tm1 and tm2′ (Figure 1B), possibly indicating cooperativity between the monomers. In the E. coli aspartate receptor, tm2 has been shown to move “down” (toward the cytoplasm) in response to attractant (Chervitz and Falke, 1996). However, ligand-induced motions of tm2 in both directions have been observed in structural studies of the NarX (Cheung and Hendrickson, 2009) and TorS (Moore and Hendrickson, 2009) receptors, which are similar in secondary structure and organization to the Tar aspartate receptor. Similar rearrangements have been observed for transmembrane receptors based on PAS domains. For example, crystallographic studies of the periplasmic domain of the CitA histidine kinase receptor revealed a large contraction of the PAS domain upon binding citrate, which causes the central β-sheet to repack, moving tm2 “down” (toward the cytoplasm) (Sevvana et al., 2008).
Similarly, our model for low pH-induced changes in urea ligation within TlpB suggests a loosening of the urea binding site and/or partial unfolding of the PAS domain, due to protonation of Asp114 and consequent steric interference with bound urea. The proposed rearrangement of the PAS domain is likely to result in movement of tm2 relative to tm1 (Figure 6). Consistent with this latter scenario, single molecule studies of the PYP blue light photoreceptor suggested partial unfolding and expansion of the PAS domain is response to absorption of a photon, which acts as a repellent (Zhao et al., 2006).
The emerging models of chemoreceptor signaling pose that local conformational changes, such as protonation events or ligand binding, lead to global conformational changes that relay signals. Our data suggest a model that urea acts as a pH-sensing cofactor to confer structural stability within TlpB which is consistent with current models and provides new insight into how chemoreceptors can sense their environment.
Experimental Procedures
Protein purification methods (WT, D114N, K166Q, Y140F, K166R)
TlpB full length gene was PCR amplified from genomic SS1 H. pylori (Lee et al., 1997) DNA and cloned into pBH4 (Prehoda et al., 1999), a derivative of the pET19-b vector (Novagen). The periplasmic portion (amino acids 33–211) of TlpB was subcloned from the above plasmid into pBH4.
TlpB 33–211 (TlpBpp) was cloned using the forward primer 5′ AAA CCGGGA TCC AAG GTT ATG CAA AAA GAT GTG 3′ (which contains a Bam site, GGATCC) and the reverse primer with stop codon 5′ AAA CCG CTC GAG TTA TAG TCT TGT GGT GTT TTC 3′ (which contains a Xho site, CTCGAG) into a Bam/Xho-linearized pBH4 plasmid. The mutants were made using primers containing the mutation and PCR. The plasmid was transformed into BL21 (DE3) E. coli cells. The E. coli culture was grown at 37°C until OD600 0.5, then moved to 16°C until culture reached an OD600 0.6–0.9. N-terminal, 6X His-tagged protein expression was induced with 0.25mM IPTG and grown 16hrs at 16°C. All subsequent steps were performed at 0–4°C. Cells were lysed in lysis buffer (10mM KH2PO4, 300mM NaCl, 10mM imidazole, 1mM DTT, pH 7.8–8.0), sonicated and debris pelleted. The supernatant was mixed with 5–6mL Ni-NTA resin that was pre-washed with lysis buffer. The supernatant and resin were incubated at 4°C for 1.5–2hrs followed by centrifugal washing of the resin three times with 10X bed volume of lysis buffer. The resin was transferred to a plastic column, washed with 10X bed volume of lysis buffer, then 10X bed volume of lysis buffer containing 30mM imidazole, and finally eluted with lysis buffer containing 300mM imidazole. The high absorbance (280nm) fractions were pooled and cleaved with TEV protease for 16hrs during dialysis into 10mM KH2PO4, 300mM NaCl and 2mM DTT. The His tag was removed by passing the dialyzed sample through a second Ni-NTA column. Purified TlpBpp was stored in 300mM NaCl, 10mM KH2PO4, pH 7.8 at 4°C.
Wild type TlpB and K166R are soluble while D114N, Y140F and K166Q are less soluble (<50% found in the supernatant compared to the pellet during purification).
