Abstract
Histone tails and their post-translational modifications (PTM) play important roles in regulating the structure and dynamics of chromatin. For histone H4, the basic patch K16R17H18R19 in the N-terminal tail modulates chromatin compaction and nucleosome sliding catalyzed by ATP-dependent ISWI chromatin remodeling enzymes while acetylation of H4 K16 affects both functions. The structural basis for the effects of this acetylation is unknown. Here we investigated the conformation of histone tails in the nucleosome by solution NMR. We found that backbone amides of the N-terminal tails of histones H2A, H2B, and H3 are largely observable due to their conformational disorder. However, only residues 1–15 in H4 can be detected, indicating that residues 16–22 in the tails of both H4 histones fold onto the nucleosome core. Surprisingly, we found that K16Q mutation in H4, a mimic of K16 acetylation, leads to a structural disorder of the basic patch. Thus, our study suggests that the folded structure of the H4 basic patch in the nucleosome is important for chromatin compaction and nucleosome remodeling by ISWI enzymes while K16 acetylation affects both functions by causing structural disorder of the basic patch, K16R17H18R19.
DNA molecules in eukaryotic cells are super long negatively charged biopolymers. To be accommodated within the small (~10 μM in diameter) nucleus, DNA molecules are packaged into chromatin through association with small positively charged histone molecules to form nucleosomes, the structural unit of chromatin,1 which consists of an octameric histone core (two copies of each histone H2A, H2B, H3 and H4) wrapped around by ~146 bp DNA. At the first level of the compaction, chromatin forms a beads-on-a-string structure with the nucleosome as the beads and the linker DNA between them as the strings.2 At the next level of compaction, in the presence of linker histone H1 and/or Mg2+, chromatin forms a structure referred to as the 30-nm fibers.3,4 The structures of the nucleosomes with histones from various species have been determined at near atomic resolution using X-ray crystallography.5,6 In the crystal structure, the histone fold domain (HFD) of each histone is well defined, whereas some of the tail regions N-terminal to the HFDs are undefined or show interactions with neighboring nucleosomes in the crystal lattice. The structure of the 30-nm chromatin fiber has not been determined. Instead, two types of models have been proposed; one is a one-start helix formed with consecutive nucleosomes arranged like a solenoid3 and the other is a twisted two-start helix assembled from a zigzag ribbon of nucleosomes.7 Having an alternative arrangement for the linker DNA,8,9 a compact tetra-nucleosome structure determined at 9 Å resolution using X-ray crystallography supports the two-start model.
Histone tails play important roles in the formation of higher-order chromatin structures,10,11 which are often regulated by their post-translational modifications and ATP-dependent chromatin remodeling. For example, it has been shown that deletion of the H4 tails, including residues 1–19, prevents the formation of higher-order structures. Deletion of residues 1–13 has no such effect, indicating that residues 14–19, which contain a basic patch (K16R17H18R19), play an essential role in the formation of compact chromatin structures.12 In addition, based on the packing interactions of the basic H4 tail patch and an acidic patch of histones H2A-H2B observed between two neighboring nucleosomes in the crystal of nucleosome core particles, it has been suggested that such electrostatic interactions may also stabilize the higher-order chromatin structure.5,8 Moreover, it has been shown that the basic patch is also required for the ATP-dependent nucleosome modeling by the ISWI enzymes.13–19 Interestingly, both formation of higher-order chromatin structures and nucleosome remodeling are hindered by the acetylation of K16 in histone H4.18,20,21 How acetylation of K16 affects all of the above functions is not fully understood.
In this work, we examined the histone tail conformations in the nucleosome using solution NMR. We found that the basic patch in both H4 histones has a folded structure in the nucleosome while residues 1–15 have a disordered structure. Surprisingly, we found that K16Q mutation, a mimic of K16 acetylation, can disrupt the folding of the H4 basic patch and prevent the nucleosome array from forming higher-order structures, suggesting a novel structural mechanism for the effect of acetylation of K16 on chromatin compaction and nucleosome remodeling.
