SUMMARY
Helicases move on DNA via an ATP binding and hydrolysis mechanism coordinated by well-characterized helicase motifs. However, the translocation along ssDNA and the strand separation of dsDNA may be loosely or tightly coupled. Dda is a phage T4 SF1B helicase with sequence homology to the Pif1 family of helicases that tightly couples translocation to strand separation. The crystal structure of the Dda-ssDNA binary complex reveals a domain referred to as the ‘pin’ which was previously thought to remain static during strand separation. The pin contains a conserved phenylalanine that mediates a transient base-stacking interaction that is absolutely required for separation of dsDNA. The pin is secured at its tip by protein-protein interactions through an extended SH3 domain thereby creating a rigid strut. The conserved interface between the pin and the SH3 domain provides the mechanism for tight coupling of translocation to strand separation.
INTRODUCTION
Helicases are ATP-dependent motor proteins that play key roles in DNA and RNA metabolism (Delagoutte and von Hippel, 2003; Lohman et al., 2008; Patel and Donmez, 2006; Pyle, 2008). Classically, helicases unwind double-stranded substrates, but they can also modulate nucleic acid architecture in processes such as recombination, chromatin remodeling and RNA transport. The fact that ~2% of the yeast genome encodes helicase-like proteins reflects their importance. Many diseases, typically associated with cancer and/or premature aging, are linked to mutations in helicase genes and defective DNA repair (Bohr, 2008; Brosh et al., 2000; Ellis, 1997; Stevnsner et al., 2008). Helicases have also emerged as important therapeutic targets for the treatment of cancer (Aggarwal and Brosh, 2009) and viral infections (Frick, 2007; Lescar et al., 2008).
Helicases are classified into four superfamilies based on the presence of helicase motifs (Gorbalenya and Koonin, 1993; Singleton et al., 2007). The monomeric SF1 and SF2 helicases share an architecture based on paired RecA-like domains, while the SF3 and SF4 helicases form hexameric rings. SF1 is the largest class and is sub-divided into the SF1A and SF1B enzymes that translocate 3′-to-5′ and 5′-to-3′, respectively, on ssDNA (Singleton et al., 2007). This report focuses on the SF1B subfamily which includes members that are conserved from bacteria to humans. Examples include Dda from bacteriophage T4, RecD2 from bacteria (Saikrishnan et al., 2009), Pif1 from eukaryotes (Bochman et al., 2010) and human DNA helicase B (Gu et al., 2004). Although key information has recently become available (Saikrishnan et al., 2008; Saikrishnan et al., 2009), SF1B helicases are poorly understood compared to their SF1A and SF2 counterparts despite their emerging importance in key biological functions. Notably, human DNA helicase B is involved in DNA repair and replication (Gu et al., 2004; Taneja et al., 2002), and the Pif1 helicase has roles in telomere regulation (Boule et al., 2005; Boule and Zakian, 2010) and mitochondrial DNA repair (Cheng et al., 2007).
The Dda helicase from phage T4 is an ideal model system for understanding the basic mechanism of the SF1B class, although its biological role is still somewhat obscure. Dda knockouts are fully viable, and the helicase is not needed for phage replication, recombination or repair (Behme and Ebisuzaki, 1975). Nonetheless, Dda-deficient phage show a delay in origin usage and overall DNA replication, and replication is compromised during infections with mutants deficient in both Dda and gp59 which loads the replicative helicase gp41 (Gauss et al., 1994). These results suggest that Dda has an overlapping function with gp59 in loading gp41 at T4 replication origins (Gauss et al., 1994). In vitro experiments have suggested additional roles for Dda that have yet to be verified in vivo. First, Dda can promote DNA polymerase strand switching that could play a key role in survival after DNA damage (Kadyrov and Drake, 2004). Second, Dda can displace proteins from DNA suggesting that it helps clear the DNA template for replication (Bedinger et al., 1983; Byrd and Raney, 2006). Third, Dda has been shown to both inhibit and stimulate recombination strand invasion reactions (Kodadek, 1991; Kodadek and Alberts, 1987) suggesting a role in homologous recombination. Consistent with this, Dda binds both UvsX (the T4 recombinase) and gp32 (the T4 ssDNA-binding protein) (Formosa and Alberts, 1984; Hacker and Alberts, 1992). Dda single mutants show no obvious recombination phenotype (Behme and Ebisuzaki, 1975) but phenotypes do appear in double knockout phages. Most notably, a Dda/gp61 (the T4 primase) double knockout phage was found to be lethal due to poor DNA packaging (Belanger, 1997). Primase-deficient infections are known to generate excessive ssDNA which, in turn, promotes recombination and the overproduction of DNA branches that inhibit packaging. In this scenario, it has been suggested that Dda acts as an anti-recombinase that facilitates packaging by reducing the levels of DNA branches.
In contrast, the enzymology of the Dda helicase has been well characterized. Dda unwinds DNA as a monomer (Nanduri et al., 2002) in a 5′-to-3′ direction at a rate of ~ 250 bp/s (Eoff and Raney, 2006), and can efficiently displace proteins in its path (Morris and Raney, 1999). In the absence of impediments, Dda molecules function independently (Eoff and Raney, 2010), but when impediments are present, displacement efficiency is augmented by multiple copies of Dda working in tandem via protein-protein interactions (Byrd and Raney, 2004). This is true for the displacement of streptavidin (Byrd and Raney, 2004), the removal of proteins (Byrd and Raney, 2006), and movement past chemical perturbations in the DNA (Eoff et al., 2005). Helicases are described as being active or passive depending on whether the enzyme actively separates the base pairs or simply sequesters ssDNA that forms due to thermal fraying (Manosas et al., 2010). The ratio of the rate of unwinding dsDNA to the rate of translocation on ssDNA distinguishes between the two; a helicase is considered highly active when the ratio is close to 1 and passive with ratios less than 0.25 (Manosas et al., 2010). Recently, we determined that Dda translocates on ssDNA at nearly the same rate as it unwinds dsDNA, making it one of the most “active” helicases yet studied (Byrd et al. 2012).
Although Dda shares the same helicase motifs as other superfamily 1 helicases, the highly active nature of Dda suggests that alternative or perhaps additional mechanisms are present to explain the extraordinary efficiency for dsDNA unwinding. Here, we present the crystal structure of the Dda-ssDNA binary complex. The structure, together with biochemical and mutagenesis data, supports the proposed 5′-to-3′ translocation mechanism of the SF1B helicases (Saikrishnan et al., 2009) and reveals a new mechanism by which translocation is directly coupled to strand separation of duplex DNA. The results provide the structural basis for distinguishing a highly active helicase from a passive helicase.
RESULTS
Structure Determination
Dda is difficult to express in a soluble form, but the active site point mutant K38A could be readily expressed and purified. The mutation does not affect the DNA-binding properties of Dda, and a purified complex with a dT8 oligonucleotide produced crystals that diffract to 3.5 Å. The structure was determined using SAD phasing with selenomethionine-substituted protein. There are three molecules in the C2 asymmetric unit (ASU). We subsequently grew crystals that diffract to 3.3 Å, and refined the structure to Rwork and Rfree values of 0.204 and 0.258, respectively. Data collection and refinement statistics are shown in Table 1.
