Abstract
Cameroon is a West African country where high genetic diversity of HIV-1 has been reported. The predominant CRF02_AG is involved in the emergence of more complex intersubtype recombinants. In this study, we sequenced the full-length genome of a novel unique recombinant form (URF) of HIV-1, 02CAMLT04 isolated in blood donors in urban Cameroon. Phylogenetic tree and bootscan analysis showed that 02CAMLT04 was complex and appeared to be a secondary recombinant derived from CRF02_AG and CRF22_01A1. The genomic composition of 02CAMLT04 strain showed that it is composed of three segments; twenty four percent of the genome is classified as CRF02_AG, spanning most of the envelope gene. The remaining seventy six percent of the genome is classified as CRF22_01A1. In addition, the sequence analysis of 13 full-length sequences from HIV-1 positive specimens received from Cameroon between 2002 and 2010 indicated that five specimens are pure CRF22_01A1 viruses, and six others have homology with CRF22_01A1 sequences in either gag, pol or env region where as 6% of strains contain portions of CRF22_01A1. Further study demonstrated that CRF22_01A1 is a primary prevalence strain co-circulating in Cameroon and is involved in complex intersubtype recombination events with subtypes (D or F), subsubtypes (A1 or F2) and CRFs (CRF01_AE or CRF02_AG). Our studies show that novel recombinants between CRF22_01A1 and other clades and recombinant forms may be emerging in Cameroon that could contribute to the future global diversity of HIV-1 in this region and world wide.
Keywords: HIV-1, recombinant, genetic diversity, phylogenetic analysis, CRF22_01A1, CRF02_AG, Cameroon
INTRODUCTION
The genetic diversity of HIV-1 is very broad in Cameroon where all group M clades and several circulating recombinant forms CRFs (in particular CRF01_AE, CRF02_AG, CRF06_cpx, CRF09_cpx, CRF11_cpx, CRF13_cpx, CRF22_01A1, CRF36_cpx and CRF37_cpx), in addition to groups O, N and P viruses, have been identified1–10. Among the broad genetic diversity of HIV-1 strains, CRF02_AG represents over 65% of HIV infections with an additional 26% classified as unique recombinant forms (URFs)8, 10. Co-circulation of different subtypes and CRFs in Cameroon results in the continuing emergence of new intersubtype recombinants or even M/O recombinants11. Therefore, Cameroon is an ideal area to investigate the genetic diversity of HIV-1 and its impact on the global pandemic. In 2002, we initiated a molecular and serologic epidemiology survey in blood donors in Douala and Yaoundé, in Cameroon to investigate HIV-1 genetic diversity, study virologic and immunologic characteristics of the viruses, evaluate the performance of US FDA licensed HIV-1 assays in the presence of numerous HIV-1 variants, and identify samples to serve as candidate reference reagents for diagnostics. We found that several group M subtypes (A, B, C, D, F2 and G) and CRFs (CRF02_AG, CRF06_cpx, CRF11_cpx, CRF13_cpx, CRF19_cpx and CRF22_01A1) were circulating in this population. A new URF strain, 02CAMLT04, had CRF22_01A1gag-CRF02_AGenv genotype12, and a novel strain, 02CAMLT72 isolated in Douala, was designated as pure CRF22_01A113.
CRF22_01A1 strain was initially identified in Cameroon in 20014 and circulating in this country for many years. Brennan’s studies reported that the percentage of CRF22_01A1 in concordant specimens was 6.6% (1996–1999), 4.7% (2000–2002), and 6.6% (2003–2004) over the 9-year period14, however, molecular, epidemiological and evolutionary study of CRF22_01A1 phylogenetic association with different HIV-1 subtypes was not described. Here, we report 14 full-length genomic sequences of HIV-1 specimens isolated in Cameroon from 2002–2003 and 2006–2010. Of these 14 specimens, 5 were pure CRF22_01A1, 6 were determined to be recombinants of CRF22_01A1 with CRF02_AG. These new data support involvement of CRF22_01A1 in the emerging diversity of HIV-1 in Cameroon.
