Abstract
Bla g allergens are major targets of IgE responses associated with cockroach allergies. However, little is known about corresponding T cell responses, despite their potential involvement in immunopathology and the clinical efficacy of Specific ImmunoTherapy (SIT). Bioinformatic predictions of the capacity of Bla g 1, 2, 4, 5, 6, and 7 peptides to bind HLA DR, DP and DQ molecules, and PBMC responses from 30 allergic donors, identified 25 T cell epitopes. Five immunodominant epitopes accounted for over half of the response. Bla g 5, the most dominant allergen, accounted for 65% of the response, and Bla g 6 accounted for 20%. Bla g 5 induced both IL-5 and IFN-γ responses, while Bla g 6 induced mostly IL-5 and, conversely, Bla g 2 induced only IFN-γ. Thus, responses to allergens within a source are independently regulated, suggesting a critical role for the allergen itself, and not extraneous stimulation from other allergens or co-presented immunomodulators. In comparing antibody with T cell responses for several donor/allergen combinations we detected IgE titers in the absence of detectable T cell responses, suggesting that unlinked T-B help might support development of IgE responses. Finally, SIT resulted in IL-5 down-modulation, which was not associated with development of IFN-γ or IL-10 responses to any of the Bla g derived peptides. In summary, the characteristics of T cell responses to Bla g allergens appear uncorrelated with IgE responses. Monitoring these responses may therefore yield important information relevant to understanding cockroach allergies and their treatment.
INTRODUCTION
Allergy to cockroach, a significant health problem worldwide, is associated with urban development, and often inner city environments, and clinical consequences. A number of cockroach proteins are potent environmental aeroallergens. There is evidence that early life exposure (1) to cockroach allergens leads to allergic sensitization to cockroaches, which has been shown to have a strong correlation with the incidence of asthma (2), particularly in children (3–5), and asthma exacerbations (3, 6). Childhood sensitization to cockroach allergens also has been associated with an increased risk for persistent asthma and bronchial hyper-responsiveness and with a greater loss of lung function (7). In general, cockroach allergens are an important cause of asthma exacerbations in many parts of the world (8, 9).
At the immunological level, cockroach allergies are mediated by both humoral and cellular responses (10–14). Regarding humoral responses, IgE levels (measured by RAST or skin test) against cockroach allergens are highly correlated with clinical allergic status, and commonly utilized in the diagnosis of cockroach allergies. By comparison, much less is known regarding the role of T cells in allergy and asthmatic reactions to cockroach antigens (15).
Both German (Blattella germanica) and American (Periplaneta americana) cockroach species can induce allergic responses, although the German cockroach (CR) is most frequently associated with severe clinical allergy in the USA, and American cockroach is associated with allergies in tropical areas (4). Allergen proteins expressed by the two species are highly homologous. Several different allergens have been identified on the basis of their reactivity with IgE from allergic patients, and their sequences have been determined (4, 16). These allergens include the Bla g 1, 2, 4, 5, 6, and 7 allergens. Indeed, the study of the patterns of serological reactivity to these antigens has contributed to definition of their relevance in cockroach allergy, which can aid in the design of diagnostics and immunotherapeutics (17–23).
By contrast to this wealth of information on antibody responses, no T cell epitopes have been defined for any of the Bla g allergens, and the frequency, phenotype and specificity of T cell responses is unexplored. Specifically, the pattern of immunodominance of T cell responses is unknown, and it is also unknown whether T cell responses correlate with IgE responses. These knowledge gaps are particularly relevant because of the potential role of T cells in both the development of cockroach allergies and in the efficacy of cockroach specific immunotherapy (SIT).
A key issue, still the object of much debate, is whether both generation of IgE responses and SIT is mediated by linked or unlinked T-B cooperation at the level of individual allergenic proteins. That is, whether induction of T cell responses against one particular allergen can provide help for IgE responses directed against a different allergen, or is help restricted to the IgE response to the same allergen. Modulation of T cell responses may be able to act in an unlinked mode, if the two allergens are both present in the same allergy inducing substance. Recent clinical trials have reported some successful results from SIT regimens utilizing one or few recombinant antigens for the treatment of allergic symptoms caused by complex allergens, suggesting that unlinked mechanisms may indeed play a role in SIT clinical efficacy (24). In the context of cockroach allergies, it is unknown whether the same allergens are recognized by T cell and humoral responses, and whether it is necessary that IgE producing B cells receive help from T cells specific for the same allergens, or whether unlinked help also contributes to the generation of responses.
Cockroach immunotherapy is not commonly used and reports on its effectiveness are very limited (25, 26). Little is known regarding the immunological basis for its clinical efficacy. In the case of other allergens, several non mutually exclusive mechanisms have been proposed, including: 1) induction on IgG antibodies that can prevent the allergenic effects caused by IgE or block IgE facilitated allergen uptake and presentation and 2) inhibition of Th2 responses by modulation of T cell responses, either by altering the Th1/Th2 balance, or induction of IL10-producing regulatory T cells (27, 28). Inhibition of Th2 responses would lead to eventual decrease in IgE titers. Indeed, it has been proposed that induction of IL10 producing Tregs by the subcutaneous (s.c.) administration of allergen extract might be responsible for the clinical benefit (29, 30). Furthermore, in the timothy grass (TG) Phl p 1 system (31) the majority of allergen-specific T cell clones raised before SIT revealed a Th2-like pattern of cytokine production, while those established after SIT revealed Th1 characteristics. Previous work from our group in the TG allergen system delineated frequently recognized epitopes associated with ten major known TG allergens (32). When individuals that had undergone SIT were compared to individuals that were allergic to TG, but were not SIT treated, we detected a generalized decrease in Th2 responses and no increase in either Th1 or IL-10 responses. In this context the study of Bla g specific T cell responses is of obvious interest to examine whether these potential mechanisms are also associated with successful SIT treatment for cockroach allergies.
In the present study we identify T cell epitopes derived from Bla g antigens, and use them to characterize T cell responses in allergic individuals before SIT treatment and after reaching the SIT maintenance phase. The results reveal that Bla g T cell responses are associated with strong patterns of immunodominance and immunoprevalence, with the Bla g 5 and Bla g 6 allergens being most dominantly and prevalently recognized. Furthermore, different allergens are associated with unique patterns of lymphokine polarization, with Bla g 2 being associated with responses strongly polarized toward Th1-, and Bla g 6 with responses strongly polarized toward Th2. Interestingly, the pattern of T cell reactivity to specific Bla g proteins at the level of individual donors frequently did not correlate with IgE responses, suggesting that T cell responses might regulate antibody responses in an unlinked fashion. Finally, longitudinal analysis of samples before and after establishment of the SIT treatment revealed a marked downregulation of Th2 responses to the Bla g allergens, which was not associated with a concomitant increase in Th1 or IL10 producing T cells. In conclusion, these data suggest that T cell responses to Bla g allergens have important distinguishing features from IgE responses to the same allergens, and that more in depth studies of these responses might yield important information relevant to our understanding of cockroach allergies.
MATERIALS AND METHODS
Patient donor population
Patient recruitment for this study was performed under three IRB protocols. The first two were conducted at Johns Hopkins University, one as part of the NIAID-funded Inner City Asthma Consortium (ICAC) (NIAID Protocol Number ICAC-18), and the second NIAID sponsored, but separate from ICAC. All participants came from the Baltimore area, were aged 18–55 and had a history of allergic rhinitis and/or asthma, and sensitivity to cockroach. Thirty individuals provided 100 ml of blood for PBMCs and 20 ml serum samples. A subset of 9 study participants provided samples both prior to and 6 months after the initiation of subcutaneous immunotherapy for German cockroach (SCITCO), after receiving biweekly dose escalations for 11–12 weeks followed by 14 weeks of weekly maintenance injections. Clinical case histories and other information were collected and recorded by the local clinical investigators. IgE specific for German CR extract was used to determine sensitivity to German CR. For a subset of patients that received immunotherapy, skin test reactivity to German CR was also performed by standard methods and both wheal (mm) and flare (mm) reactions were measured.
The third group of study participants (n=4) were from the greater San Diego area, and were recruited under LIAI protocol VD1-059-0311, with Institutional Review Board approval (Federal Wide Assurance #00000032). Informed consent, study ID numbers, clinical case histories and other information were collected and recorded by clinical investigators. Skin test reactivity to a panel of extracts from 32 common allergens, including German CR, was determined by standard methods. Both wheal (mm) and flare (mm) reactions were measured. All volunteers were asked to provide a 5 ml serum sample and 400 ml peripheral blood. IgE specific for German CR extract was also determined in this patient cohort.