Table 1. Data Collection and Refinement Statistics.
| Dda/K38A(SeMet) | ||
|---|---|---|
| SAD (peak) | Higher resolution | |
| Data Collectiona | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 223.9, 106.7, 85.3 | 225.5, 107.3, 85.5 |
| α, β, γ (°) | 90.0, 94.3, 90.0 | 90.0, 94.1, 90.0 |
| Wavelength (Å) | 0.9793 | 1.0 |
| Resolution (Å) | 50.0-3.80 (3.87-3.80) | 40.0- 3.30 (3.42-3.30) |
| No. of reflections | ||
| Measured | 120,589 | 208,930 |
| Unique | 19,895 | 29,389 |
| Completeness (%) | 99.5(98.4) | 95.8(71.4) |
| Redundancy | 6.1(4.9) | 7.1(4.5) |
| I/σ(I) | 16.6(2.6) | 19.82(2.05) |
| Rsym (%) | 11.7(48.2) | 9.2(47.3) |
|
| ||
| Phasing | ||
| Resolution (Å) | 3.8 | |
| FOM | 0.29 | |
| FOM after density modification | 0.63 | |
|
| ||
| Refinement | ||
| Resolution (Å) | 32.3–3.3 | |
| No. reflections | 27,990 | |
| Completeness (%) | 91.1 | |
| Rwork (%) | 20.4 | |
| Rfree (%)b | 25.8 | |
| RMSDs | ||
| Bonds lengths(Å) | 0.004 | |
| Bonds angles(°) | 0.916 | |
| No. protein residues in A.U. | 1317 | |
| No. dT in A.U. | 24 | |
| Water | 10 | |
| Average B-factor (Å2) | 110 | |
| Ramachandran plot (%) | ||
| Most favored regions | 89.3 | |
| Additional allowed regions | 10.0 | |
| Generously allowed regions | 0.7 | |
| Disallowed regions | 0.0 | |
Highest resolution shell is shown in parenthesis.
Rfree is the R value obtained for a test set of reflections consisting of randomly selected 5% subset of the data set excluded from refinement.
Description of the Structure
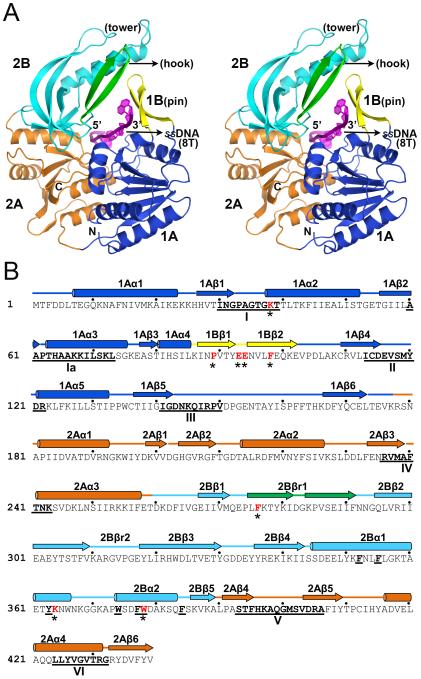
The Dda structure is shown in Figure 1A and it is most similar to the SF1B helicase RecD2 (Saikrishnan et al., 2009). At the heart of the molecule are the two signature RecA-like domains 1A (residues 1-173) and 2A (182-257 and 391-439) spanned by the ssDNA. Within domain 1A is an extended β-ribbon (86-102) that is referred to as the ‘pin’ or domain 1B. Within domain 2A is an SH3-like β-barrel domain 2B (260-389) comprising five β-strands that matches a similar domain in RecD2 (Saikrishnan et al., 2009). However, unlike RecD2, the Dda SH3 domain contains two insertions, a second β-ribbon (275-291) that we refer to as the ‘hook’ and a β-ribbon/two-helix substructure (305-322 and 345-383) that we refer to as the ‘tower’. The Dda sequence is shown in Figure 1B with the secondary structure elements labeled to reflect the domains structure, and the helicase motifs are highlighted (Blair et al., 2009).
Figure 1. Structural overview of the Dda-ssDNA complex.
(A) Secondary structure representation of the complex showing the key structural and functional domains and motifs in stereo view. The 1A and 2A RecA-like domains are blue and orange, respectively, the SH3 domain (2B) including the tower is cyan, the β-ribbon pin (1B) is yellow, and the β-ribbon (hook) extension from 2B is green. The ssDNA is shown in magenta passing through the central arch. (B) The primary structure of Dda with the secondary structures labeled and colored as in (A). The helicase motifs are underlined in bold and labeled. Residues mutated in the study are shown in red with an asterisk. Key hydrophobic residues in the tower region (Figure 2) are bold and underlined.
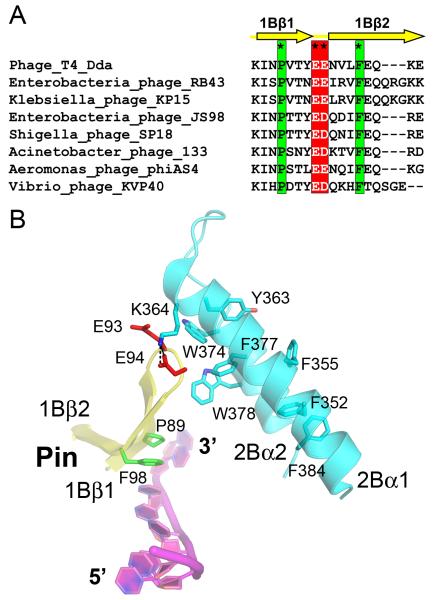
The pin and tower interact at their distal ends, creating an arch through which the ssDNA passes. Sequence comparisons with other T4-like phages reveal that the pin and the tower are conserved features of Dda (Figure 2A). Most prominent are seven aromatic residues (Phe352, Phe355, Tyr363, Trp374, Phe377, Trp378 and Phe384) that stabilize the tower substructure (Figure 2B). Three of these residues (Trp374, Phe377 and Trp378) together with salt bridges between Glu93, Glu94 and Lys364 create the pin-tower interface (Figure 2B).
Figure 2. Details of the interaction between the pin and the tower in the Dda-ssDNA complex.
(A) The pin residues are highly conserved in Dda proteins from T4-like phages. Completely conserved is a pair of acidic residues within the tight turn at the end of the pin (asterisks and red box) that mediate an electrostatic interaction with the tower. Also completely conserved are a proline and a phenylalanine (asterisks and green box) that form a stacking interaction with a base of the translocating ssDNA. (B) A close up of the pin-tower interface highlighting key charged and aromatic residues described in the text. E93, E94, K364 and W378 are the residues that have been mutated to probe the functional importance of this interface. In the figure, the pin is shown in yellow, the tower is cyan and the bound ssDNA is magenta.
