Summary
The Mre11–Rad50–Nbs1 (MRN) complex tethers, processes and signals DNA double strand breaks, promoting genomic stability. To understand the functional architecture of MRN, we determined the crystal structures of the Schizosaccharomyces pombe Mre11 dimeric catalytic domain alone and in complex with a fragment of Nbs1. Two Nbs1 subunits stretch around the outside of Mre11’s nuclease domains, with one subunit additionally bridging and locking the Mre11 dimer via a highly conserved asymmetrical binding motif. Our results reveal that Mre11 forms a flexible dimer and suggest that Nbs1 is not only a checkpoint adaptor, but also functionally impacts on Mre11-Rad50. Clinical mutations in Mre11 are located along the Nbs1 interaction sites and weaken the Mre11–Nbs1 interaction. However, they differentially affect DNA repair and telomere maintenance in Saccharomyces cerevisiae, potentially providing insight into their different human disease pathologies.
Introduction
Genome stability is constantly threatened by environmental influences and cellular metabolism. DNA double strand breaks (DSBs), perhaps the most hazardous DNA lesions arise from failures in DNA replication, ionizing radiation (IR) or genotoxic agents1–3. Cells have evolved a complex damage response utilizing two major pathways to efficiently repair DSBs: homologous recombination (HR) and non–homologous end–joining (NHEJ)4–6. In HR DSBs are repaired in a relatively error free manner by using a sister–chromatid as a template7, whereas the two DNA ends are directly ligated together in the more error–prone NHEJ pathway. Beside classical NHEJ, alternative NHEJ pathways such as micro–homology mediated end joining (MMEJ), where DNA ends are trimmed and joined via micro–homologies8–10 may be utilized.
The Mre11–Rad50–Nbs1 (MRN) complex and its Saccharomyces cerevisiae counterpart Mre11–Rad50–Xrs2 (MRX) play a central role in various DNA end associated processes, including HR, NHEJ, MMEJ, meiosis and telomere maintenance10–15. MRN consists of an Mre11 endo/exonuclease dimer, two Rad50 ATP binding cassette proteins, and the Nbs1 protein. Whereas Mre11 and Rad50 are present in all three domains of life, Nbs1/Xrs2 is only found in eukaryotes. Mre11 and Rad50 together form an ATPregulated nuclease that senses DSBs and tethers DNA via long Rad50 coiled–coil domains16,17. MRN and bacterial MR are 3’–5’ dsDNA exonucleases as well as ATP dependent endonucleases18–20. The endonuclease activity of the complex, which is additionally supported by Sae2/CtIP in eukaryotes, liberates short oligonucleotides from the 5’ end of DSBs21. This process is suggested to remove Spo11 from meiotic breaks and promote subsequent processive 5’ resection by other nucleases22–25.
Perhaps the mechanistically least understood function of MRN is checkpoint signaling and ATM (ataxia telangiectasia mutated kinase) activation. Nbs1 contains a structured N–terminus with a FHA (fork head associated) domain followed by dual BRCT (breast cancer associated 1 C–terminus) domains26,27. This region binds phosphorylated substrates and helps to organize repair foci. However, DSB signaling has been associated with the C–terminus of Nbs1, which contains highly conserved and functionally critical motifs. This domain is predicted to be of an extended structure and does not contain recognizable structural folds, but helps recruit and activate ATM/Tel127–30. However, ATM can also be activated by MR alone in vitro and requires the C–terminus of Mre11 as well as the signature motif of Rad5031–33. On the other hand, a peptide comprising the Nbs1 C–terminus can activate ATM in Xenopus egg extracts34.
Activated ATM phosphorylates a large set of DNA damage response factors including Nbs1 itself35–37. Following Nbs1 phosphorylation, MRN also functions downstream of ATM and participates in the damage checkpoint and DNA repair by recruiting other factors involved in the DNA damage response26,27.
The peculiar nature of the association between Nbs1 and ATM with Mre11 and Rad50 is exemplified by the closely related disease syndromes linked with mutations in their corresponding genes: Ataxia telangiectasia (A–T) is caused by disruption of ATM, while A–T like disease (A–TLD), Nijmegen breakage syndrome (NBS) and NBS–like disease are caused by hypomorphic mutations in Mre11, Nbs1 and Rad50, respectively38–42. Patients exhibit radiation hypersensitivity, chromosome instability, cancer predisposition and distinct neuropathologic phenotypes. Differences between neuropathology phenotypes were proposed to result from different impacts of the different mutations on DNA DSB signaling and apoptosis43, although the precise molecular bases for the similarities and differences between A–T, A–TLD and NBS are yet poorly understood. For instance, a recent report described an Mre11 mutation, in close sequence proximity to an A–TLD mutation, that causes an NBS–like disease (NBSLD) instead of A–TLD44.
To understand the human disease syndromes caused by MRN defects, it is crucial to reveal how Nbs1 interacts with Mre11. Here, we report structures of the catalytic domain of S. pombe Mre11 (Mre11cd) and its complex with the Mre11 interacting region of Nbs1 (Nbs1mir). Our structures and biochemical studies define A–TLD and NBSLD mutation sites and we addressed the molecular defects of corresponding mutations in yeast by in vitro and in vivo assays. Unexpectedly, we discovered that the Mre11 dimer is flexible and is stabilized by Nbs1. Our results indicate that ATP– and DNA–dependent conformational changes in MR may be coupled to the Mre11–Nbs1 interface, suggesting a global structural switch as a basis for DSB signaling.
Results
Structure of the catalytic domain of S. pombe Mre11
We first used a limited proteolysis approach to screen for stable fragments of S. pombe (Sp)Mre11 that were suitable for crystallization and determined the X–ray crystal structure of SpMre1115–413 at 3.0 Å resolution (Table 1). SpMre1115–413 encompasses the whole catalytic core of eukaryotic Mre11 and is further referred to as SpMre11cd (catalytic domain).
Table 1.