Crystallization, derivatization and diffraction data collection
See Supplementary Information. The coordinates for all four structures have been deposited in the RCSB Protein Data Bank. TlpBpp with urea bound is 3UB6, TlpBpp with hydroxyurea bound is 3UB9, TlpBpp with acetamide bound is 3UB7 and TlpBpp with formamide bound is 3UB8.
Data reduction and structure determination
See Supplementary Information and Table 1.
Reassessment of Protein Data Bank entry 2QHK
See Supplementary Information and Figure S1. The new coordinates for the reassessed 2QHK structure have been deposited in the RCSB Protein Data Bank with an ID code of 4EXO.
Determining an apparent binding affinity of urea to TlpBpp using equilibrium dialysis
Thermal stability assays and secondary structure analysis using circular dichroism
Circular dichroism (CD) was done using a Jasco model 810 spectrometer equipped with a PDF-425S thermal controller. CD spectra were measured to identify secondary structure for TlpBpp (20μg/mL protein, 10mM KH2PO4, pH 2.5–7.8) using a quartz cuvette (QS-111 from Hellma). Spectra were recorded for each sample from 260-200nm at 1.4–2.8°C using a bandwidth of 1nm and a response time of 16sec at a scan speed of 10nm/min.
CD monitored thermal denaturation profiles were collected at 220nm using a J-810 spectrometer and a QS-111 cell with both in-cell stirring and an in-cell thermal probe. The samples were heated at 2°C/min from 1.2–86°C. The CD signal was recorded every 0.2°C. TlpBpp and TlpD samples were at 20μg/mL in 300mM NaCl, 10mM NaH2PO4, pH 3–7, with or without 5mM urea, acetone, formamide, or acetamide.
The semi-melt and urea re-melt experiments were also performed using a J-810 spectrometer. Briefly, 20μg/mL of TlpBpp was heated at 2°C/min from 1.2°C to 28°C (partially melted). Urea was spiked in to a final concentration of 1mM and the CD signal at 220nm was recorded for 1600sec at a constant temperature of 28°C. Then, urea was spiked in to a final concentration of 2mM and the CD signal at 220nm was recorded until the CD signal reached a plateau. Urea was spiked in to a final concentration of 5mM, cooled to 2°C and then the sample was heated at 2°C/min from 2–86°C. The CD signal at 220nm and the temperature of the solution was recorded every 0.2°C.
In vitro urea binding assay
To determine the % urea bound per mole of TlpB, we used a colorimetric assay (U. S. patent # 4,273,556 (Gindler, 1981)). Briefly, reagent A (0.195g o-phthaladehyde, 91.9mL water, 8.1mL concentrated H2SO4) and reagent B (1.64mL concentrated H2SO4, 2.63g boric acid,1.64g chromotropic acid) were made fresh and mixed 1:1. A urea standard curve was generated from 0–350μM (see Figure S2A) and measured simultaneously with the protein samples. Urea standards and protein samples (100μL) were mixed with 1.0mL of A+B mixture, incubated at 37°C for 1hr, then briefly centrifuged to clarify. The absorbance at 450nm was read for each sample. The mole of urea per mole of TlpB monomer was calculated based on the standard curve and presented as percentages (see Figure S2A and S2B).
Western Blots
H. pylori strains (G27 wildtype (Covacci et al., 1993), mG27 tlpB::cat (Williams et al., 2007), G27 tlpBY140F, G27 tlpBD114N, G27 tlpBK166Q, G27 tlpBK166R) were grown from plate in BB10 (Brucella Broth, 10% FBS) for 16hrs shaking at 37°C in 10% CO2 to an OD600 > 1.0. The strains were diluted to OD600 1.0, loaded onto a standard 10% SDS PAGE gel and transferred to PVDF. The membrane was blocked and then probed with a rabbit polyclonal antibody to GST-TlpA22 to the common CheW binding region among TlpA, B, C and D at 1:2000 dilution for 16hrs (Williams et al., 2007).