The Interaction with ssDNA
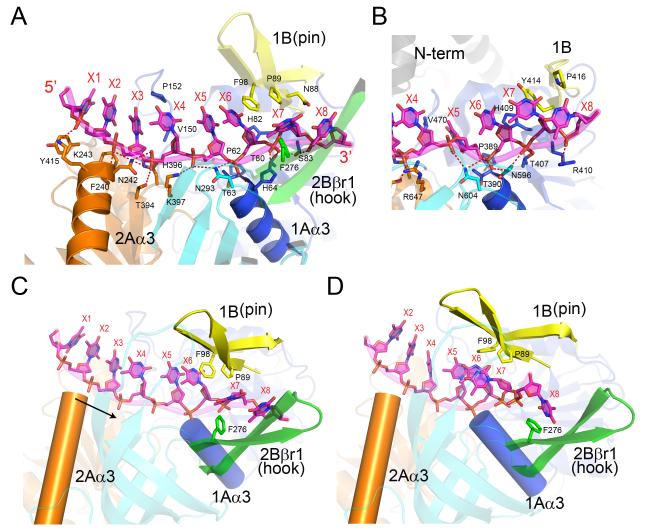
The ssDNA traverses the 2A and 1A domains in a 5′-to-3′ direction. As viewed in Figure 1A, the ssDNA would move away from the viewer by the action of Dda. The protein-ssDNA interactions principally involve the sugar-phosphate backbone that makes hydrogen-bonding and ionic interactions with the side chains in the ssDNA-binding groove that are conserved amongst the Dda-like helicases (Figure 3A). The register of the ssDNA with respect to domains 1A and 2A may be partly controlled by the positive helix dipoles at the N-termini of two topologically equivalent α-helices (Kerr et al., 2007), which are 1Aα3 and 2Aα3 in Dda. These two helices engage the 5’ phosphate groups of the ssDNA at positions X6 and X3, respectively. The bases are mostly stacked and do not contribute to the protein-ssDNA interface apart from positions X3-X4 and X6-X7.
Figure 3. The ssDNA-binding interactions in Dda.
(A) The ssDNA traverses the RecA-like domains 1A and 2A, and P89 and F98 form a triple stack interaction with the DNA base at position X6. Note that the N-termini of helices 1Aα3 (domain 1A) and 2Aα3 (domain 2A) point directly at the DNA 5′ phosphates at positions X6 and X3, respectively. The coloring scheme matches that shown in Figure 1. (B) The open conformation of the RecD2-ssDNA complex (Saikrishnan et al., 2009) centered on position X6. To facilitate the comparison with (A), the two structures are aligned and equivalently colored and labeled. Note that the bases X5, X6 and X7 are in different orientations in the two structures with X5 stacked onto X6 in Dda but rotated into a pocket in RecD2. (C) The observed open conformation structure of the Dda-ssDNA complex highlighting the locations of the pin (yellow), the hook (green), and helices 1Aα3 (blue cylinder) and 2Aα3 (orange cylinder) relative to ssDNA. The arrow indicates the direction in which α-helix 2Aα3 moves relative to ssDNA during domain closure. (D) A model of the closed Dda-ssDNA complex based on the structure of the RecD2 ternary complex (Saikrishnan et al., 2009). Due to domain closure and rotation, Phe98 no longer stacks onto the base at X6, and the hook becomes aligned along the axis of the ssDNA towards the incoming duplex with Phe276 forming a new stacking interaction with the base at position X8. Note that helix 2Aα3 has moved closer to helix 1Aα3 by one phosphate group compared to (C).
Domains 1A and 2A interact almost exclusively with nucleotides X5-X8 and X1-X4, respectively (Figure 3A). Within domain 1A, His64 interacts with the phosphate groups of X6 and X7, Thr80 with X7, and Ser83 with X7 and X8. The ribose groups of X6 and X7 stack against the side chains of His82 and Asn88, respectively. Pro89 and Phe98 from opposite sides of the pin stack onto the base at X6 to create a three-stack structure that introduces a kink in the ssDNA. Within domain 2A, Lys243, Asn242 and Thr394 engage the phosphates at positions X2, X3/X4 and X4, respectively. Phe240 forms van der Waal interactions with the X2 and X3 ribose groups and His396 stacks against the ribose of X3. This bipartite interaction pattern is not strictly followed; Val150 interacts with the base-ribose at X4 and disrupts the base stacking at X3-X5, and Asn293 and Lys397 interact with the phosphates of X5/X6 and X5, respectively.
Comparison with the RecD2-ssDNA Complex
The Dda-ssDNA and RecD2-ssDNA binary complexes display many similarities. In terms of overall architecture, the 1A, 2A, 1B (pin) and 2B (SH3) sub-domains are in equivalent positions with respect to their primary structures. The ‘N-term’ domain and the C-terminal extension in RecD2 are not present in Dda and, conversely, the tower in Dda is not present in RecD2. The hook structure in Dda is replaced by a short β-ribbon in RecD2. Both proteins interact with ssDNA in a similar fashion, traversing the 1A and 2A domains and engaging the SH3 β-barrel at the 5′ end. Most of the residues in Dda that mediate its interaction with ssDNA have direct counterparts in RecD2. However, there is one significant difference that involves the bases at positions X5 and X6. In Dda, Pro89 and Phe98 form a triple stack with the base at X6 and the adjacent base at X7 is pushed out of alignment by an edge-to-face interaction with Pro89 (Figure 3A). This interaction is not seen in RecD2 (Figure 3B) where the triple stack is absent and the base at X5 is instead rotated into a small hydrophobic pocket (Saikrishnan et al., 2009).
The Dda-ssDNA-ATP Ternary Complex
We have not been able to crystallize the Dda-ssDNA-nucleotide ternary complex, but we constructed a model based on the structure of the equivalent RecD2 complex (Saikrishnan et al., 2009). In the RecD2 structure, domains 1A and 2A clamp down on the ATP-binding site at the interface and the linker region changes conformation to accommodate the bound nucleotide. When the Dda structure is adjusted to adopt these movements, an almost identical ATP-binding site is generated. Gln10 (Gln343 in RecD2) is the so-called ‘Q motif’ that forms a hydrogen-bonding interaction with the adenine ring, the adjacent threonines 39 and 40 (367 and 368) hydrogen bond to the β and α phosphates, respectively, and Arg178 (493) and Arg431 (679) make salt bridge interactions with the phosphate groups. Each of the helicase motifs associated with binding ATP is in the expected location. Motif I (the Walker A motif) creates the phosphate binding loop, and Lys38 (366) within this loop is the residue that we mutated to facilitate crystallization.
The three molecules in the Dda ASU have very similar conformations, but there is some variability in the orientations of domain 1A versus the tightly associated domains 2A/2B. When modeling the Dda ternary complex, we therefore assumed that 2A and 2B move as a rigid body with respect to 1A. This reveals two features (Figures 3C and 3D). First, the interaction between the tips of the pin and the tower must act as a pivot to maintain this interaction. Second, the hook engages the incoming ssDNA, points towards the incoming duplex and interacts with the base at X8 via a stacking interaction with Phe276. An equivalent stacking interaction is observed in the RecD2 structure involving Tyr598 (Saikrishnan et al., 2009).
Functional Studies – The Pin-DNA Interaction
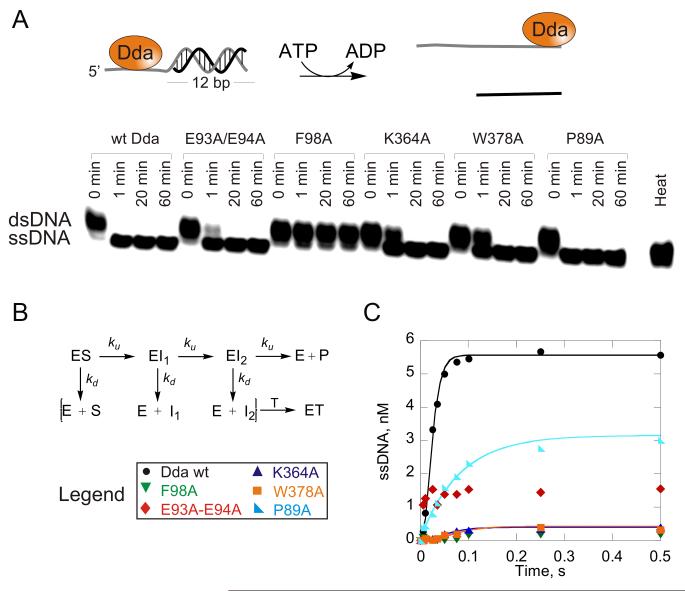
The roles of the RecA domains 1A and 2A in the inchworm model for translocation (Velankar et al., 1999) have been examined for several helicases (Singleton et al., 2007), and their modified roles in SF1B 5′-3′ translocation have been established (Saikrishnan et al., 2009). We therefore focused on the less well characterized roles of domains 1B and 2B, and generated mutants within these two regions and biochemically characterized each of them (Figure S1).