Data collection and refinement statistics
| Nbs1mir-Mre11cd SeMet |
Nbs1mir-Mre11cd | Nbs1mir-Mre11cd +50mM Mn2+ |
Unfused Mre11cd-Nbs1mir +50mM Mn2+ |
Mre11cd | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) | 58.9, 79.3, 218.5 | 60.3, 78.9, 222.7 | 59.3–79.0 223.0 |
59.1, 80.0, 220.9 | 76.3, 82.3, 164.3 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90. | 90.0, 90.0, 90 |
| Resolution (Å) | 50–2.8 | 47.9–2.4 | 50–2.5 | 47.5–2.2 | 46–3.0 |
| Rsym or Rmerge | 5.5 (31.3) | 5.0 (36.2) | 4.6 (29.7) | 5.3 (44.5) | 6.1 (45.8) |
| I / σI | 18.06 (3.74) | 27.41 (4.36) | 20.89 (4.2) | 24.96 (3.93) | 16.02 (2.87) |
| Completeness (%) | 96.4 (87.6) | 96.4 (87.9) | 97.7 (93.8) | 95.8 (85.9) | 92.4 (82.3) |
| Redundancy | 3.68 | 7.05 | 3.84 | 7.63 | 3.64 |
| Refinement | |||||
| Resolution (Å) | 47.9–2.4 | 47.4–2.5 | 47.5–2.2 | 45.6 – 3.0 | |
| No. reflections | 43364 | 36562 | 53872 | 20978 | |
| Rwork / Rfree | 22.2/24.2 | 21.3/25.7 | 23.3/24.4 | 22.2/ 28.9 | |
| No. atoms | |||||
| Protein | 6702 | 6627 | 6491 | 5895 | |
| Ligand/ion | 22 | 4 | 4 | 4 | |
| Water | 210 | 145 | 208 | - | |
| B-factors | |||||
| Protein | 51.8 | 46.8 | 50.2 | 76.3 | |
| Ligand/ion | 49.6 | 41.0 | 40.4 | 62.1 | |
| Water | 46.9 | 47.9 | 46.2 | - | |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.012 | 0.009 | 0.014 | 0.008 | |
| Bond angles(°) | 1.795 | 1.133 | 1.766 | 1.393 | |
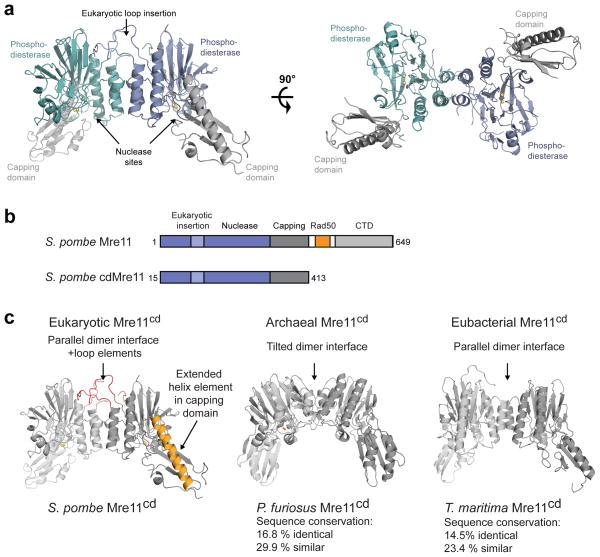
SpMre11cd dimerizes via phosphodiesterase domains flanked by a α/β “capping” domain, creating a U–shaped particle with a broad DNA/Rad50 binding cleft. SpMre11cd harbors two nuclease sites analogous to bacterial and archaeal Mre11 dimers (Fig. 1) and retains its ssDNA endo/exo nuclease activity (Supplementary Fig. 1a). Two Mn2+ ions are coordinated in each of the two apparently functional nuclease sites ( Supplementary Fig. 1b).
Figure 1. Structure of S. pombe apo–Mre11cd and comparison with homologous Mre11 structures from P. furiosus and T. maritima.
(a) Mre11cd dimer (cyan/blue phosphodiesterase and grey capping domains), shown as ribbon representation from side and top view.
(b) Domain maps of full length S. pombe Mre11 and the crystallized Mre11cd construct. CTD, Mre11 C–terminal domain; Rad50, Rad50–interacting region.
(c) Comparison of the eukaryotic S. pombe Mre11cd structure with archaeal P. furiosus Mre11 (PDB 1II7) and eubacterial T. maritima Mre11 (PDB 2Q8U). The percent identity and similarity are relative to S. pombe Mre11.
The structure of SpMre11cd displays prominent differences to prokaryotic Mre11 (Fig. 1c). Notably, SpMre11cd has a large loop insertion – unique to eukaryotes – that extends the dimer interface distal to the DNA/Rad50 binding cleft. Mutations found in A–TLD 3/4 and NBSLD map to this loop element, linking it to pathologies caused by MRN defects. In the capping domain, S. pombe Mre11 possesses an extended α–helix relative to prokaryotic Mre11 (αF, residues 332 to 358; Fig. 1c and Supplementary Fig. 1c). Previous analysis indicates that αF binds the Rad50 NBD in the absence of ATP 19. An internal deletion in αF Del(340–366) is associated with A–TLD, suggesting that this region is critical for Mre11 function and/or stability42. These structural differences likely contribute to the additional functions of eukaryotic Mre11 in DNA damage signaling.
Structure of the Nbs1mir–Mre11cd complex
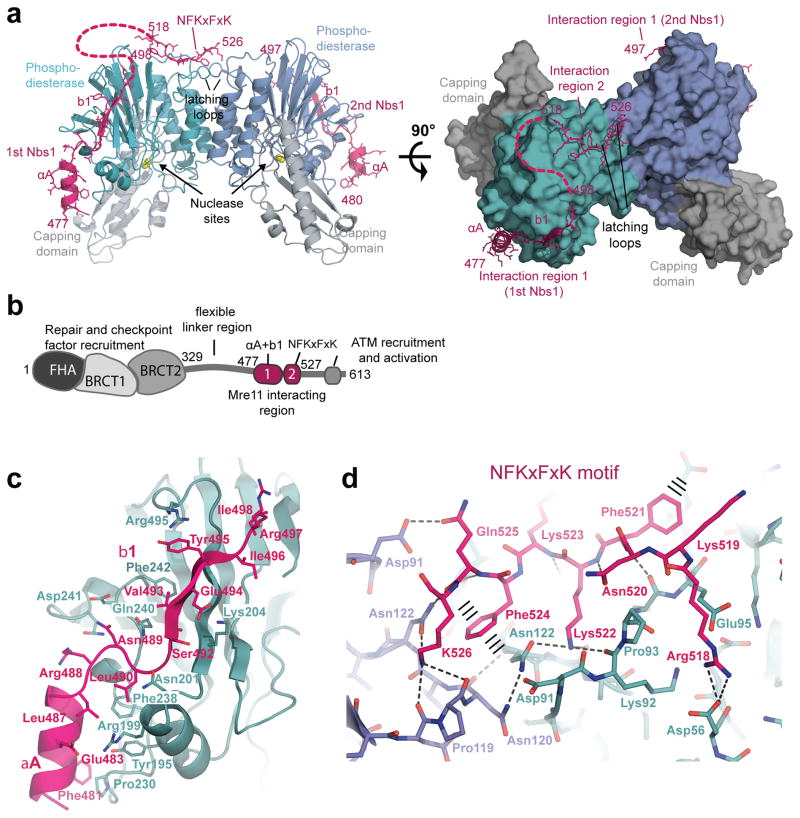
To crystallize a complex between Nbs1 and Mre11cd, we first narrowed down the Mre11 interaction site of S. pombe Nbs1 to residues 474–531 (Supplementary Fig. 2a)45, referred to as Nbs1mir (for Mre11 interacting region). Co–purifying Mre11cd with Nbs1 variants revealed complexes with approximate equimolar stoichiometry (Supplementary Fig. 2b) that were too sensitive to higher ionic strengths to allow for stringent purification. To obtain preparations suitable for structural studies, it was necessary to covalently fuse Nbsmir to Mre11cd via a peptide linker (Supplementary Fig. 2c). This allowed us to determine the X–ray crystal structure of the Nbsmir–Mre11cd fusion protein at 2.4 Å resolution (Fig. 2a, Table 1). To rule out structural influences of the fusion, we also generated a reversed and cleavable fusion protein (Mre11cd 7–413 fused to Nbsmir 474–531) (Supplementary Fig. 2d). After proteolytic cleavage of the linker peptide, the X–ray crystal structure of the unfused complex of Mre11cd and Nbsmir was determined at 2.2 Å resolution. Both structures are highly similar, arguing against influences of fusion and construct design (Supplementary Fig. 2e). All crystallization conditions for fused or unfused Nbs1mir–Mre11cd contained citrate ions, which chelates one Mn2+ ion from the Mre11 active site. To yield crystals of Nbs1mir–Mre11cd with two Mn2+ ions per active site, we added 50 mM MnCl2 to the crystallization condition (Table1, Supplementary Table 2).
