Abstract
Leucyl-tRNA synthetase (LeuRS) produces error free leucyl-tRNALeu by coordinating translocation of the 3′ end of (mis-)charged tRNAs from its synthetic site to a separate proof-reading site for editing. Here we report co-crystal structures of the Escherichia coli LeuRS-tRNALeu complex in the aminoacylation or editing conformations and show that translocation involves correlated rotations of four flexibly linked LeuRS domains. This pivots the tRNA to guide the charged tRNA 3′ end from the closed aminoacylation state to the editing site. The editing domain unexpectedly stabilizes the tRNA during aminoacylation while a large rotation of the leucine-specific domain positions the conserved KMSKS loop to bind the 3′ end of the tRNA, promoting catalysis. Our results give new insight into the structural dynamics of a molecular machine that is essential for accurate protein synthesis.
Introduction
Leucyl-tRNA synthetase (LeuRS) is a large, multi-domain, class Ia aminoacyl-tRNA synthetase (aaRS) whose essential function in all organisms is to synthesize Leu-tRNALeu for use in protein synthesis. Like several other synthetases, notably the other two class 1a synthetases, valyl- (ValRS) and isoleucyl- (IleRS) tRNA synthetases, LeuRS possesses an error correction mechanism to enhance the specificity of aminoacylation and thus the accuracy of protein synthesis. This post-transfer editing mechanism hydrolytically deacylates tRNALeu that has been mischarged with non-cognate amino acids similar to leucine, such as isoleucine, methionine or non-canonical norvaline1.
LeuRS comprises a main enzyme body (Rossmann-fold catalytic domain and class 1a anticodon binding domain) and four flexibly linked additional domains, denoted zinc (ZN1), editing, leucine-specific and C-terminal (Fig.1a). Proof-reading requires that the 3′ end of the tRNA, which is initially charged (or mischarged) in the so-called synthetic site of the enzyme, translocates to the editing site, located ~35 Å away in an independently folded editing, or CP1, domain2. Previously we and others have characterized various structural and biochemical features of the editing state of LeuRS. These include determination of the crystal structure of the Thermus thermophilus LeuRS (LeuRSTT) tRNALeu complex with the 3′ end of the tRNA bound in the editing site3, elucidation of the structural basis for the amino acid specificity of the LeuRS editing site that binds non-cognate amino acids, but rejects cognate leucine2, and a theoretical analysis of the hydrolytic mechanism4. Recently, a series of benzoxaborole compounds were shown to bind specifically in the LeuRS editing site in a tRNA-dependent fashion5. These compounds form a long lifetime covalent adduct with the tRNA 3′ terminal ribose hydroxyls in the editing site, thus trapping the tRNA on the enzyme and inhibiting aminoacylation5. Benzoxaboroles have potential to be potent antibiotics and are under development against fungal, bacterial and parasitic pathogens5–8.
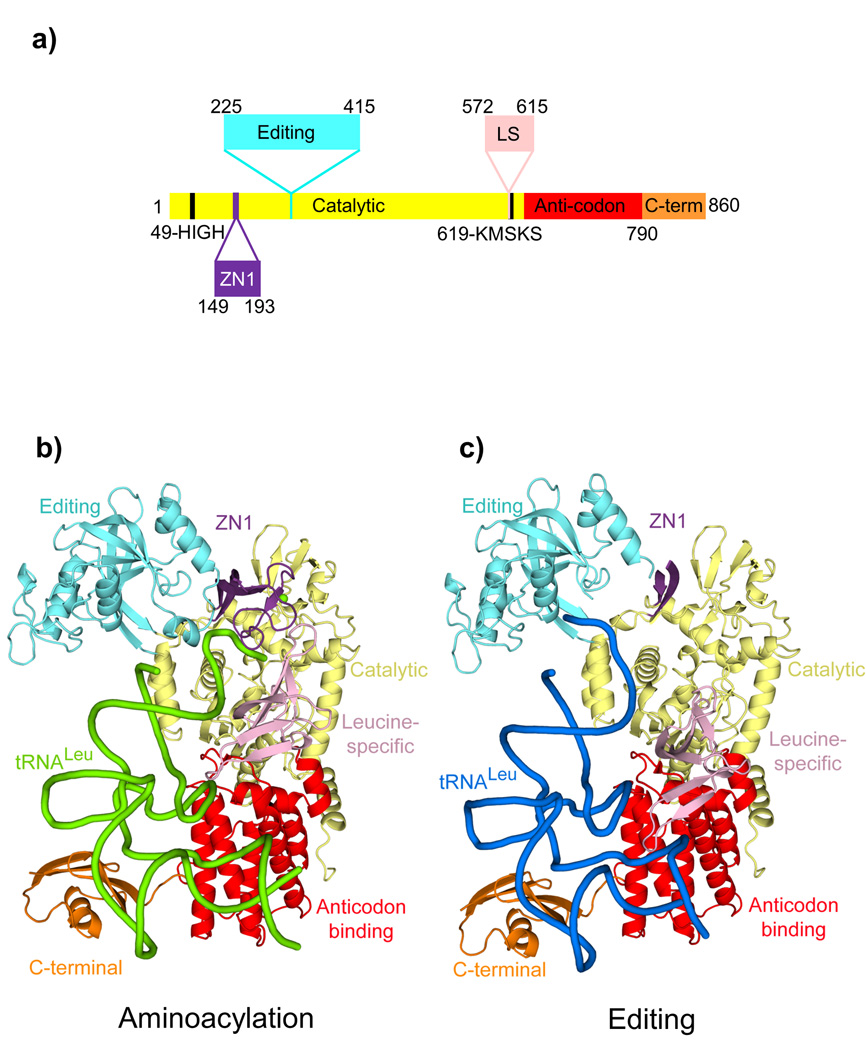
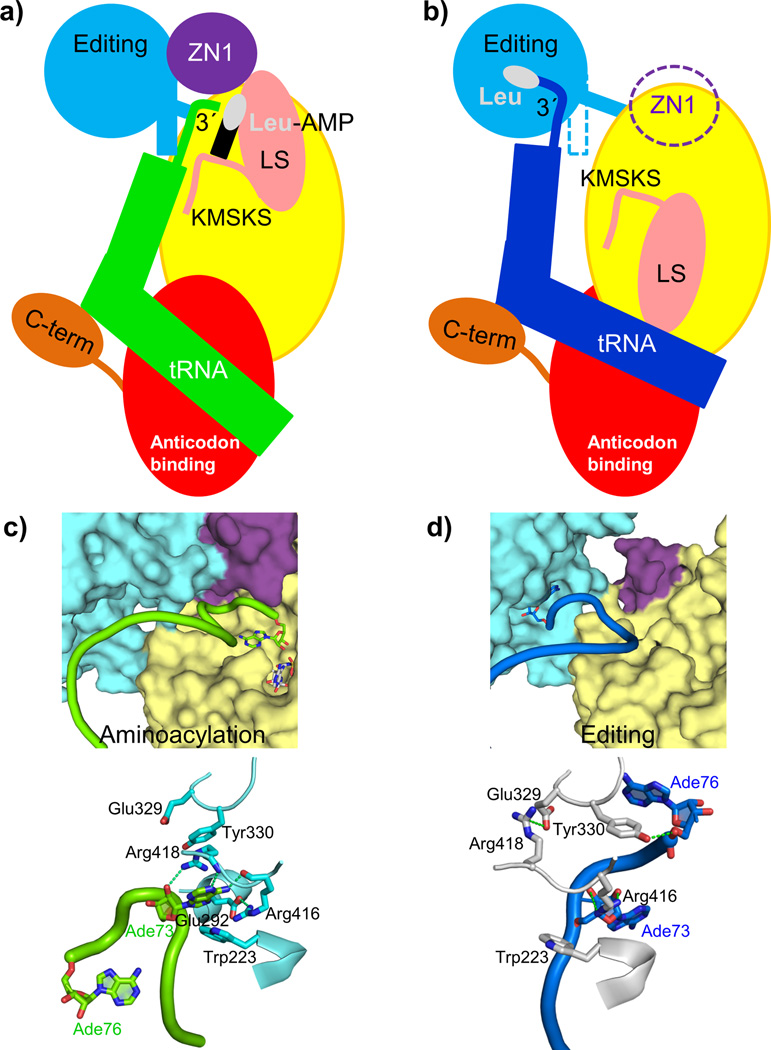
Figure 1. Structures of the E. coli LeuRS-tRNALeu complex in the aminoacylation and editing states.
a. The domain structure of LeuRSEC. Residue numbers indicate domain boundaries. The color code used throughout this paper for the various domains is catalytic (yellow), zinc (ZN1) (purple) with the zinc ion in green, editing (cyan), leucine-specific (pink), anticodon-binding (red) and C-terminal (orange).