Bioinformatic analyses
Nine Bla g allergen sequences, including isoforms, were considered and scanned for unique 15-mer peptides (UniProt ID: O96522, P54958, P54962, O18598, Q9NG56, Q9UAM5, Q1A7B3, Q1A7B2, Q1A7B1). Additional variants of these allergens are known, especially for Bla g 4, which has very frequent sequence variations that are quite disparate in discrete regions, with 0–32 substitutions (82.4–100% identity) (33, 34). However, we have limited the present analysis to include only those sequences found in the IUIS database. Each peptide was predicted for the capacity to bind to a panel of 20 HLA class II alleles (DPA1*0103/DPB1*0201, DPA1*0201/DPB1*0101, DPA1*0201/DPB1*0501, DPA1*0301/DPB1*0402, DQA1*0101/DQB1*0501, DQA1*0301/DQB1*0302, DQA1*0401/DQB1*0402, DQA1*0501/DQB1*0301, DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0405, DRB1*0701, DRB1*0802, DRB1*1101, DRB1*1302, DRB1*1501, DRB3*0101, DRB4*0101, DRB5*0101) using the consensus prediction described by Wang et al. (35). Peptides with predicted binding scores in the top 20% for a given allele were considered potential binders, and the number of HLA molecules each peptide was predicted to bind was enumerated. All peptides predicted to bind 7 or more HLA molecules were selected for synthesis and further study.
Peptide synthesis
Peptides used for screening studies were purchased from Mimotopes (Clayton, Victoria, Australia) and/or A and A (San Diego, CA) as crude material on a small (1 mg) scale. Peptides utilized as radiolabeled ligands were synthesized on larger scale, and purified (>95%) by reversed phase HPLC.
HLA binding assays
Assays to quantitatively measure peptide binding to MHC class II molecules are based on the inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules, and have been described in detail elsewhere (36). Briefly, MHC molecules were purified from EBV transformed homozygous cell lines by monoclonal Ab-based affinity chromatography. HLA-DR, DQ and DP molecules were captured by repeated passage of lysates over LB3.1 (anti-HLA-DR), SPV-L3 (anti-HLA-DQ) and B7/21 (anti-HLA-DP) columns.
For inhibition experiments, 0.1–1 nM of radiolabeled peptide was co-incubated at room temperature or 37°C with 1 μM to 1 nM of purified MHC in the presence of a cocktail of protease inhibitors and various amounts of inhibitor peptide. Following a 2 to 4 day incubation, the percent of MHC bound radioactivity was determined by capturing MHC/peptide complexes on LB3.1 (DR), L243 (DR), HB180 (DR/DQ/DP), SPV-L3 (DQ) or B7/21 (DP) Ab coated Optiplates (Packard Instrument Co., Meriden, CT), and bound cpm measured using the TopCount (Packard Instrument Co.) microscintillation counter. Inhibitor peptides were tested in at least three independent assays at six different concentrations covering a 100,00-fold dose range. Under the conditions utilized, where [label]<[MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (37, 38).
PBMC isolation and HLA typing
PBMC were obtained by density gradient centrifugation (Ficoll-Hypaque, Amerhsam Biosiences, Uppsala, Sweden) from one unit of blood (450 ml), according to manufacturer’s instructions, and cryo-preserved for further analysis. HLA typing was performed according to standard methods (Blood system Laboratories, Tempe, AZ, USA).
In vitro expansion of Bla g-specific T cells
PBMC were cultured in RPMI 1640 (V Scientific, Tarzana, CA) supplemented with 5% human serum (Cellgro, Herndon, VA) at a density of 2×106 cells/ml in 24-well plates (BD Biosciences, San Jose, CA) and stimulated with 25 μg/ml German cockroach (Blatella germanica; Bla g) extract (Greer, Lenoir, NC), or individual peptides. Cells were cultured at 37°C in 5% CO2 and additional IL-2 (10 U/ml; eBioscience, San Diego, CA) was added every 3 days after initial antigenic stimulation. On day 14, cells were harvested and screened for reactivity against Bla g-specific peptide pools or individual peptides.
ELISPOT assays
The production of IL-5, IFN-γ, and IL-10 was analyzed in ELISPOT assays. Flat-bottom 96-well nitrocellulose plates (Millipore, Bedford, MA) were prepared according to manufacturer’s instructions and coated with either 10 μg/ml anti-human IL-5 (Clone TRFK5; Mabtech, Cinncinati, OH), anti-human IFN-γ (Clone 1-D1K; Mabtech), or anti-human IL-10 (Clone 9D7, Mabtech). Cells were then incubated at a density of 1×105/well either with peptide pools or individual peptides (10 μg/ml), German cockroach extract (25 μg/ml), PHA (10 μg/ml), or medium (containing 1% DMSO corresponding to the percentage of DMSO in the pools/peptides) as a control. After 24 hours, cells were removed, and plates were incubated with either 2 μg/ml biotinylated anti-human IL-5 Ab (Clone 5A10, Mabtech) and 1:200 HRP-conjugated anti-human IFN-γ Ab (Clone 7-B6-1, Mabtech) or 2 μg/ml biotinylated anti-human IL-10 Ab (Clone 12G8, Mabtech) at 37°C. After 2 hours, spots corresponding to the biotinylated Abs (IL-5, IL-10) were developed by incubation with Alkaline Phosphatase-Complex (Vector Laboratories, Burlingame, CA) followed by incubation with Vector Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories) according to the manufacturer’s instructions. Spots corresponding to the HRP-conjugated Ab (IFN-γ) were developed with 3-amino-9-ethylcarvazole solution (Sigma-Aldrich, St. Louis, MO). Spots were counted by computer-assisted image analysis (Zeiss, KS-ELISPOT reader, Munich, Germany).
Each assay was performed in triplicate. The level of statistical significance was determined with a Student’s t-test using the mean of triplicate values of the response against relevant pools or individual peptides versus the response against the DMSO control. Criteria for peptide pool positivity were 100 spot-forming cells (SFCs)/106 PBMC, p ≤ 0.05 and a stimulation index (SI) ≥ 2, while criteria for individual peptide positivity were ≥ 20 SFC/106 PBMC, p ≤ 0.05, and a SI ≥ 2.
HLA restriction
To determine the HLA locus restriction of identified epitopes, mAb inhibition assays were performed. After 14 days of stimulation with German cockroach extract (50 μg/ml) or specific peptide (10 μg/ml), for locus or allele restriction assays respectively, PBMCs were incubated with 10 μg/ml of mAbs (Strategic Biosolutions, Windham, ME) against HLA-DR (LB3.1), DP (B7/21) or DQ (SVPL3) 30 min prior to peptide addition. Cytokine production against positive peptides was then measured in ELISPOT assays as described above. The pan MHC class I Ab (W6/32) was used as a control. A restricting locus was identified by ≥50% inhibition of the response by the corresponding mAb.
To determine the specific HLA allele restriction, donor derived T cells were expanded for 10 days using a single epitope peptide, and these cells were then subsequently incubated with peptide pulsed EBV cell lines and/or fibroblasts expressing known HLA molecules also expressed in the donor from whom the T cells were derived. Cytokine specific ELISPOT assays were performed as described above to determine cytokine production and allele restriction determined by analyzing a matrix of negative and positive cytokine responses with the HLA expressing EBV lines and fibroblasts used.
Serological determinations
Sera were analyzed for specific IgE antibody binding to rBla g 1, rBla g 2, rBla g 4, rBla g 5 and rPer a 7 using allergen-coated streptavidin-ImmunoCAPs. Recombinant allergens were expressed in Pichia pastoris (rBla g 1, rBla g 2, rBla g 4, rPer a 7) or Escherichia coli (rBla g 5) and purified by affinity chromatography. Purified allergens were biotinylated and bound to streptavidin-coated ImmunoCAPs (Phadia US Inc., Portage, MI) at an optimized amount of 1 μg per ImmunoCAP. Specific IgE antibody binding to extracts from Blattella germanica (i6), Dermatophagoides pteronyssinus (d1) and D. farinae (d2), and total IgE antibody were also measured by ImmunoCAP analysis.
RESULTS
Heterogeneity and immunodominance in T cell response to German cockroach allergens in allergic donors
We have recently described a general strategy to identify T cell epitopes derived from common allergens based on the observation that while responses to complex allergens in humans are very heterogeneous and involve recognition of a large number of epitopes, a relatively small number of the most dominant and prevalent responses encompass a significant fraction of the total response (32). We have further shown that these epitopes can be predicted on the basis of their capacity to bind a panel of HLA class II molecules representative of the most frequent alleles expressed at the DR, DP and DQ loci. Here, we took advantage of this approach to identify T cell epitopes derived from German cockroach allergens, denominated Bla g.
