Abstract
T-cell acute lymphoblastic leukemias and lymphomas (T-ALL) are aggressive hematologic cancers frequently associated with activating mutations in NOTCH1. Early studies identified NOTCH1 as an attractive therapeutic target for the treatment of T-ALL through the use of γ-secretase inhibitors (GSIs). Here, we characterized the interaction between PF-03084014, a clinically-relevant GSI, and dexamethasone in preclinical models of glucocorticoid-resistant T-ALL. Combination treatment of the GSI PF-03084014 with glucocorticoids induced a synergistic antileukemic effect in human T-ALL cell lines and primary human T-ALL patient samples. Mechanistically PF-03084014 plus glucocorticoid treatment induced increased transcriptional upregulation of the glucocorticoid receptor and glucocorticoid target genes. Treatment with PF-03084014 and glucocorticoids in combination was highly efficacious in vivo, with enhanced reduction of tumor burden in a xenograft model of T-ALL. Finally, glucocorticoid treatment effectively reversed PF-03084014-induced gastrointestinal toxicity via inhibition of goblet cell metaplasia. These results warrant the analysis of PF-03084014 and glucocorticoids in combination for the treatment of glucocorticoid-resistant T-ALL.
Introduction
The NOTCH1 receptor is a class I single-pass transmembrane protein involved in cell-fate decisions during development. Interaction of NOTCH1 with Delta-like or Jagged ligand molecules expressed on the surface on a neighboring cell induces the proteolytic cleavage of the receptor by ADAM10 at the cell surface and then by the gamma-secretase complex in the transmembrane domain (1). This double proteolytic processing releases the intracellular portion of NOTCH1 from the plasma membrane and triggers its translocation to the cell nucleus where it activates the transcription of NOTCH-target genes. Activating mutations in the NOTCH1 gene are present in over 50% of T-cell acute lymphoblastic leukemia (T-ALL) cases, and constitutive activation of NOTCH1 signaling plays a major role in the pathogenesis of this disease (2, 3). Given the high prevalence of NOTCH1 mutations and the strict requirement of the gamma-secretase complex for effective NOTCH1 signaling, small molecule gamma-secretase inhibitors (GSIs) have been proposed as anti-NOTCH targeted therapy in T-ALL. However, early work testing GSIs in the treatment of human leukemia showed limited antitumor response and overt gastrointestinal toxicity (4).
Glucocorticoids are essential drugs in the treatment of T-ALL due to their ability to induce apoptosis and cell cycle arrest in leukemia lymphoblasts. In the absence of ligand, the glucocorticoid receptor protein (NR3C1) is located in the cytoplasm in an inactive complex with heat shock proteins (5). Glucocorticoid binding induces activation of the receptor and triggers its translocation to the nucleus where it binds to DNA and activates a broad gene expression program resulting in cell cycle arrest and induction of apoptosis in T-ALL cells (6–8). The importance of glucocorticoids in the treatment of T-ALL is highlighted by the poor prognosis associated with limited initial response to glucocorticoid therapy and the frequent development of secondary glucocorticoid resistance in patients at relapse (9, 10).
Our previous work found that Compound E and dibenzazapine (DBZ), two generic gamma secretase inhibitors, can reverse glucocorticoid resistance in T-ALL (11). Moreover, glucocorticoid treatment antagonizes the intestinal toxicity associated with systemic inhibition of NOTCH signaling with GSIs. Here, we describe preclinical studies characterizing the interaction between glucocorticoids and PF-03084014, a clinically-relevant GSI. Our results show a synergistic antitumor response to PF-03084014 and glucocorticoids in primary human T-ALL samples and cell lines and demonstrate effective protection from GSI-induced gut toxicity in animals treated with PF-03084014 and glucocorticoids in combination.
Materials and Methods
Inhibitors and drugs
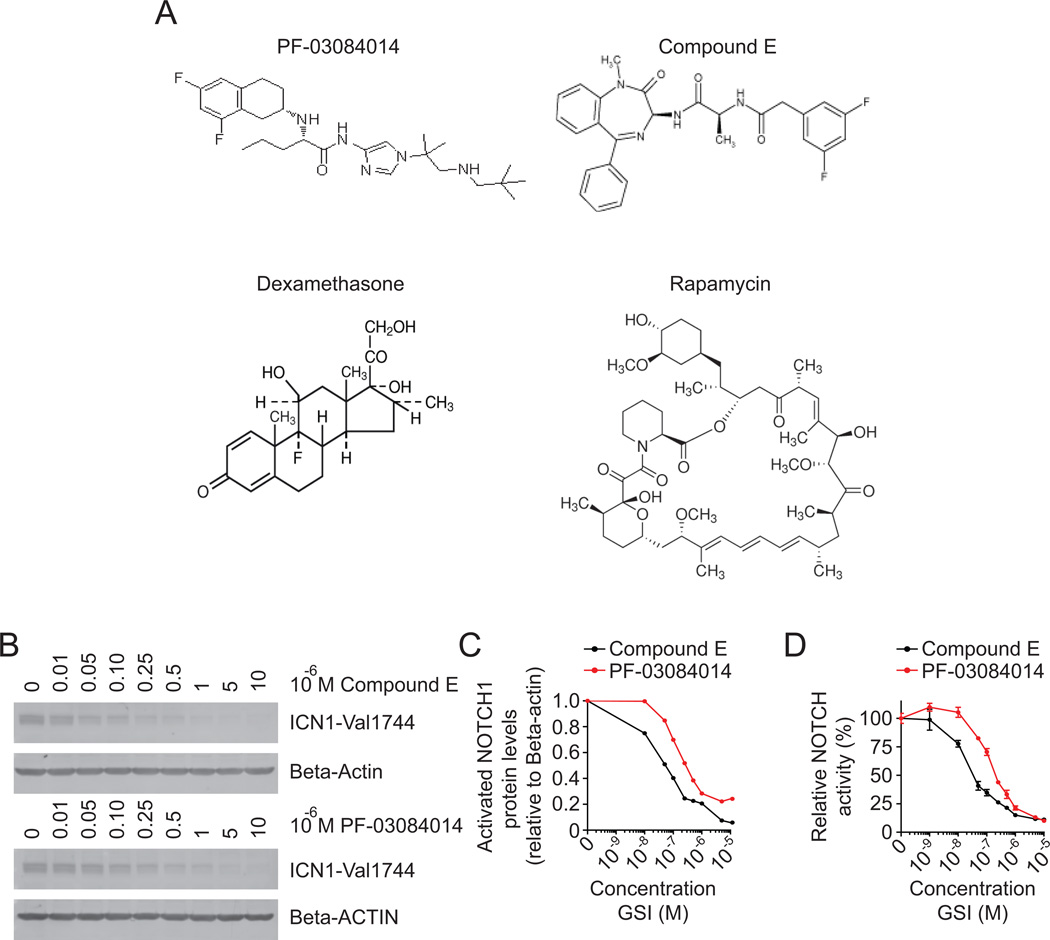
Compound E was purchased from Enzo Life Sciences, PF-03084014 [(S)-2-((S)-5,7-difluoro-1,2,3,4-tetrahydronaphthalen-3-ylamino)-N-(1-(2-methyl-1-(neopentylamino)propan-2-yl)-1H-imidazol-4-yl)pentanamide] was synthesized at Pfizer, Groton, CT. Dexamethasone, etoposide, methotrexate, vincristine, and rapamycin were all purchased from Sigma-Aldrich. L-asparaginase was purchased from Roche. Imatinib mesilate was a gift from Dr. David Sternberg (Mount Sinai School of Medicine, New York, NY). Chemical structures for PF-03084014, Compound E, dexamethasone and rapamycin are reported in Figure 1A.