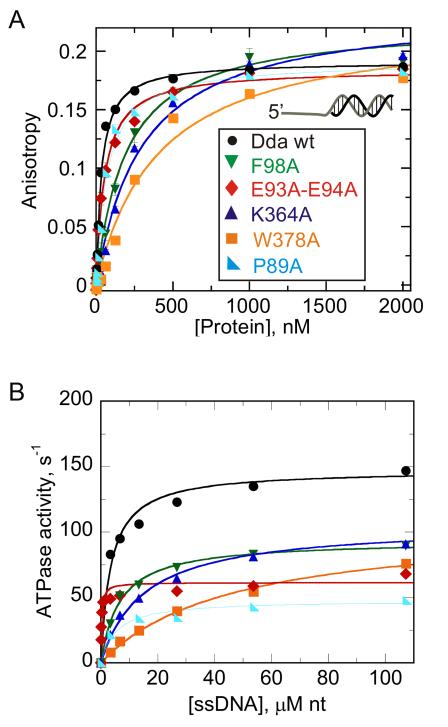
Consistent with its location with respect to the bound ssDNA, the pin has been proposed to split the incoming duplex during the 5′-to-3′ unwinding of DNA in RecD2. The Dda structure further reveals that Phe98 within the pin, buttressed by Pro89, forms a stacking interaction on the 3′-side of base X6 (Figures 3A and 3C). We postulate that this interaction serves to facilitate the unwinding reaction. Although the RecD2 structure does not show this interaction (Figure 3B), Tyr414 is perfectly positioned to be the functionally equivalent residue to Phe98. To investigate this, Phe98 was mutated to alanine to remove the stacking interaction. F98A bound to a ss/dsDNA partial duplex with lower affinity than the wild type (wt) enzyme with a ~6-fold higher equilibrium dissociation constant, KD, and the ssDNA-stimulated ATPase activity was reduced by less than 2-fold (Figure 4A, Table 2). Hence, replacement of Phe98 with alanine reduces the affinity of Dda for ssDNA, but does not greatly reduce the inherent ability of the enzyme to hydrolyze ATP in a ssDNA-stimulated manner. The stacking interaction between F98 and Pro89 was investigated by creating P98A. The resulting DNA binding and ATPase activities were reduced, but only by 2-3 fold (Figure 4, Table 2).
Figure 4. DNA binding and single-stranded DNA-stimulated ATPase activity.
(A) Binding was measured by using fluorescence anisotropy of F-dT7:16bp DNA duplex in the presence of wild type Dda, F98A, E93A/E94A, K364A, W378A, and P89A. Data were fit to the quadratic equation to obtain KD values (listed in Table 2). (B) ATP hydrolysis of wt and mutant forms of Dda were measured using a coupled spectrophotometric assay in which ATP hydrolysis is coupled to oxidation of NADH through the enzymes pyruvate kinase (PK) and lactate dehydrogenase (LDH). Rates of hydrolysis were determined in the presence of increasing concentrations of denatured calf thymus DNA. Data were fit to a hyperbola to obtain ATPase constants kcat and Kact where the latter constant is the concentration of denatured calf thymus DNA (in nucleotides) needed to reach half the maximum ATP hydrolysis rate (listed in Table 2). See also Figure S1.
Table 2. Activities of wild-type and variant Dda enzymes.
| Enzyme | DNA binding |
ATPase activity | DNA unwinding | ssDNA translocase |
Ratio of unwinding to translocation rates |
||
|---|---|---|---|---|---|---|---|
| KD (nM) | kcat (s−1) | Kact (μM, nt) | ku (bp s−1) | amplitude (nM) |
ktrans (nt s−1) | ku/ktrans | |
| wtDda | 34 ± 3 | 147 ± 3 | 3.5 ± 0.5 | 258 ± 17 | 5.6 ± 0.1 | 233 ± 27 | 1.10 ± 0.15 |
| F98A | 192 ± 26 | 94 ± 2 | 6.7 ± 0.6 | N.D.a | N.D. | 249 ± 23 | N.D. |
| E93A/E94A | 55 ± 5 | 61 ± 2 | 0.26 ± 0.05 | N.D. | N.D. | 77.0 ± 4.9 | N.D. |
| K364A | 298 ± 38 | 106 ± 2 | 15.1 ± 1.0 | 47 ± 9 | 0.39 ± .05 | 212 ± 7 | 0.22 ± 0.04 |
| W378A | 409 ± 72 | 107 ± 2 | 46.5 ± 3.9 | 36 ± 8 | 0.43 ± .05 | 157 ± 11 | 0.23 ± 0.05 |
| P89A | 72 ± 8 | 48 ± 1 | 5.4 ± 1.1 | 96 ± 13 | 3.0 ± 0.2 | 149 ± 6 | 0.64 ± 0.09 |
Those activities listed as ‘not determined’ (N.D.) indicate too little product formed to reliably determine a rate constant, amplitude, or binding constant.
DNA unwinding data are from experiments conducted at 1 μM enzyme and 10 nM substrate under single turnover conditions. See also Table S1.
DNA unwinding was measured under multiple-cycle conditions to determine whether any unwinding was observed for the Dda variants (Figure 5A). A substrate containing a short duplex (12 bp) was chosen to detect even low levels of strand separation. F98A was devoid of unwinding activity but P89A was able to unwind the short substrate. To determine the rates for unwinding by the variant forms of Dda, single-cycle DNA unwinding experiments were conducted using a well-characterized partial duplex substrate containing 7 nt of ssDNA and 16 bp (Figures 5B and 5C). Wild type Dda unwound the substrate at a rate of 258 ± 17 bp/s. No product was observed with F98A Dda, as expected, but P89A Dda unwound the substrate in a single kinetic step at a rate of 96 ± 13 bp/s. Hence, whereas loss of stacking interaction between P89 and F98 reduces efficiency of DNA unwinding, removal of the stacking interactions between F98 and the nucleobases completely disrupts DNA unwinding. This has not been accounted for in any helicase mechanism thus far proposed.
Figure 5. DNA unwinding activity.