METHODS
Specimens
For HIV characterization studies, 441 blood samples from HIV-1-seropositive individuals were collected at Douala (LT), Bamenda (LPH), Buea (BDHS), Limbe (LB), Yaounde (ARC) and a few villages in Cameroon (NYU) in 2002–2010. A total of 208 viruses were isolated and genotyped based on partial sequence analysis, 63% of them were CRF02_AG, 26% were URFs, and 6% were CRF22_01A1 containing recombinants12,15. In this study, 13 viruses containing CRF22_01A1 genome and 3 URF strains were selected for full-length sequence analysis, however nucleic acid was available for only 14 of them which were completely sequenced and characterized.
RNA Extraction, RT-PCR and Nearly Full-Length Genomic Sequencing
Viral RNA was extracted from the plasma samples using QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA). Then, reverse transcription, polymerase chain reaction (PCR) and sequencing were performed as previously described6, 7. The nearly full-length genomic sequences reported in this study have been deposited in the GenBank database (accession numbers: EU743964, JN864047-JN864059).
Phylogenetic and Recombination Sequence Analysis
Nucleotide sequences were aligned with HIV-1 reference strains of different subtypes, subsubtypes and CRFs from Los Alamos HIV database (A1_SE8538, A1_PS1044, A1_92UG037, A1_Q23 17, A1SE7535, A1_92RW008; A2_97CDKTB48, A2_94CY017; B_HXB2 LAI IIIB, B_671_00T36; C_BR025, C_ETH2220; D_ELI, D_94UG114; F1_VI850, F1_93BR020_1; F2_MP255, F2_02CM.0016BBY; G_DRCBL, G_92NG083; H_VI997, H_VI991; J_SE7887, J_SE7022; K_EQTB11C, K_MP535; 01_90CF4071, 01_90CF11697, 01_93TH051, 01_93TH253, 01_CM240; 02_pBD6_15, 02_IBNG)16 using Clustal W method17 as implemented in Vector NTI. The phylogenetic reconstructions were also performed with MEGA 4 software package using the neighbor-joining method18. To analyze the recombinant structure of the new viruses, the SimPlot 3.5.1 software was used to determine the percentage of similarity between selected pairs of sequences and to calculate bootscan plots19. The regions that did not cluster with any of the known subtypes were submitted to BLAST analysis (http://hiv-web.lanl.gov/BASIC-BLAST) to find the closely related sequences of other HIV-1 strains. The viral nucleotide positions correspond to the reference strain HXB2 (GenBank Accession Number K03455) used throughout this manuscript.