NMR chemical shift assignments of histone tails in the nucleosome
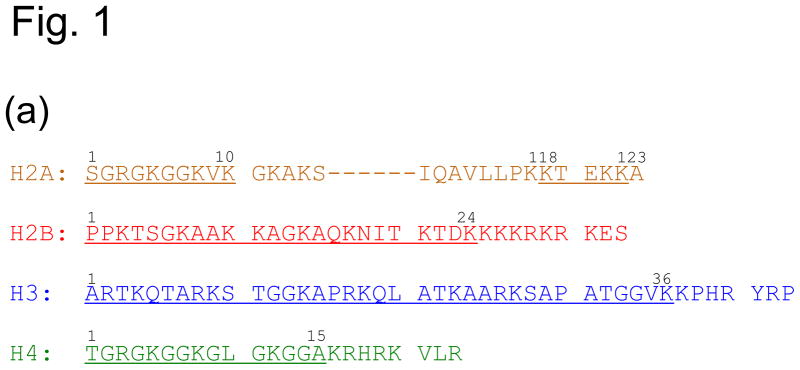
To investigate the histone tail conformation in the nucleosome, we reconstituted the nucleosome with recombinant Drosophila histones and 167 base pair of DNA containing the Widom ‘601’ sequence in the middle.8,22 For chemical shift assignments, we labeled one histone type with 15N and 13C for each nucleosome sample and observed a limited number of peaks in the two-dimensional 1H-15N HSQC spectra. Such results are expected since the nucleosome has a molecular weight larger than 200 KDa. The NMR signals of the backbones in the folded region of the nucleosome should not be observable by the conventional methods due to its slow tumbling rate. The disordered regions such as flexible histone tails, however, have fast local dynamic motions and should be observable. Indeed, the assignments show that the observed peaks are from residues in the histone tails. They include residues 2–10 at the N-terminal tail and 118–122 at the C-terminal tail of H2A, 3–24 at the N-terminal tail of H2B, 3–36 at the N-terminal tail of H3, and 1–15 at the N-terminal tail of H4 (Fig. 1(a)-(c)).
Fig. 1. Chemical shift assignment of histone tails in the nucleosome.
(a) Amino acid sequences of the Drosophila histone tails. Residues assigned by NMR are underlined. (b) 1H-15N HSQC spectra and chemical shift assignment. (c) Different views of the nucleosome crystal structure, illustrating the positions of histone tails. The numbers indicate the first or last residues that are observable in the histone tails in the crystal structure (pdb ID: 2PYO). Histones H2A, H2B, H3, and H4 are colored in orange, red, blue and green, respectively. Production of isotope-labeled histones, DNA, and reconstitution of nucleosomes are described in our earlier work.27 The chemical shifts of the histone tail residues were assigned using the triple resonance NMR methods on Bruker 500, 600, and 700 MHz instruments.29 Mutation of H4 K16Q was introduced by site-directed mutagenesis using the QuikChange™ kit (Stratgene, CA) and were subsequently verified by DNA sequencing. NMR data is processed using NMRPipe30 and analyzed using NMRView.31
The two H2A histones in the crystal structure of the Drosophila nucleosome include residues 11–121 and 13–120, respectively (pdb ID: 2PYO).23 We found that residues 2–10 and 118–122 are disordered in solution. Thus, for the N-terminal tail of H2A, our NMR data is consistent with the crystal structure. However, we observed two sets of peaks for residues R3, K5, and G7, indicating asymmetrical conformations for the two N-terminal tails of H2A. In addition, the observation of the region 118–122 by NMR indicates that this C-terminal region of H2A, which is ordered in the crystal structure, is actually flexible in solution. For H2B, the N-terminal regions (1–27) are absent in the crystal structure for both histones. We observed residues 3–24 by NMR. The missing of the first two residues is likely due to fast exchange rates of the amide protons. The three residues 25–27 are either folded in the nucleosome in solution or have dynamic motions on the millisecond time scale, rendering their signals unobservable by NMR. For H3, the residues 1–36 are absent in the nucleosome structure. We assigned residues 3–36 in the 1H-15N correlation spectra (Fig. 1(b)). For H4, residues 1–14 are absent in one H4 molecule while residues 1–22 are absent in the other H4 molecule in the crystal structure. We observed residues 1–15 in the 1H-15N HSQC spectra. Since only one set of peaks for residues 1–15 in H4 is observed, we conclude that the lack of defined structure for the residues 16–22 in one of the two H4 molecules in the crystal structure is caused by crystal packing.