Figure 1. Inhibition of NOTCH1 activation and activity by PF-03084014.
(A) Chemical structures of PF-03084014, Compound E, dexamethasone, and rapamycin. (B) Western blot analyses of activated NOTCH1 protein in CUTLL1 lymphoma cells treated with CompE or PF-03084014; β-Actin is shown as loading control. (C) Activated NOTCH1 protein levels (ICN1-Val1744) relative to β-Actin. (D) Luciferase reporter analysis of NOTCH1 transcriptional activity in 293T cells transfected with an activated mutant form of NOTCH1 (ΔE NOTCH1) treated with CompE or PF-03084014. Data are represented relative to vehicle only (DMSO) treated cells.
Cell lines and pediatric leukemia samples
The CUTLL1 cell line derived from a glucocorticoid resistant T-cell acute lymphoblastic lymphoma patient at relapse was generated, validated and fingerprinted and characterized in the Ferrando laboratory at Columbia University (12). KOPTK1, TALL1, ALL-SIL and RPMI-8402 T-ALL cells were purchased from American Type Culture Collection and the Deutsche Sammiung von Mikroorganismen und Zellkulturen. Hairpin oligonucleotide sequences targeting either the NR3C1 gene or a non-silencing control were expressed in the pGIPZ lentiviral vector. Oligonucleotide sequences for shRNAs targeting the PTEN or luciferase gene were expressed in the pLKO-GFP lentiviral vector. Lentivirus production and spin infection of CUTLL1 cells were performed as previously described (13). Primary T-ALL lymphoblast samples were provided by collaborating institutions in the USA (Department of Pediatrics, Columbia Presbyterian Hospital, New York), the Hospital Central de Asturias (Oviedo, Spain) and the Eastern Cooperative Oncology Group (ECOG). All samples were collected with informed consent and analyzed under the supervision of the Columbia University Medical Center Institutional Review Board.
Antibodies and western blotting
Antibodies against activated NOTCH1 (Val1744, Cell Signaling); PTEN (clone 6H2.1, Cascade Biosciences), beta-ACTIN (C-11, Santa Cruz Biotechnology), and NR3C1 (E-20, Santa Cruz Biotechnology) were used for western blot analysis according to standard procedures. Protein expression was visualized by chemifluorescence using the Typhoon Trio Variable Mode Imager (GE Healthcare). ICN1-Val1744 band intensity relative to beta-Actin was calculated using ImageJ software (National Institutes of Health).
Luciferase assay
We co-transfected 293T cells in triplicate with pCS2-ΔE-NOTCH1; pGA-luc, a reporter containing six tandem CSL-binding sites upstream of the firefly luciferase gene (a gift from Dr. Honjo at Kyoto University, Kyoto, Japan); and pRL, a plasmid expressing the Renilla luciferase gene under the control of the cytomegalovirus (CMV) promoter used as an internal control. We performed transfections using FuGENE 6 (Roche) following the manufacturer’s protocol. Cells were treated for 24 hours with increasing doses of either Compound E or PF-03084014, and we performed luciferase assays using the Dual-Luciferase Reporter Assay System and a Modulus II microplate reader (Promega). The range of concentrations of GSI used in these experiments was 10−9 to 10−5 M. Statistical significance was calculated by one-tailed Student’s t-test using GraphPad Prism software.