RESULTS
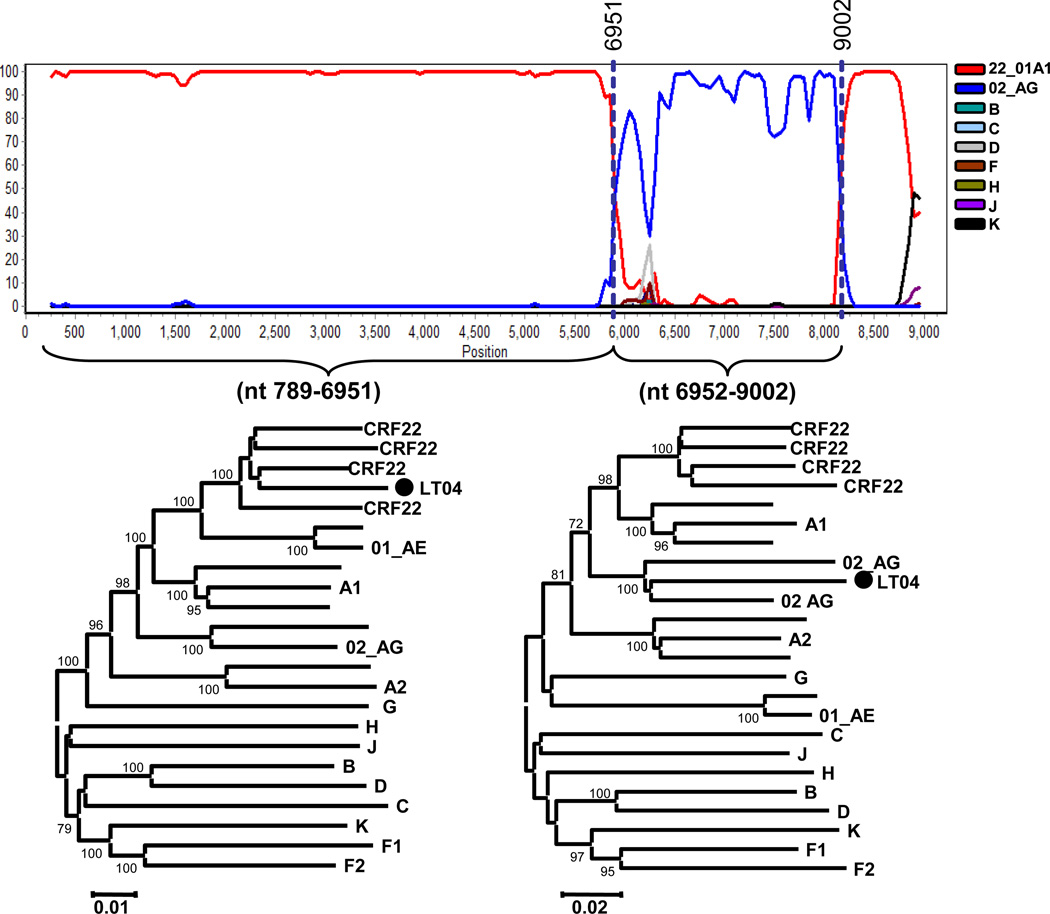
Near full-length genome sequencing was completed for 14 viruses of interest. One virus, NYU488, was collected from Bapile located in the East Province of Cameroon, and thirteen viruses were collected from the Southwest Province of Cameroon (Figure 1A). Phylogenetic analysis of these sequences confirmed their relatedness, as they all clustered together in the CRF22_01A1, CRF02_AG or subtype G radiation with a high bootstrap value (BV). As shown in Figure 2, six strains (02CAMLT04, LPH27MF, LB005, LB011, LB013 and LB054) were deeply clustered in the CRF22_01A1 radiation (BV, 100%) revealing a close relationship, 3 strains (LB045, NYU488 and ARC087) clustered between CRF22_01A1 and CRF01_AE suggesting they are CRF22_01A1 recombinants. 4 strains (ARC007, LB052, BDSH129 and LT66) clustered with CRF02_AG (BV, 93%) and LT31 clustered with subtype G (BV, 100%). These sequences were further analyzed by bootscanning to determine the phylogenetic mosaic structure. The 02CAMLT04 sequence was plotted against references restricted to the subsubtype A1, F2, subtype B, C, H, K, CRF22_01A1 and CRF02_AG. The genome of 02CAMLT04 was divided into three segments at the two breakpoints of nt 6951 and nt 9002, and the neighbor-joining trees were further built for each segment (Figure 3). The first segment extended from the gag to the beginning of the env gene (nt 789–6951), and clustered with CRF22_01A1 (BV, 100%). The second segment representing most of the env gene (nt 6952–9002) clustered with CRF02_AG radiation (BV, 100%) and the rest of the segments covering the nef gene (nt 9003–9408) clustered with CRF22_01A1. A significant portion (76%) of the 02CAMLT04 genome was in perfect alignment with CRF22_01A1 reference sequences except that most of the env gene clustered with CRF02_AG. These results demonstrated that the 02CAMLT04 virus was generated by the recombination of CRF22_01A1 and CRF02_AG. To our knowledge, this is the first report of a novel URF of CRF22_01A1 recombinant with CRF02_AG in full-length sequence.