To identify dominant Bla g T cell epitopes, we obtained PBMC donations from 34 different allergic donors. Allergic status was defined as a positive skin test reaction (>3 mm) and RAST IgE to Bla g extract >0.35kU/L, and a history of allergic rhinitis and/or asthma. The sequences of six previously described Bla g allergens (Bla g 1, 2, 4, 5, 6 and 7) were selected for analysis, including known isoallergens described in the WHO/IUIS Allergen Nomenclature Database (www.allergen.org). These sequences were scanned with predictive algorithms specific for 20 different common HLA DR, DP and DQ molecules, representative of the most common molecules encountered in the general population, irrespective of ethnicity (39). Peptides ranking in the top 40% of predicted affinities for 7 or more of 20 HLA class II alleles were selected. This prediction strategy was aimed at identifying peptides potentially capable of binding to multiple HLA class II molecules, and thereby most likely to be prevalently recognized.
A total of 195 peptides from the Bla g 1, 2, 4, 5, 6, and 7 allergens were synthesized (Supplemental Table I), and arranged into 13 pools containing 12–18 peptides each. These pools were tested with extract-stimulated PBMC cultures for production of IL-5, as a prototype Th2 lymphokine, and IFN-γ, as a prototype Th1 lymphokine. Positive pools were deconvoluted to identify specific epitopes.
Of the 34 donors tested, 32 responded to stimulation with the allergen extract, and of these 32, individual peptide responses were obtained in 19 (a total of 21 responded to the peptide pools). In all, 41 peptides were identified that elicited a positive response in at least one donor. As discussed in more detail in the following sections, the fact that some individuals did not respond to the peptides is not likely a reflection of the computational analysis not identifying all relevant peptides. As also discussed in the following results section, it is possible that T cell responses are directed against additional proteins not analyzed herein, and that also the relatively weak sensitization of the patient cohort studied contributed to this phenomenon.
Some peptides were highly homologous because they were derived from isoforms of the same allergen, or were derived from the same allergen protein and represented nearly identical overlapping sequences and donor responses. After removal of these redundancies, a total of 32 unique peptide responses were identified (Supplemental Table II). Further consolidation of largely overlapping contiguous epitopes allowed the definition of 25 distinct antigenic regions of 15–20 amino acids in length (Table I). These results highlight the high degree of heterogeneity of human responses to the Bla g allergens studied.
Table I.
25 distinct Bla g regions are recognized by T cell responses from allergic individuals
| Antigen | Position | Region ID | Sequence | Donors responding | Total SFC | % Total response | Cumulative response |
|---|---|---|---|---|---|---|---|
| Bla g 5 | 181 | 17 | ALREKVLGLPAIKAWVAKRP | 8 | 4953 | 20.3 | 20.3 |
| Bla g 5 | 66 | 12 | VAISRYLGKQFGLSG | 2 | 2443 | 10.0 | 30.3 |
| Bla g 5 | 16 | 10 | GEPIRFLLSYGEKDFEDYRF | 5 | 2342 | 9.6 | 39.9 |
| Bla g 5 | 96 | 13 | ISDFRAAIANYHYDA | 2 | 2103 | 8.6 | 48.5 |
| Bla g 5 | 156 | 15 | YFVAILDYLNHMAKE | 2 | 1730 | 7.1 | 55.6 |
| Bla g 6 | 66 | 20 | EEFCTLASRFLVEED | 2 | 1433 | 5.9 | 61.5 |
| Bla g 6 | 6 | 23 | PEQIQLLKKAFDAFD | 2 | 1245 | 5.1 | 66.6 |
| Bla g 1 | 331 | 2 | LIDDVLAILPLDDLK | 1 | 1157 | 4.7 | 71.4 |
| Bla g 5 | 166 | 16 | HMAKEDLVANQPNLKALREK | 3 | 1140 | 4.7 | 76.0 |
| Bla g 5 | 131 | 14 | TKKFDEVVKANGGYLAAGKL | 2 | 927 | 3.8 | 79.8 |
| Bla g 5 | 46 | 11 | SMPFGKTPVLEIDGK | 2 | 874 | 3.6 | 83.4 |
| Bla g 2 | 11 | 5 | FAVATITHAAELQRV | 1 | 855 | 3.5 | 86.9 |
| Bla g 6 | 11 | 18 | EQISVLRKAFDAFDREKSGS | 2 | 757 | 3.1 | 90.0 |
| Bla g 6 | 71 | 19 | EFVTLAAKFIIEEDS | 1 | 443 | 1.8 | 91.8 |
| Bla g 1 | 40 | 3 | PEFQSIVQTLNAMPEYQNLL | 2 | 327 | 1.3 | 93.2 |
| Bla g 1 | 281 | 4 | PELQNFLNFLEANGL | 1 | 323 | 1.3 | 94.5 |
| Bla g 6 | 31 | 24 | MVGTILEMLGHRLDD | 2 | 317 | 1.3 | 95.8 |
| Bla g 2 | 321 | 9 | HFFIGDFFVDHYYSE | 1 | 250 | 1.0 | 96.8 |
| Bla g 2 | 26 | 6 | PLYKLVHVFINTQYA | 1 | 240 | 1.0 | 97.8 |
| Bla g 1 | 351 | 1 | FETIVVTVDSLPEFK | 1 | 167 | 0.7 | 98.5 |
| Bla g 2 | 296 | 8 | ISSQYYIQQNGNLCY | 1 | 153 | 0.6 | 99.1 |
| Bla g 6 | 140 | 22 | SGTVDFDEFMEMMTG | 1 | 57 | 0.2 | 99.4 |
| Bla g 2 | 46 | 7 | GNQNFLTVFDSTSCN | 1 | 55 | 0.2 | 99.6 |
| Bla g 6 | 86 | 21 | EAMEKELREAFRLYD | 1 | 53 | 0.2 | 99.8 |
| Bla g 6 | 101 | 25 | GYITTNVLREILKEL | 1 | 50 | 0.2 | 100.0 |
Conversely, we also noted a clear hierarchy of immunoprevalence and immunodominance. At the level of immunoprevalence, some T cell epitopes were recognized in only one donor, while others were recognized in multiple donors. At the level of immunodominance, we noted that the strength of the responses varied over 1000-fold. Indeed, the top 5 peptides accounted for over half (55.6%) of the response, and the top 9 and 13 accounted for 76% and 90% of the total response, respectively (Table I).
One of the epitope reactivities (Epitope region 5) was directed against a peptide contained within the leader sequence. Since the natural Bla g 2 N-terminal sequencing (40) showed that the mature protein starts at residue 25, this result is unexpected and might reflect recognition of a minor isoform where the signal sequence is cleaved at an alternate position.
Taken together these results indicate that while responses to Bla g allergens in humans are very heterogeneous and involve recognition of a large number of T cell epitopes, a small number of epitopes that elicit the most dominant and prevalent responses encompass a significant fraction of the total response in this population of donors. Similar results have been reported in the timothy grass system (32).
The T cell epitopes identified account for a significant fraction of the response
We next evaluated the thoroughness of the epitope identification studies by three different types of analyses. First, since the candidate epitopes were identified on the basis of predicted HLA binding we wanted to exclude that a large fraction of T cell epitopes might have been missed by the predictions.
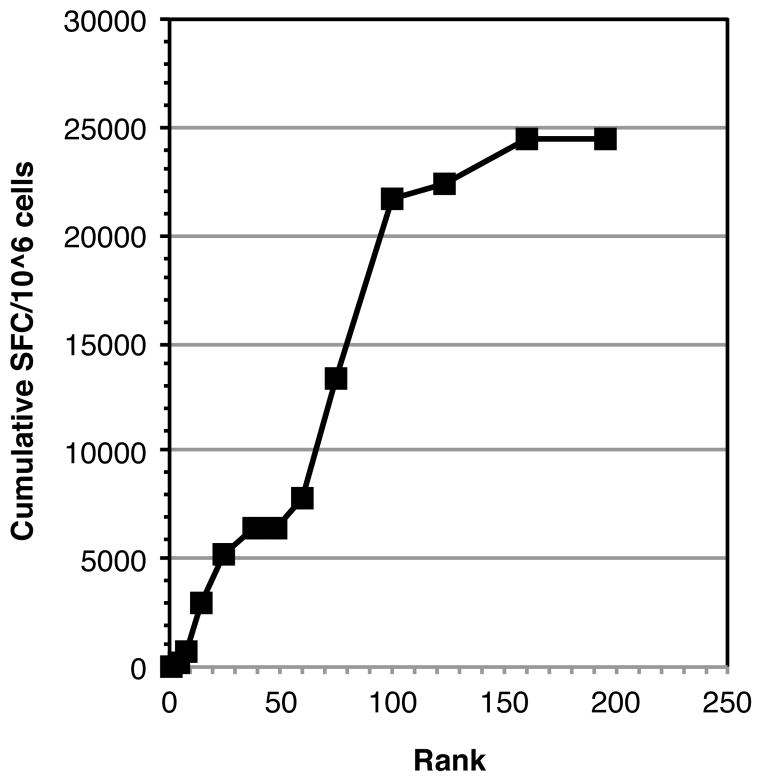
First we note that because of the relatively low stringency used in the prediction a large fraction of the sequence of each of the Bla g proteins would be covered by the predicted peptides tested. Indeed, as shown in Supplemental Table III, an average of 69% of the overall sequences were covered, corresponding to about 42% of unique 15-mers, considering a ten-residue overlap, spanning the entire sequence. Second, we considered having missed a large fraction of the epitopes unlikely, based on previous timothy grass studies (32), which had shown that predictions of this level of stringency would identify approximately 75% of the total response detected with complete sets of overlapping peptides. In this context we reasoned that if the predictions were reasonably effective, most of the response would be associated with the peptides ranking high in predicted binding capacity. If the predictions are exhaustive, lower ranking peptides would be associated with diminishing success, and the curve of prediction success versus rank would start to level off. The data shown in Figure 1 shows that this is indeed the case.