In vitro cell viability assays
We cultured cells for 48 hours (KOPTK1) or 72 hours (CUTLL1, TALL1, RPMI-8402, ALL-SIL) in the presence of indicated drugs. Cell growth ratios were calculated using the MTT Cell Proliferation Kit I (Roche Applied Science; Indianapolis, IN). The range of concentrations used in these experiments were: GSI (10−9 to 10−5 M), dexamethasone (10−10 to 10−5 M), vincristine (10−13 to 10−7 M), rapamycin (10−12 to 10−6 M), methotrexate (10−10 to 10−4 M), L-asparaginase (10−11 to 10−5 M), etoposide (10−10 to 10−4 M), and imatinib (10−12 to 10−6 M). For the analysis of primary T-ALL patient samples, we cultured cells in MEMα medium supplemented with 10% fetal bovine serum, 10% human heat inactivated AB+ serum, 1% penicillin/streptomycin, 1% GlutaMAX, human IL-7 (10ng/mL), human SCF (50ng/mL), human FLT3-ligand (20ng/mL), and insulin (20nM) on a feeder layer of MS5 stromal cells overexpressing the NOTCH ligand Delta-like 1, as previously described in (14). In these experiments, T-ALL lymphoblasts were cultured in triplicate and treated with either 1µM Compound E or PF-03084014 in the presence or absence of dexamethasone (1nM or 10nM). We harvested cells after 72 hours and analyzed cell viability using the BD cell viability kit with liquid counting beads (BD Bioscience) in combination with APC-conjugated anti-CD45 staining to gate out stromal cells. We acquired data using a FACSCanto II flow cytometer (BD Bioscience) and analyzed it using FlowJo software (Tree Star, Inc.). We analyzed apoptosis using the Annexin V PE Apoptosis Detection Kit I and cell cycle using the APC BrdU Flow Kit (BD Bioscience) following 48 hours (KOPTK1) and 72 hours (CUTLL1 and TALL1) of treatment with PF-03084014 plus dexamethasone. Statistical significance for cell viability assays was calculated by one-tailed Student’s t-test using GraphPad Prism software.
DNA microarray analysis
RNA was isolated from CUTLL1 cells treated for 48 hours with DMSO, 1µM PF-03084014, 1µM dexamethasone, or PF-03084014 plus dexamethasone, and samples were labeled and hybridized to Affymetrix Human U133 Plus 2.0 microarrays as previously described (11). Normalization was performed with GC-RMA using the open-source Bioconductor project within the statistical programming language R (15, 16). Differentially expressed genes between PF-03084014 plus dexamethasone and dexamethasone treatment (fold change > 1.25) were ranked based on Spearman correlation with an upregulated by dexamethasone and synergistically upregulated by PF-03084014 plus dexamethasone arbitrary vector (P < 0.05). Microarray data is available in Gene Expression Omnibus (GEO) with accession code GSE33562.
Quantitative real-time PCR
Total RNA was extracted from CUTLL1 cells using the RNeasy mini kit (Qiagen). We synthesized cDNA using the SuperScript First Strand Synthesis System (Invitrogen) and performed quantitative real-time PCR using SYBR Green PCR Master Mix and the 7300 Real-Time PCR System (Applied Biosystems). Relative expression levels were normalized with GAPDH expression used as a reference control. Sequences of primers are: HES1 forward: 5’-CTGGAAATGACAGTGAAGCACCT-3’; HES1 reverse: 5’-ATTGATCTGGGTCATGCAGTTG-3’; DTX1 forward: 5’-AAGAAGTTCACCGCAAGAGGATT-3’; DTX1 reverse: 5’-CTAGGTAGCTAGCGTCCGGGTAG-3’; NR3C1 forward: 5’-GGCAATACCAGGTTTCAGGA-3’; NR3C1 reverse: 5’-ACACAGCAGGTTTGCACTTG-3’; BCL2L11 forward: 5’-CACAAACCCAAGTCCTCCTT-3’; BCL2L11 reverse: 5’-TTCAGCCTGCCTCATGGAA-3’; GAPDH forward: 5’-GAAGGTGAAGGTCGGAGT-3’; and GAPDH reverse: 5’-GAAGATGGTGATGGGATTTC-3’. Statistical significance for quantitative PCR analysis was calculated by one-tailed Student’s t-test using GraphPad Prism software.
Mice and animal procedures
We kept all mice in specific pathogen–free animal facilities at Columbia University Medical Center. Mouse procedures were reviewed, approved, and performed under the supervision of the Columbia University Medical Center Institutional Animal Care and Use Committee. We carried out toxicity experiments in 6–8-week-old C57BL/6 female mice (Jackson Laboratory). To analyze the effects of dexamethasone in GSI-induced toxicity, we treated mice with vehicle (DMSO in 0.5% Methocel E4M/0.1% Tween-80), dexamethasone (1mg/kg, 5mg/kg, or 15 mg/kg), PF-03084014 (150mg/kg) and dexamethasone plus PF-03084014 for 5 days. Dexamethasone was administered by once daily intraperitoneal injection and PF-03084014 was administered BID by oral gavage. At the end of the treatment, mice were sacrificed and tissues were collected and processed for histological and immunohistochemical analysis following overnight fixation in 10% neutral buffered formalin. We performed xenograft experiments using 6–8-week-old NOD-SCID female mice (Taconic Farms) as recipients. CUTLL1 FUW-LUC cells were generated as previously described (11). For subcutaneous xenograft experiments, we injected 5×106 CUTLL1 FUW-LUC cells embedded in Matrigel basement membrane matrix (BD Bioscience) subcutaneously into the flank of NOD-SCID mice. After one week, we segregated mice into four groups (vehicle, dexamethasone, PF-03084014, and dexamethasone+PF-03084014) and treated them with vehicle, dexamethasone, PF-03084014, or dexamethasone+PF-03084014 as described above for four days. For imaging studies, we anesthetized mice by isoflurane inhalation and injected them intraperitoneally with D-Luciferin at 50 mg/kg (Caliper Life Sciences). We imaged photonic emission with the IVIS Molecular Imaging System (Caliper Life Sciences) with a collection time of 1 min and quantified tumor bioluminescence using the Living Image software package (Caliper Life Science). Statistical significance for subcutaneous xenograft experiments was calculated by one-tailed paired t-test using GraphPad Prism software.
Immunohistochemistry
We performed anti-Ki67 (Dako) and anti-lysozyme (Dako) immunohistochemistry on formalin-fixed paraffin-embedded tissue sections after antigen retrieval by microwave heating in citrate buffer (pH 6.0) for Ki67 and by proteinase K for lysozyme. After epitope recovery, slides were incubated with antibody (anti Ki67 1:50, anti lysozyme 1:500) overnight at room temperature before antigen detection with diaminobenzidine (DAB).