(A) DNA Unwinding under multiple-cycle conditions. DNA, 10 nM dT8:12bp substrate was incubated with helicase (100 nM) and the reaction was initiated by mixing with ATP, Mg2+, phosphoenol pyruvate, pyruvate kinase/lactate dehydrogenase, and DNA trap. ssDNA was separated from dsDNA on a 20% native polyacrylamide gel and visualized by phosphorimager analysis. (B) A 16 bp DNA substrate is unwound by Dda in a three-step unwinding scheme which was used to fit data using KinTek Explorer (Johnson et al., 2009). Starting with enzyme (E) and substrate (S) prebound, the enzyme can convert the substrate to product in a series of three identical steps, defined by rate constant (ku). At each step, the enzyme can also dissociate from the substrate with rate constant kd and bind to the DNA trap (T). (C) DNA unwinding under single-cycle conditions. Unwinding of 10 nM dT7:16bp substrate by 1 μM Dda. Enzyme and substrate were pre-incubated, and the reaction was initiated by mixing with ATP, Mg2+, DNA trap, and protein trap. Data were fit to the three-step sequential mechanism shown in B for wt, W378A and K364A forms of Dda. A single exponential was used to fit unwinding data for the P89A variant. The rate constants and amplitudes of unwinding are listed in Table 2. See also Figure S2.
Functional Studies – The Pin-Tower Interaction
Three variant proteins were produced to test the importance of the pin-tower interface (Figure S1), and these were examined for DNA binding, ssDNA-stimulated ATPase activity, and DNA unwinding activity. These data reveal that the pin-tower interaction is critical for the tight coupling of ATP hydrolysis to DNA unwinding in Dda.
E93A/E94A removes the electrostatic interaction with K364 (Figure 2B), and DNA binding and DNA-stimulated ATPase activity were only slightly reduced compared to wtDda (Figure 4A, Table 2). E93A/E94A was able to unwind DNA under multiple-cycle conditions (Figure 5A). In contrast, single-cycle DNA unwinding produced an unusual result in which a small quantity of ssDNA product appeared during the first few milliseconds of the reaction but remained constant over the remaining time-frame (Figure 5C). This pattern of ssDNA appearance is consistent with ATP-independent unwinding reactions, although such activity is not observed with wtDda under these conditions. Control experiments supported the conclusion that the small degree of unwinding observed for E93A/E94A was indeed due to ATP-independent melting (Figure S2). The electrostatic interactions appear to serve an important role in transmitting ATP hydrolysis energy to DNA unwinding. This was confirmed by the K364A mutant that showed reduced DNA binding and moderately reduced ATPase activity (Figure 4B, Table 2). Although K364A was able to unwind DNA under multiple-cycle conditions, single-cycle conditions revealed significantly reduced DNA unwinding activity (Figure 5C, Table 2).
The hydrophobic interaction between the pin and the tower was investigated by mutating Trp378. Although W378A showed reduced DNA binding, the kcat for ATPase activity was only moderately reduced (Figure 4A, Table 2). In addition, the DNA unwinding activity of W378A was observed under multiple-cycle conditions, but single-cycle unwinding indicated that both the amplitude of product formed and the rate constant for product formation were reduced by about 10-fold compared to wtDda. These results indicate that disruption of the pin-tower interaction significantly decouples ATP binding and hydrolysis from DNA unwinding.
Functional Studies – The Hook Region
To test the prediction that the extended hook region mediates interactions with the incoming duplex via Phe276 (Figure 3D), we generated the point mutant F276A and the 6-amino acid deletion mutant 279-284 that truncates the end of the β-ribbon. Neither the mutation nor the deletion had a strong impact on the DNA unwinding activity (supplemental Fig. 2C). The unwinding rate was similar for the deletion mutant and was reduced by less than 2-fold for F276A. The amplitude was reduced by ~ 2-fold for each variant. Hence, the role of the hook region is not critical for DNA unwinding activity.
Translocation on ssDNA is Only Modestly Reduced by Mutations in the Pin-Tower Coupling Path
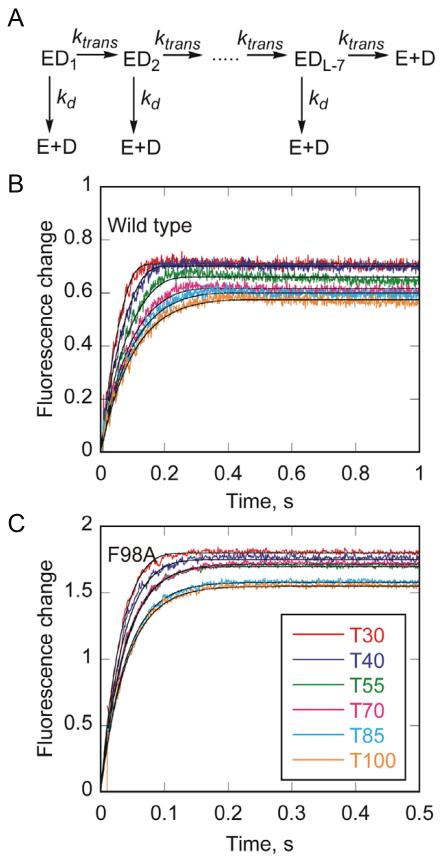
We sought to determine whether the newly identified coupling path through the pin-tower interaction was necessary for translocation on ssDNA. Translocation was determined by measuring dissociation of Dda from increasing length oligonucleotides by stopped-flow fluorescence (Rajagopal et al., 2010). Longer oligonucleotides require longer dissociation times because Dda must translocate further to reach the end of the track (Figure 6A). Figure 6B shows dissociation data of wtDda from oligonucleotides of increasing length. Fitting the data to a model for translocation provided a translocation rate constant of 233 ± 27 nt s−1 (see supplement for detailed description of translation data analysis). Dissociation of F98A Dda was determined from the same oligonucleotides (Figure 6C). Interestingly, the rate constant for translocation was 249 ± 23 nt s−1, which was essentially the same as wtDda. Thus, the F98A variant was incapable of unwinding dsDNA, but exhibited rapid translocation on ssDNA. Furthermore, the other variant forms of Dda all exhibited robust translocation rate constants (Figure S3, Table 2). Hence, translocation on ssDNA is not dependent on the newly identified coupling path between the tower region and the pin. However, efficient strand separation requires this interaction.
Figure 6. ssDNA translocase activity.
(A) A translocation scheme was used to fit data from DNA translocation experiments using KinTek Explorer (Johnson et al., 2009). The enzyme was equally distributed among all DNA binding sites (ED1 to EDL-7) where L is the length of ssDNA to allow for random binding to the substrate and account for the binding site size of 8 nucleotides based on the structure. The enzyme can translocate on the substrate in a series of identical steps, defined by rate constant (ktrans). At each step, the enzyme can also dissociate from the substrate with rate constant kd and bind to the dextran sulfate trap. (B) Change in fluorescence of Dda upon dissociation of wild type Dda from T30, T40, T55, T70, T85, and T100 oligonucleotides. Enzyme and DNA were pre-incubated, and the reaction was initiated by mixing with ATP, and dextran sulfate. Data were fit to the mechanism shown in (A). The rate constants for translocation and dissociation are 233 ± 27 nt/s and 5.75 ± 1.60 s−1, respectively. (C) Change in fluorescence upon dissociation of F98A Dda from T30, T40, T55, T70, T85, and T100 oligonucleotides. Enzyme and DNA were pre-incubated, and the reaction was initiated by mixing with ATP and dextran sulfate. Data were fit to the mechanism shown in (A). The rate constants for translocation and dissociation are 249 ± 23 nt/s and 20.4 ± 2.3 s−1, respectively. See also Figure S3.