Barrier formation assay
The barrier assays were performed as described in Rader et. al (Croxen et al., 2006; Rader et al., 2011), with the following changes. G27 wildtype, mG27 tlpB::cat, G27 tlpBY140F, G27 tlpBD114N, G27 tlpBK166Q and G27 tlpBK166R strains were grown from plate in BB10 for 6hrs, shaking at 37°C in 10% CO2. 8μL of culture were spotted onto the center of a glass slide and covered with a 1mm thick, 22 × 22mm cover slip. Three sides were sealed with clear finger nail polish and the fourth side was sealed until approximately a 10mm opening remained. 8μL of DPD (100μM in BB10, Omm Scientific), BB10, or HCl (100mM in BB10) was gently ejected into the open side of the cover slip. After 5–10min of incubation, to allow slight mixing to cease, slides were visualized and still images were taken.
Video chemotaxis assay
The stopping rate of motile bacteria was calculated from 10sec videos of HCl treated and untreated cultures of H. pylori. H. pylori were grown in BB10 from plate for 3–5hrs shaking in a 37ºC/10% CO2 incubator. 5μL of H. pylori culture was added to 5μL of 50mM HCl in BB10 or BB10 on parafilm and mixed by pipeting up and down, and 2μL was transferred to a 10-well microscope slide (MP Biomedicals). A cover slip was placed on top and after a 3min incubation, videos were taken through a 20X objective at 15 frames per sec using a Cohu CCD camera (model 4912) and USBVision Capture software. Videos were analyzed using custom particle tracking programs written in MATLAB. Analysis was performed on inverted image frames (white bacterial particles on black background) of 702 × 480pixels where 1pixel equaled 0.86μm. Background and non-motile particles were digitally removed by subtracting the average image intensity, integrated over the entire movie, from each frame. Tracked particles were identified as local intensity maxima in high-pass filtered and thresholded images, with a filter size of 8pixels and an intensity threshold of 95%. Each particle’s position was assigned to be the centroid of the thresholded object. The bacterial length (approx. 3μm) spans several pixels, and so centroid-finding provides a measure of localization that is sufficiently precise relative to bacterial size. Objects were linked into tracks by identifying the nearest object in the subsequent frame of the movie. Tracks were analyzed for stops only if they were greater than 1sec in duration and had a frame-to-frame variance greater than 1pixel. A bacterium was considered to have “stopped” if its velocity dropped below 1.25pixels/frame for two consecutive frames, while otherwise having a velocity of at least 1.75pixels/frame for at least two consecutive frames. The overall stopping rate (stops/sec) was assessed over the duration of the movie. This approach was tested by comparing computer determined stops with those identified using the judgment of an experienced H. pylori experimenter. The videos for each strain and treatment combination were performed with cultures from a minimum of three different days and no more than three videos taken per culture. The average stops/sec were graphed using Prism (Graphpad). The statistics were performed using ANOVA with Tukey’s pairwise comparison (Alpha = 0.01). Statistical analyses were performed using the JMP Pro 9 software package.
Supplementary Material
Highlights.
The crystal structure of TlpB reveals a periplasmic urea bound PAS domain
Urea binds TlpB with high affinity and stabilizes TlpB in a pH dependent manner
Mutants with defective urea binding by TlpB have decreased responses to acid in vivo
A urea-mimetic PAS domain mutant retains acid-responsiveness
Acknowledgments
We would like to thank Dr. Brian Matthews and Dr. Walter Baase for CD expertise and use of the J-810 spectrometer. We would also like to thank Dr. Karen Ottemann for reagents and Dr. Khoosheh Gosink and Karen Kallio for useful advice on experimental design and suggestions on the manuscript. Dr. Rick Faber was instrumental in designing a high-throughput method for analyzing the chemotaxis videos. Finally, we would like to thank the anonymous reviewers whose insightful comments have significantly improved the manuscript. This work was supported by Public Health Service grant R01 DK075667 (to K.G.) from the National Institutes of Health and National Research Service Award A1091098-02 (to E.G.S.) from the National Institutes of Health and MCB 208731 (to S.J.R.) from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bowie JU, Pakula AA, Simon MI. The three-dimensional structure of the aspartate receptor from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1995;51:145–154. doi: 10.1107/S0907444994010498. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celej MS, Dassie SA, Freire E, Bianconi ML, Fidelio GD. Ligand-induced thermostability in proteins: thermodynamic analysis of ANS-albumin interaction. Biochim Biophys Acta. 2005a;1750:122–133. doi: 10.1016/j.bbapap.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Celej MS, Fidelio GD, Dassie SA. Protein Unfolding Coupled to Ligand Binding: Differential Scanning Calorimetry Simulation Approach. J Chem Ed. 2005b;82:87–92. [Google Scholar]
- Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Pokkuluri PR, Szurmant H, Schiffer M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. Journal of bacteriology. 2010;192:1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. Journal of bacteriology. 2006;188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S, Dorrell N, Ward SJ, Stabler RA, McColm AA, Rycroft AN, Wren BW. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindler EM. Determination of Urea. Sherwood Medical Industries Inc; St. Louis, Mo: 1981. U.S. Patent, ed. [Google Scholar]
- Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J Biol Chem. 2010;285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DY. Efficient identification and evaluation of effective Helicobacter pylori therapies. Clin Gastroenterol Hepatol. 2009;7:145–148. doi: 10.1016/j.cgh.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Ligand Binding PAS Domains in a Genomic, Cellular, and Structural Context. Annu Rev Microbiol. 2010 doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. ChePep Controls Helicobacter pylori Infection of the Gastric Glands and Chemotaxis in the Epsilonproteobacteria. MBio. 2011:2. doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M, Macnab RM. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. Journal of bacteriology. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Lee A, O’Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- Lowenthal AC, Simon C, Fair AS, Mehmood K, Terry K, Anastasia S, Ottemann KM. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology. 2009;155:1181–1191. doi: 10.1099/mic.0.021857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BJ, Goodwin CS, Warren JR, Murray R, Blincow ED, Blackbourn SJ, Phillips M, Waters TE, Sanderson CR. Prospective double-blind trial of duodenal ulcer relapse after eradication of Campylobacter pylori. Lancet. 1988;2:1437–1442. doi: 10.1016/s0140-6736(88)90929-4. [DOI] [PubMed] [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun. 2005;73:1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science (New York, NY. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Moore JO, Hendrickson WA. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure. 2009;17:1195–1204. doi: 10.1016/j.str.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Muller S, Gotz M, Beier D. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One. 2009;4:e6930. doi: 10.1371/journal.pone.0006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr A piston model for transmembrane signaling of the aspartate receptor. Science (New York, NY. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Simon MI. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc Natl Acad Sci U S A. 1992;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003;278:49523–49529. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellant via the chemoreceptor TlpB. Microbiology. 2011 doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Robertson AD. Some thermodynamic implications for the thermostability of proteins. Protein Sci. 2001;10:1187–1194. doi: 10.1110/ps.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- Sachs G, Weeks DL, Melchers K, Scott DR. The gastric biology of Helicobacter pylori. Annu Rev Physiol. 2003;65:349–369. doi: 10.1146/annurev.physiol.65.092101.142156. [DOI] [PubMed] [Google Scholar]
- Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A. 2004;101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Shanks AM, El-Omar EM. Helicobacter pylori infection, host genetics and gastric cancer. J Dig Dis. 2009;10:157–164. doi: 10.1111/j.1751-2980.2009.00380.x. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney EG, Guillemin K. A gastric pathogen moves chemotaxis in a new direction. MBio. 2011:2. doi: 10.1128/mBio.00201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K, Williams SM, Connolly L, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresguerres M, Buck J, Levin LR. Physiological carbon dioxide, bicarbonate, and pH sensing. Pflugers Arch. 2010;460:953–964. doi: 10.1007/s00424-010-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura T, Matsumoto Y, Ohnishi K, Homma M, Kawagishi I. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J Biol Chem. 2002;277:1593–1598. doi: 10.1074/jbc.M109930200. [DOI] [PubMed] [Google Scholar]
- Watts KJ, Johnson MS, Taylor BL. Different conformations of the kinase-on and kinase-off signaling states in the Aer HAMP domain. Journal of bacteriology. 2011;193:4095–4103. doi: 10.1128/JB.01069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JM, Lee H, Nome RA, Majid S, Scherer NF, Hoff WD. Single-molecule detection of structural changes during Per-Arnt-Sim (PAS) domain activation. Proc Natl Acad Sci U S A. 2006;103:11561–11566. doi: 10.1073/pnas.0601567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.