Figure 1. Peptide binding predictions allowed efficient identification of a preponderance of the Bla g specific T cell response.
Bla g allergen sequences were scanned with a panel of bioinformatics algorithms predicting binding to 20 common HLA class II molecules, as described in the materials and methods. Peptides were ranked on the basis of predicted binding promiscuity, and all peptides predicted to bind 7 or more molecules were selected for synthesis and tested for recognition in allergic donors. Then cumulative Bla g specific response (total SFC) as function of peptide rank was tabulated. Saturation of responses was noted at a rank of about 160, corresponding to approximately the top 35% scoring peptides, and over 75% of the response was associated with the top 100 (corresponding to the top 22%) predicted peptides.
Finally, to have a crude estimate whether the epitopes identified accounted for a significant fraction of the response, we compared the total response observed against the cockroach extract to the sum total of the epitope specific response. By this analysis the sum total of epitope responses corresponded to 90% of the sum total extract response (data not shown). These values should not be taken as directly comparable, since an optimal amount of peptide epitope is used in the assay, while cockroach extracts contain an unknown amount of each allergen. So, this percentage could vary very significantly depending on different extract preparations or techniques. Nevertheless, the above considerations strongly suggest that the identified epitopes likely represent a very large fraction of the T cell epitopes contained in the Bla g allergens studied. However, in 13 of the donors that were positive for cockroach extract IgE reactivity, and for which a significant response to extract stimulation was observed, no epitope derived from the Bla g 1, 2, 4, 5, 6, and 7 allergens could be identified. This suggests that additional, as yet undefined, proteins might be recognized by T cell responses in these donors.
Diverse HLA locus restriction of Bla g epitopes
The HLA locus restriction of the 13 most frequently recognized epitopic regions (Table I; regions recognized by more than one donor) was determined by inhibition experiments utilizing DR, DP and DQ specific monoclonal antibodies. The results are presented in Table II. Overall, 20 locus restrictions were determined. DR accounted for the most (11/20) restrictions analyzed, but restriction by DQ molecules was also relatively frequent (6/20). By contrast, the DP locus restricted only three epitopes.
Table II.
HLA restriction of Bla g responses
| Region ID | Donor | Locus | Donor class II alleles bound | Tentative restriction |
|---|---|---|---|---|
| 17 | XT0025 | DR | DRB1*0101, DRB1*0401, DRB4*0103 | DRB1*01 |
| XT0021 | NA | DRB1*0101, DRB1*1301 | ||
| XT0023 | DR | DRB1*0102, DRB1*0804 | ||
| XT0029 | DR | DRB1*0102, DRB1*1302 | ||
| XT0034 | DR | DRB1*0102, DRB1*1102 | ||
| XT0013 | DR | DRB1*1201, DRB1*1316 | ||
| XT0012 | NA | DRB1*0804, DRB1*1503, DRB5*0101 | ||
| XT0041 | NA | DRB1*0701, DRB1*1503, DRB5*0101 | ||
|
| ||||
| 12 | XT0023 | DP | DPB1*0201 | DPB1*0201 |
| XT0030 | NA | DPB1*0201 | ||
|
| ||||
| 10 | XT0011 | DQ | DQB1*0501 | DQB1*0501 |
| XT0021 | NA | DQB1*0501 | ||
| XT0023 | NA | DQB1*0501 | ||
| XT0030 | NA | DQB1*0501 | ||
| XT0041 | NA | DQB1*0303 | ||
|
| ||||
| 13 | XT0024 | DQ | DQB1*0301, DQB1*0504, DRB1*0101, DRB1*0804 | DQB1*0301/DQB1*05 |
| XT0025 | DQ DR |
DQB1*0301, DQB1*0501 DRB1*0101, DRB1*0401, DRB4*0103 |
||
|
| ||||
| 15 | XT0024 | DQ | DQB1*0504 | DQB1*0504 |
| XT0041 | NA | |||
|
| ||||
| 20 | XT0015 | DR | DRB1*1302 | DRB1*1302 |
| XT0024 | NA | |||
|
| ||||
| 23 | U00023 | NA | ||
| XT0024 | NA | |||
|
| ||||
| 2 | XT0003 | DP | DPB1*0101 | DPB1*0101 |
|
| ||||
| 16 | XT0029 | DR | DRB1*1302 | DRB1*13 |
| XT0013 | DR | DRB1*1316 | ||
| XT0021 | NA | DRB1*1301, DRB1*0101 | ||
|
| ||||
| 14 | XT0023 | DQ | DQB1*0301 | DQB1*0301 |
| XT0034 | DQ | DQB1*0301 | ||
|
| ||||
| 11 | XT0041 | DR | ||
| XT0021 | NA | |||
|
| ||||
| 5 | U00023 | NA | ||
|
| ||||
| 18 | XT0024 | DR | DRB1*0101, DRB1*0804 | DRB1*0101/DRB1*0804 |
| XT0022 | DP | |||
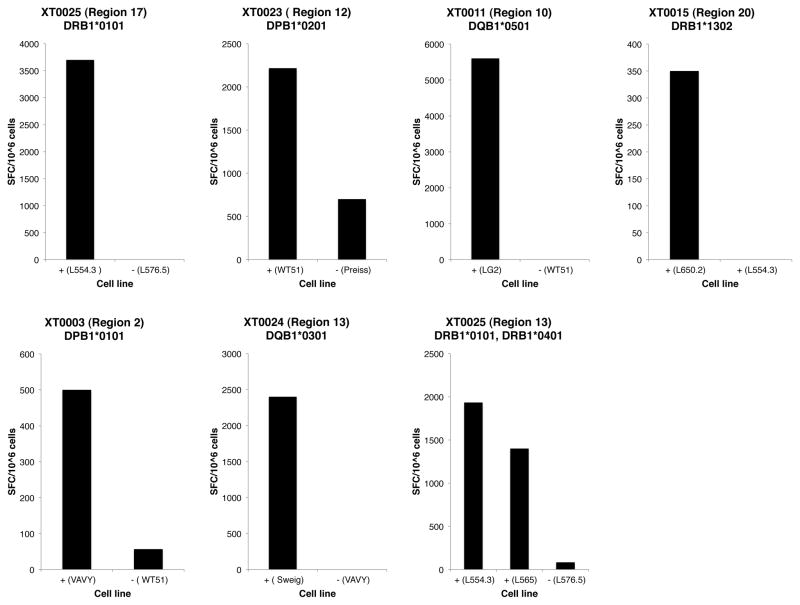
These Bla g epitopes were also tested for their capacity to bind a panel of 35 different DR, DP and DQ molecules (39, 41–43) representative of the most common allelic variants worldwide. This HLA binding information was then utilized to infer potential HLA allelic restrictions for each patient/epitope combination. For each instance in which the restricting locus was determined by antibody inhibition experiments, the HLA types expressed at that locus by the corresponding donor were considered. The binding data was utilized to further narrow the potential restricting molecule by eliminating molecules that were shown to not be able to bind the epitope in in vitro assays utilizing purified HLA. Whether several allergic donors responding to the same epitope shared particular HLA molecules that bind the epitope, at a locus shown to restrict epitope, was also considered. In many cases these data allowed inference of the likely restricting HLA molecule (see last column of Table II). In selected cases the inferred likely restriction was confirmed by the use of transfected cell lines expressing single HLA class II molecules and/or the use of HLA matched/mismatched EBV transformed cell lines. In particular, by this approach we could demonstrate that epitope regions 17, 12, 10, 20 and 2 are restricted by DRB1*0101, DPB1*0201, DQB1*0501, DRB1*1302, and DPB1*0101, respectively. Region 13, was promiscuous in its restriction, being restricted by DQB1*0301 in donor XT0024 and both DRB1*0101 and DRB1*0401 in donor XT0025 (Figure 2).
Figure 2. Inferred HLA restriction of T cell responses to Bla g epitopes.