Location and structure of the two H4 tails in the nucleosome
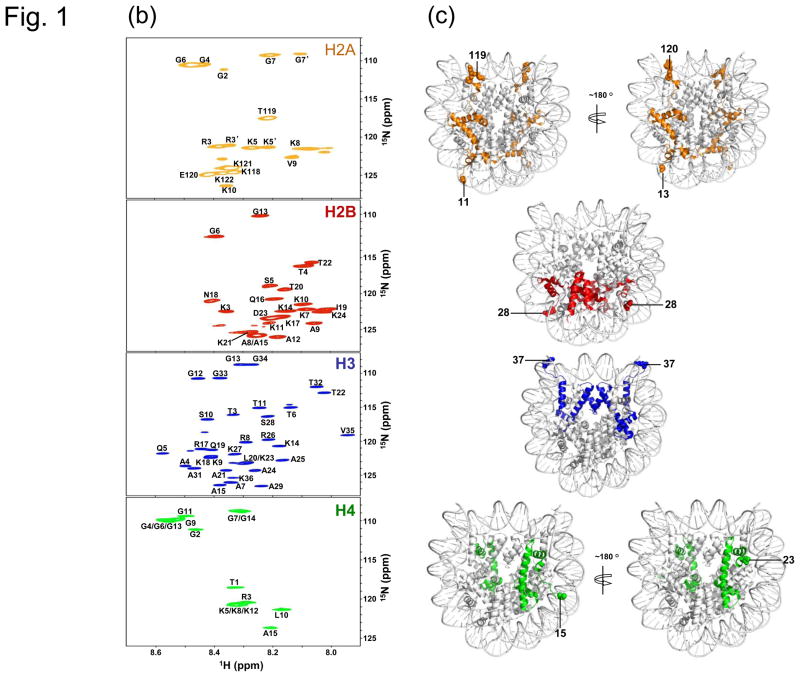
To test if the regions 16–22 in both H4 tails have the same folded structure in solution as one of the H4 tails determined in the crystal structure, we used paramagnetic relaxation enhancement (PRE)24 and methyl-TROSY methods to examine their locations in the nucleosome.25 We reconstituted a nucleosome in which residue A15 in H4 was mutated to Cys and then linked to MTSL (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrroline-3-yl)methyl methanesulfonothioate) while the methyl groups of Ile, Leu, and Val in histone H3 were labeled with 13C and all hydrogen atoms other than those in these methyl groups were deuterated.26 By taking the 1H-13C methyl-TROSY spectra under oxidized and reduced conditions (Fig. 2(a), (b)), we calculated the ratios of the peak intensities for the methyl groups in H3 (C110A mutant) using the assignments made in our earlier work.27 We then plotted the intensity ratio as a function of the distance of the Cβ atom of A15 to the carbon atoms in the methyl groups of H3 (Fig. 2(c)). We fitted the data to the formal equation for describing the effect of PRE as described in [1]24 (Fig. 2(c)):
| [1] |
Fig. 2. Verification of the location of H4 tails in the nucleosome by methyl-TROSY NMR and PRE.