b. Aminoacylation conformation with the tRNA in green.
c. Editing conformation with the tRNA in blue. In this state, the ZN1 domain is partially disordered.
While the editing state of bacterial class 1a LeuRS3, ValRS9 and IleRS10 is well characterized, the only published structure of any class 1a synthetase with tRNA bound in the aminoacylation active site is that of an archaeal LeuRS from Pyrococcus horikoshii (LeuRSPH)11. However, archaeal and eukaryotic cytoplasmic LeuRSs are architecturally distinct from bacterial LeuRS12,13. Therefore, the available structures of the editing and aminoacylation states, respectively from bacteria and archaeal systems, are not directly comparable. Furthermore, the LeuRSPH-tRNALeu aminoacylation complex lacks any bound small substrates and does not represent the enzymatically functional aminoacylation state. Indeed, surprisingly few class 1 synthetase-tRNA complexes have been determined in this functional state, the only examples being the class 1b GlnRS14–16 and GluRS17,18 aminoacylation complexes. In other class I co-crystal structures the tRNA is either directed to the editing site9,10 or the 3′ end is disordered or incorrectly bound19–24 or the adenylate is not present25.
Here we present the crystal structure of the functional aminoacylation complex of E. coli LeuRS (LeuRSEC). In this ternary complex structure, the 3′ end of E. coli tRNA5Leu(UAA) is bound in the synthetic site and poised to interact with leucyl-adenylate (present as a non-hydrolysable analogue), the enzyme-bound activated intermediate of the two-step aminoacylation reaction (Fig. 1b). In addition we present high resolution structures of LeuRSEC with the tRNA 3′ end bound in the editing site (Fig. 1c) and leucine or leucyl-adenylate analogue bound in the synthetic site, as well as two different crystal forms of the complex with the tRNA trapped in the editing site by the simplest benzoxaborole compound, 1-hydroxy-3H-2,1-benzoxaborole. Comparison of all these structures enables us to describe the substantial domain and active site rearrangements that accompany tRNA translocation between the two functional conformations, providing new insight into the mechanism of aminoacylation and proof-reading in class 1a synthetases.
Results
Summary of structures determined
Complexes of LeuRSEC with a tRNA5Leu(UAA) transcript (Supplementary Fig. 1ab) and various combinations of small molecule substrates and inhibitors such as leucine, leucinol, ATP, AMP, LeuAMS (the sulphamoyl-analogue of leucyl-adenylate, LeuAMP) and 1-hydroxy-3H-2,1-benzoxaborole, were subjected to extensive crystallization screens with the aim of obtaining crystals of different functional states of the enzyme. Two different crystal forms, both diffracting to up to 2 Å resolution, were obtained with the tRNA directed towards or bound in the editing site. These also grew with or without 1-hydroxy-3H-2,1-benzoxaborole bound in the editing site (where it makes a covalent adduct with the 3′ end of the tRNA5) and with or without leucine or LeuAMS bound in the synthetic site. A different condition gave crystals diffracting to 2.5 Å resolution of the LeuRS-tRNALeu(UAA)-LeuAMS ternary complex with the tRNA in the aminoacylation conformation (Supplementary Fig. 1c). Details of the structure determination are given in the Online Methods and Table 1.
Table 1.