Functional Surfaces of Dda
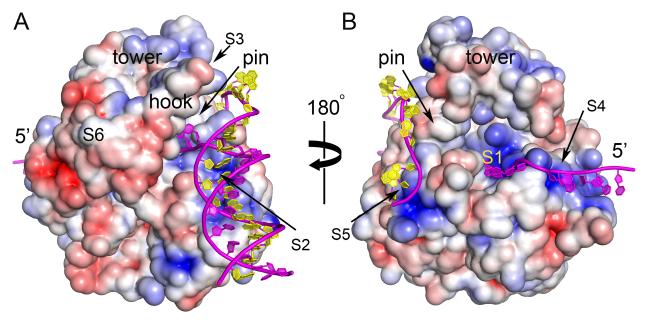
Figure 7 shows the surface electrostatic potential of Dda which reveals that the path of the translocated ssDNA through the arch has the appropriate positive potential (surface S1). Four additional positively charged surfaces can be identified (S2-S5) which suggest the binding sites for the incoming duplex (S2 or S3), the exiting translocating strand (S4) and the displaced strand (S5). Modeling cannot distinguish whether surface S2 or S3 is the binding site for the incoming duplex because either is consistent with the role of the pin and the path of the translocating ssDNA strand. However, we prefer the model shown in Figure 7 because it mirrors the binding mode of duplex DNA in PcrA (Velankar et al., 1999) and UvrD (Lee and Yang, 2006) where the 90° turn is suggested to facilitate unwinding. The model also maximizes the proposed interactions with the hook region. Surfaces S4 and S5 suggest that Dda maintains contact with both the translocating and the displaced DNA strands, to prevent reannealing and/or to direct them towards UvsX and gp32 that interact with Dda (Formosa and Alberts, 1984; Hacker and Alberts, 1992). Surface S4 leads towards a conserved hydrophobic patch on the SH3 domain (surface S6), and this may represent the docking site for these interacting partners.
Figure 7. Surface electrostatic potential of Dda and substrate binding.
(A) The view corresponding to Figure 1A and (B) is rotated by 180°. Blue is positive potential, red is negative and white is neutral/hydrophobic (±5 kT/e electrostatic potential). Key surfaces are labeled S1-S6. S1 engages the translocating strand. S2 and S3 are putative alternate binding sites for the incoming duplex. S4 and S5 are putative binding sites for the exiting translocated strand and the displaced strand, respectively. S6 is a conserved hydrophobic patch on the SH3 domain that is well positioned to bind T4 UvsX and/or gp32. Superimposed on the figure is a model of the full DNA substrate based on these sites. S2 is preferred over S3 for binding the duplex in this model. See also Movie S1.
DISCUSSION
The SF1 and SF2 helicases comprise a core motor of paired RecA-like domains, which undergo an ATP-driven cyclical conformational change that brings the domains together upon binding ATP and moves them apart after ATP hydrolysis. Crystallographic and functional studies have led to the ‘inchworm’ model (Velankar et al., 1999) or the revised ‘wrench and inchworm’ model (Lee and Yang, 2006) for how this conformational change is coupled to nucleic acid translocation. Single-stranded nucleic acid traverses the RecA-like domains that alternately bind and release the sugar-phosphate backbone during the conformational change, moving the nucleic acid by one nucleotide per cycle. SF1A and SF1B helicases translocate in 3′-to-5′ or 5′-to-3′ directions, respectively, and structural studies have revealed that the key to directionality is the location of amino acid side chains that act as ratchets which favor nucleic acid movement in one direction over the other. Thus, the two RecA-like domains in the SF1A and SF1B helicases bind and release ssDNA during the conformational change in reversed order.
Dda and RecD2 are both SF1B helicases, and their structures and mode of binding to ssDNA in the non-ATP bound form are very similar. Our Dda structure supports the 5′-to-3′ translocation mechanism (Saikrishnan et al., 2009) but it provides new insights into the ratchet mechanism. In Dda, Phe98 within the pin forms a stacking interaction with the X6 nucleobase (Figures 2B, 3A and 3C). This interaction is not seen in the RecD2 structure even though an equivalent Tyr414 is available (Figure 3B). Conversely, in RecD2, the base at X5 is bound within a pocket created by Pro389, Met439 and Val470, and this is not seen in Dda although an equivalent pocket comprising Pro62, Met119 and Val150 is available. We propose that both interactions serve as ratchets during translocation, but they have been independently observed in the two structures. The ratchets appear to be alternate conformations that cannot exist simultaneously; the Phe98-X6 interaction stabilizes the stacking of the adjacent X5 base and prevents its rotation into the pocket, whereas rotation of X5 into the pocket would destabilize the Phe98-X6 interaction. The RecD2 ternary complex reveals that the transition between the open and closed conformations involves a twisting motion, and our model of the Dda ternary complex (Figure 3D) shows that this is sufficient to remove the Phe98 stacking interaction and permit the passage of ssDNA. This suggests that our open complex of Dda reflects the conformation just prior to domain closure and that the RecD2 open complex is the conformation that precedes this. In the RecD2 ternary complex, the base at X5 is flipped out of the pocket similar to Dda, supporting this model. It has been proposed (Saikrishnan et al., 2009) that Val470 in RecD2, and therefore Val150 in Dda, may ‘sense’ the nucleotide status of the helicase during the conformational change because it is at the C-terminus of motif III which directly links the nucleotide and nucleic acid binding sites. Nucleotide binding may be the trigger that flips the X5 base from the pocket and initiates domain closure.
SF1 and a subset of SF2 helicases perform strand separation (Jankowsky and Fairman, 2007), and a common feature of these enzymes is a β-ribbon ‘pin’ that inserts into and splits the incoming duplex such that one strand passes through the motor while the second strand is displaced (Buttner et al., 2007; Kim et al., 1998; Korolev et al., 1997; Lee and Yang, 2006; Pike et al., 2009; Theis et al., 1999; Velankar et al., 1999). In the 3′-to-5′ helicases thus far studied, the pin is associated with the 2A RecA domain to facilitate interaction with the incoming duplex. In contrast, the pin in Dda and RecD2 is part of the 1A RecA domain to accommodate the entry of the duplex from the opposite direction. In the RecD2 ternary complex (Saikrishnan et al., 2009), Tyr598 within a short loop in the SH3 domain acts as an additional ratchet role to prevent ‘backsliding’ of the ssDNA as the 1A domain repositions to the ‘open’ conformation, and Phe276 is equivalently placed to perform this same role in the extended hook region. However, mutations of Phe276 and the hook region reveal that these are not essential for unwinding, consistent with its lack of conservation among T4-like Dda sequences.
The biological roles of Dda during T4 infection have yet to be firmly established, but a number of in vitro studies suggest that its function is related to the ability to efficiently unwind dsDNA roadblocks (Byrd and Raney, 2004, 2006; Eoff et al., 2005). This is consistent with our kinetic data which show that Dda is essentially an optimally active helicase, meaning that it separates dsDNA at the same rate as it translocates on ssDNA (ku/ktrans ≈ 1, Table 2) (Manosas et al., 2010). Our structure of the Dda-ssDNA binary complex suggests that the elongated pin and its tight interaction with the tower region at its tip and the ssDNA at its base are the key features that contribute to this efficiency. The tower region within domain 2B is rigidly connected to domain 2A and appears to be specifically designed for the task of supporting the extended pin. We suggest that 2A and 2B move as one unit during the ATP-driven translocation of ssDNA while maintaining contact with the pin. In this scenario, the pin-tower interaction can be considered as an additional ‘transmission’ site that serves to more efficiently couple the energy from ATP binding and hydrolysis to the unwinding of dsDNA. Movie S1 shows a model of the proposed Dda translocation/unwinding mechanism. Point mutations designed to break this energy transmission pathway support this model.