Methyl-TROSY spectra of the nucleosome with methyl-labeled H3 (C110A) and MTSL-labeled H4 at position A15 in (a) oxidized and (b) reduced form. (c) Plot showing changes in peak intensities for the methyl groups in H3 as a function of the distances between the Cβ atom of A15 in H4 and the carbon atoms in the methyl groups. Methyl groups that show large changes in peak intensities are labeled in the plot. A bar graph showing the peak intensity change is shown in the supplementary figure. (d) Structural illustration for the effects of PRE. The MTSL-labeling at position A15 of H4 follows the procedure of Racki et al.32
Here parameters A and B include combined terms related to the physical properties of the spin.24 Parameter d is used to approximately correct the different positions between the Cβ atom of Ala15 and the unpaired electron in the MTSL. A reasonably good fitting is obtained (Fig. 2(c)). In particular, all of the methyl groups that displayed large decreases in their peak intensities (> 50%) including residues Leu65, Leu70, Val71, Leu74, Leu82, and Val89 in H3, are close to residue Ala15 in the H4 in the crystal structure of the nucleosome. Thus, we conclude that the regions (16–22) in both H4 tails fold onto the nucleosome like the structure of one of the histone tails determined by the crystallographic method.
K16Q mutation leads to structural disorder of the basic patch
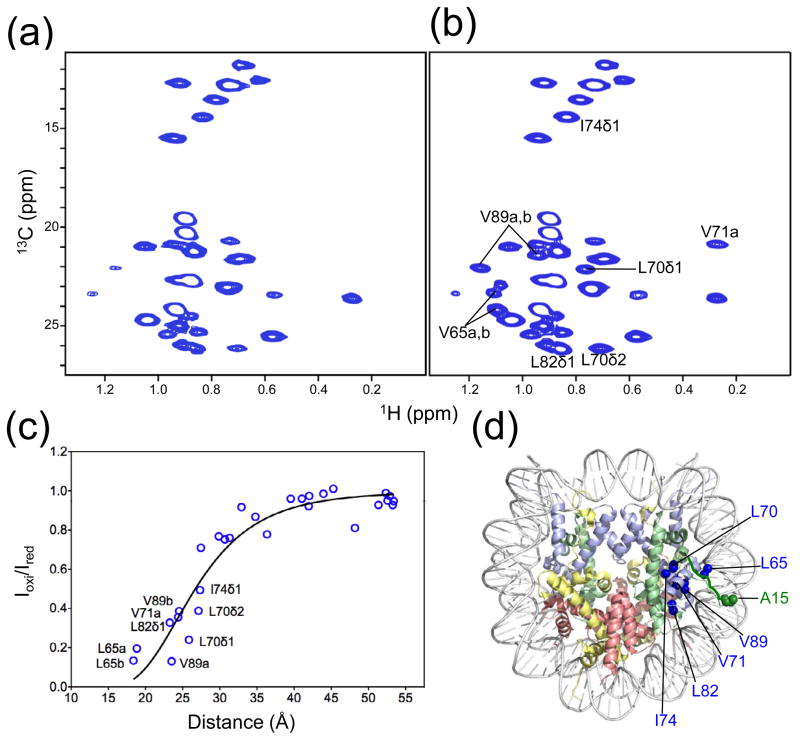
Since the side chain of K16 in H4 interacts with the DNA through electrostatic interactions (Fig. 3(a)), acetylation of K16 in H4 may abrogate this interaction and cause structural changes in the H4 tails. To test such a possibility, we used NMR to examine the H4 tail with a K16Q mutation in the nucleosome. This particular mutation was chosen as it partially mimics the acetylation effect of K16. We found that this H4 K16Q mutation led to the appearance of four extra peaks in the 1H-15N NMR spectra of the nucleosome, in addition to residues 1–15 observed with wild-type H4. We assigned these extra peaks using 15N/13C-labeled H4 nucleosome and found that they are from residues Q16, R17, H18, and R19 (Fig. 3(b)). The observation of residues 16–19 in the 1H-15N HSQC spectra indicates that the K16Q mutation indeed disrupts the structure of the basic patch in the nucleosome.
Fig. 3. Unfolding of the H4 basic patch upon K16Q mutation.
(a) Illustration of the conformation for one of the two H4 tails in the crystal structure of the nucleosome. (b) 1H-15N HSQC spectrum of the nucleosome with H4 K16Q mutation.