Figure 2. Structure of the Nbs1mir–Mre11cd complex.
(a) Structure of the two Nbs1mir (magenta) bound to the Mre11cd dimer (cyan/blue phosphodiesterase and grey capping domains), shown as ribbon representation with highlighted secondary structures. Nbs1 binds with “interaction region 1” around the outside of the phosphodiesterase domain. One of the two Nbs1 additionally binds with “interaction region 2” to two “signaling loops” at the Mre11 dimer interface.
(b) Molecular surface representation of the Mre11 dimer with bound Nbs1 molecules highlights the asymmetric bridging of the Mre11 dimer by Nbs1 “interaction region 2”.
(c) Domains and motifs of S. pombe Nbs1.
(d) Mre11 interaction region 1 of Nbs1 (magenta) binds to the outside of Mre11’s phosphodiesterase domain (cyan) with two secondary structure elements (αA and b1) and partially polar, partially hydrophobic interface. Key residues from both interaction partners are annotated.
(e) Interaction region 2 of Nbs1 (magenta) contains the highly conserved NFKxFxK motif and binds asymmetrically across the Mre11 dimer (blue/cyan) via a network of hydrogen bonds and π–stacking interactions (highlighted). The structure is rotated by 180° around its central vertical axis in comparison to Figure 2a.
The Nbs1mir–Mre11cd structure reveals a complex with 2:2 stoichiometry, but with striking internal asymmetry (Fig. 2a). Two Nbs1mir molecules wrap around the outside of the two Mre11 phosphodiesterase domains, each binding in a highly extended conformation via a α–helix–β strand motif (Interaction region 1). However, one of the two Nbs1mir molecules additionally binds across the Mre11 dimer interface, forming a second interaction opposite the nuclease cleft (Interaction region 2) (Fig. 2a,b). Consequently, a highly conserved “NFKxFxK motif” (residues 518–526) from one Nbs1mir bridges both copies of the eukaryote–specific loop insertion in the Mre11 dimer. Binding of the second NFKxFxK motif from the other Nbs1 molecule is sterically excluded at this site, “breaking” the symmetry of the Mre11 dimer. Binding of the NFKxFxK motif at the Mre11 dimer interface is mediated by the eukaryote–specific insertion loops (one from each Mre11 protomer). As a result, these loops undergo a substantial structural change and a disorder to order transition, leading us to name these as “latching loops”. The peculiar interaction of Nbs1 with the Mre11 dimer suggests that Nbs1 plays an active role in the functional architecture of the MRN complex.
Details of the Nbs1mir–Mre11cd interfaces
Interaction motif 1 is polar with a few conserved hydrophobic anchor points. Helix αA (Nbs1 Asp477–Arg486) attaches to the Mre11 loop emerging from the metal–coordinating active site motif IV (Fig. 2c and Supplementary Fig. 3a). This orients the N–terminal region of Nbs1 (absent in our structure) towards the entry/exit of Mre11’s nuclease cleft, placing repair and checkpoint factors that interact with the Nbs1 FHA and BRCT domains near DNA26,27. Nbs1 residues Leu487–Gly491 bind as a loop across the short Mre11 helix element αD, creating a hydrophobic interaction between the highly conserved residues Mre11 Phe238 and Nbs1 Leu490.
Interaction motif 2 (“NFKxFxK”) binds via a hydrogen bonding and π–stacking network across both latching loops of Mre11 (Fig. 2d, Supplementary Fig. 3b,c). It is itself pseudosymmetric and pseudosymmetrically binds both latching loops in the Mre11 dimer via similar contacts. Residues of the “NFKxFxK” motif that make direct contact to Mre11 are almost invariant among Nbs1 species, suggesting the high functional relevance of this interaction (Supplementary Fig. 3b).
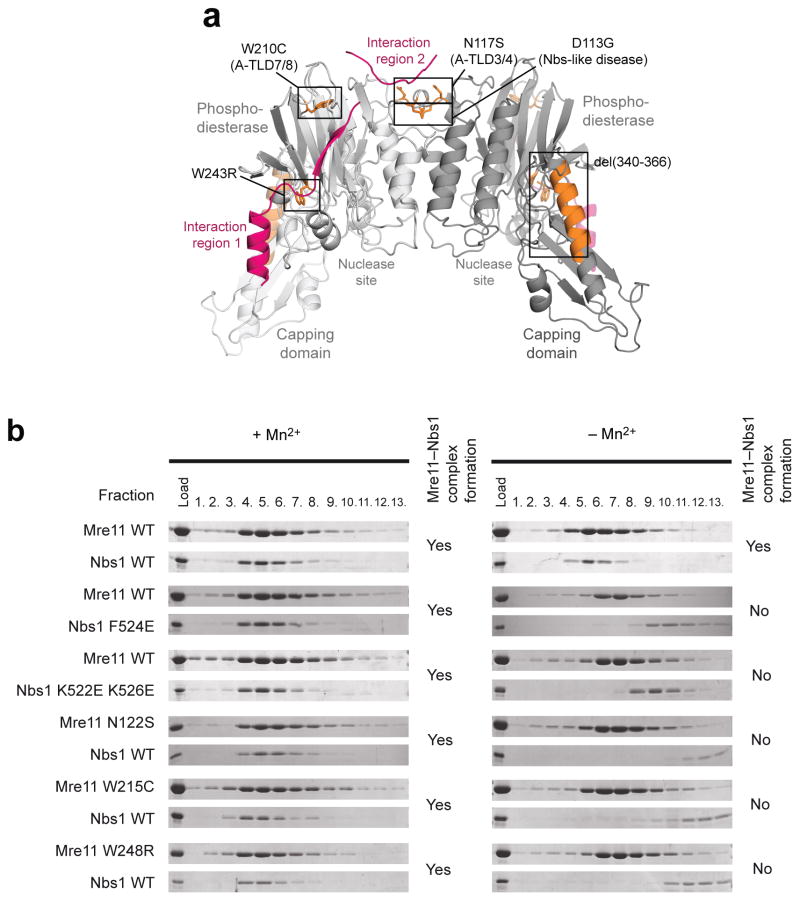
Analysis of A–T like and NBS like disease mutations
Our results provide a structural framework for the molecular pathologies of several human A–TLD and NBSLD mutations (Fig. 3a and Supplementary Fig. 4). Most remarkably, the human equivalent of SpMre11Asn122 – the residue that sandwiches the invariant Nbs1 phenylalanine finger in the Mre11 dimer interface – is mutated in A–TLD3/4 (ref. 41) (human Mre11 N117S, see Fig. 3a and Supplementary Fig. 4a). Human Mre11 W210C leads to A–TLD 7/8 (ref. 46), and the equivalent residue SpMre11 Trp215 caps the three stranded shared β–sheet between Mre11 and Nbs1 (Fig. 3a and Supplementary Fig. 4b). Thus, A–TLD 7/8 likely affects Nbs1 interaction at interaction region 1. The same structural region is mutated in S. cerevisiae mre11(ts) (human Mre11 P162S), which also exhibits compromised MRX complex formation47. More recently, it was shown that the compound heterozygous mutation of human W243R with the deletion mutation Del(340–366) is associated with A–TLD, the latter mutation mapping to the central long helix of the capping domain42 (Fig. 3a). Such a severe truncation probably destabilizes the protein, explaining the decreased Mre11 levels in these patients, but is also expected to affect the precision of interaction with Rad50. The point mutation W243R in the other allele maps to SpMre11 Trp248, which forms the hydrophobic core of the structural region linking the nuclease active site and contact site to SpNbs1 αA (Supplementary Fig. 4c). Thus, human Mre11 W243R could affect both nuclease activity as well as Nbs1 binding.