Disruption of the pin-tower interface in K364A and W378A reduces the processivity for DNA unwinding by more than 10-fold and partially uncouples the translocase and helicase activities. However, Phe98 plays a more important role in unwinding. Phe98 is required for DNA unwinding presumably due to its ability to engage the duplex through stacking interactions with an incoming base. The role of the pin-tower interface appears to allow the pin to form a more rigid body through interaction with two domains of the enzyme. The pin is thereby made responsive to ATP driven conformational changes that modulate the ability of Phe98 to actively engage the duplex. Disruption of the pin-tower interaction reduces this activity whereas mutation of Phe98 to Ala eliminates this activity. Translocation of F98A remains intact because splitting the DNA is not necessary for translocation of ssDNA. The fact that E93A/E94A has a greater effect on ssDNA translocation than F98A may be surprising. However, translocation measurements may be affected by the enhanced, ATP-independent sliding activity of E93A/E94A, which is not exhibited by F98A. Architecturally, Dda resembles the HCV NS3 RNA helicase which also contains an extended pin attached at its distal end to a helical domain (Kim et al., 1998), and mutations at the interaction surface produced similar results to ours (Lam et al., 2003).
Two positively charged surfaces are appropriately located to act as transient binding sites for the two emerging DNA strands to prevent re-annealing and, potentially, to contribute to unwinding efficiency. Dda, like RecD2, contains an SH3 domain that typically mediates protein-protein interactions, and the SH3 surface may be the docking site for the T4 recombinase UvsX and/or the T4 ssDNA-binding protein gp32 (Formosa and Alberts, 1984; Hacker and Alberts, 1992). A hydrophobic patch on the SH3 domain could mediate these interactions, and one of the positively-charged surfaces is appropriately positioned to channel the translocated strand of DNA towards this patch (Figure 7). Our model of the Dda closed ternary complex (Figure 3D) also suggested that the hook region and Phe276 at the base of the hook might contribute to unwinding efficiency, but mutagenesis studies did not support this. However, the hook is well positioned to facilitate the displacement of bound proteins from the incoming dsDNA, which is a well characterized property of Dda (Bedinger et al., 1983; Byrd and Raney, 2006; Morris and Raney, 1999). These features are also shown in Movie S1. We are in an excellent position to characterize these potential interactions now that the structure of Dda is available.
EXPERIMENTAL PROCEDURES
Dda(K38A) Expression and Purification
The ATPase deficient Dda mutant, K38A was cloned as an N-terminally 6xHis-tagged protein in pET28b. The construct was transformed into BL21 (DE3) cells (Novagen, Madison, WI), and cells were grown in LB medium containing 20 ug/ml kanamycin at 37°C until the OD600 reached 0.4-0.6. The culture was grown at 16 °C for 1 hour before the cells were induced for 16 hours by adding isopropyl-β-D-thiogalactoside (0.5 mM). Cells were harvested by centrifugation at 2000×g for 20 minutes. The pellet was resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5% glycerol, 1mM imidazole, 5 mM β-mercaptoethanol, 0.4 mg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride supplemented with EDTA-free proteinase inhibitor cocktail (Roche) and lysed using a microfluidizer. NaCl was added to a final concentration of 400 mM, and the suspension was centrifuged at 154,000×g for 1 hour before application to a Ni2+ chelation column (GE Healthcare) in buffer A (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 1mM imidazole, 5 mM β-mercaptoethanol). Dda(K38A) was eluted with a 1 to 200 mM imidazole gradient in buffer A. Cleanup was performed by gel filtration on a HiLoad® 16/60Superdex® 75 column (Pharmacia) run in 0.5 mM EDTA, 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol. For crystallographic phasing, a selenomethionine-substituted version of Dda(K38A) was expressed in the E. coli methionine auxotroph strain B834 (Novagen) as described (Price et al., 2001).
Crystallization, Data Collection and Crystallography
Poly-dT8 and Dda(K38A) in a 2:1 molar ratio were concentrated to ~10 mg/ml using 10,000 MWCO Amicon® Ultra Centrifugal Filters (Millipore). Crystallization trials were performed at 18 °C using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, CA), and the best crystals were grown in 4.5% (w/v) PEG8000, 8% (v/v) ethylene glycol, 0.1 M Hepes, pH 6.5, 20 mM DTT. Crystals were cryo-protected in 25% glycerol, and a single-wavelength (peak) SAD dataset was collected at the SERCAT ID line at the Advanced Photon Source to 3.8Å. SAD phasing followed by density modification generated an electron density map that revealed three Dda molecules in the asymmetric unit. We subsequently collected 3.3 Å data from higher quality crystals grown in the same conditions, and fitted the amino acid sequence using iterative rounds of model building, refinement and phase combination. Data collection and refinement statistics are shown in Table 1. Data processing was performed using HKL2000 (Otwinowski and Minor, 1997) and SAD phases were calculated with PHENIX (Adams et al., 2010). Initial models were built and refined using the O program (Kleywegt and Jones, 1997) and CNS (Brunger et al., 1998), and final model building and refinement used COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010). Structural figures and surface electrostatic potentials were generated using PyMol (DeLano, 2002), and the structural statistics were calculated using PROCHECK (Laskowski et al., 1993). The movie was generated using UCSF Chimera (Pettersen et al., 2004) and PyMol (DeLano, 2002).
ATPase Activity Assays
ATP hydrolysis was measured using a spectrophotometric assay in which ATP hydrolysis is coupled to oxidation of NADH through pyruvate kinase (PK) and lactate dehydrogenase (LDH). The assay buffer contained 25 mM HEPES pH 7.5, 10 mM KOAc, 2 mM β-mercaptoethanol, 0.1 mg/mL BSA (bovine serum albumin) 10 mM Mg(OAc)2, 5 mM ATP,0.8 mM phosphoenol pyruvate, 17.05 U/mL PK, 24.75 U/mL LDH, and 0.7 mg/mL NADH. Dda (50 nM) was added, followed by denatured calf thymus DNA. The change in absorbance associated with NADH conversion to NAD+ was monitored at 380 nm and was directly correlated to ATP hydrolysis.
Fluorescence Anisotropy
Binding assays were performed in assay buffer (25 mM Hepes pH 7.5, 10 mM KOAc, 0.1 mM EDTA, 2 mM β-mercaptoethanol, 0.1 mg/ml BSA, and 10 mM Mg(OAc)2). An oligonucleotide substrate (dT7 single-stranded overhang with a 16 bp duplex, dT7:16bp) containing a 5′-fluorescein label was used for studies. Enzyme was incubated with 1 nM DNA for 10 minutes at 25° C before measuring fluorescence anisotropy on a Perkin Elmer 1420 Multilabel Counter Victor3V instrument and analyzing using KaleidaGraph software.
DNA unwinding assays
Multiple cycle DNA unwinding was performed using a 12bp duplex. The substrate was prepared by annealing a radio-labeled dT812mer to a 12mer to form a dT8:12bp substrate. Dda (100 nM) was pre-incubated with 10 nM DNA for 10 min at 25 °C in assay buffer without Mg(OAc)2. The reactions were initiated by mixing with 5 mM ATP, 10 mM Mg(OAc)2, and a DNA trap (300 nM compliment 12mer). Phosphoenol pyruvate (4 mM) and PK (10 U/mL) and LDH (14.5 U/mL) were added with the ATP to prevent depletion of ATP during the course of the reaction. All concentrations listed are final, after mixing. Reactions were quenched with 400 mM EDTA.