Data collection and refinement statistics
|
E. coli LeuRS + E. coli tRNA (UAA) |
Editing + Leucine |
Editing + benzoxaborole |
Editing + benzoxaborole |
Aminoacylation + LeuAMS |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P21 | C2 |
| Cell dimensions | ||||
| a, b, c (Å) | 77.08, 119.37, 141.10 |
76.18, 118.94, 141.03 |
89.68, 77.11, 91.14 | 158.66, 69.20, 228.84 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
90.00, 90.00, 90.00 | 90.00, 102.24, 90.00 |
90.00, 104.35, 90.00 |
| Resolution (Å)* | 50–2.00 (2.00 – 2.10) |
50–2.08 (2.08–2.15) | 50–2.02 (2.02–2.09) | 50–2.5 (2.50–2.60) |
| Rsym | 7.1 (71.6) | 5.6 (36.7) | 6.6 (68.8) | 8.4 (51.5) |
| I / σI | 10.5 (2.0) | 20.1 (3.1) | 14.2 (1.6) | 11.1 (2.1) |
| Completeness (%) | 99.3 (99.1) | 96.7 (74.2) | 98.2 (84.5) | 90.3 (76.6) |
| Redundancy | 3.90 (3.76) | 3.91 (2.80) | 3.65 (2.81) | 2.89 (2.16) |
| Refinement | ||||
| Resolution (Å) | 2.0 | 2.08 | 2.02 | 2.5 |
| No. reflections work/free |
83506/4411 | 71134/3759 | 74330/3977 | 71885/3826 |
| Rwork / Rfree | 0.210 / 0.257 | 0.199 / 0.244 | 0.208 / 0.249 | 0.188 / 0.250 |
| No. atoms | ||||
| Protein | 6465 | 6517 | 6476 | 2 × 6836 |
| tRNA | 1516 | 1711 | 1777 | 2 × 1653 |
| Ligand | 9 (Leucine) | 9 (AN2679) | 9 (AN2679) | 64 (2 × LeuAMS) |
| Mg2+ | 1 | 2 | 1 | 2 |
| Water/other | 415 | 325 / 4 glycerol | 402 | 169 |
| B-factors | ||||
| Protein | 42.1 | 30.4 | 42.3 | 46.2[A]b/62.5[D] |
| tRNA | 68.6 | 55.6 | 80.9 | 51.3[B]/74.1[E] |
| Ligand | 35.5 | 29.9 | 31.9 | 23.9[F]/36.4[G] |
| Mg2+ | 62.1 | 47.9 | 50.1 | 61.7[M] |
| Water | 45.1 | 31.4 | 39.4 | 41.1[Z] |
| R.M.S. deviations | ||||
| Bond lengths (Å) | 0.014 | 0.011 | 0.013 | 0.014 |
| Bond angles (°) | 1.68 | 1.50 | 1.64 | 1.77 |
Values in parentheses are for highest-resolution shell.
Values are for each molecule in the asymmetric unit with chain indicator given in square brackets.
The LeuRS-tRNALeu-LeuAMS aminoacylation complex structure
The E. coli LeuRS-tRNALeu-LeuAMS ternary complex (Fig. 1b) shows the 3′ end of the tRNA bending back into the synthetic active site, bringing the 2′OH of the Ade76 ribose into the required position to attack the non-reactive leucyl-adenylate analogue; thus the structure closely mimics the functional aminoacylation state. Significantly, only in this state are all four flexibly linked domains of the enzyme fully ordered due to a complex network of mutually stabilizing inter-domain and protein-tRNA interactions. Due to extra contacts between bases 69–76 of the tRNA 3′ strand to multiple domains of LeuRSEC, the tRNA makes considerably more contacts with the synthetase in the aminoacylation state compared to the editing state (Fig 2a, Supplementary Figs. 1a,b and Supplementary Table 1). These extra contacts not only ensure correct selection of cognate tRNAs, but also control the domain re-arrangements that create the catalytically active aminoacylation state. In the editing state, discrimination of cognate tRNA is no longer essential and the key Ade73 identity element26,27 is not recognised by the protein.
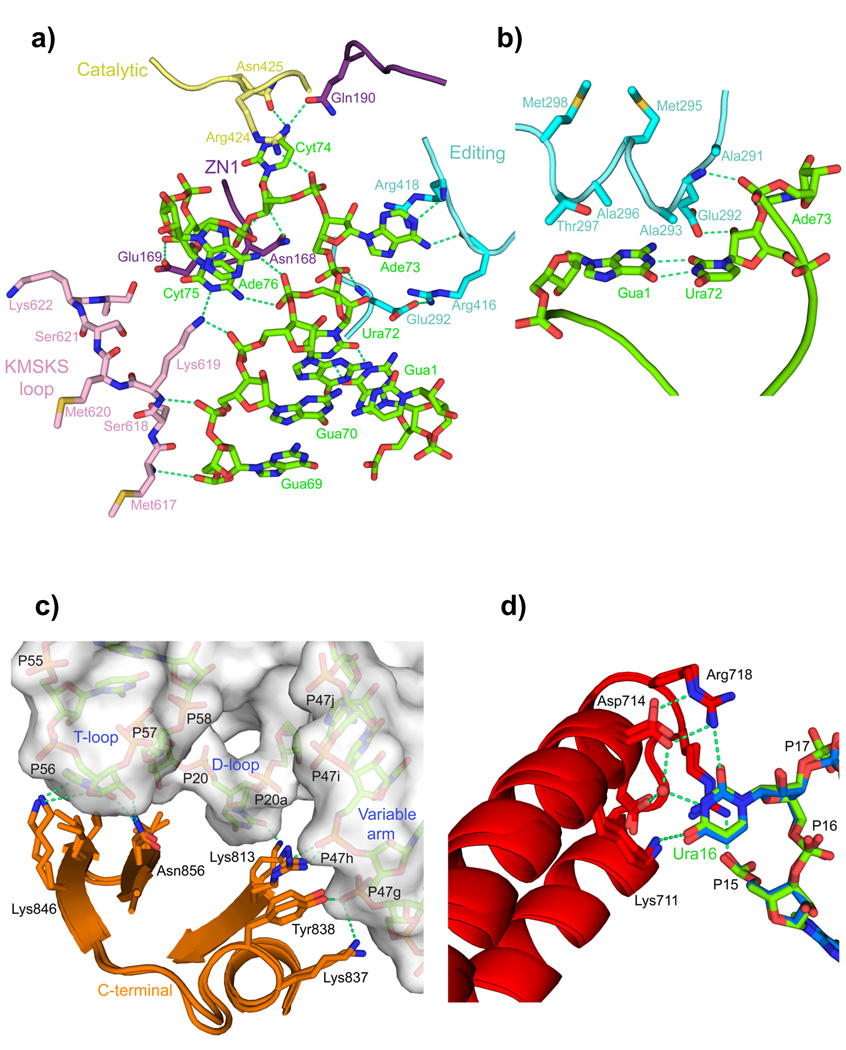
Figure 2. LeuRS-tRNALeu interactions in the aminoacylation complex.
a. Several domains (color coded as in Fig. 1) of LeuRS are involved in binding and stabilizing the conformation of nucleotides 69 to 76 of the 3′ end of tRNALeu (green). The base of Gua71 is omitted for clarity.
b. The α-helix 291–298 of the editing domain stacks on the G1-U72 base pair and contacts the backbone of Ade73.
c. Interactions of the C-terminal domain with the T-loop, D-loop and long-variable arm of the tRNA (surface representation) are conserved in the editing and aminoacylation states (overlaid).
d. A network of interactions from the anti-codon binding domain, conserved between the two states (overlaid), specifically recognizes the base of Ura16 (see also Supplementary Figs. 4a,b).