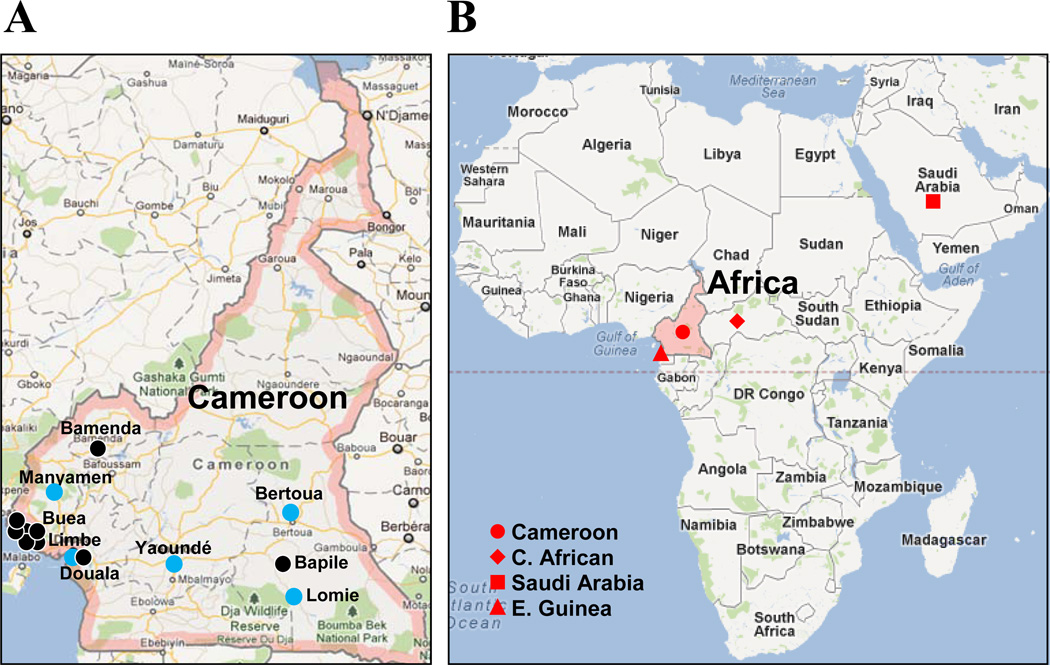
Figure 1. CRF22_01A1 distribution in Cameroon (A) and Africa (B).
Approximate locations of the rural villages or cities from which the specimens were obtained are named. Circle indicates the specimens in Cameroon; light blue circle indicates CRF22_01A1 reference strains described in previous study13, black circle indicates new CRF22_01A1 containing strains characterized in this study. Specimens isolated in Africa and Saudi Arabia are indicated as red symbol.
Figure 2. Phylogentic analysis of the nearly full-length genome sequence.
Analysis was performed using the neighbor-joining methods with Kimura’s two-parameter method and bootstrap analysis (1000 replicates). The reference subsubtypes and CRFs were used to construct the tree, the five strains 02CAMLT72, 01CM.1867LE, 01CM.0001BBY, 02CM.3097MN and 02CM.1917LE were used as CRF22_01A1 references, some references have been omitted for clarity. Bootstrap values above 70% are shown, numbers at the nodes of the tree represent maximum parsimony bootstrap values. The scale bar represents 1% genetic distance. The new identified HIV-1 strains are indicated as red “●”, and blast searched HIV-1 strains are indicated as blue “Δ”.
Figure 3. Recombination in full-length of 02CAMLT04 sequence.