Figure 3. Structural basis for A–TLD and NBS–like disease mutations.
(a) A–TLD mutation sites (orange sticks) are found throughout the Mre11 dimer (light/dark grey cartoon model). All point mutations are located in places that are critical for the interaction with Nbs1 (magenta cartoon model).
(b) Interaction studies of SpNbs1428–613 with SpMre11cd, analyzed by co–migration on gel filtration. Shown are SDS–PAGE bands from gel filtration fractions for both SpMre11cd and SpNbs1428–613.
Another recent patient study reported that a compound heterozygous mutation of human Mre11 D113G is linked with a so far unreported NBSLD44. As described above, the equivalent SpMre11 Asp118 forms a salt bridge across Mre11 protomers (Fig. 3a and Supplementary Fig. 4d). Thus, differential impacts on Mre11 dimer functionality and Nbs1 interaction may account for the observed phenotypic differences.
In gel filtration analyses testing the effect of A–TLD mutations and mutations in the Nbs1 NFKxFxK motifs, SpNbs1 containing F524E or K522E K526E mutations interacted with SpMre11 in the presence but not in absence of Mn2+ (Fig. 3b), reflecting a compromised but not abolished interaction. Similarly, A–TLD like mutations SpMre11 N122S, W215C and W248R interact with Nbs1428–613 in the presence but not absence of Mn2+. Although a direct structural link between Nbs1 binding and metal binding to the Mre11 active sites is unexpected, it is consistent with a mutation in the Mre11 phosphodiesterase motif that was found to disrupt MRX in vivo48.
Impact of Nbs1 binding on Mre11 dimer conformation
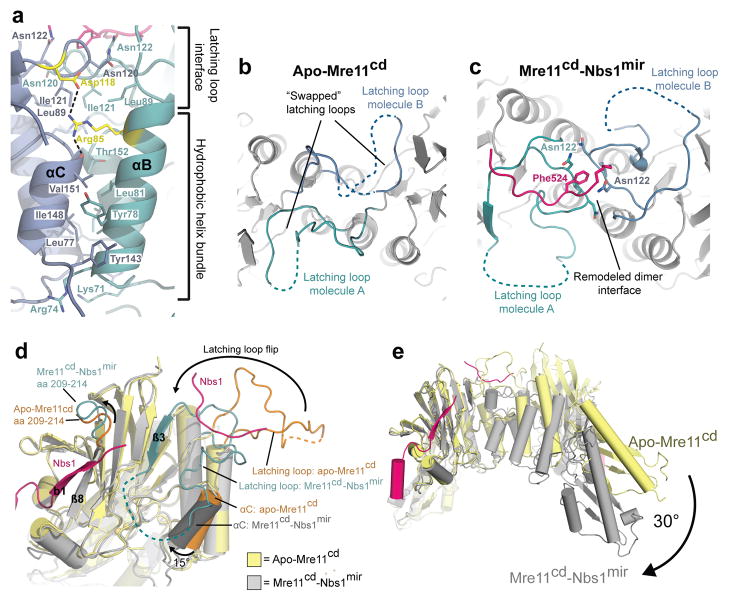
The latching loops not only bind Nbs1, but substantially extend the Mre11 dimer interface, which can be split into two functional parts (Fig. 4a). The first part, found in Mre11 from all phylogenetic domains, is a 4–helix bundle between αB and αC from each protomer. The second part, unique to eukaryotic Mre11, comprises the latching loops that interact with each other via an extended loop in the presence (but not absence) of Nbs1. As seen from the structures of Nbs1mir–Mre11cd and apo–Mre11cd, Nbs1 induces a disorder–to–order transition and geometric rearrangement of the latching loops (Fig. 4b–d), flipping them almost 180° towards the basal phosphodiesterase core. The Mre11 dimer interface helix αC rotates by 15° towards a more tilted orientation in respect to the Mre11 dimer axis. A second eukaryotic loop insertion (residues 209–214) moves to cap the beta sheet interaction between β8 of Mre11 and β1 of Nbs1mir.
Figure 4. Conformational impact of Nbs1 binding on the Mre11 dimer configuration.
(a) Details of the Mre11 dimer interface in the Nbs1mir–Mre11cd structure. The Mre11 monomers are colored in blue and cyan, while Nbs1mir is colored in pink. The dimer interface can be distinguished into two different regions, the hydrophobic four–helix and the latching loop interface. Both motifs are connected with each other via salt bridges between the conserved residues SpMre11 Arg85 and Asp118 (colored in yellow). The structure is rotated by 180° around its central vertical axis in comparison to Figure 2a.
(b) A top view of apo–Mre11cd along the Mre11 dimer axis shows the latching loops indicated in cyan (molecule A) and blue (molecule B).
(c) A top view of Nbs1mir–Mre11cd similar to (B) on the latching loop conformation in presence of the bound Mre11 interaction region 2 of Nbs1.
(d) Nbs1 binding causes several conformational rearrangements in Mre11. Shown is an overlay of Mre11 monomers from apo–Mre11cd and Nbs1mir–Mre11cd structures.
(e) An overlay of apo–Mre11cd and Nbs1mir–Mre11cd by aligning of Nbs1mir–Mre11cd to just one apo–Mre11cd protomer reveals a distinct macromolecular change. The dimer angle of Nbs1mir–Mre11cd is rotated by 30°, in comparison to apo–Mre11cd, towards a more compact and closed conformation.
Nbs1 also induces a 30° rotation in the dimer angle towards a conformation with a narrower nuclease cleft (Fig. 4e). The highly conserved arginine SpMre11 Arg85 appears to be a critical element of the Mre11 dimer conformation, reaching across the dimer interface and binding to the C–terminus of helix αC at the other protomer, thereby stabilizing the geometry of the 4–helix bundle (Fig. 4a). Arg85 is coordinated in this conformation by a salt bridge with SpMre11 Asp118 of the latching loop; this interaction is missing in the absence of Nbs1, enabling the 4–helix bundle to shift.
Notably, this buried interface salt bridge between SpMre11 Arg85 and Asp118 is evolutionarily conserved and a recent clinical study linked the compound heterozygous mutation of human Mre11 D113G with NBSLD44. It is remarkable that hypomorphic mutations that are in close spatial proximity and are located in the same functional loop such as human Mre11 D113G and N117S (A-TLD 3/4, ref. 41) lead to different patient phenotypes. However, our observation that the latching loop is not only important for Mre11–Nbs1 interaction but also stabilizes Mre11 dimer conformation offers a plausible explanation for this observation. Thus, if the A–TLD and NBSLD mutations differentially affect Nbs1 interaction and Mre11 dimer geometry and stability, this could differentially impact on repair and checkpoint functions of MRN and hence disease phenotypes.