K16Q mutation affects nucleosome array compaction
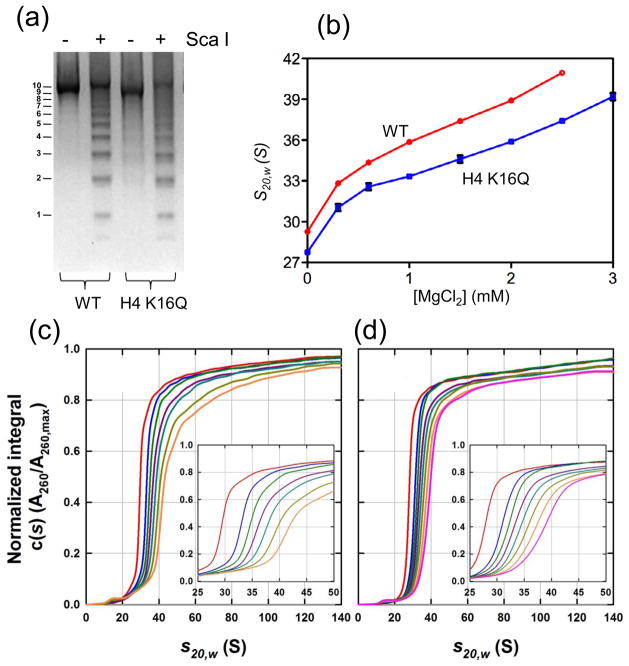
To examine the effect of K16Q mutation on the formation of higher-order chromatin structure, we reconstituted nucleosome arrays containing wild-type histones and histone H4 with K16Q mutation. The nucleosome array contains 10 nucleosomes that are positioned with the Widom ‘601’ sequence (147 bp) and linked with 61 bp DNA. Saturation of all 10 nucleosome sites was confirmed by limited and complete digestion of the array with restriction enzyme Sca I (Fig. 4(a)). We then performed sedimentation experiments at different Mg2+ concentrations. The sedimentation coefficient of the wild type nucleosome array reached 36S at 1 mM Mg2+ before forming aggregates at higher concentration of Mg2+, which is consistent with earlier results using a similar array.28 In contrast, the sedimentation coefficient of the nucleosome array containing the H4 K16Q mutation is lower at 1 mM Mg2+ and reaches ~36S at ~2.0 mM Mg2+ before it starts forming aggregates at higher concentrations of Mg2+. Similar behavior is reported for the nucleosome arrays with H4 K16Q mutation21 or deletion of the H4 tail that includes the basic patch in previous studies in the range of 0 to 1 mM Mg2+.12 Our results show that a higher concentration of Mg2+ is required to compact an H4 K16Q nucleosome array to the same extent as that comprising the wild-type histones. We note that the sedimentation coefficients reported here are for the soluble fraction at higher concentrations of Mg2+. This is because the wild-type nucleosome array starts forming aggregates at Mg2+ concentrations higher than 1.0 mM while the nucleosome array with the H4 K16Q mutation nucleosome array starts aggregating at Mg2+ concentrations higher than 2.5 mM. In addition, the compaction of the nucleosome arrays as a function of Mg2+ seems to be non-cooperative and the formation of nucleosome array aggregates (inter-nucleosome packing) does not necessarily require a maximal intra-nucleosome compaction.
Fig. 4. K16Q mutation hinders nucleosome array compaction.