Bootscan plot were performed using SimPolt 3.5.1 software configured with 500 bootstrap replicates, 500 bp window, and a step size of 50 bp. The x-axis shows all 8771 aligned nucleotides of the sequence analyzed, and the y-axis shows the bootstrap value. The 02CAMLT04 was plotted against subtypes/CRF02_AG and a group of CRF22_01A1 references including 02CAMLT72, 02CM.1867LE, 01CM.0001BBY, 02CM.3097MN and 02CM.1917LE as indicated at the top panel. Breakpoints are indicated above the plot. Regions of the sequence alignment were extracted according to the indicated breakpoints. The reference subsubtypes and CRFs were fetched from Los Alamos HIV Sequence Database and initially used to construct the trees, some references have been omitted for clarity. Each segment was analyzed separately by neighbor-joining with the bootstrap above 70%. Representative trees are illustrated for regions a through e, Loci of genome segments are based on the HXB2 numbering engine. Scale bars represent 1–2% distance.
Bootscan analysis of these viruses revealed the presence of CRF22_01A1 homology, the overall structures of these CRF22_01A1 containing viruses are diagrammed in Figure 4. Five sequences (LPH27MF, LB005, LB011, LB013 and LB054) clustered phylogenetically with CRF22_01A1 references throughout the entire genome demonstrating that these viruses share the same genomic mosaic and can be designated as pure CRF22_01A1. Two viruses (ARC087 and NYU488) shared identical recombinant breakpoints and were largely CRF22_01A1 (80% of whole genome spanning mostly pol to nef genes) and 20% for CRF02_AG. LB045 virus was divided into 4 segments with a complex CRF02/CRF22/CRF02/CRF22 recombinant, about 65% of the LB045 genome was CRF22_01A1. BDSH129 virus was almost entirely CRF02_AG (84%) with a small piece of CRF22_01A1 (16%) spanning the gag gene and the beginning of the pol gene. LB052, a triple CRF02/CRF22/CRF02 recombinant, was also predominantly CRF02_AG (76%) with a small portion of CRF22_01A1 fragment spanning the most of env gene from the beginning. ARC007 virus was a subtype B/CRF02 recombinant strain. LT66 and LT31 viruses were a complex F2/CRF02/F2/CRF02/F2 and CRF01/G/CRF02/G recombinant, respectively (data not shown).
Figure 4. Diagram of the genomic structure of CRF22_01A1 containing recombinants.
HBX2 genomic regions are indicated at the top of the plot as reference structure. Breakpoint locations are based on the HXB2 number engine. Each of the resulting genomes was analyzed by SimPlot and Genotyping separately; recombinant structures of each strain were determined and listed. Red bar indicates the CRF22_01A1 regions, open bar indicates subtype D, F, subsubtype A1, F2, CRF01_AE (01) and CRF02_AG (02) as indicated.
To further understand the involvement of CRF22_01A1 in genetic recombination, the blast search of HIV sequence database against 6162 nt of 02CAMLT04 segment (nt 789–6951) yielded four high search score HIV-1 strains 01CM.0130NY, 01CM.1152NG, 02CM.3163MN and 01CM.4008HAN which were originally identified in Cameroon and classified as 01AF2U, A1U, AF2 and 01DU, respectively20. Phylogenetic analysis showed that 01CM.0130NY virus clustered with high confidence to CRF22_01A1 radiation (BV, 100%, Figure 2), 02CM.3163MN and 01CN.1152NG clustered with other CRF22_01A1 containing recombinant(BV, 85%). 01CM.4008HAN was a unique strain clustered between CRF01_AE and subsubtype A1. Further bootscanning analysis showed that they were recombinants of CRF22_01A1 with HIV-1 subtypes F, D, subsubtype A1, F2 or CRFs, all of these strains had a unique mosaic pattern combining CRF22_01A1 (Figure 4). The detailed phylogenetic composition of CRF22_01A1 containing recombinants described above is listed in table (Supplemental Content).