The hairpin structure of the tRNA 3′ end is stabilised both protein-tRNA and intra-tRNA interactions, the latter including base-base stacking and phosphate-base hydrogen bonds. A rotamer flip of the class 1a conserved tyrosine28, Tyr43, into an open position avoids a steric clash with the position of the bound ribose of Ade76. This tyrosine acts as a lid that can either pack down on the hydrophobic amino acid substrate, preventing premature hydrolysis of the leucyl-adenylate, or be open, to allow Ade76 binding and the aminoacylation reaction to proceed.
The discriminator base Ade73 is stacked between Trp223 and Arg416, making base-specific hydrogen bonds to the main-chain of Arg416 and Arg418, which are part of the highly conserved (see below) 416-RLRDWGVSRQRYWG-429 motif linking the editing domain to the catalytic domain 3,11. Other residues from this motif also interact with the tRNA, whereas Cyt74 is flipped out into a pocket formed between the ZN1 domain and the catalytic domain (Fig. 2a). The ZN1 domain, ordered in the aminoacylation but not the editing state (see below), has a mutually stabilizing interface with the first four residues of the editing domain loop 286–298, also disordered in the editing state. Unexpectedly, a short helix within this loop stacks upon the G1-U72 base-pair of the tRNA, which remains unbroken (Fig 2b). Glu292, within this helix, hydrogen bonds to the phosphate of Ade73 and the 2′-OH of Ura72 and also forms a salt bridge with Arg416, part of the motif mentioned above. These interactions respectively block the acceptor stem in place and form a physical barrier separating the single-stranded region 73–76 from the acceptor stem helix (Fig. 2a). In order for the 3′ end of the tRNA to translocate from the synthetic site into the editing site the Glu292-Arg416 salt bridge would need to be broken. The unexpected observation that the CP1 editing domain loop 286–298 is directly involved in positioning the tRNA for aminoacylation provides an explanation for the required integrity of this loop for efficient aminoacylation by LeuRSEC29,30. This may also explain the preserved editing domain in metazoan mitochondrial LeuRS, even though the editing active site is defunct31.
Comparison of the LeuRSEC aminoacylation complex with that of archaeal LeuRSPH11 shows that the mode of binding of the tRNA 3′ end is similar although there are marked differences in the orientations of the 74-Cyt-Cyt-Ade bases (Supplementary Fig. 1d). The ZN1 domain is in the identical position in the two complexes but there is no equivalent to the leucine-specific domain in the archaeal enzyme. Additionally there is no leucyl-adenylate in the archaeal complex, and the KMSKS loop, discussed in more detail below, is in an open, inactive conformation.
Phylogenetic analysis of 27 LeuRS sequences from representative species covering the whole breadth of bacterial clades supports the importance of key residues and interactions cited here and elsewhere in the text (Supplementary Figure 2a-e). For instance, the 416-R/KLRDWGVSRQRYWG-429 motif is very highly conserved (Supplementary Figure 2c), as are ZN1 domain residues Asn168, Glu169 and Gln190 that interact with the tRNA (not shown). The alignment also suggests that the editing domain loop containing the helix that stacks on the first base-pair of the tRNA is structurally and likely functionally conserved, although the sequence can diverge (Supplementary Figure 2b). A Glu or Asp is found at position 292 in about half the sequences, suggesting that the 292–416 salt bridge is not essential.
Structure of the E. coli LeuRS-tRNALeu editing complex
The overall structure of the E. coli LeuRS-tRNALeu editing complex, determined at nearly 2 Å resolution in the orthorhombic form, is shown in Figure 1c. The orthorhombic and monoclinic forms of the complex have only minor differences in domain orientations and the occurrence of similar structures in two different crystal forms suggests that they faithfully represent the post-transfer editing state of the enzyme. This is reinforced by the global similarity (apart from slight changes in the orientation of the flexibly linked editing and C-terminal domains) of the LeuRSEC editing complex to the previously published structure of the LeuRSTT editing complex (PDB entry 2BYT) (Supplementary Fig. 3). However, there are some notable differences between the two editing complexes. Firstly, the ZN1 domain is well-ordered and closes over the leucine binding site in the LeuRSTT complex, whereas there is only weak, un-interpretable electron density for the corresponding region in the LeuRSEC editing complex. This flexibly linked domain plays a more active role in the LeuRSEC aminoacylation complex as described above. Secondly, the LeuRSEC leucine-specific domain is larger and has a different topology than in LeuRSTT. Only in the LeuRSEC editing complex does the leucine-specific domain directly interact with the tRNA. This occurs via an extended beta hairpin, absent from LeuRSTT, which contacts bases 10 and 27 via Arg595 and Arg600 (Supplementary Fig. 3). Due to the high variability in the sequence of the leucine specific domain (and its absence in some cases), these contacts are likely to be idiosyncratic for E. coli and other closely related bacterial LeuRS enzymes.
A second notable difference in protein-tRNA interactions concerns tRNA base 16, which is not contacted in the T. thermophilus system. However in both the LeuRSEC aminoacylation and editing complexes, Ura16 is involved in a network of direct- and water-mediated interactions with Lys711 (K), Asp714-Asp715 (DD), Arg718-Arg719 (RR), referred to as the K/DD/RR motif (Fig.2d and Supplementary Fig. 4a). Mutations of these residues reduce tRNA binding and catalytic efficiency for aminoacylation, and substitution of Ura16 by guanine or cytosine eliminates aminoacylation (Supplementary Figs. 4b,c). This suggests that Ura16 is a previously overlooked identity element, at least for E. coli LeuRS and related bacteria where the K/DD/RR motif is conserved (Supplementary Fig. 2d). Phylogenetic analysis shows that Ura16 is conserved not only in all E. coli tRNALeu isoacceptors, but also in those of most bacteria. Uridine is more favored than cytosine, which occurs occasionally, by the interaction of its O4 with well-conserved Lys711 (Supplementary Fig. 4a).