(a) Limited Sca I digestion indicates full saturation of the nucleosome array. The numbers on the left of the gel indicate the number of nucleosomes in the corresponding bands. (b) Measurement of sedimentation coefficients of nucleosome arrays with wild-type histones (circles) and with H4 K16Q mutation (squares) at different Mg2+ concentrations. (c) Normalized, integral c(s) distributions for the array with wild-type histone H4 in 0 (red), 0.3 (blue), 0.6 (green), 1.0 (maroon), 1.5 (cyan), 2.0 (light green) and 2.5 (orange) mM Mg2+ and 20 mM Tris-HCl (pH 8.0), 1 mM DTT. (d) Normalized, integral c(s) distributions for the array with H4 K16Q mutation in 0 (red), 0.3 (blue), 0.6 (green), 1.0 (maroon), 1.5 (cyan), 2.0 (light green), 2.5 (orange) and 3.0 (pink) mM Mg2+ and 20 mM Tris-HCl (pH 8.0), 1 mM DTT. The fully saturated nucleosome array was reconstituted from the recombinant histones and the DNA fragment containing 10 tandem repeats of 208 bp ‘601’ sequences by step-wise salt dialysis in 10 mM Tris-HCl (pH 8.0) according to the procedure of earlier established procedures.8 To ensure the saturation of all 10 repeats with histone octamers, the ratio of octamers to ‘601’ DNA sites was kept at 1.25. After dialysis, the sample was centrifuged at 15,000 rpm for 15 min to remove any insoluble components. The reconstituted arrays were selectively precipitated at 5 mM MgCl2. The purity and octamer/DNA ratio of the sample was checked on a 2% agarose gel. The saturation of all 10 nucleosome sites in the array was confirmed by DNA digestion with Sca I that cleaves the linker DNA region of the array. Sedimentation experiments were performed in duplicate at 20.0°C on a Beckman-Coulter ProteomeLab XL-I analytical ultracentrifuge at rotor speeds of 18,000 or 15,000 rpm. Absorbance data collected at 260 nm were analyzed in terms of a continuous c(s) distribution in SEDFIT33 using a range of 0 – 150 S, a resolution of 300 and a confidence interval of 68%. A partial specific volume of 0.65 cm3g−1 was used,34 solution densities and viscosities were calculated in SEDNTERP35 and sedimentation coefficients were corrected to standard conditions s20,w. The sedimentation coefficients shown in (b) represent the value for which the differential c(s) distribution is at a maximum.
In summary, our solution NMR studies of the nucleosome reveal that the basic patch in the tails of both H4 histones fold onto the nucleosome core with a structure identical to that determined in the crystal structure of the nucleosome. K16Q mutation, a mimic of K16 acetylation, of H4 leads to structural disorder of the basic patch. Our results suggest a new structural mechanism by which K16 acetylation of H4 inhibits chromatin compaction and remodeling by causing structural disorder of the basic patch, K16R17H18R19 of H4.
Supplementary Material
We assigned chemical shifts of histone tails in the nucleosome.
Residues 16–23 in H4 tails are structured in the nucleosome.
K16Q in H4 causes conformational disorder of its basic patch K16R17H18R19.
Disorder of the basic patch may affect chromatin compaction and remodeling.
Acknowledgments
We thank Drs. Carl Wu and Tim Richmond for providing the plasmids of histones and the 601 DNA, respectively. The work is supported by the intramural research programs of NIH: NCI (B.Z, H.F., H.K. and Y.B.), NIDDK (R.G.) and NHLBI (J. G.)
References
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Olins AL, Olins DE. Spheroid chromatin units (n bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 3.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 9.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 10.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 11.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 12.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 13.Hamiche A, Kang JG, Dennis C, Xiao H, Wu C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc Natl Acad Sci U S A. 2001;98:14316–14321. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazzio TG, Gelbart ME, Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol Cell Biol. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shogren-Knaak M, Ishii H, Sun J, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu C, Nordenskiold L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2010;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Clapier CR, Chakravarthy S, Petosa C, Fernandez-Tornero C, Luger K, Muller CW. Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer. Proteins. 2008;71:1–7. doi: 10.1002/prot.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 25.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 26.Tugarinov V, Hwang JE, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 27.Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal proten 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander EW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci U S A. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bax A. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 30.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausio J, Borochov N, Seger D, Eisenberg H. Interaction of chromatin with NaCl and MgCl2. Solubility and binding studies, transition to and characterization of the higher-order structure. J Mol Biol. 1984;177:373–398. doi: 10.1016/0022-2836(84)90291-2. [DOI] [PubMed] [Google Scholar]
- 35.Cole JL, Lary JW, Moody TP, Laue TM. Analytical ultracentrifugation:sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.