In vivo functions of Mre11 latching loops
To understand the function of the Mre11 latching loops and to clarify the different phenotypes of A–TLD and NBSLD mutations in this region, we studied the consequences of certain mutations in vivo using S. cerevisiae as a model organism. In particular, we analyzed the charged interface residue ScMre11 Arg76 at the helix bundle and ScMre11 N113S, equivalent to the A–TLD3/4 mutation of human Mre11 N117S41 (Arg85 and Asn122 in S. pombe, Fig. 4a). We also analyzed ScMre11 Asp109 at the latching loop (Asp118 in S. pombe, Supplementary Fig. 3a), mutated in human NBSLD44 (human Mre11 D113G).
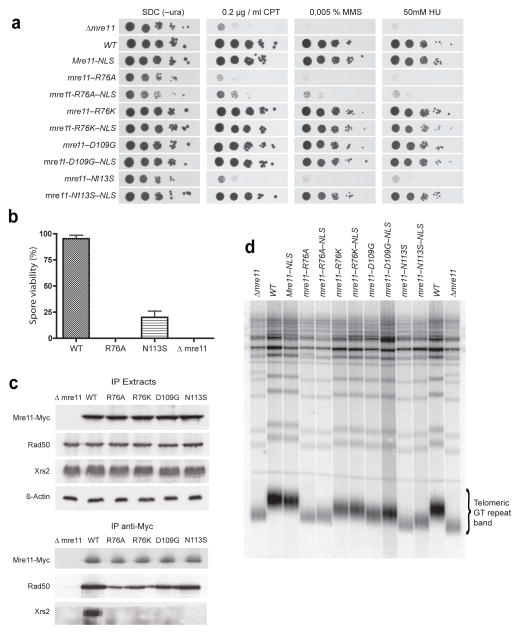
We utilized a plate survival assay to monitor the consequences of the above mutations in response to various genotoxic agents. mre11–R76A cells are as sensitive as mre11Δ cells to methyl methanesulfonate (MMS), hydroxyurea (HU) and the Topoisomerase 1 inhibitor camptothecin (CPT), whereas the conservative R76K mutation causes no detectable hypersensitivity phenotype (Fig. 5a). A comparable sensitivity is observed for mre11–N113S cells, while the NBSLD analogous D109G mutation does not show any visible defect. Consistent with the plate survival assay, homologous recombination is impaired in mre11–R76A and –N113S strains when analyzed in a mating type switch assay49 (Supplementary Fig. 5a). Thus, ScMre11 R76 and N113 are crucial for the mitotic repair functions of Mre11 in S. cerevisiae.
Figure 5. In vivo characterization of Mre11 latching loop compromising mutations in S. cerevisiae.
(a) Plate survival assays by serial dilutions reveal sensitivity of latching loop targeting mutations to camptothecin (CPT), methyl–methanesulfonate (MMS) and hydroxyurea (HU).
(b) Spore viability is strongly reduced in ScMre11 R76A and ScMre11 N113S diploid strains. Spore viability was calculated as the percentage of viable spores on total spores after tetrad dissection. Error bars represent s.e.m. spore viability obtained in two independent experiments, with 64 spores analyzed for each strain in each experiment.
(c) Mre11–Rad50–Xrs2 complex formation defects of Mre11 latching loop targeting mutations tested by co–immunoprecipitation. Cell extracts were immunoprecipitated with an anti–Myc antibody against Mre11-Myc and proteins were visualized by Western blotting with anti–Myc (Mre11), anti–Rad50, and anti–Xrs2 antibodies respectively. β–Actin was included as a loading control.
(d) Telomere lengths of Mre11 latching loop targeting mutations. Southern blot of XhoI–digested yeast DNA probed with a poly(GT)20 oligonucleotide specific for telomeric repeats is shown. The bracket indicates the telomeric GT repeat band derived from Y′ element–containing chromosomes.
Due to Mre11 functions in meiosis, we also compared the meiotic phenotypes of the mutants that displayed growth defects in the plate survival assays. Both N113S and R76A mutations strongly reduce spore viability (Fig. 5b), but whereas the mre11–N113S mutant generates viable spores at reduced levels compared to wild–type cells, we could not observe any viable mre11–R76A spores. Thus, the Mre11 dimer interface mutation ScMre11 R76A resembles an mre11 null phenotype regarding both mitotic repair and meiotic functions.
We next monitored if the phenotype of the mutants was due to defects in the cellular localization of Mre11. Both N113S and R76A mutations strongly decrease the nuclear accumulation of Mre11 (Supplementary Fig. 5b). The DNA damage sensitivity caused by XRS2 deletion can be rescued by adding a nuclear localization sequence (NLS) to Mre1150, so we generated Mre11 variants that contain an additional SV40 large T–antigen NLS at the C–terminus. Interestingly, although all NLS–tagged Mre11 mutants relocated to the nucleus, the NLS only rescued the repair defects of mre11–N113S but not of mre11–R76A strains in plate survival and mating type switch assays (Fig. 5a and Supplementary Fig. 5a,b).
Since nuclear mislocalisation caused by a defect in Xrs2 interaction cannot explain the severe phenotype of mre11–R76A, we examined the integrity of the MRX complex for this and other Mre11 mutations. Protein levels of Mre11, Rad50 and Xrs2 were similar in cell lysates from wild–type and mutant cells, ruling out general misfolding and degradation as the main cause for the observed phenotypes (Fig. 5c). Mre11 R76K, N113S and D109G form stable complexes with Rad50 but the interaction of Mre11 R76A with Rad50 is reduced (Fig. 5c). In contrast, all Mre11 mutants have impaired interaction with Xrs2, suggesting that the reduced affinity for Xrs2 does not necessarily lead to repair defects and that Mre11 R76K and D109G must retain a residing ability to bind Xrs2 in order to localize in the nucleus.
ScMre11 Arg76 maps to the dimer interface of Mre11, and R76A is not rescued by an NLS, indicating that it may impact dimerization. To test this, we introduced a second plasmid, coding for a C–terminally HA–tagged Mre11, into the myc–tagged mre11 shuffle strain to create “mixed” dimer complexes. We then immunoprecipitated Myc–tagged Mre11 and probed the dimer integrity by using an antibody against HA–tagged Mre11 (Supplementary Fig. 5c). Mre11 dimer stability was reduced in the mre11–R76A strain compared to WT, indicating that the Mre11 latching loops not only play a crucial role in binding to Nbs1/Xrs2 but also in stabilizing Mre11 dimers in S. cerevisiae.
Finally we tested how mutations targeting the latching loops affect the function of Mre11 in telomere length maintenance (Fig. 5d). This process depends on all components of the MRX complex, with null mutations of any of these factors resulting in telomeres of shorter but stable length51. Our analyses revealed that, similar to cells totally disrupted for Mre11 function (mre11Δ), both the R76A mutant and the A–TLD analogous mre11–N113S mutant strain possess significantly shorter telomeres than wild–type cells, while the NBSLD analogous D109G mutation displayed shortened telomeres of intermediate length between those in wild–type and mre11Δ cells. In contrast, no significant telomere shortening could be observed in strains bearing the conservative R76K mutation. Interestingly, addition of the NLS had no or only very minor rescue effects on the telomere lengths of N113S and D109G mutant strains.
Taken together, these results show that the mitotic repair and recombination deficiency of mre11–N113S can be explained by a nuclear localization defect. Indeed, this mutant is still competent in the Xrs2 independent mitotic repair functions and its sensitivity to genotoxic agents can be rescued by addition of a NLS to Mre11. In contrast, our data suggest that telomere maintenance depends on a stable interaction between the NFKxFxK motif of Xrs2/Nbs1 and the latching loops of Mre11, which is probably still compromised in mre11–N113S–NLS. A summary of all observed Mre11 mutant strain phenotypes is shown in Supplementary Table 3.