Results
PF-03084014 inhibits NOTCH1 activation and function
PF-03084014 is a selective tetralin amino imidazole inhibitor of gamma-secretase currently in phase I clinical trials for the treatment of relapsed and refractory T-ALL (17, 18). PF-03084014 inhibits NOTCH1 activation by preventing the proteolytic release of the intracellular, active domain of NOTCH1 from the plasma membrane, which blocks its translocation to the nucleus and the activation of NOTCH-target genes. To test the effects of gamma-secretase inhibition with PF-03084014 in T-ALL we first analyzed the effects of this GSI in NOTCH1 in CUTLL1 cells, a glucocorticoid-resistant human T-ALL cell line that expresses high levels of NOTCH1 as a result of the t(7;9)(q34;q34) translocation (12). Treatment of CUTLL1 cells with increasing doses of PF-03084014 or Compound E (CompE), a well characterized generic GSI, resulted in dose-dependent reduction of activated NOTCH1 protein (Figure 1B, 1C). To establish the ability of PF-03084014 and CompE to inhibit NOTCH1 signaling, we analyzed transcriptional activity of NOTCH1 using a CSL-dependent luciferase reporter assay. In these experiments, we treated NOTCH1-transfected 293T cells with increasing doses of these GSIs, and found both to be potent inhibitors of NOTCH1 transcriptional activity, with 50% inhibitory concentrations (IC50) of 141nM for PF-03084014 and 30nM for CompE (Figure 1D). In these experiments, maximal inhibition of NOTCH activation and activity was achieved using 1µM PF-03084014, so this dose was selected for use in future cell viability assays.
PF-03084014 reverses glucocorticoid resistance in human T-ALL cell lines and primary lymphoblasts
Despite the prominent role of aberrant NOTCH1 signaling in T-cell transformation GSIs seem to exert only a limited antitumor effect in T-ALL (2, 19). These results question the clinical relevance of GSIs in the treatment of human leukemia and suggest that combination of NOTCH1 inhibitors with glucocorticoids and chemotherapy may be needed to potentiate the antileukemic effects of these drugs (11, 20, 21). To examine the interaction between PF-03084014 and glucocorticoids, we treated CUTLL1 cells with PF-03084014 (1µM) and increasing concentrations of dexamethasone (ranging from 10−10 to 10−5M). These experiments showed a synergistic decrease in cell viability in this glucocorticoid-resistant cell line treated with PF-03084014 and dexamethasone in combination (P < 0.001) (Figure 2A). The synergistic antileukemic effects of PF-03084014 and dexamethasone were reproducible in two additional glucocorticoid-resistant cell lines, KOPTK1 and TALL1 (Figure 2A). Similar results were obtained using a fixed concentration of dexamethasone (1µM) and increasing concentrations of PF-03084014 (ranging from 10−9 to 10−5M) (Supplementary Figure 1).
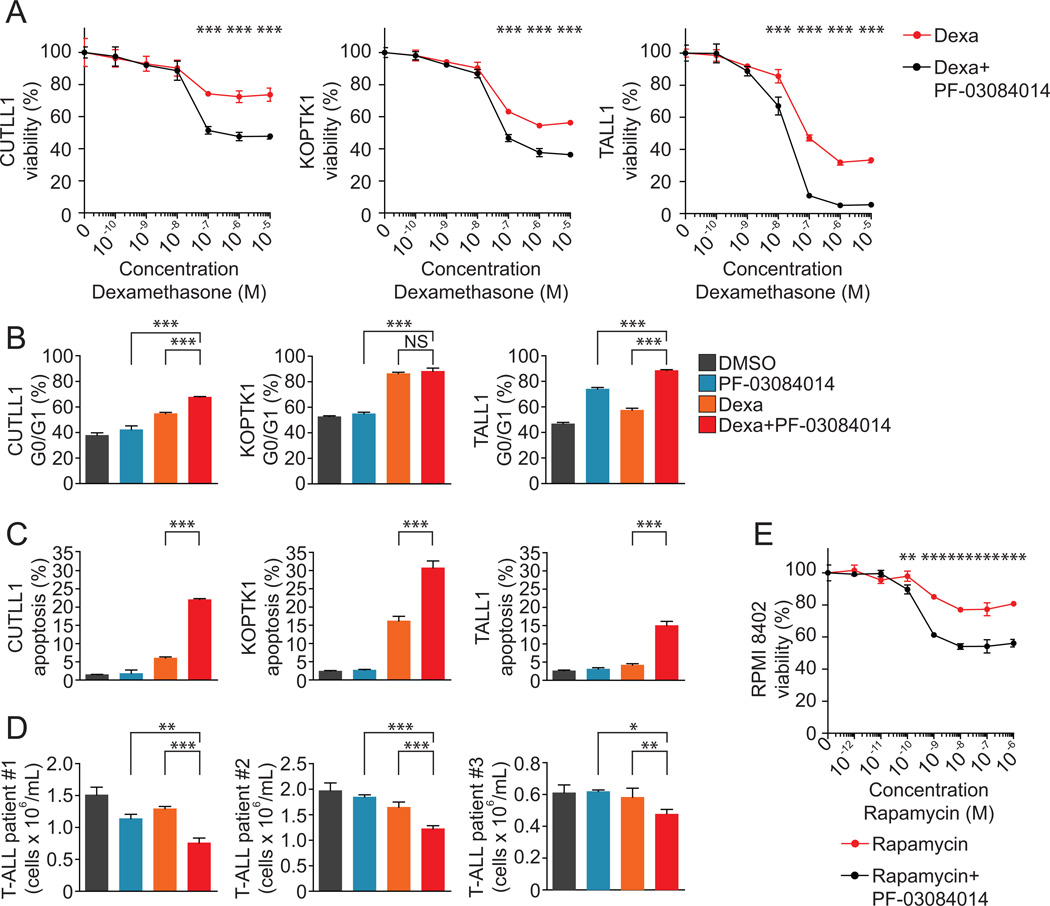
Figure 2. PF-03084014 reverses glucocorticoid resistance in human T-ALL cells.