The HLA restriction of donor responses to Bla g epitopes was determined in selective cases using cell lines transfected with a single HLA class II molecule and/or the use of HLA matched (+)/mismatched (−) EBV transformed cell lines. Representative data are shown.
Differential dominance and polarization of Bla g allergens for T cell responses
The data was next analyzed in terms of the specific antigen from which the various epitopes were derived (Table III). It was found that the Bla g 5 allergen was most dominant, by far, in comparison to the other Bla g allergens analyzed, in that it alone accounted for 67.7% of the total response. Bla g 6 was second in terms of the immunodominance hierarchy, accounting for 17.9% of the response, and Bla g 1 was third, accounting 8.1%. Little or no response was detected for the Bla g 2, 4, and 7 allergens. This dominance profile was not merely due to size differences between the allergens, nor was it correlated to the number of peptides predicted and tested. Indeed the number of peptides tested for either Bla g 5 or 6 was far less than the number tested for the less frequently and less strongly recognized Bla g 1 and 2.
Table III.
Differential polarization of T cell responses to Bla g allergens
| Protein | Total SFC | % Total response | Peptides tested | IL-5 SFC | IFN-γ SFC | Ratio1 |
|---|---|---|---|---|---|---|
| Bla g 1 | 1973 | 8.1 | 85 | 1083 | 890 | 1.22 |
| Bla g 2 | 1553 | 6.4 | 37 | 0 | 1553 | <0.01 |
| Bla g 4 | 0 | 0.0 | 14 | 0 | 0 | - |
| Bla g 5 | 16512 | 67.7 | 20 | 9637 | 6875 | 1.40 |
| Bla g 6 | 4355 | 17.9 | 24 | 4045 | 310 | 13.0 |
| Bla g 7 | 0 | 0.0 | 15 | 0 | 0 | - |
|
| ||||||
| Total | 24394 | 100.0 | 195 | 14765 | 9629 | 1.53 |
IL-5/IFNγ
As mentioned above, we assayed for IL-5 and IFN-γ production, as prototype Th2 and Th1 lymphokines, respectively. Furthermore, we analyzed the data in terms of the ratio of the IL-5 to IFN-γ responses detected for the various allergens (Table III). As expected, the overall IL-5 production exceeded IFN-γ. However, a surprisingly wide variation was observed in terms of the individual allergen proteins. In the case of the most dominant Bla g 5 antigen, both IL-5 and IFN-γ responses were detected, with the IL-5 response only slightly more vigorous than that of IFN-γ. In the case of Bla g 6, there was a clear preponderance of IL-5. Conversely, in the case of Bla g 2, responses were detected only for IFN-γ, and not for IL-5.
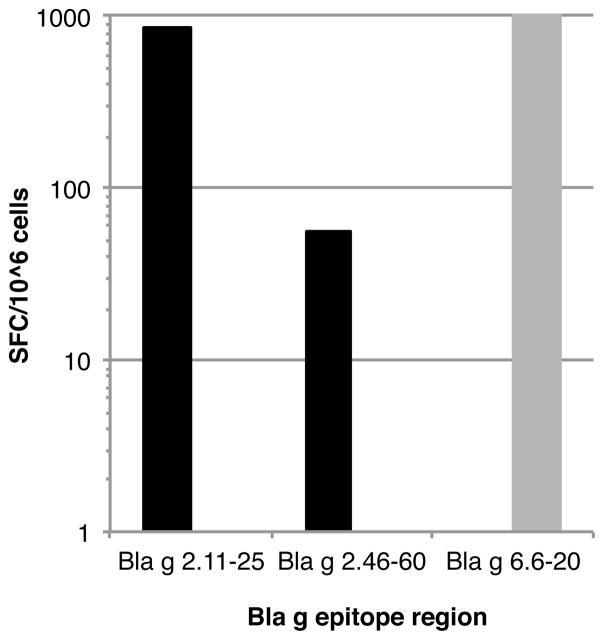
Strikingly, even within an individual donor, responses to different allergens could be differentially polarized, with responses to one allergen dominated by Th1 responses, and to a different allergen dominated by Th2 responses. An example of this type of situation was observed in donor U00023, who responded to epitope regions 5 and 7 from Bla g 2 and region 23 from Bla g 6. As shown in Figure 3, the T cell response to Bla g 6 was associated only with IL-5 production; conversely, the response to the two Bla g 2 epitopes produced IFN-γ, but no IL-5. Taken together, these results suggest that different allergen proteins might be associated with different patterns of polarization of the responding T cell subsets.
Figure 3. Polarized T cell responses to Bla g antigens within an individual donor.
T cell responses against Bla g allergens are differentially polarized at the level of individual donors. IFNγ (black bars) and IL-5 (gray bars) responses in donor U00023 to epitopes derived from Bla g 2 (epitope region 5, Bla g 2.11–25, and epitope region 7, Bla g 2.46–60) and Bla g 6 (epitope region 23, Bla g 6.6–20) are shown. The T cell response to Bla g 6 was associated only with only IL-5 production, while the response to the two Bla g 2 epitopes was only associated with IFN-γ.
Lack of correlation between prevalence of IgE and T cell responses to individual Bla g proteins
In parallel with the determination of the T cell responses, IgE titers to specific cockroach allergens were measured by streptavidin-ImmunoCAP assays. Total IgE and specific IgE antibodies to cockroach and two species of mite allergens were also determined. This allowed correlating, in the same donor population, the prevalence of IgE and T cell responses against the specific allergens. As shown in Table IVa, there is a trend (statistically not significant) towards the IgE response being more broadly directed against the various allergens. In agreement with previous reports (23), Bla g 5 was the most frequently recognized allergen in both T cell and IgE assays, with no clear dominance in IgE reactivity.
Absolute values of specific IgE antibody binding against specific allergens were low for most of the sera tested. The average of total IgE was 427.8 kU/L (range 35.6 – 2,152; n=34), and for cockroach specific IgE was 13.69 (range <0.35–21.3 kU/L; n=34). Absolute values of IgE antibody binding to specific allergens were low for most of the sera tested. Reactivity to mite extracts from D. pteronyssinus and D. farinae was also measured, with an average of 11.41 and 15.2, respectively (range of <0.35–>100, and n=34, for both). There was no correlation between reactivity to mite extracts and cockroach tropomyosin. Therefore, reactivity to mite tropomyosin would not be responsible for the reactivity to cockroach tropomyosin (Per a 7) observed.
The correlation between T cell responses and IgE responses was further investigated by examining for each donor which allergens were recognized by IgE and T cell responses (Table IVb and Supplemental Table IV). No significant correlation was detected for any of the antigens, suggesting that distinct mechanisms may govern responsiveness against the various allergens at the IgE and T cell level. Importantly, in the case of several donors we found that significant IgE titers were observed against a given antigen, while T cell responses to the same allergen were undetectable, but were vigorous and readily detected against a different Bla g protein. This observation suggests that, in these instances, unlinked T-B help might support the development of IgE responses.
Patterns of T cell reactivity following SIT treatment
For a select number of donors, we were able to obtain blood donations before, and six months after, initiation of SIT treatment. Six months is a time period routinely associated with reaching the “maintenance” phase, and it is believed that regulatory events associated with modulation of T cell responses probably occur within this time frame (44, 45). For four individual donors who responded to Bla g epitopes before SIT treatment, responses in PBMC cultures were also studied 6 months after initiation of SIT treatment. T cells producing IL-5, IFN-γ, and IL-10 were measured by standard ELISPOT assays. Results are shown in Table V, and the overall data can be summarized as follows. First, in no case was SIT treatment associated with development of reactivity against new epitopes (data not shown). Second, SIT resulted in a profound decrease in IL-5 producing cells, despite normal PHA responses. Third, this loss of IL-5 producing cells was not associated with development of increased responses to either IFN-γ or IL-10 against any of the peptides derived from the Bla g allergens considered.
Table V.