The R76A mutation impairs not only Xrs2 interaction, but also more directly impacts the Xrs2–independent repair functions of MR by compromising the Mre11 dimer structure. However, purified SpMre11cd R85A still forms dimers and interacts with Nbs1428–613, although the Nbs1 interaction is Mn2+–sensitive (Supplementary Fig. 5d), indicating that it is weakened. Thus, it is unlikely that the equivalent ScMre11 R76A mutation completely disrupts Mre11 dimers.
Discussion
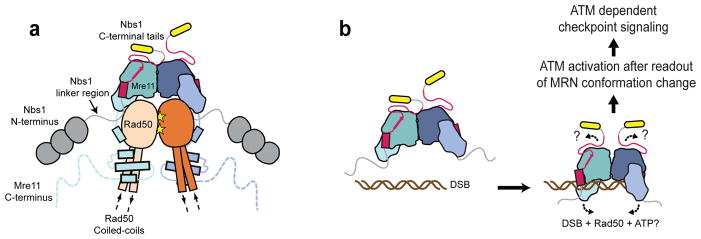
Our results provide a structural framework for eukaryotic Mre11 and its complex with Nbs1, revealing the molecular basis of a core part of the MRN complex (Fig. 6a). Nbs1 wraps as an extended chain around the Mre11 phosphodiesterase domain with 2:2 (M:N) stoichiometry, but only one of the two Nbs1 completely binds to Mre11 via the NFKxFxK motif as the observed asymmetric binding mode sterically excludes binding of the second Nbs1 NFKxFxK motif to the latching loops. Although we have crystallized only a small portion of Nbs1, the observed complex likely harbors most if not all of the interaction sites between Nbs1 and Mre1150. However, it is thought that there exists, at least for the human MRN complex, an additional interface between Nbs1 and Rad50, which might map to the region N–terminal to the co–crystallized fragment52.
Figure 6. Models for the principle architecture of eukaryotic MRN and MRN dependent DNA double–strand break signaling.
(a) Proposed model for the principle architecture of the eukaryotic MRN complex. Shown is the ATP bound conformation observed in the homologous archaeal and eubacterial crystal structures20,57.
(b) Speculative model for DNA double–strand break signaling by a DNA– and Rad50–induced structural switch in MRN. Such a switch could alter the Nbs1 binding geometry at the Mre11 dimer interface and hence reposition the adjacent ATM binding motifs (yellow). ATM could be activated after sensing the DNA bound MRN conformation via a multi–contact interface with MRN including binding to the C–terminal Nbs1 region. Activated ATM then promotes downstream checkpoint signaling by its kinase activity.
Recently, a crystal structure of human Mre11 was published that differs significantly from the dimeric conformation of the S. pombe Mre11 structure presented here53, despite an otherwise identical fold (Supplementary Fig. 6a). Whereas in S. pombe the canonical four helix bundle forms the dimer interface19,54, the human Mre11 dimer is connected by a disulfide bond between helix aC from each monomer (Supplementary Fig. 6b,c). The responsible cysteines are not conserved between H. sapiens and S. pombe, or even all vertebrates (e.g. Xenopus laevis). Thus, at this time, it is unclear what functional state the conformation of the human Mre11 structure in the absence of Nbs1 displays.
The extended interface of Nbs1 along the phosphodiesterase domains of Mre11 gives insights into the molecular pathology of A–TLD. A–TLD associated point mutations along these interfaces validate the observed interactions and show that binding between Mre11 and Nbs1 is mediated by several distributed, independent interaction sites. (Fig. 4). Thus, single point mutations are unlikely to completely disrupt the complex, explaining the hypomorphic phenotype of different A–TLD variants. Furthermore, the mitotic repair and recombination functions of MR are largely unaltered by the ScMre11 N113S mutation, if nuclear levels of MR are recovered by an NLS on Mre1141 (Fig. 5). This argues for proficient Mre11 nuclease and Rad50 ATP binding capabilities in of MRN in ATLD3/4. However, the NLS scarcely rescued telomere shortening in the mre11–N113S, indicating that a stable interaction between the NFKxFxK motif of Xrs2/Nbs1 and the latching loops of Mre11 may be crucial for MRX–mediated telomere maintenance. This is in agreement with a former study in which an acidic point mutation in the Xrs2 NFKxFxK motif led to shortened telomere length50. One major role of the MRX complex in telomere maintenance is to recruit the checkpoint kinase Tel1 to short telomeres55, so the phenotypes observed may represent defects in Tel1 recruitment or activation.
In contrast, the arginine finger mutation (Arg76 in S. cerevisiae and Arg85 in S. pombe) cannot be rescued by NLS-tagging, suggesting that this mutation induces a fundamental defect in Mre11. As SpMre11cd R85A still formed dimers and interacted with Nbs1mir in vitro, and prokaryotic Mre11 lack this motif altogether, it is surprising that the arginine finger mutation has such a dramatic phenotype19,54. Thus, we propose a role of this arginine in orienting the Mre11 dimer in a particular conformation that is important for MRN function. In addition, this arginine finger coordinates a conserved aspartate in the latching loop (Asp109 in S. cerevisiae and Asp118 in S. pombe), mutated in NBSLD, and the analogous mutation in S. cerevisiae resulted in significantly shortened telomeres, We assume that this stems from a partly destabilized Mre11 latching loop coordination and Xrs2 interaction in mre11–D109G, which impairs the Xrs2 dependent telomere maintenance functions of the complex.
The asymmetric bridging of Mre11 dimers by a single Nbs1 subunit, paired with the uncovered intrinsic flexibility of the eukaryotic Mre11 dimer, is perhaps the most significant and unexpected finding of our structural and functional analyses. Nbs1 side chains in direct contact with Mre11 at the latching loops are more or less invariant across species41,56, so this asymmetric bridging appears to be a conserved feature of the Nbs1–Mre11 interaction. We do not know what the other “free” NFKxFxK motif does in the complex. One possibility is that one Nbs1 controls the functional architecture of MR while the other may interact with other repair proteins or DNA. This could provide an asymmetry that may reflect the necessarily asymmetric protein interactions at a DNA end.
Because the Mre11 dimer structure is bridged by the pseudo–symmetric NFKxFxK motif, it is tempting to speculate that conformational changes in the Mre11 dimer and Mre11–Nbs1 interaction are important for MRN function. In support of this, the dimer interface residues of bacterial and archaeal Mre11 undergo conformational changes upon Rad50 dependent ATP binding 19,20,57,58. Additionally, DNA bound archaeal Mre1159 and S. pombe Mre11 structures have different angles in the Mre11 dimer, which might represent different functional states of the protein (Supplementary Fig. 6d,e). An Mre11 dimer angle rotation, controlled by Nbs1 on one side and by DNA and/or Rad50 plus ATP on the opposing side of Mre11, might be sensed by ATM via the C–terminal tails of Nbs1, adjacent to the NFKxFxK motif (Fig. 6b). This region of Nbs1 contains an ATM interaction motif30 which directly stimulates ATM in Xenopus egg extracts34,60.
While the mechanistic link between Rad50 and DNA binding to Mre11 requires further studies, our data suggest that Mre11 dimer flexibility and its control by Nbs1 could be an important part of MRN function.