The oligonucleotide substrate for single cycle DNA unwinding studies was a dT7:16bp substrate. Enzyme (1 μM) and DNA(10 nM) were allowed to incubate in assay buffer without Mg(OAc)2 at 25°C for 10 minutes before initiating the reaction by mixing with 5 mM ATP and 10 mM Mg(OAc)2. A protein trap (75 μM nt, T150) and a DNA trap (300 nM compliment 16mer) were included with the ATP to ensure single cycle conditions with respect to the DNA substrate. The reaction was quenched with 400 mM EDTA. For some experiments, Dda was mixed with DNA, ATP, and Mg(OAc)2 to initiate the reaction. For these experiments, the protein and DNA traps were added with the quench. Samples were mixed with gel loading buffer (0.1% xylene cyanol, 0.1% bromophenol blue, 10% glycerol), separated on a 20% native gel, visualized with a Typhoon Trio phosphorimager (GE Healthcare), quantified using ImageQuant software (GE Healthcare), and fit to the scheme shown in Figure 5B using KinTek Explorer (Johnson et al., 2009).
Translocation Assays
100 nM Dda was pre-incubated with 6 μM ssDNA (in nucleotides) in buffer without bovine serum albumin in an SX.18MV stopped flow reaction analyzer (Applied Photophysics). Reactions were initiated at 25 °C by mixing with a saturating concentration of ATP (600 μM) (Byrd et al., 2012) and 12 mg/ml dextran sulfate. The change in tryptophan fluorescence was monitored after a 320 nm cutoff filter (Newport Optical Filter #FSQ-WG320) with excitation at 280 nm through 1 mm slits. The fluorescence data was fit to the mechanism in Figure 6A using KinTek Explorer (Johnson et al., 2009) to obtain the rate constant for translocation.
Supplementary Material
HIGHLIGHTS.
New insights into the mechanism of SF1B helicases.
Insights into how a monomeric helicase is optimized to perform strand separation.
Uncoupling of the unwinding and translocation activities of a helicase.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM066934 (to SWW and KNK) and GM098922 (to KDR), UL1RR029884 from the NCRR, Cancer Center core grant CA21765 and the American Lebanese Syrian Associated Charities (ALSAC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract No. W-31-109-Eng-38. We thank Rebecca DuBois and Darcie Miller for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Refined coordinates and structure factors have been deposited in the Protein Databank as 3UPU.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and one table and one movie can be found with this article online.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Brosh RM., Jr. Hitting the bull’s eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J Cell Biochem. 2009;106:758–763. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P, Hochstrasser M, Jongeneel CV, Alberts BM. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Behme MT, Ebisuzaki K. Characterization of a bacteriophage T4 mutant lacking DNA-dependent ATPase. J Virol. 1975;15:50–54. doi: 10.1128/jvi.15.1.50-54.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger KG. PhD Thesis Dissertation. Duke University Medical Center; 1997. Origin-dependent DNA replication in bacteriophage T4: A mechanism for initiation. [Google Scholar]
- Blair LP, Tackett AJ, Raney KD. Development and evaluation of a structural model for SF1B helicase Dda. Biochemistry. 2009;48:2321–2329. doi: 10.1021/bi801919s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- Boule JB, Zakian VA. Characterization of the helicase activity and anti-telomerase properties of yeast Pif1p in vitro. Methods Mol Biol. 2010;587:359–376. doi: 10.1007/978-1-60327-355-8_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr., Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. The Journal of biological chemistry. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AK, Matlock DM, Bagchi D, Aarattuthodilyil S, Harrison D, Croquette V, Raney KD. Dda helicase tightly couples translocation on single-stranded DNA to unwinding of duplex DNA: Dda is an optimally active helicase. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.04.007. doi:10.1016/j.jmb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q Rev Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- Ellis NA. DNA helicases in inherited human disorders. Curr Opin Genet Dev. 1997;7:354–363. doi: 10.1016/s0959-437x(97)80149-9. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- Eoff RL, Raney KD. Kinetic mechanism for DNA unwinding by multiple molecules of Dda helicase aligned on DNA. Biochemistry. 2010;49:4543–4553. doi: 10.1021/bi100061v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoff RL, Spurling TL, Raney KD. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- Formosa T, Alberts BM. The use of affinity chromatography to study proteins involved in bacteriophage T4 genetic recombination. Cold Spring Harb Symp Quant Biol. 1984;49:363–370. doi: 10.1101/sqb.1984.049.01.043. [DOI] [PubMed] [Google Scholar]
- Frick DN. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr Issues Mol Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- Gauss P, Park K, Spencer TE, Hacker KJ. DNA helicase requirements for DNA replication during bacteriophage T4 infection. J Bacteriol. 1994;176:1667–1672. doi: 10.1128/jb.176.6.1667-1672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases - Amino-Acid-Sequence Comparisons and Structure-Function-Relationships. Current Opinion in Structural Biology. 1993;3:419–429. [Google Scholar]
- Gu J, Xia X, Yan P, Liu H, Podust VN, Reynolds AB, Fanning E. Cell cycle-dependent regulation of a human DNA helicase that localizes in DNA damage foci. Mol Biol Cell. 2004;15:3320–3332. doi: 10.1091/mbc.E04-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker KJ, Alberts BM. Overexpression, purification, sequence analysis, and characterization of the T4 bacteriophage dda DNA helicase. The Journal of biological chemistry. 1992;267:20674–20681. [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Drake JW. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. The Journal of biological chemistry. 2004;279:35735–35740. doi: 10.1074/jbc.M403942200. [DOI] [PubMed] [Google Scholar]
- Kerr ID, Sivakolundu S, Li Z, Buchsbaum JC, Knox LA, Kriwacki R, White SW. Crystallographic and NMR analyses of UvsW and UvsW.1 from bacteriophage T4. The Journal of biological chemistry. 2007;282:34392–34400. doi: 10.1074/jbc.M705900200. [DOI] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Model building and refinement practice. Methods in Enzymology. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- Kodadek T. Inhibition of protein-mediated homologous pairing by a DNA helicase. The Journal of biological chemistry. 1991;266:9712–9718. [PubMed] [Google Scholar]
- Kodadek T, Alberts BM. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987;326:312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. The Journal of biological chemistry. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci U S A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patel SS, Donmez I. Mechanisms of helicases. The Journal of biological chemistry. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci U S A. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AC, Zhang YM, Rock CO, White SW. Structure of beta-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry. 2001;40:12772–12781. doi: 10.1021/bi010737g. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Rajagopal V, Gurjar M, Levin MK, Patel SS. The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase. The Journal of biological chemistry. 2010;285:17821–17832. doi: 10.1074/jbc.M110.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja P, Gu J, Peng R, Carrick R, Uchiumi F, Ott RD, Gustafson E, Podust VN, Fanning E. A dominant-negative mutant of human DNA helicase B blocks the onset of chromosomal DNA replication. The Journal of biological chemistry. 2002;277:40853–40861. doi: 10.1074/jbc.M208067200. [DOI] [PubMed] [Google Scholar]
- Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.