(A) Cell viability assays of glucocorticoid-resistant T-ALL cell lines (CUTLL1, KOPTK1, TALL1) treated with PF-03084014 (1µM) plus increasing concentrations of dexamethasone. Data are represented relative to treatment with PF-03084014 plus vehicle control. (B) Cell cycle analysis following 48h (KOPTK1) or 72h (CUTLL1 and TALL1) treatment with DMSO, PF-03084014 (1µM), dexamethasone (1µM), or PF-03084014 plus dexamethasone. Data are represented as percentage of cells within G0/G1 following flow cytometry analysis of BrdU incorporation and 7-AAD staining. (C) Percent apoptotic cells following 48h (KOPTK1) or 72h (CUTLL1 and TALL1) treatment with DMSO, PF-03084014 (1µM), dexamethasone (1µM), or PF-03084014 plus dexamethasone. Data are represented as percentage of cells staining as apoptotic (AnnexinV+7-AAD−) by flow cytometry. * indicates a P value < 0.05, ** indicates a P value < 0.01, *** indicates a P value < 0.001. (D) Analysis of viability of primary human T-ALL patient samples treated with DMSO or 1µM PF-03084014 in the presence or absence of dexamethasone (1–10nM). (E) Cell viability assay of RPMI-8402 cells treated with PF-03084014 (1µM) plus increasing concentrations of rapamycin. Data are represented relative to treatment with PF-03084014 plus vehicle control.
Cell cycle analysis through BrdU incorporation and 7-AAD staining revealed that treatment with PF-03084014 and dexamethasone resulted in G0/G1 cell cycle arrest, with a greater than 20% increase in percent cells in G0/G1 when compared to dexamethasone alone in two of the three glucocorticoid-resistance cell lines examined (Figure 2B). Treatment with PF-03084014 in combination with dexamethasone also resulted in increased apoptosis, with an 11-fold increase in apoptosis when compared to GSI alone (22% versus 2% Annexin V+7-AAD−), and a 4-fold increase when compared to dexamethasone alone (22% versus 6% Annexin V+7-AAD−) (Figure 2C). These results were reproducible in the glucocorticoid-resistant cell lines KOPTK1 and TALL1 (Figure 2C). Importantly, analysis of human primary T-ALL samples showed synergistic antileukemic effects when treated with PF-03084014 and dexamethasone in comparison to PF-03084014 or dexamethasone alone in three out of five primary patient samples examined (Figure 2D). Loss of the PTEN tumor suppressor gene, and consequent constitutive activation of the PI3K-AKT-mTOR signaling pathway, has been associated with GSI resistance in leukemia cell lines (19). Notably, shRNA knockdown of PTEN or the glucocorticoid receptor (NR3C1) in CUTLL1 cells rendered them resistant to induction of apoptosis by PF-03084014 and dexamethasone, suggesting that glucocorticoid treatment cannot overcome GSI resistance in these cells (Supplemental Figure 2). However, rapamycin, an mTOR inhibitor, has been shown to enhance the growth suppression of GSIs in mouse models of T-ALL and human leukemic cell lines (20, 22). Consistently, we observed a synergistic antileukemic interaction between rapamycin and PF-03804014 which was most prominent in the RPMI-8402 T-ALL cell line. In these experiments, the rapamycin IC50 for RPMI-8402 cells was 6.4×10−8 M, which was decreased 2.5-fold upon treatment with PF-03084014 (Figure 2E).
Combination multi-agent chemotherapy including etoposide, a topoisomerase II inhibitor; L-asparaginase, which blocks protein synthesis by depriving cells of asparagine; methotrexate, an anti-folate and vincristine an inhibitor of microtubule assembly is commonly used in the treatment of T-ALL. To examine the interaction of PF-03084014 with other chemotherapeutic agents, we treated CUTLL1 cells with etoposide, L-asparaginase, methotrexate and vincristine, in the presence or absence of Compound E or PF-03084014. Additionally, we examined PF-03084014 in combination with imatinib, an inhibitor of the NUP214-ABL1 tyrosine kinase oncoprotein expressed in the ALL-SIL cell line (23). Overall, these experiments demonstrated the specificity of the interaction between PF-03804014 and glucocorticoids as none of these drugs showed increased antileukemic response in combination with PF-03804014 (Supplementary Figure 3).
PF-03084014 enhances the glucocorticoid gene expression signature
To analyze the possible mechanisms mediating the interaction of PF-03804140 and glucocorticoids, we performed gene expression profiling using oligonucleotide microarrays and RNA from CUTLL1 cells treated for 48 hours with vehicle (DMSO), PF-03084014 (1µM), dexamethasone (1µM), or PF-03084014 plus dexamethasone. Analysis of gene expression changes induced by PF-03084014 plus dexamethasone showed a robust gene expression signature associated with enhancement of the glucocorticoid response, including increased upregulation of known glucocorticoid-target genes such as RUNX2, PFKFB2, BCL2L11, BMF, and TSC22D3 (Figure 3A). Treatment with PF-03084014 induced downregulation of known NOTCH1-target genes such as HES1, DTX1, and NRARP (Figure 3B). Finally, and consistent with previous reports, cells treated with PF-03084014 and dexamethasone showed increased upregulation of the glucocorticoid receptor (NR3C1) concomitant with a 3-fold increase in the apoptotic factor BIM (BCL2L11) compared to cells treated with dexamethasone alone (Figure 3A, 3B). Synergistic upregulation of NR3C1 was validated by western blot analysis, with a nearly 5-fold induction in NR3C1 levels following treatment with PF-03084014 and dexamethasone compared to DMSO treatment (Figure 3C).
Figure 3. Enhancement of the glucocorticoid response by combination treatment with PF-03084014 and dexamethasone.
(A) Heat map representation of top differentially expressed genes associated with PF-03084014 plus dexamethasone compared with dexamethasone treatment alone. Relative expression levels are color-coded as indicated at the bottom. (B) Quantitative RT-PCR analysis of HES1, DTX1, NR3C1, and BCL2L11 genes in CUTLL1 cells treated with DMSO, PF-03084014, dexamethasone, or PF-03084014 plus dexamethasone. Data are represented relative to DMSO-treated cells. *** indicates a P value < 0.001. (C) Western blot analysis of NR3C1 protein levels following 24 hours treatment of the CUTLL1 cell line with DMSO, 1µM PF-03084014, 1µM dexamethasone, or PF-03084014 plus dexamethasone; β-Actin is shown as a loading control.