Overall pattern of responses pre- and post-SIT treatment
| Donor | Region | Antigen | PRE -SIT | POST -SIT | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| IFNg (SFC/106) | IL-5 (SFC/106) | IL-10 (SFC/106) | IFNg (SFC/106) | IL-5 (SFC/106) | IL-10 (SFC/106) | |||
| XT0003 | 2 | Bla g 1.0101 | 160 | 997 | 0 | 0 | 0 | 0 |
| 3 | Bla g 1.0101 | 153 | 87 | 0 | 0 | 0 | 0 | |
| PHA | 1083 | 2650 | 450 | 1837 | 2463 | 73 | ||
|
| ||||||||
| XT0008 | 24 | Bla g 6.0201 | 0 | 50 | 0 | 0 | 0 | 0 |
| 25 | Bla g 6.0201 | 0 | 50 | 0 | 0 | 0 | 0 | |
| PHA | 1907 | 2017 | 167 | 1807 | 2160 | 127 | ||
|
| ||||||||
| XT0011 | 10 | Bla g 5 | 0 | 1537 | 0 | 0 | 0 | 0 |
| PHA | 1507 | 2987 | 900 | 1427 | 2820 | 1653 | ||
|
| ||||||||
| XT0012 | 17 | Bla g 5 | 197 | 0 | 0 | 0 | 0 | 0 |
| PHA | 1613 | 1540 | 147 | 1440 | 3093 | 273 | ||
DISCUSSION
Herein we describe the first T cell epitopes to be identified from six Blattella germanica allergens that have been implicated in cockroach allergy, a major health problem that is increasing in frequency, particularly in urban and inner city settings. The epitopes identified were utilized to determine the quality, immunodominance and immunoprevalence of T cell responses, to explore their relation to IgE responses, and to probe the evolution of T cell responses associated with SIT treatment with cockroach extracts.
The epitopes were identified utilizing a prediction schema previously validated in the context of timothy grass allergens (32) and further validated in the present study. The data presented herein shows that bioinformatics predictions can be used to reduce the number of peptides needed to be tested, while still identifying a majority of the epitopes. These strategies are of relevance as they simultaneously target the most common allelic variants expressed at the human HLA class II DR, DP and DQ loci (39, 41–43). Predictive strategies are of particular interest in cases where limiting amounts of human samples are available. Alternatively, bioinformatic predictions can be utilized to allow efficient performance of large-scale screening of comprehensive panels of antigens. Indeed our group has recently initiated a large-scale screening effort to map allergen epitopes from over two-dozen different common human allergens. However, the data also show that about 20% of all possible peptides need to be examined to identify the majority of epitope sequences. Not all possible peptides have been tested, and it is likely, that additional epitopes that were not selected by the bioinformatics approach we have taken might yet be identified. Further improvement in prediction strategies might incorporate additional factors, such as the influence of antigen processing (46).
A total of 25 epitopic regions were identified from the six Bla g proteins analyzed, underlining the heterogeneity of human allergen-specific T cell responses. These data are in good agreement with what has been observed previously in the case of the timothy grass system (32) and in other systems (47–61). However, as in the case of timothy grass, we also found that a rather limited number of epitopes could account for the majority of responses. This is of relevance for potential diagnostic or therapeutic applications, as it demonstrates that a finite number of molecular structures can be used to recapitulate the heterogeneity observed in human populations of allergic individuals.
A particular caveat with the data herein is that while 32 of the 34 donors with a history of cockroach sensitivity had positive responses to the extract, only 19 of them had responses to the peptides identified by bioinformatics analysis. Further, 6 of the 19 responders had a response to a single peptide, and half of these responses were less than 200 SFC/10^6 cells. At the same time, only four donors (XT0021, U00023, XT0024 and XT0041), which responded to 4 or more regions, accounted for 21 of the 48 (44%) unique donor/region responses (see Supplemental Table 1). Thus, the range of informative data is narrow. It is possible that T cell responses are directed against additional proteins or isotype sequences not analyzed herein. Future studies will be directed at the identification and characterization of possible novel cockroach allergens. It is also possible that the relatively weak sensitization of the patient cohort studied has contributed to this phenomenon. For the present study, we were largely limited to the donors available through the ICAC. In future work we hope to expand our studies to include additional donors, and preferably those that are more highly sensitized.
In the present study we have utilized in vitro expansion of PBMC by stimulation with cockroach allergen extract. We recognize that this approach can create significant bias. However, this approach is also commonly used in the literature describing allergens, as it allows the study of low frequency responses. Allergen extracts can vary significantly in terms of the relative concentrations of various components, as well as in relation to what variant is actually inhaled and/or is causative of the allergic reaction. In this respect, further studies are clearly warranted. For example, it would be informative to perform stimulations with recombinant allergens, especially those for which few or no responses were obtained following stimulation with Bla g extracts.
As mentioned above, the prediction schema utilized is based on the most common DR, DP and DQ allelic variants. When the epitopes identified were examined in terms of the HLA locus restriction, we indeed found a diverse breadth of restrictions, demonstrating that strategies only targeting the most often studied DR locus might be unwise and yield an incomplete representation of the epitopic landscape. The locus restriction data, together with binding data and HLA typing of the responding donors was used in a number of instances to predict likely allelic restrictions, which were verified in several instances by the use of transfected cell lines and/or matched and mismatched homozygous EBV transformed cell lines. The resulting data will be of potential interest in terms of assisting in the generation of HLA tetrameric staining reagents, valuable for more detailed characterization of specific responses.
An interesting observation that was derived from the experiments relates to the fact that different allergens appear to elicit patterns of responses that are differentially polarized in terms of their Th1/Th2 balance, at least as judged by IL-5 (Th2) or IFN-γ (Th1) production. Hales et al. have also noted differences in the balance of IL-5 and IFN-γ responses to the Der p 1 and Der p 7 allergens (62). Strikingly, even within an individual donor, responses to different allergens could be differentially polarized, with responses to one allergen dominated by Th1 responses, and to a different allergen by Th2 responses. The molecular basis for this effect is presently unknown, and might reflect differences in the relative concentrations and accessibility in the natural state of the allergens when inhaled, their processing and presentation, and potentially the presence of distinct costimulatory signals associated with each allergen. The study of this mechanism might suggest avenues to influence or alter the lymphokine balance of Th responses, and thus to potentially alter the outcome of responses in terms of IgE titers.
A few caveats must also be noted with respect to the significance of the cytokine bias data. Specifically, the Bla g 2 responses, showing a Th1 bias, are from a total of only 3 donors. At the same time, while 5 donors responded to Bla g 6 regions (representing a Th2 bias), the preponderance of the total Bla g 6 response is dominated by responses from a single donor. Further examination of the responses to Bla g 5 data on a per donor basis, which overall reflected a fairly balanced Th1/Th2 response, reveals that 3 of the 11 donors had a strong Th1 bias (ratio >5-fold), 3 had a strong Th2 bias, and only 5 could be deemed balanced responses.
Another striking finding originating from our study is the lack of correlation, at the level of individual donors and individual Bla g proteins, between IgE titers and T cell responses. Specifically, we found that in a given donor significant IgE titers could be observed against a given antigen, while T cell responses in the same donor to the same allergen could be undetectable, while vigorous and readily detected T cell responses occurred against a different allergen. This finding is similar to what was observed previously in the timothy grass system, and is most readily interpreted by postulating that antibody responses to a given allergen can be “helped” and modulated by T cell responses to a different antigen (unlinked cognate T-B cooperation). The molecular mechanism for such unlinked T-B cooperation is unknown but may be caused by 2 or more allergens being present in the same physical structure, such as micron-sized particles. Furthermore, this observation is also consistent with the fact that for 13 of the donors that had elevated IgE titers and that responded to extract stimulation, not a single epitope derived from the Bla g 1, 2, 4, 5, 6, and 7 allergens was identified, which might suggest that additional, as yet undefined, proteins might be recognized by T cell responses in these donors and that these responses could provide help for the antibody response to the Bla g allergens.
Finally, the definition of the Bla g epitopes allowed us to follow in a longitudinal pattern the magnitude and specificity of T cell responses to the main Bla g allergens as a function of SIT treatment. These results are of relevance in the light of previous studies that indicate that SIT treatment might be associated with deviation of Th2 responses towards a Th1 phenotype, and/or induction of regulatory IL-10 producing T cells. Our results do not support this notion, at least in the case of Bla g allergens and cockroach extracts. Indeed, SIT treatment appeared to be associated with a generalized down regulation of T cell responses, in the absence of new or increased IFN-γ and IL-10 production. We are aware that the in vitro expansion step we have used to characterize responses could alter the pattern of Th subsets detected. However, direct ex vivo experiments with either extract or epitopes did not yield detectable/reproducible responses either before or after SIT treatment, thus precluding ex vivo analysis. Thus, the results obtained in the case of SIT treatment, might favor the hypothesis that SIT efficacy might be associated with T cell responses directed against different T cell antigens, and/or development of IgG responses competing with the IgE recognizing the known allergens.
In conclusion, our experiments provide a characterization of Bla g-specific T cell responses. The results highlight that these responses are associated with a unique pattern of immunodominance, and T cell responses are differentially polarized at the level of the different allergens. The observed pattern of immunodominance is distinct from that observed at the level of IgE responses, and suggests the possibility that unlinked T-B cooperation contributes to shape IgE responses, or that very low IgE responses might not need cognate help. Finally, T cell responses are generally downregulated by SIT treatment, without evidence of induction of Tregs or deviation towards Th1 responses.