In the LeuRSTT-tRNA editing complex, a tRNALeu was used with a truncated long variable arm comprising two base-pairs and a tetraloop3. In both LeuRSEC conformations, the long variable arm is as wild-type (four base-pairs and a tetraloop) and contacts the C-terminal domain via the variable stem bases 47F-47I (Fig. 2c, Supplementary Fig. 1). In all LeuRSEC complex structures the extremity of the anti-codon stem-loop does not contact the synthetase and is poorly disordered, as previously observed for the T. thermophilus system3.
Structures of E. coli LeuRS with the tRNA in the editing conformation have been obtained either in the absence of other substrates, or with leucine or LeuAMS in the synthetic site or with the benzoxaborole covalent adduct in the editing site. In the absence of benzoxaborole, the 3′ CCA end of the tRNA is less tightly bound in the editing active site and the last four base-pairs of the acceptor stem and discriminator base, which are not in contact with protein, are poorly ordered due to flexibility. Formation of the benzoxaborole-tRNA adduct strongly stabilizes tRNA binding in the editing site and improves the ordering of the tRNA acceptor stem, although the mobility (as judged by the crystallographic B-factors) of the two last base-pairs and discriminator base is still high. The interactions of the 1-hydroxy-3H-2,1-benzoxaborole in the editing active site are identical to that previously described for similar compounds5,6. Binding of leucine or LeuAMS results in little change of the structure apart from the closure of Tyr43, which is otherwise in an open configuration, over the substrate leucine (see above).
Global comparison of the editing and aminoacylation states
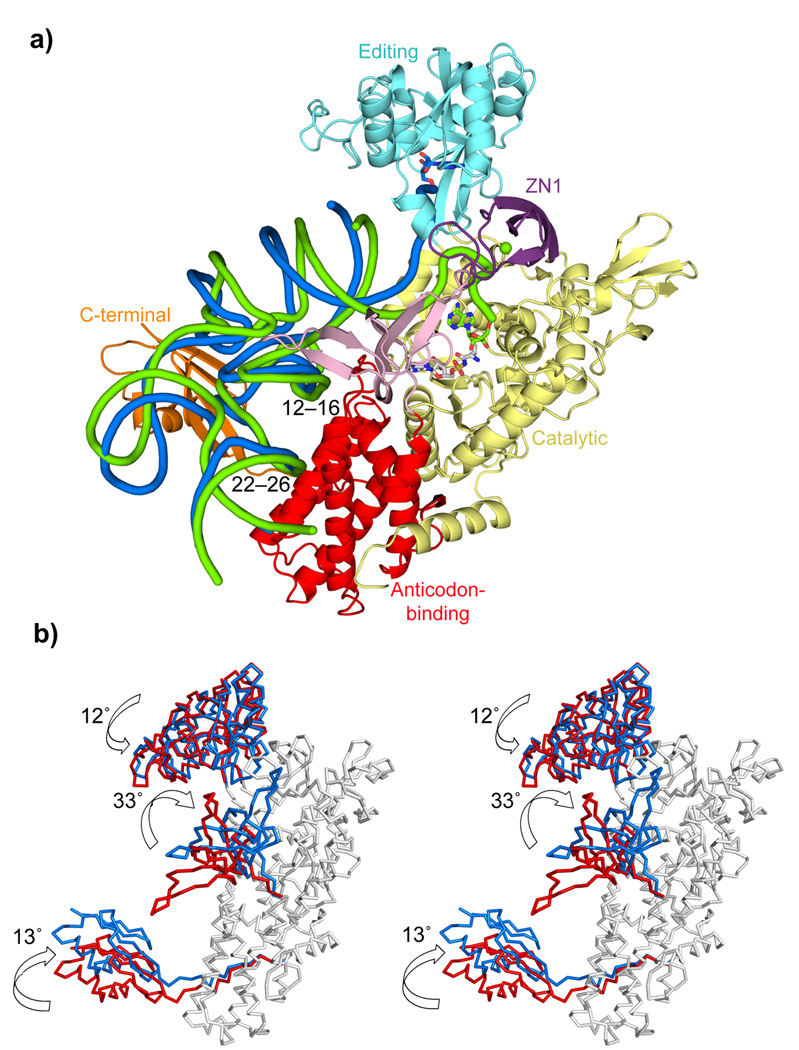
Both the tRNA and the flexibly linked domains undergo dramatic changes in their orientation during the transition between the aminoacylation state to the editing state (Fig 3a, b and Supplementary Video 1). A major feature of the aminoacylation complex compared to the editing conformation is that the entire leucine-specific domain rotates 33° towards the synthetic active site. This brings the beta-hairpin 577–583 into contact with the ZN1 domain, at the same time breaking the contact that occurs in the editing state between the 595–601 hairpin and the tRNA. The entire tRNA pivots by 15° around conserved contacts with the anti-codon binding domain (Fig. 3a) tipping it towards the synthetic active site. The pivoting tRNA is accompanied by the C-terminal domain, maintaining its conserved interactions with the T-loop and long-variable arm (Fig. 2c, Supplementary Fig. 2e). The editing domain rotates by 12° to open up a passage for translocation of the 3′ end of the tRNA from the otherwise closed aminoacylation state. Although the ZN1 domain is only ordered in the aminoacylation state in the E. coli system, in structures of LeuRSTT with bound adenylate (PDB 1H3N12) or with tRNA in the editing state (PDB 2BYT3), the ZN1 domain is packed over the leucyl-adenylate in a position completely incompatible with tRNA 3′ end binding3 (Supplementary Fig. 3c). This position may help to prevent premature hydrolysis of the adenylate, but the domain then needs to rotate by about 44° into a position that is compatible with 3′ end binding (Supplementary Fig. 3d,e). More detailed implications of these domain motions are discussed below.
Figure 3. Comparison between the aminoacylation and the editing configurations.
a. Global comparison of the two states. The LeuRSEC-tRNALeu aminoacylation complex (tRNA green tube) is shown together with the tRNA in the editing conformation (tRNA blue tube) after superimposition of the catalytic and the anticodon-binding domains of the two complexes.
b. Stereo diagram showing rotations of the flexibly linked domains between the editing (red) and aminoacylation states (blue), after superposition of the body of the enzyme (grey).