Increased antitumor efficacy of PF-03084014 and dexamethasone in vivo
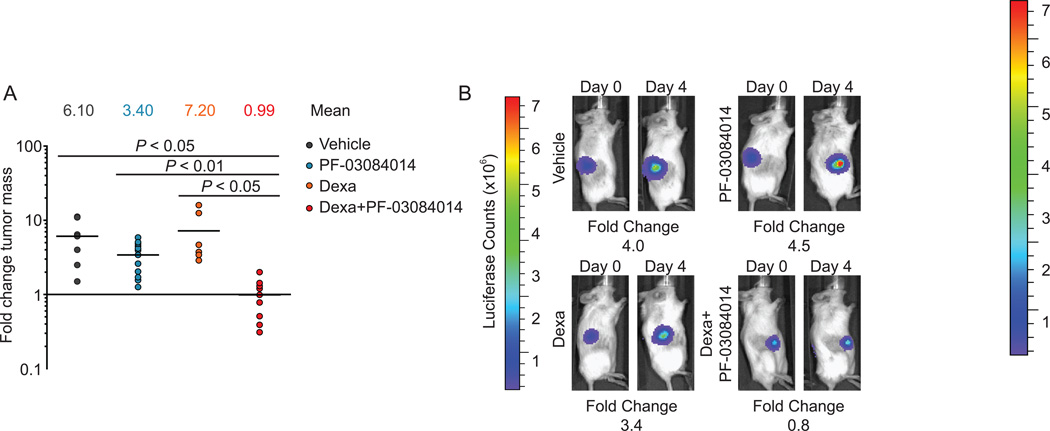
Following the establishment of a synergistic interaction between PF-03084014 and dexamethasone in vitro, we sought to determine if this drug combination can demonstrate improved efficacy in vivo using a xenograft model of glucocorticoid-resistant T-cell lymphoblastic lymphoma. In these experiments we injected luciferase-expressing CUTLL1 cells in the flank of immunodeficient NOD-SCID mice and verified tumor engraftment and disease progression by in vivo bioimaging in animals treated with vehicle only, dexamethasone (15mg/kg), PF-03084014 (150mg/kg), or dexamethasone plus PF-03084014. Mice treated with vehicle, dexamethasone, or PF-03084014 showed a four- to seven-fold increase in tumor burden compared to day zero (Figure 4). In contrast, combination treatment with PF-0384014 and dexamethasone effectively abrogated tumor growth and induced leukemic regression in four of the nine tumors analyzed (Figure 4).
Figure 4. Antitumor effects of PF-03084014 and glucocorticoids in vivo.
(A) Bioimaging quantification of tumor mass changes in subcutaneous CUTLL1 lymphoma xenografts in mice treated with vehicle, 15mg/kg dexamethasone, 150mg/kg PF-03084014, or 150mg/kg PF-03084014 plus 15mg/kg dexamethasone for four days. (B) Bioluminescence images depicting mice with tumor burdens closest to the median showing changes in tumor growth after treatment with vehicle, dexamethasone, PF-03084014, or PF-03084014 plus dexamethasone.
Glucocorticoids reverse PF-03084014-induced gastrointestinal toxicity
Systemic inhibition of NOTCH signaling induces goblet cell metaplasia characterized by the aberrant differentiation of intestinal progenitors into secretory cells (24). Notably, co-treatment with glucocorticoids can abrogate intestinal metaplasia associated with inhibition of NOTCH signaling in mice (11). To test the effects of PF-03084014 alone and in combination with glucocorticoids in the gut we treated C57BL/6 mice with vehicle, dexamethasone (15mg/kg), PF-03084014 (150mg/kg), or PF-03084014 plus dexamethasone for five days. In this experiment, treatment with PF-03084014 induced a marked increase in the number of goblet cells in the distal ileum compared to vehicle or dexamethasone alone, which was concomitant with a loss of the Ki67+ proliferative compartment of the intestinal crypts (Figure 5). In contrast, animals treated with dexamethasone plus PF-03084014 at a 15mg/kg dose showed no change in the number of goblet cells or proliferative Ki67+ cells in their intestinal crypts compared to vehicle-only treated cells (Figure 5). Animals treated with dexamethasone alone showed an accumulation of lysozyme-positive Paneth cells at the crypt base as compared to vehicle-treated controls with no other alterations in the architecture of the intestine (Figure 5).
Figure 5. Glucocorticoids reverse PF-03084014-induced goblet cell metaplasia.
Histological and immunohistochemical analysis of distal ileum sections from mice treated for 5 days with vehicle, 15mg/kg dexamethasone, 150mg/kg PF-03084014, or dexamethasone plus PF-03084014. Scale bars represent 50µm.