Taken together these results enhance our understanding of T cell responses in cockroach allergy and their potential role in SIT therapy. They demonstrate that T cell responses to Bla g allergens have important distinguishing features from IgE responses to the same allergens and suggest that more in-depth studies of these responses might significantly enhance our understanding of cockroach allergies and their treatment.
Supplementary Material
Table IV.
Correlation between IgE and T cell responses against Bla g allergens.
| a. Prevalence of IgE and T cell responses
| ||
|---|---|---|
| Allergen | Donors responding (%)
|
|
| T cell | IgE | |
| Bla g 1 | 4 (12) | 4 (12) |
| Bla g 2 | 3 (9) | 8 (24) |
| Bla g 4 | 0 (0) | 4 (12) |
| Bla g 5 | 11 (32) | 15 (44) |
| Bla g 6 | 5 (17) | NA1 |
| Bla g 7/Per a 7 | 0 (0) | 4 (12) |
| b. Correspondence between IgE and T cell responses
| |||||
|---|---|---|---|---|---|
| Allergen | Donors with responses to:
|
p value | |||
| Both | T cell only | IgE only | Neither | ||
| Bla g 1 | 1 | 3 | 3 | 27 | 0.35 |
| Bla g 2 | 1 | 2 | 7 | 24 | 0.43 |
| Bla g 4 | 0 | 0 | 4 | 30 | 1.00 |
| Bla g 5 | 7 | 4 | 8 | 15 | 0.087 |
| Bla g 7 | 0 | 0 | 4 | 30 | 1.00 |
Reagents unavailable
Acknowledgments
We thank Scott Southwood, Carrie Moore, Kate Bradley, and Duy Le for performing the MHC binding assays, and Jaye Lohman for measuring IgE antibody levels. La Jolla Institute for Allergy and Immunology & Kyowa Hakko Kirin California publication number 1467.
Footnotes
This study was supported by Nation Institutes for Health (NIH) - National Institute Of Allergy And Infectious Diseases (NIAID) contracts HSN272200700048C (AS, LIAI) and HHSN272200900052C (BW, JHU), NIH grant NO1-AI-25482 (BW, JHU) and NIAID grants R01AI077653 (AP) and U19AI100275 (AS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health.
References
- 1.Arshad SH. Indoor allergen exposure in the development of allergy and asthma. Current allergy and asthma reports. 2003;3:115–120. doi: 10.1007/s11882-003-0023-8. [DOI] [PubMed] [Google Scholar]
- 2.Litonjua AA, V, Carey J, Burge HA, Weiss ST, Gold DR. Exposure to cockroach allergen in the home is associated with incident doctor-diagnosed asthma and recurrent wheezing. The Journal of allergy and clinical immunology. 2001;107:41–47. doi: 10.1067/mai.2001.111143. [DOI] [PubMed] [Google Scholar]
- 3.Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, Mitchell H, McNiff-Mortimer K, Lynn H, Ownby D, Malveaux F. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. The New England journal of medicine. 1997;336:1356–1363. doi: 10.1056/NEJM199705083361904. [DOI] [PubMed] [Google Scholar]
- 4.Arruda LK, Vailes LD, Ferriani VP, Santos AB, Pomes A, Chapman MD. Cockroach allergens and asthma. The Journal of allergy and clinical immunology. 2001;107:419–428. doi: 10.1067/mai.2001.112854. [DOI] [PubMed] [Google Scholar]
- 5.Gruchalla RS, Pongracic J, Plaut M, Evans R, 3rd, Visness CM, Walter M, Crain EF, Kattan M, Morgan WJ, Steinbach S, Stout J, Malindzak G, Smartt E, Mitchell H. Inner City Asthma Study: relationships among sensitivity, allergen exposure, and asthma morbidity. The Journal of allergy and clinical immunology. 2005;115:478–485. doi: 10.1016/j.jaci.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Visness CM, Calatroni A, Gergen PJ, Mitchell HE, Sampson HA. Effect of environmental allergen sensitization on asthma morbidity in inner-city asthmatic children. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2009;39:1381–1389. doi: 10.1111/j.1365-2222.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platts-Mills TA, Rakes G, Heymann PW. The relevance of allergen exposure to the development of asthma in childhood. The Journal of allergy and clinical immunology. 2000;105:S503–508. doi: 10.1016/S0091-6749(00)90051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss KB, Gergen PJ, Crain EF. Inner-city asthma. The epidemiology of an emerging US public health concern. Chest. 1992;101:362S–367S. doi: 10.1378/chest.101.6.362s. [DOI] [PubMed] [Google Scholar]
- 9.Perzanowski MS, Platts-Mills TA. Further confirmation of the relevance of cockroach and dust mite sensitization to inner-city asthma morbidity. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2009;39:1291–1293. doi: 10.1111/j.1365-2222.2009.03327.x. [DOI] [PubMed] [Google Scholar]
- 10.Pollart SM, Chapman MD, Fiocco GP, Rose G, Platts-Mills TA. Epidemiology of acute asthma: IgE antibodies to common inhalant allergens as a risk factor for emergency room visits. The Journal of allergy and clinical immunology. 1989;83:875–882. doi: 10.1016/0091-6749(89)90100-0. [DOI] [PubMed] [Google Scholar]
- 11.Huss K, Adkinson NF, Jr, Eggleston PA, Dawson C, Van Natta ML, Hamilton RG. House dust mite and cockroach exposure are strong risk factors for positive allergy skin test responses in the Childhood Asthma Management Program. The Journal of allergy and clinical immunology. 2001;107:48–54. doi: 10.1067/mai.2001.111146. [DOI] [PubMed] [Google Scholar]
- 12.Antony AB, Tepper RS, Mohammed KA. Cockroach extract antigen increases bronchial airway epithelial permeability. The Journal of allergy and clinical immunology. 2002;110:589–595. doi: 10.1067/mai.2002.127798. [DOI] [PubMed] [Google Scholar]
- 13.Bhat RK, Page K, Tan A, Hershenson MB. German cockroach extract increases bronchial epithelial cell interleukin-8 expression. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2003;33:35–42. doi: 10.1046/j.1365-2222.2002.01481.x. [DOI] [PubMed] [Google Scholar]
- 14.Slater JE, James R, Pongracic JA, Liu AH, Sarpong S, Sampson HA, Satinover SM, Woodfolk JA, Mitchell HE, Gergen PJ, Eggleston PA. Biological potency of German cockroach allergen extracts determined in an inner city population. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2007;37:1033–1039. doi: 10.1111/j.1365-2222.2007.02751.x. [DOI] [PubMed] [Google Scholar]
- 15.Finn PW, Boudreau JO, He H, Wang Y, Chapman MD, Vincent C, Burge HA, Weiss ST, Perkins DL, Gold DR. Children at risk for asthma: home allergen levels, lymphocyte proliferation, and wheeze. The Journal of allergy and clinical immunology. 2000;105:933–942. doi: 10.1067/mai.2000.106546. [DOI] [PubMed] [Google Scholar]
- 16.Arruda LK, Chapman MD. The role of cockroach allergens in asthma. Curr Opin Pulm Med. 2001;7:14–19. doi: 10.1097/00063198-200101000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Pomes A, Melen E, Vailes LD, Retief JD, Arruda LK, Chapman MD. Novel allergen structures with tandem amino acid repeats derived from German and American cockroach. The Journal of biological chemistry. 1998;273:30801–30807. doi: 10.1074/jbc.273.46.30801. [DOI] [PubMed] [Google Scholar]
- 18.Arruda LK, Vailes LD, Hayden ML, Benjamin DC, Chapman MD. Cloning of cockroach allergen, Bla g 4, identifies ligand binding proteins (or calycins) as a cause of IgE antibody responses. The Journal of biological chemistry. 1995;270:31196–31201. doi: 10.1074/jbc.270.52.31196. [DOI] [PubMed] [Google Scholar]
- 19.Arruda LK, Vailes LD, Mann BJ, Shannon J, Fox JW, Vedvick TS, Hayden ML, Chapman MD. Molecular cloning of a major cockroach (Blattella germanica) allergen, Bla g 2. Sequence homology to the aspartic proteases. The Journal of biological chemistry. 1995;270:19563–19568. doi: 10.1074/jbc.270.33.19563. [DOI] [PubMed] [Google Scholar]
- 20.Arruda LK, Vailes LD, Platts-Mills TA, Hayden ML, Chapman MD. Induction of IgE antibody responses by glutathione S-transferase from the German cockroach (Blattella germanica) The Journal of biological chemistry. 1997;272:20907–20912. doi: 10.1074/jbc.272.33.20907. [DOI] [PubMed] [Google Scholar]
- 21.Hindley J, Wunschmann S, Satinover SM, Woodfolk JA, Chew FT, Chapman MD, Pomes A. Bla g 6: a troponin C allergen from Blattella germanica with IgE binding calcium dependence. The Journal of allergy and clinical immunology. 2006;117:1389–1395. doi: 10.1016/j.jaci.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Asturias JA, Gomez-Bayon N, Arilla MC, Martinez A, Palacios R, Sanchez-Gascon F, Martinez J. Molecular characterization of American cockroach tropomyosin (Periplaneta americana allergen 7), a cross-reactive allergen. Journal of immunology. 1999;162:4342–4348. [PubMed] [Google Scholar]