Active site interactions of the KMSKS loop
Comparison of the E. coli LeuRS aminoacylation and editing complexes shows that the class 1 conserved and catalytically important 619-KMSKS loop moves as an integral part of the leucine-specific domain. The movement brings it some 6–7 Å closer into the active site in the aminoacylation state allowing it to make critical interactions with the 3′ strand that correctly position the tRNA extremity for the transfer reaction (Figs. 2a and 4a,b). In contrast, in the editing conformation, there are no contacts to the 3′ strand of the acceptor stem (Supplementary Fig. 1b). In the closed aminoacylation state, Met568 flips to stack against the center of the adenine base of the LeuAMS (Fig. 4a). This correlates with a rotamer switch of Glu532 allowing it, the N3 of Ade76, and the LeuAMS sulphate to coordinate a water molecule, or possibly a magnesium ion, that could be important in the catalytic mechanism (Figs. 4b,c, see below). One result of this tightening of the active site is that the adenylate itself is slightly compressed into a strained conformation in comparison to the more extended conformation observed in structures without the tRNA in the aminoacylation state (Figs. 4a,b). Whereas the leucyl moiety is unchanged, the adenylate ribose is closer to the carbonyl group, partly due to the maintenance of the interaction of its 2′OH with Glu532, which, as mentioned above, has changed side chain conformation (Fig. 4b). In contrast, in the editing conformation with leucine or LeuAMS in the synthetic site, the KMSKS loop is in an open, relaxed conformation (Fig. 4a). In this case, the adenine moiety of the LeuAMS is much less tightly bound with no base-specific contacts to the backbone of Val569 and Met620 and no stacking with Met568 (Figs. 4a,b).
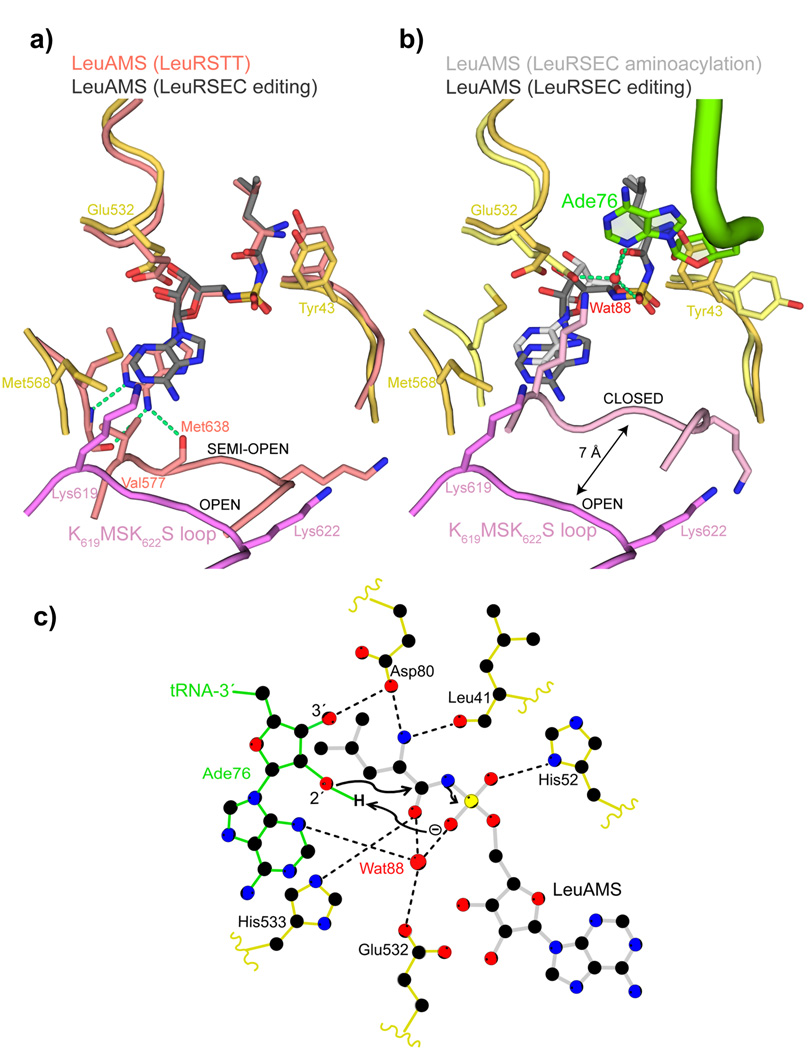
Figure 4. tRNA 3′ end binding induces full closure of the KMSKS loop.
a. Comparison between the synthetic sites of the LeuRSEC-LeuAMS-tRNALeu complex with the tRNA in the editing configuration (catalytic domain yellow, fully open KMSKS loop purple, LeuAMS dark grey) and the LeuRSTT-LeuAMS complex (PDB 1H3N) with the KMSKS loop in the semi-open conformation (all chains and LeuAMS salmon). Hydrogen bonds between the semi-open KMSKS loop conformation and the adenine base of the LeuAMS are indicated by green dotted lines but are absent in the open conformation.
b. Comparison between the synthetic sites of the LeuRS-LeuAMS-tRNALeu complex in the aminoacylation configuration (catalytic domain pale yellow, closed KMSKS loop light purple, LeuAMS light grey, tRNA green) and editing configuration (as in (a)). In the aminoacylation state, the presence of Ade76 necessitates a side-chain flip of Tyr43 and a water molecule (Wat88, red sphere) is coordinated by Ade76 N3, the re-orientated Glu532 carboxylate, a LeuAMS sulphate oxygen and the carbonyl-oxygen of the substrate leucine as indicated by green dotted lines (see text and (c)).
c. Schematic diagram of the LeuRSEC active site indicating the presumed substrate assisted mechanism of aminoacylation (see text).
These observations are consistent with a previous suggestion based on TyrRS structures that the KMSKS loop could be in three states: open, semi-open and closed32. With the tRNA in the editing conformation and either without synthetic site substrates or with bound leucine or LeuAMS, the KMSKS loop is in the fully open conformation and unable to form base-specific interactions to the adenine (Fig. 4a). With adenylate bound in the absence of tRNA (LeuRSTT-LeuAMS complex12, PDB 1H3N) the KMSKS loop is in a semi-open conformation with base-specific interactions to the adenine base (Fig. 4a). The fully closed conformation of the KMSKS loop has only been observed with the leucyl-adenylate bound together with the 3′ of the tRNA (Fig. 4b). However a similar state might occur for the preactivation state when ATP and leucine are both bound, as observed for instance for TyrRS19 (PDB 1H3E) or GlnRS14, even though the only known structure of a class 1a enzyme with an ATP analogue bound, but no amino acid substrate, that of ArgRSPH24 (PDB 2ZUE) has the KMSKS loop in the semi-open state. As suggested for the tyrosyl-system, the semi-open post-activation state with bound LeuAMP (as observed with LeuRSTT, Fig. 4a) may be required to permit initial binding of the tRNA 3′ end prior to re-closure, induced by 3′ end interactions, to allow the aminoacylation reaction.