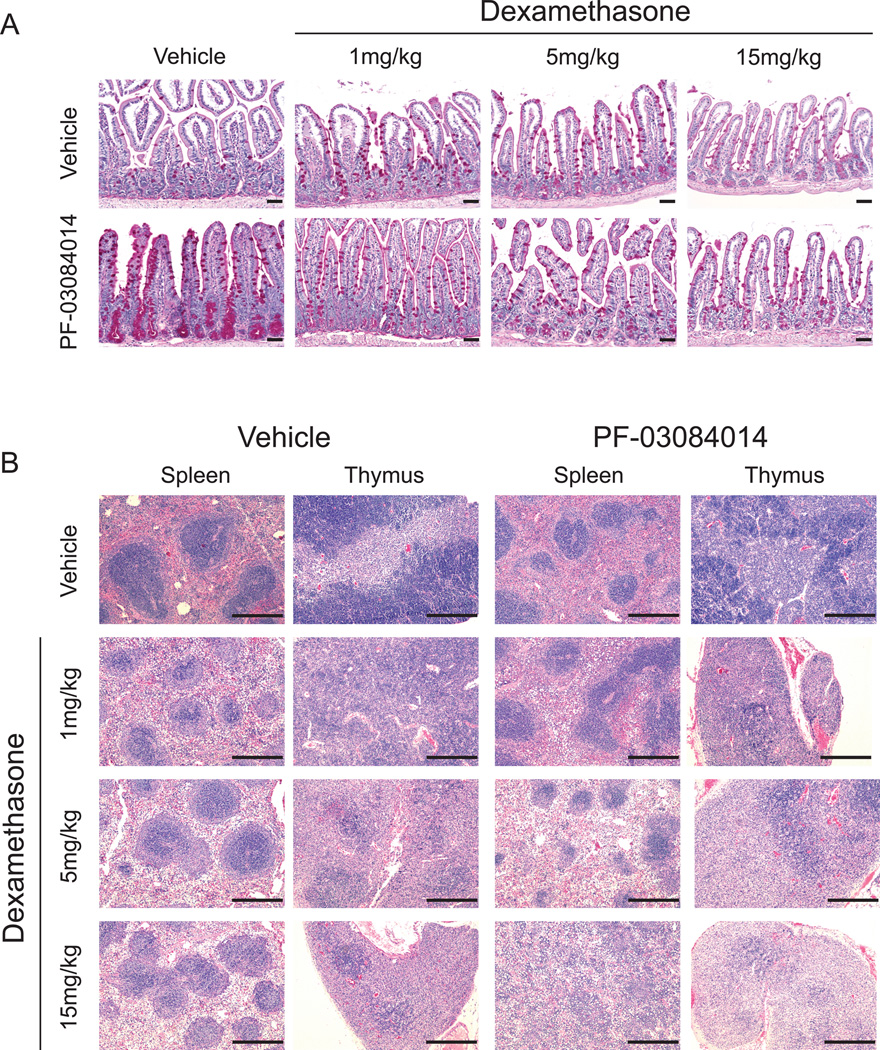
Chronic treatment with glucocorticoids is associated with adverse side effects, including immunosuppression associated with atrophy of the spleen and thymus (25). To test if reduced, less toxic, doses of dexamethasone could retain the enteroprotective effects of glucocorticoids against GSI-induced gut toxicity, we performed a dexamethasone descalation experiment in which we treated mice with 150mg/kg PF-03804014 in the presence or absence of 1mgkg, 5mg/kg, or 15mg/kg dexamethasone. In this experiment, doses as low as 5mg/kg and 1mg/kg dexamethasone were able to reduce the goblet cell metaplasia induced by PF-03084014 in the distal ileum (Figure 6A). Moreoever, mice treated with 1mg/kg of dexamethasone also had a reduction in the degree of glucocorticoid-induced atrophy of the spleen and thymus, suggesting that a reduced, less toxic, glucocorticoid regimen may retain the enteroprotective effects of dexamethasone against GSI-induced goblet cell metaplasia. (Figure 6B).
Figure 6. Dose reduction of dexamethasone for the reversal of PF-03084014-induced gastrointestinal toxicity.
(A) Periodic acid-Schiff (PAS) stain of distal ileum sections harvested from mice treated for 5 days with vehicle, dexamethasone (1mg/kg, 5mg/kg, or 15mg/kg), 150mg/kg PF-03084014, or dexamethasone plus PF-03084014. (B) Hematoxylin and eosin (H&E) stain of spleens and thymii harvested from mice in (A). Scale bars represent 50µm (intestines) or 400µm (spleen, thymus).
Discussion
Activating mutations in the NOTCH1 gene are present in over 50% of human T-ALL cases, making NOTCH1 the most prominent oncogene specifically involved in the pathogenesis of this disease. Importantly, cleavage by the gamma-secretase complex is required for the activity of NOTCH1, bringing small-molecule inhibitors of gamma-secretase to the forefront of molecularly-targeted therapies for the treatment of T-ALL. However, the translation of gamma-secretase inhibitors into the clinic has been hindered by a lack of cytotoxic anti tumor responses and by severe gastrointestinal toxicity associated with inhibition of NOTCH signaling in the intestinal epithelium. To date, the main mechanism around this toxicity has been the use of strategic dosing approaches to minimize goblet cell metaplasia based on the turnover rate of the intestinal epithelium (22). In this context, we have recently shown that co-treatment of dexamethasone and the generic azepine-class gamma-secretase inhibitors Compound E and DBZ results in increased antileukemic effects and reversal of GSI-mediated gastrointestinal toxicity. Based on these results, we examined here whether PF-03084014, a structurally unique and clinically-relevant GSI, had a synergistic interaction with dexamethasone for the treatment of glucocorticoid-resistant T-ALL.
Previous studies have found that seven days of continuous treatment with PF-03084014 was required for maximal induction of cell cycle arrest, thought in most cell lines, there was a minimal increase in apoptosis (18). We found a statistically significant decrease in cell viability of glucocorticoid-resistant T-ALL cell lines and primary T-ALL lymphoblasts following only three days of treatment with dexamethasone and PF-03084014. In vitro, treatment with PF-03084014 enhanced the antileukemic effects of dexamethasone in three GSI-sensitive and glucocorticoid resistant cell lines. Reduced cell viability was characterized by increased apoptosis and/or increased G0/G1 cell cycle arrest.
This synergistic interaction was specific to glucocorticoids, with a minimal increase in therapeutic advantage when combining PF-03084014 with etoposide, methotrexate, vincristine, L-asparaginase, or imatinib. A synergistic interaction between gamma secretase inhibitors and rapamycin has been shown in the past, whereby combination treatment resulted in decreased cell viability and increased apoptosis in both mouse and human models of T-ALL (20, 22). Consistently, rapamycin treatment enhanced the antileukemic effects of PF-03084014 in the RPMI-8402 T-ALL cell line.