DISCUSSION
We identified five pure CRF22_01A1 and six CRF22_01A1 containing URFs strains in Cameroon. We also reclassified four CRF22_01A1 containing recombinants from the HIV Sequence GenBank (Figure 4). Nine unique recombination patterns were identified; breakpoints were found throughout the genome, the most definitive breakpoints were at the beginning of the pol, accessory or beginning of the env region. CRF22_01A1 fragments may be identified in any region across the HIV whole genome. Table 1 summarizes the subtype/CRF assignments of recombinants containing CRF22_01A1 fragments evaluated in samples collected from 2000–2010. Since the identification of CRF22_01A1 in Cameroon, it may be emerging as a dominant HIV-1 variant in this population that could act as a parental subtype for recombination events. The emergence of more complex intersubtype recombinants containing fragments of CRF22_01A1, such as CRF36_cpx, have been reported in Cameroon7. Reanalysis of the CRF36 genome including CRF22_01A1 reference sequences showed that the CRF36_cpx contains the fragment of CRF22_01A1, CRF01_AE and CRF02_AG. Brennan et al found that 22.4% of HIV-1 URFs were CRF22_01A1 in the blood donor samples collected in 1996–2004 in Cameroon14. Our recent study of samples collected from 2006 to 2008 in Cameroon also identified three CRF02gag/CRF22pol/CRF22env strains12,13. These studies highlight the complexity of recombinants between CRF22_01A1 and CRF02_AG in Cameroon (Table 1). CRF02_AG appeared to be the dominant virus in the population and accounted for 60–68% of HIV-1 infections with an additional 26% classified as URFs2, 21–23. This high degree of recombination could be due to preferential founder effects, and the subsequent introduction of the other genetic form, such as CRF22_01A1 or CRF02_AG, frequently resulting in recombinants containing fragments of CRF22_01A1 or CRF02_AG.
Table 1.
Subtype/CRF assignments of recombinants containing CRF22_01A1 fragment.
| Isolates ID |
collected year |
country /village |
patient information | Genotype | GenBank No | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| age | sex | mar. status | sexual partner | gag | pol | env | ||||
| 02CAMLT04 | 2002 | Cam/Douala | ND | ND | ND | ND | CRF22 | CRF22 | CRF02 | EU743964 |
| NYU488 | 2006 | Cam/Bapile | 68 | F | widow | heterosexual | CRF02 | CRF22 | CRF22 | JN864047 |
| ARC087 | 2006 | Cam/Yaounde | 38 | F | married | heterosexual | CRF02 | CRF22 | CRF22 | JN864048 |
| LPH27MF | 2006 | Cam/Bamenda | 37 | F | married | multiple | CRF22 | CRF22 | CRF22 | JN864049 |
| BDSH129 | 2008 | Cam/Buea | 32 | F | married | heterosexual | CRF22 | CRF02 | CRF02 | JN864052 |
| LB005 | 2010 | Cam/Limbe | 32 | F | single | multiple | CRF22 | CRF22 | CRF22 | JN864050 |
| LB011 | 2010 | Cam/Limbe | 23 | F | married | multiple | CRF22 | CRF22 | CRF22 | JN864051 |
| LB013 | 2010 | Cam/Limbe | 56 | M | married | multiple | CRF22 | CRF22 | CRF22 | JN864058 |
| LB045 | 2010 | Cam/Limbe | 36 | F | single | heterosexual | CRF02 | CRF22 | CRF22 | JN864053 |
| LB052 | 2010 | Cam/Limbe | 33 | F | married | heterosexual | CRF02 | CRF02 | CRF22 | JN864054 |