Online Methods
Expression constructs
Constructs were generated using standard techniques and verified by DNA sequencing. Cloning details are provided in the Supplementary Methods. All primers used for cloning in this study are listed in Supplementary Table 1.
Expression and purification of recombinant proteins
All proteins, except selenomethione labelled Nbsmir–Mre11cd, were expressed in Escherichia coli Rosetta DE3 (Stratagene), grown in Luria–Bertani medium with 250 μM IPTG by shaking at 18 °C overnight. Harvested cells were lysed in a buffer containing 50 mM Hepes pH 7.5, 300 mM NaCl (Mre11) or 500mM NaCl (Nbs1), 2mM EDTA, 2 mM β–mercaptoethanol and purified by Glutathion–Sepharose affinity chromatography (GE Healthcare). Afterwards the GST–Tag was proteolytically cleaved by adding Prescission protease (GE Healthcare) or Tobacco Etch Virus (TEV) protease depending on the fusionprotein. GST was removed by additional Glutathione–Sepharose affinity chromatography. The flowthrough was collected and further purified by size exclusion chromatography on a S200 column (GE Healthcare). All buffers used for size exclusion chromatography can be found in Supplementary Table 2. Expression of selenomethionine containing Nbsmir–Mre11cd fusion protein was performed in Escherichia coli B834 Rosetta (DE3) in a shaking culture of minimal medium 1 containing 50 mg/L selenomethionine with 250 μM IPTG at 18 °C overnight. Nbsmir–Mre11cd SeMet was purified similar to native protein but in the presence of 5mM β–mercaptoethanol.
Protein crystallization
All proteins were crystallized at 20°C using the hanging drop vapour diffusion technique and mixing 1 μl protein solution with 1 μl reservoir solution. The preparation buffers, protein concentrations, screen compositions and cryo protectants for all crystallized proteins are listed in Supplementary Table 2. Cryocooling was achieved by soaking the crystals for 30 seconds in mother liquor solution containing the cryo protectant and flash freezing in liquid nitrogen.
X-ray Diffraction Data Collection, Phasing and Refinement
Nbsmir-Mre11cd was crystallized in space group P212121 with two molecules per asymmetric unit. Data to 2.8 Å resolution were collected at the Swiss light source (Villigen, Switzerland) and the European synchrotron radiation facility (Grenoble, France) and phased by selenium single anomalous dispersion using selenomethionine derivatized protein. After model building, the structure was refined against 2.4 Å resolution native data. Mre11cd and unfused Nbsmir plus Mre11cd were both crystallized in space group P212121, and their 3.0 Å and 2.2 Å resolution structures, respectively, were determined by molecular replacement using the Nbsmir-Mre11cd as search model. Data collection and refinement statistics are listed in Table 1. Further details are described in the Supplementary Methods.
Analytical size exclusion chromatography
10 nmol Mre11 and 7.5 nmol Nbs1 proteins were mixed and dialysed against the chromatography running buffer containing 20 mM Hepes pH 7.5, 100 mM KCl, 5% glycerol, 2mM β–mercaptoethanol and 1mM MnCl2 or 2mM EDTA respectively. We loaded 350 μl protein sample onto a Superdex S200 10/300 GL column (GE Healthcare) with a flow rate of 0.5 ml/min. Elution fractions were analysed for Mre11 and Nbs1 by SDS–PAGE and Coomassie staining.
Nuclease activity assay
Mre11 nuclease activity was tested using a 6–FAM– 5′ labelled 60mer poly(dT) oligonucleotide. For each reaction, 10 nM DNA was incubated with 5 μM of Mre111–413 or Mre111–413 H134S proteins in 10 μl buffer containing 20 mM Hepes pH 7.5, 100 mM KCl, 5% glycerol, 5mM MnCl2 and 2mM mercaptoethanol for 2h at 37°C. The reaction was stopped by addition of 3 μl loading buffer (10mM Tris, pH 8.0–16.6% formamide, 16.6% glycerol, 5mM EDTA) and incubated for 5min at 95°C. Reaction products were resolved on a denaturating 18% acrylamide gel in 1× TBE buffer containing 8M Urea. Gels were imaged with a Typhoon 9400 fluorescence scanner (GE Healthcare) using the green–exited (532nm) fluorescence mode.
S. cerevisiae strains for in vivo studies
All assays were carried out using a S. cerevisiae W303 Δmre11 strain (Genotype: mre11delta::KanMX4, Mata/α; ura3–1, trp1–1, his3–11,15; leu2–3, 112; ade2–1; can1–100) 2. The respective Mre11 variant was introduced by transformation of a pRS416 Mre11–13Myc plasmid.
Plate survival assays
One loop of freshly growing cells from a plate was resuspended in 1 ml H2O. Five 10 fold dilutions were prepared and 6 μl of each dilution spotted onto the corresponding plates: SDC (–Ura), SDC (–Ura) + 0.2 μg/ml Camptothecin (CPT), SDC (–Ura) + 0.005% methyl methanesulfonate (MMS), SDC (–Ura) + 50 or 200 mM hydroxyurea (HU).
Spore viability assay
Homozygous diploids mre11Δ/mre11Δ were transformed either with the empty plasmid (pRS416) or with a plasmid carrying the MRE11, mre11R76K, or mre11N113S alleles. Transformants were grown on selective medium plates in order to maintain the plasmid and subsequently moved on sporulation plates at 25°C for 2 days to allow sporulation. The spores of the resulting tetrads were separated using a tetrad dissector and incubated at 30°C for 2 days. Spore viability was then scored as the ability of spores to form a colony.
Mating type switching assay
mre11Δ cells were transformed either with the empty plasmid (pRS416) or with a plasmid carrying the MRE11, mre11R76A, mre11R76A–NLS, mre11N113S, or mre11N113S–NLS alleles. The obtained strains were transformed with a pGAL–HO plasmid and grown on double selection medium for the whole experiment. Exponentially growing cells in SCraff–Ura–Leu (raf) were transferred to medium containing galactose to induce HO expression. After one hour cells were transferred to medium containing glucose to allow repair (time zero). StyI–BamHI digested genomic DNA was subjected to Southern blot analysis with a MAT probe that detects 0.9–kb fragments (MATa) in the absence of HO–cut, while HO–induced DSB formation results in generation of an 0.7–kb fragment corresponding to an HO–cut fragment, which can be eventually repaired by HR with donor sequence HMR or HML, generating MATa (0.9–kb) and MATalpha (1.8–Kb) repair products, respectively. The coordinates of the MAT probe are 201082 to 201588 of chromosome III
Co–Immunoprecipitation
All Co–Immuniprecipitation experiments were carried out as decribed before, exept for the use of Protein G beads (Stratagene) instead of Protein A Sepharose 3. The following buffer was used for cell lysis and all washing steps: 50mM Hepes pH 7.5, 140mM NaCl, 10% Glycerin, 0.5% NP–40, 1mM PMSF, 2mM benzamidine hydrochloride, 2 μM Pepstatin, 0.5 μM Leupeptin, 3.3 μM Chymostatin. Samples were detected by SDS–PAGE and subsequent western blot analysis. For western blot analysis of IP inputs, whole cell extracts were prepared using TCA–mediated protein precipitation as described before 4. A monoclonal anti–Myc antibody (Sigma–Aldrich, clone 9E10, produced in mouse) or a monoclonal anti–HA antibody (Abcam, Clone 12CA5, produced in mouse) were used to immunoprecipitate and detect Myc–or HA–tagged Mre11, respectively. Rad50 and Xrs2 were detected using polyclonal rabbit antisera. (generous gifts from J. Petrini) and β–Actin with an monoclonal mouse antibody (Abcam, ab8224).