The critical role of the KMSKS loop in activation and aminoacylation and its coupling with the leucine-specific domain explain why a gross deletion of this domain (569–618 replaced by 3 or 5 alanines) abolishes aminoacylation activity of E. coli LeuRS33. However the leucine-specific domain is highly variable in size and sequence and some bacterial LeuRS lack it completely, e.g. Campylobacter jejuni, Helicobacter pylori, and the second LeuRS (denoted B2) from Agrobacterium radiobacter, which is resistant to the natural antibiotic TM84 (an analogue of LeuAMP)34. Interestingly, Mycoplamsa mobile has evolved a minimal LeuRS, lacking both the leucine-specific domain and also, uniquely, the entire editing domain35. It remains to be seen how the absence of the leucine-specific domain affects the function of these bacterial enzymes, but one might hypothesize that the KMSKS loop is more dynamic as it does not have the inertial mass of the rest of the leucine-specific domain to carry with it.
Aminoacylation catalytic mechanism
For class I synthetases the aminoacylation reaction requires the nucleophilic attack of Ade76 ribose O2′ on the carbonyl carbon of the aminoacyl-adenylate. The reaction is most likely substrate assisted, with the phosphate of the adenylate acting as the general base to abstract the proton from the 2′OH in a concerted mechanism14,36 (Fig. 4c). This mechanism is potentially universal as it does not depend on nearby, non-conserved residues, as proposed by some authors15. The LeuRSEC aminoacylation complex structure is generally consistent with this mechanism since the O2′ is 3.6 Å from the sulphate oxygen of the leucyl-adenylate analogue LeuAMS, comparable to 3.8 Å for the equivalent distance in the GlnRS ternary complex15 (PDB 1QTQ). However, these distances are substantially longer than a hydrogen bond suggesting that the energy barrier for deprotonation might be too high for the direct reaction. This could reflect the fact that the structures do not correspond precisely to the real situation (e.g. the sulphamoyl-analogue lacks the negative charge of leucyl-adenylate) or, that additional elements might be required to explain the observed low energy barrier for the aminoacylation reaction. One fully conserved LeuRS residue that could play a role in the aminoacylation reaction is Glu532, which is strategically placed in the aminoacylation complex (Figs. 4b,c). To test the role of this residue, an E532Q mutant of LeuRSEC was expressed and the temperature dependence of the pyrophosphate (PPi) exchange and overall leucylation reactions was measured (Supplementary Figs. 4c). The E532Q mutant has similar activation energies for the first step and the overall reaction (93 and 94.9 kJ mol−1 respectively) in contrast to wild type, which had corresponding values of 25.2 and 55.0 kJ mol−1. This suggests that the rate limiting step for the mutant E532Q has shifted to the ATP-dependent activation of leucine (Supplementary Figs. 4d), consistent with an important role for Glu532 in activation, perhaps by coordinating a magnesium ion. Indeed Wat88, coordinated by Glu532, could occupy the magnesium position (Fig. 4b,c). In the presence of the authentic, negatively charged LeuAMP it is plausible that this site could still bind a magnesium ion, and this might facilitate the aminoacylation reaction, even though the E532Q mutant does not reveal this. Combined quantum-molecular dynamics simulations will likely be required to determine the most plausible mechanism for the aminoacylation reaction in LeuRS.
Discussion
In the aminoacylation state the tRNA 3′ end is deeply buried in the catalytic domain. How does aminoacyl transfer with concomitant formation of AMP lead to product release and translocation? With reference to the schematic diagram of the two functional states (Figs 5a,b) we propose the following mechanism for the functional cycle of E. coli LeuRS. We hypothesize that the aminoacylation state is under strain with regard to the unusual conformation of the 3′ end of the tRNA, the distorted state of the adenylate and the fully closed conformation of the KMSKS loop. Formation of this state is possible due to the presumably high binding energy of the tRNA 3′ end. Once the covalent integrity of the adenylate is broken by completion of the aminoacylation reaction, the strain is released by relaxation of the KMSKS loop into its open conformation, accompanied by the dissociation of AMP and the flip of the entire leucine-specific domain, whose inertia will prevent easy reversion to the closed state. Concomitant with this, the KMSKS loop interactions with the tRNA are broken, destabilizing the binding of the entire acceptor end of the now charged tRNA. This leads to broader cooperative destabilization of the interactions holding the ZN1 domain and the editing domain in contact with each other and with the tRNA. The release of constraints allows the tRNA 3′ end to relax to its preferred conformation with the discriminator base stacked on the 1:72 base-pair and the CCA end now able to enter the editing active site. Locally, this is accompanied by a re-arrangement of the interactions of Glu292, Arg416, Arg418 and Tyr330 with the tRNA, each of which has distinct roles in the two states (Fig. 5c,d). More globally, the editing domain slightly rotates as the tRNA pivots around its fixed contact points on the anti-codon binding domain and the C-terminal domain moves synchronously, maintaining its contacts to the tRNA. In this conformation the tRNA is already partially disassociated from the catalytic domain (Supplementary Figs. 1a,b and Supplementary Table 1) favoring total tRNA release after proof-reading in the case of correctly charged Leu-tRNALeu or possibly direct re-aminoacylation, without full release, of hydrolyzed mischarged tRNA.
Figure 5. Dynamics of the functional cycle of bacterial LeuRS.
a, b. Schematic diagram of the structural changes between the aminoacylation and proof-reading conformations of the LeuRSEC-tRNALeu complex (see text). Color code for each domain is as in Figure 1, leucine is represented as a white oval and AMP as a black rectangle.
c, d. Dynamic rearrangements in the interface between the editing, ZN1 and catalytic domain that allow translocation of the tRNA 3′ end from the synthetic site (c) to the editing site for proof-reading (d). Some elements of the catalytic domain have been removed for clarity. Top panels show the cavities of the catalytic and editing sites in surface representation, and bottom panels the residues important for the translocation of the tRNA (rotated 180 ° respect to the top panels for clarity).