- 23.Satinover SM, Reefer AJ, Pomes A, Chapman MD, Platts-Mills TA, Woodfolk JA. Specific IgE and IgG antibody-binding patterns to recombinant cockroach allergens. The Journal of allergy and clinical immunology. 2005;115:803–809. doi: 10.1016/j.jaci.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Pree I, Shamji MH, Kimber I, Valenta R, Durham SR, Niederberger V. Inhibition of CD23-dependent facilitated allergen binding to B cells following vaccination with genetically modified hypoallergenic Bet v 1 molecules. Clin Exp Allergy. 2010;40:1346–1352. doi: 10.1111/j.1365-2222.2010.03548.x. [DOI] [PubMed] [Google Scholar]
- 25.Kang BC, Johnson J, Morgan C, Chang JL. The role of immunotherapy in cockroach asthma. J Asthma. 1988;25:205–218. doi: 10.3109/02770908809071367. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava D, Gaur SN, Arora N, Singh BP. Clinico-immunological changes post-immunotherapy with Periplaneta americana. European journal of clinical investigation. 2011;41:879–888. doi: 10.1111/j.1365-2362.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- 27.Akdis M, Verhagen J, Taylor A, Karamloo F, Karagiannidis C, Crameri R, Thunberg S, Deniz G, Valenta R, Fiebig H, Kegel C, Disch R, Schmidt-Weber CB, Blaser K, Akdis CA. Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J Exp Med. 2004;199:1567–1575. doi: 10.1084/jem.20032058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross F, Metzner G, Behn U. Mathematical modeling of allergy and specific immunotherapy: Th1-Th2-Treg interactions. J Theor Biol. 2011;269:70–78. doi: 10.1016/j.jtbi.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Soyer OU, Akdis M, Akdis CA. Mechanisms of subcutaneous allergen immunotherapy. Immunol Allergy Clin North Am. 2011;31:175–190. vii–viii. doi: 10.1016/j.iac.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Jutel M, Akdis M, Budak F, Aebischer-Casaulta C, Wrzyszcz M, Blaser K, Akdis CA. IL-10 and TGF-beta cooperate in the regulatory T cell response to mucosal allergens in normal immunity and specific immunotherapy. Eur J Immunol. 2003;33:1205–1214. doi: 10.1002/eji.200322919. [DOI] [PubMed] [Google Scholar]
- 31.Ebner C, Siemann U, Bohle B, Willheim M, Wiedermann U, Schenk S, Klotz F, Ebner H, Kraft D, Scheiner O. Immunological changes during specific immunotherapy of grass pollen allergy: reduced lymphoproliferative responses to allergen and shift from TH2 to TH1 in T-cell clones specific for Phl p 1, a major grass pollen allergen. Clin Exp Allergy. 1997;27:1007–1015. doi: 10.1111/j.1365-2222.1997.tb01252.x. [DOI] [PubMed] [Google Scholar]
- 32.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong KY, Yi MH, Jeong KJ, Lee H, Hong CS, Yong TS. Sequence diversity of the Bla g 4 cockroach allergen, homologous to lipocalins, from Blattella germanica. International Archives of Allergy and Immunology. 2009;148:339–345. doi: 10.1159/000170388. [DOI] [PubMed] [Google Scholar]
- 34.Jeong KY, Lee H, Shin KH, Yi MH, Jeong KJ, Hong CS, Yong TS. Sequence polymorphisms of major German cockroach allergens Bla g 1, Bla g 2, Bla g 4, and Bla g 5. International Archives of Allergy and Immunology. 2008;145:1–8. doi: 10.1159/000107460. [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidney J, Southwood S, Oseroff C, Del Guercio MF, Sette A, Grey H. Current Protocols in Immunology. John Wiley & Sons, Inc; 1998. Measurement of MHC/Peptide Interactions by Gel Filtration; pp. 18.13.11–18.13.19. [DOI] [PubMed] [Google Scholar]
- 37.Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 39.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arruda LK, Vailes LD, Benjamin DC, Chapman MD. Molecular cloning of German cockroach (Blattella germanica) allergens. International Archives of Allergy and Immunology. 1995;107:295–297. doi: 10.1159/000237006. [DOI] [PubMed] [Google Scholar]
- 41.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. 2010;185:4189–4198. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. [DOI] [PubMed] [Google Scholar]
- 44.Passalacqua G, Durham SR. Allergic rhinitis and its impact on asthma update: allergen immunotherapy. The Journal of allergy and clinical immunology. 2007;119:881–891. doi: 10.1016/j.jaci.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 45.Bohle B. T cell responses during allergen-specific immunotherapy of Type I allergy. Frontiers in bioscience : a journal and virtual library. 2008;13:6079–6085. doi: 10.2741/3139. [DOI] [PubMed] [Google Scholar]
- 46.German RN, Castellino F, Han R, Reis e Sousa C, Romagnoli P, Sadegh-Nasseri S, Zhong GM. Processing and presentation of endocytically acquired protein antigens by MHC class II and class I molecules. Immunol Rev. 1996;151:5–30. doi: 10.1111/j.1600-065x.1996.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 47.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, Sette A. A Quantitative Analysis of the Variables Affecting the Repertoire of T Cell Specificities Recognized after Vaccinia Virus Infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 49.Botten J, Alexander J, Pasquetto V, Sidney J, Barrowman P, Ting J, Peters B, Southwood S, Stewart B, Rodriguez-Carreno MP, Mothe B, Whitton JL, Sette A, Buchmeier MJ. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J Virol. 2006;80:8351–8361. doi: 10.1128/JVI.00896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotturi MF, Botten J, Sidney J, Bui HH, Giancola L, Maybeno M, Babin J, Oseroff C, Pasquetto V, Greenbaum JA, Peters B, Ting J, Do D, Vang L, Alexander J, Grey H, Buchmeier MJ, Sette A. A Multivalent and Cross-Protective Vaccine Strategy against Arenaviruses Associated with Human Disease. PLoS Pathog. 2009;5:e1000695. doi: 10.1371/journal.ppat.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mothe BR, Stewart BS, Oseroff C, Bui HH, Stogiera S, Garcia Z, Dow C, Rodriguez-Carreno MP, Kotturi M, Pasquetto V, Botten J, Crotty S, Janssen E, Buchmeier MJ, Sette A. Chronic Lymphocytic Choriomeningitis Virus Infection Actively Down-Regulates CD4+ T Cell Responses Directed against a Broad Range of Epitopes. J Immunol. 2007;179:1058–1067. doi: 10.4049/jimmunol.179.2.1058. [DOI] [PubMed] [Google Scholar]
- 52.Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia Virus-Specific CD4+ T Cell Responses Target a Set of Antigens Largely Distinct from Those Targeted by CD8+ T Cell Responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 53.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 54.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A. Dissociation between Epitope Hierarchy and Immunoprevalence in CD8 Responses to Vaccinia Virus Western Reserve. J Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic Analysis of the Repertoire of T-Cell Specificities for Influenza A Virus in Humans. J Virol. 2008;82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blythe M, Zhang Q, Vaughan K, de Castro R, Salimi N, Bui HH, Lewinsohn D, Ernst J, Peters B, Sette A. An analysis of the epitope knowledge related to Mycobacteria. Immunome Research. 2007;3:10. doi: 10.1186/1745-7580-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaughan K, Blythe M, Greenbaum J, Zhang Q, Peters B, Doolan DL, Sette A. Meta-analysis of immune epitope data for all Plasmodia: overview and applications for malarial immunobiology and vaccine-related issues. Parasite Immunol. 2009;31:78–97. doi: 10.1111/j.1365-3024.2008.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaughan K, Greenbaum J, Blythe M, Peters B, Sette A. Meta-analysis of all immune epitope data in the Flavivirus genus: inventory of current immune epitope data status in the context of virus immunity and immunopathology. Viral Immunol. 2010;23:259–284. doi: 10.1089/vim.2010.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zarebski LM, Vaughan K, Sidney J, Peters B, Grey H, Janda KD, Casadevall A, Sette A. Analysis of epitope information related to Bacillus anthracis and Clostridium botulinum. Expert Rev Vaccines. 2008;7:55–74. doi: 10.1586/14760584.7.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hales BJ, Shen H, Thomas WR. Cytokine responses to Der p 1 and Der p 7: house dust mite allergens with different IgE-binding activities. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2000;30:934–943. doi: 10.1046/j.1365-2222.2000.00901.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.