Immunofluorescence Microscopy
Cells from exponentially grown yeast cultures (W303 Δmre11 cells + P527 mre11–myc mutant plasmids) were fixed with 37% HCOH for 90 min and washed in spheroblasting premix (1.2 M sorbitol, 0.1 M K–phosphate buffer, pH 7.4, 0.5 mM MgCl2). Afterwards, cells were resuspended in 200μl of spheroblasting premix 0.5μg/μl 100T zymolyase and incubated for 30 min at 30°C. Next, cells were pelleted and resuspended in 10× the volume of the cells in spheroblasting premix and a drop of cell suspension loaded onto a polylysine precoated fluorescence microscopy slide. The immobilized cells were washed with blocking buffer (1% BSA powder in PBS (phosphate buffered saline, 2.7 mM KCl, 7.9 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl) and fixed by subsequent incubation with methanol and acetone at −80 °C. Slides were incubated afterwards first with blocking buffer for 10 min and then with the first antibody (mouse α–c–Myc antibody 9E10, Sigma Aldrich, diluted in 1% BSA in PBS) at RT for 2 h. Then the slides were washed with blocking buffer + 0,1 % Triton–X at RT and incubated with the second antibody (α –Mouse IgG (H+L) Alexa Fluor 488, Invitrogen, diluted in blocking buffer) for 1 h. Finally, cells were washed with blocking buffer + 0,1 % Triton–X and cell nuclei stained in SSC buffer (300 mM NaCl; 30 mM Na–citrate, pH 7.0) with DAPI (10mg/ml) for 10 min. Slides were analysed using a fluorescence microscope (Leica Microsystems, Wetzlar).
Yeast DNA extraction and analysis of telomeric DNA
Genomic DNA was isolated from 50 ml culture at OD600 of 1.0. For analysis of telomere length, genomic DNA was digested overnight using XhoI and separated on an 1% Agarose gel in 1× Tris–acetate–EDTA buffer. DNA was transferred to nylon membranes (HybondN+) by capillary blotting using 0.4 N NaOH. Detection of telomeric DNA fragments was performed as described elsewhere5.
Structure based sequence alignments
Protein sequences of Mre11 from eukaryotic organisms Schizosaccharomyces pombe (SpMre11), Saccharomyces cerevisisae, Danio rerio and Homo sapiens were aligned with ClustalW 6. The archaeal Mre11 sequence from Pyrococcus furiosus (PfMre11) was added after calculating a pairwise alignment of PfMre11 and SpMre11cd with the program FATCAT 7 using the pdb–coordinates of PfMre11 (Protein data base entry 1II7) and SpMre11cd as input files. The PfuMre11/SpMre11 alignment was further revised by comparison of the overlaid structures with Pymol (DeLano Scientific).
Supplementary Material
Acknowledgments
We are grateful to John Petrini for his gift of anti–Mre11, anti–Rad50 and anti–Xrs2 antibodies. We thank members of the Hopfner lab for technical support and discussions. We thank Matthew Bennett, Ania Rojowska, Anna Kopetzki and Christophe Jung for help with experimentation. We thank the Max–Planck crystallization facility (Martinsried) for crystallization trials, the staffs of the synchtrotron beamlines for help with data collection and processing and SLS and ESRF for generous beamtime allowance. Research in the K.–P.H. Lab was funded by grants from the German Research Council (SFBs 684, 646, and TR5), the German Excellence Initiative (CIPSM), European Commission (IP DNA repair), and NIH U19AI83025. Research in the K.S. Lab was funded by grants from the German Research Council (SFB 646) and the European Research Council (ERC Starting Grant, Project 204522). Research in the SPJ Lab is supported by grants from Cancer Research UK (C6/A11226), the European Research Council, the European Community’s Seventh Framework Program (FP7/2007-2013) under grant agreement No. HEALTH-F2-2010-259893 and by core infrastructure funding from Cancer Research UK and the Wellcome Trust. SPJ receives his salary from the University of Cambridge, supplemented by Cancer Research UK.
Footnotes
Author Contributions
C.B.S., I.G., B.C., H.F. and C.M. designed experiments, C.B.S., F.S. and A.S. cloned constructs and purified proteins, C.B.S. and F.S. crystallized proteins, C.B.S. and K.L. determined the crystal structures, I.G., C.B.S., B.C. and H.F. carried out the S.°cerevisiae assays, C.B.S. performed analytical size exclusion experiments, C.M. carried out nuclease activity assays, K.P.H. and C.B.S. wrote the manuscript, K.L., I.G. and B.C. contributed to the writing and S.P.J., H.F. and C.M. revised the manuscript, K.P.H. S.P.J. and K.S. supervised the research, K.P.H. initiated the project and designed the research.
Accession codes
The PDB submission will be finished in the next days.
References
- 1.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–6. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113–39. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutation research. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 7.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 8.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahal EA, et al. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle. 2010;9:2866–77. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faure V, Coulon S, Hardy J, Geli V. Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol Cell. 2010;38:842–52. doi: 10.1016/j.molcel.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 15.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–63. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 16.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–6. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 17.de Jager M, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–35. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 18.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–88. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lammens K, et al. The Mre11:Rad50 Structure Shows an ATP-Dependent Molecular Clamp in DNA Double-Strand Break Repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATP{gamma}S complex: understanding the interplay between Mre11 and Rad50. Genes & development. 2011;25:1091–104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nature structural & molecular biology. 2010;17:1478–85. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–4. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–7. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–11. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–21. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Difilippantonio S, et al. Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis. J Exp Med. 2007;204:1003–11. doi: 10.1084/jem.20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 32.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–6. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 33.Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nature structural & molecular biology. 2006;13:451–7. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 35.Yazdi PT, et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–82. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–4. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 37.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–81. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3:1219–25. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Waltes R, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–16. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–86. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 41.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–87. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 42.Uchisaka N, et al. Two brothers with ataxia-telangiectasia-like disorder with lung adenocarcinoma. J Pediatr. 2009;155:435–8. doi: 10.1016/j.jpeds.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 43.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–80. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto Y, et al. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair (Amst) 2011;10:314–21. doi: 10.1016/j.dnarep.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Ueno M, et al. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol Cell Biol. 2003;23:6553–63. doi: 10.1128/MCB.23.18.6553-6563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernet M, et al. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum Mol Genet. 2005;14:307–18. doi: 10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 47.Chamankhah M, Fontanie T, Xiao W. The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics. 2000;155:569–76. doi: 10.1093/genetics/155.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annual review of genetics. 1998;32:561–99. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 50.Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell. 2005;16:597–608. doi: 10.1091/mbc.E04-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–28. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 2009;37:1580–8. doi: 10.1093/nar/gkn1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 54.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–85. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 55.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–61. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shima H, Suzuki M, Shinohara M. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics. 2005;170:71–85. doi: 10.1534/genetics.104.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mockel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams GJ, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol. 2011;18:423–31. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–79. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 1.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–72. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–49. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 4.Janke R, et al. A truncated DNA-damage-signaling response is activated after DSB formation in the G1 phase of Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:2302–13. doi: 10.1093/nar/gkp1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–28. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–5. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.