Molecular characterization of the mechanism of interaction of GSI with glucocorticoids through gene expression profiling revealed upregulation of known glucocorticoid target genes, suggesting that the increase in cytotoxicity is due to a synergistic increase in glucocorticoid activity rather than enhanced gamma-secretase inhibitor efficacy. Mechanistically, we found that combination treatment of PF-03084014 plus dexamethasone resulted in enhanced upregulation of the glucocorticoid receptor and its target genes, and that the induction apoptosis was dependent on expression of the glucocorticoid receptor. These data are consistant with our previous findings showing that NOTCH1 inhibits, via HES1, glucocorticoid receptor autoupregulation, a critical positive feed-back loop required for glucocorticoid induced apoptosis (11). A synergistic interaction between PF-03084014 and dexamethasone was also recapitulated in vivo using a subcutaneous model of glucocorticoid-resistant T-ALL, with a significant increase in antitumor response compared to PF-03084014 and dexamethasone alone.
GSI-induced intestinal goblet cell metaplasia represents a major hurdle for the use of gamma secretase inhibitors as antileukemic agents. This on-target toxicity is mediated by inhibition of NOTCH1 and NOTCH2 in the intestine, which, in turn, downregulates Hes1, a transcriptional repressor of secretory cell lineage transcription factor genes such as Klf4 and Math1 (11, 24, 26, 27). Notably, GSI-mediated upregulation of Klf4 is inhibited following treatment with dexamethasone (11). However, intestinal-specific deletion of Math1, but not Klf4, is able to rescue mice from the effects of GSI treatment, suggesting a more prominent role of Math1 in the control of secretory cell fate in the gut (28, 29). An intriguing observation is the accumulation of Paneth cells at the crypt base in the intestines of dexamethasone-treated mice. Notably, Paneth cells have recently been shown to constitute the niche for intestinal stem cells in the intestinal crypt (30), suggesting a potential effect of glucocorticoids in intestinal stem cell homeostasis which could be related to the reversal of GSI-induced gut toxicity. High doses of dexamethasone are associated with systemic toxicities, which might limit the use of glucocorticoids as eneteroprorective agents against GSI-induced gut toxicity (31). The studies presented here show effective inhibition of GSI-induced goblet cell metaplasia with reduced, less toxic, doses of dexamethasone, highlighting the clinical applicability of this drug combination.
The antitumor activities of GSIs and the reversal of GSI-induced gut toxicity by glucocorticoids may go beyond the scope of T-ALL, with oncogenic roles for NOTCH family members in numerous cancers (32–36) and activating NOTCH1 mutations found in acute myeloid leukemia (37), chronic lymphocytic leukemia (38, 39), and lung cancer (40). Additionally, novel approaches to NOTCH pathway inhibition, including inhibitory NOTCH1 antibodies and peptide-mediated inhibition of the NOTCH1 transcriptional complex, are currently in development (41, 42). It will be interesting to see if the synergistic antileukemic interaction of PF-03084014 with glucocorticoids extends to these novel NOTCH inhibitors. Overall, the results presented here substantiate the clinical evaluation of PF-03084014 and dexamethasone in combination for the treatment of glucocorticoid-resistant T-ALL.
Supplementary Material
Acknowledgements
We thank Dennis Bonal and the Herbert Irving Comprehensive Cancer Center Molecular Pathology Shared Resource for assistance with immunohistochemistry and histological analysis.
Grant Support
This research was supported in part by Pfizer and grants from the National Institutes of Health (grants R01CA120196 and R01CA129382 to A.A.F), the New York Community Trust (A.A.F.), the Innovative Research Award by the Stand Up to Cancer Foundation (A.A.F.), the ECOG tumor bank, and the Leukemia & Lymphoma Society Scholar Award (A.A.F.).
References
- 1.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 2.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 3.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deangelo DJ, Stone RM, Silverman LB, Stock W, Attar EC, Fearen I, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J Clin Oncol. 2006;24:6585. [Google Scholar]
- 5.Grad I, Picard D. The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol. 2007;275:2–12. doi: 10.1016/j.mce.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Real PJ, Ferrando AA. NOTCH inhibition and glucocorticoid therapy in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:1374–1377. doi: 10.1038/leu.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Hongo T, Yajima S, Sakurai M, Horikoshi Y, Hanada R. In vitro drug sensitivity testing can predict induction failure and early relapse of childhood acute lymphoblastic leukemia. Blood. 1997;89:2959–2965. [PubMed] [Google Scholar]
- 10.Klumper E, Pieters R, Veerman AJ, Huismans DR, Loonen AH, Hahlen K, et al. In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood. 1995;86:3861–3868. [PubMed] [Google Scholar]
- 11.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 13.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong F, Brunet de la Grange P, Gerby B, Rouyez MC, Calvo J, Fontenay M, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113:1730–1740. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- 15.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 17.Lanz TA, Wood KM, Richter KE, Nolan CE, Becker SL, Pozdnyakov N, et al. Pharmacodynamics and pharmacokinetics of the gamma-secretase inhibitor PF-3084014. J Pharmacol Exp Ther. 2010;334:269–277. doi: 10.1124/jpet.110.167379. [DOI] [PubMed] [Google Scholar]
- 18.Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 19.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, Bekaert S, et al. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:533–542. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- 22.Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 24.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 25.Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11:1096–1106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C, et al. KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2009;296:G490–G498. doi: 10.1152/ajpgi.90393.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Li J, Chen H, Fu J, Ray S, Huang S, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TH, Shivdasani RA. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427–11433. doi: 10.1074/jbc.M110.188797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, et al. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aster JC. Notch targeting 2.0. Blood. 2009;113:6044–6045. doi: 10.1182/blood-2009-03-212829. [DOI] [PubMed] [Google Scholar]
- 32.Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullendore ME, Koorstra JB, Li YM, Offerhaus GJ, Fan X, Henderson CM, et al. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009;15:2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 35.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 36.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 37.Palomero T, McKenna K, J ON, Galinsky I, Stone R, Suzukawa K, et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 2006;20:1963–1966. doi: 10.1038/sj.leu.2404409. [DOI] [PubMed] [Google Scholar]
- 38.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.