Our structures provide detailed information on the end points of translocation and it might be difficult to obtain crystal structures of eventual intermediates, although an indication of such states was observed for the archaeal LeuRSPH aminoacylation complex11. Molecular dynamics simulations can potentially fill in the gap, as utilized in the analysis of the mechanism for Glu-tRNAGlu release from the synthetic site of class 1b GluRS18. In another study, normal mode analysis of LeuRSTT revealed both correlated and anti-correlated motions between various structural elements37. Correlated motions included the coupling of the KMSKS loop with the leucine-specific domain and of the editing domain loop 286–298 with the ZN1 domain, whereas between the leucine-specific and editing domains, anti-correlated motions were detected, consistent with the structure based results described here. In the future, a completed ensemble of structures (including the currently lacking pre-activation complex with bound ATP) together with molecular dynamics simulations and fast kinetic studies coupled with structure-based mutagenesis will hopefully provide further insight into the dynamics of the functional cycle of bacterial leucyl-tRNA synthetase. Interestingly, recent results have revealed that eukaryotic, cytoplasmic LeuRS acts as an intracellular leucine sensor in the amino-acid induced mTORC1 signaling pathway, which regulates cell growth38,39. It is proposed that Rag GTPases, which activate mTORC1, bind to the LeuRS editing domain and sense the presence of bound leucine via the conformational state of the enzyme. This provides a particularly striking illustration of the need for further analysis and understanding of the conformational dynamics of LeuRS.
Supplementary Material
Acknowledgments
The authors thank the ESRF-EMBL Joint Structural Biology Group for access to ESRF beamlines and the EMBL-ESRF-ILL-IBS Partnership for Structural Biology for access to structural biology instrumentation, notably the high-throughput crystallization platform.
Footnotes
Accession codes
Atomic coordinates and structure factors for the editing complex with L-leucine have been deposited in the wwPDB with code 4ARC; for the orthorhombic and monoclinic forms of the complexes with benzoxaborole under 4ARI and 4AS1 respectively; and for the aminoacylation complex with 4AQ7.
Author Contributions
TC crystallized the editing state complexes and AP crystallized the aminoacylation complex. TC, AP and SC collected X-ray data and performed the structural analysis. MV and TL performed the mutagenesis and associated biochemical studies under the supervision of SM. SC wrote the manuscript with input from all other authors.
References
- 1.Karkhanis VA, Mascarenhas AP, Martinis SA. Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J Bacteriol. 2007;189:8765–8768. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lincecum TL, Jr, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 3.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 4.Hagiwara Y, Field MJ, Nureki O, Tateno M. Editing Mechanism of Aminoacyl-tRNA Synthetases Operates by a Hybrid Ribozyme/Protein Catalyst. J Am Chem Soc. doi: 10.1021/ja9095208. [DOI] [PubMed] [Google Scholar]
- 5.Rock FL, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 6.Seiradake E, et al. Crystal structures of the human and fungal cytosolic Leucyl-tRNA synthetase editing domains: A structural basis for the rational design of antifungal benzoxaboroles. J Mol Biol. 2009;390:196–207. doi: 10.1016/j.jmb.2009.04.073. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YK, et al. Synthesis and structure-activity relationships of novel benzoxaboroles as a new class of antimalarial agents. Bioorg Med Chem Lett. 2011;21:644–651. doi: 10.1016/j.bmcl.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Ding D, et al. Design, synthesis, and structure-activity relationship of Trypanosoma brucei leucyl-tRNA synthetase inhibitors as antitrypanosomal agents. J Med Chem. 2011;54:1276–1287. doi: 10.1021/jm101225g. [DOI] [PubMed] [Google Scholar]
- 9.Fukai S, et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 10.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 11.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 12.Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. Embo J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga R, Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J Mol Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 14.Perona JJ, Rould MA, Steitz TA. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993;32:8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- 15.Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 16.Bullock TL, Uter N, Nissan TA, Perona JJ. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J Mol Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- 17.Sekine S, et al. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. Embo Journal. 2003;22:676–688. doi: 10.1093/emboj/cdg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekine S, et al. Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase. Structure. 2006;14:1791–1799. doi: 10.1016/j.str.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T, et al. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat Struct Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 21.Yang XL, et al. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen N, et al. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Research. 2008;36:1288–1299. doi: 10.1093/nar/gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauenstein S, Zhang CM, Hou YM, Perona JJ. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nature Structural & Molecular Biology. 2004;11:1134–1141. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- 24.Konno M, et al. Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP) FEBS J. 2009;276:4763–4779. doi: 10.1111/j.1742-4658.2009.07178.x. [DOI] [PubMed] [Google Scholar]
- 25.Delagoutte B, Moras D, Cavarelli J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000;19:5599–5610. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tocchini-Valentini G, Saks ME, Abelson J. tRNA leucine identity and recognition sets. J Mol Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]
- 27.Larkin DC, Williams AM, Martinis SA, Fox GE. Identification of essential domains for Escherichia coli tRNA(leu) aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 2002;30:2103–2113. doi: 10.1093/nar/30.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crepin T, et al. Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 2003;332:59–72. doi: 10.1016/s0022-2836(03)00917-3. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Guo N, Xia X, Wang ED, Wang YL. The peptide bond between E292-A293 of Escherichia coli leucyl-tRNA synthetase is essential for its activity. Biochemistry. 1999;38:13063–13069. doi: 10.1021/bi990384+. [DOI] [PubMed] [Google Scholar]
- 30.Du X, Wang ED. E292 is important for the aminoacylation activity of Escherichia coli leucyl-tRNA synthetase. J Protein Chem. 2003;22:71–76. doi: 10.1023/a:1023071928587. [DOI] [PubMed] [Google Scholar]
- 31.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi T, et al. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase. J Mol Biol. 2005;346:105–117. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 33.Vu MT, Martinis SA. A unique insert of leucyl-tRNA synthetase is required for aminoacylation and not amino acid editing. Biochemistry. 2007;46:5170–5176. doi: 10.1021/bi062078j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reader JS, et al. Major biocontrol of plant tumors targets tRNA synthetase. Science. 2005;309:1533. doi: 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 35.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci U S A. 2011;108:9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uter NT, Perona JJ. Active-site assembly in glutaminyl-tRNA synthetase by tRNA-mediated induced fit. Biochemistry. 2006;45:6858–6865. doi: 10.1021/bi052606b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weimer KM, Shane BL, Brunetto M, Bhattacharyya S, Hati S. Evolutionary basis for the coupled-domain motions in Thermus thermophilus leucyl-tRNA synthetase. J Biol Chem. 2009;284:10088–10099. doi: 10.1074/jbc.M807361200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han JM, et al. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012 doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 39.Bonfils G, et al. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.