| LB054 | 2010 | Cam/Limbe | 66 | M | ND | heterosexual | CRF22 | CRF22 | CRF22 | JN864059 |
| NYU1126 | 2000 | Cam/Mboy | - | - | - | - | CRF01 | CRF02 | CRF22 | EF087995 |
| NYU830 | 2000 | Cam/Gadji | - | - | - | - | CRF01 | CRF02 | CRF22 | EF087994 |
| 01CM.0001BBY | 2001 | Cam/Yaounde | - | - | - | - | CRF22 | CRF22 | CRF22 | AY371159 |
| CM53122 | 2001 | Cam/Bertoua | - | - | - | - | CRF22 | CRF22 | CRF22/01 |
AY037284+ AY037285 |
| 01CM.0130NY | 2001 | Cam/Nyabessang | - | - | - | - | CRF22 | CRF22 | CRF22/F2 | AY371167 |
| 01CM.1152NG | 2001 | Cam/Nyabessang | - | - | - | - | A1 | CRF22 | CRF22 | AY371163 |
| 01CM.4008HAN | 2001 | Cam/Ndikinim eki | - | - | - | - | D | CRF22 | D | AY371163 |
| CE03 | 2001 | Cam/Yaounde | - | - | - | - | CRF22 | - | 11_cpx |
DQ056985; DQ056864 |
| A1102 | 2001 | Cam/Douala | - | - | - | - | CRF22 | CRF02 | CRF02 |
EU619435; EU618346; EU618919 |
| 02CAMLT72 | 2002 | Cam/Douala | - | - | - | - | CRF22 | CRF22 | CRF22 | EU743963 |
| 02CM.3097MN | 2002 | Cam/Manyamen | - | - | - | - | CRF22 | CRF22 | CRF22 | GQ229529 |
| 02CM.1917LE | 2002 | Cam/Lomie | - | - | - | - | CRF22 | CRF22 | CRF22 | GQ229530 |
| 02CM.1867LE | 2002 | Cam/Lomie | - | - | - | - | CRF22 | CRF22 | CRF22 | AY371165 |
| 02CM.3163MN | 2002 | Cam/Manyamen | - | - | - | - | F2 | CRF22 | CRF22/F2 | AY371160 |
| 4455-7 | 2003 | Cam/Yaounde | - | - | - | - | CRF22 | CRF02 | CRF02 |
EU619276; EU618760; EU618187 |
| 06SP42-303556 | 2005-7 | E. Guinea | - | - | - | - | CRF22 | CRF22 | - |
EU342808; EU342854 |
| 08GQ346 | 2008 | E. Guinea | - | - | - | - | CRF22 | CRF22 | FN557311 | |
| J11469 | 2008 | S. Arabia | - | - | - | - | CRF22 | CRF22 | CRF22 | DG375282; DG375234; DG375289 |
| 01771M8 | 2008-9 | C. African | - | - | - | - | - | CRF22 | - | HM117941 |
| 36-0041F1 | 2008-9 | C. African | - | - | - | - | - | CRF22 | - | HM117942 |
| 8-CP001029 | 2008-9 | C. African | - | - | - | - | - | CRF22 | - | HM117944 |
| 38-0147M1 | 2008-9 | C. African | - | - | - | - | - | CRF22 | - | HM117951 |
| 14-CP001245 | 2008-9 | C. African | - | - | - | - | - | CRF22 | - | HM117957 |
Cam: Cameroon; E. Guinea: Equatorial Guinea; C. African: Certral African Republic; S. Arabia: Saudi Arabia; ND: Not Documented; A dash (-) indicates the data was not obtained. The bolded font indicates the samples characterized in this study.
One interesting finding in this study is that all of the recombinants of HIV-1 characterized displayed a fairly similar breakpoint at the accessory gene and env region which spans from vif to gp120 gene. For example, a breakpoint of 02CAMLT04 genome in nucleotide position nt 6911 in gp120 was also observed around similar position in 02CAMLT72 (nt 7015), 01CM.1152NG (nt 6963), LB052 (nt 6099), LB045 (nt 6261), 01CM.4008HAN (nt 6088) and 01CM.0130NY (nt 6668). The segment from one third of the env to the 5’ end of the nef region (nt 7016–8895) in 02CAMLT72, showed a CRF22_01A1 virus derived from subsubtype A1, but was replaced by CRF02_AG in a similar region of 02CAMLT04 virus, suggesting that this region may be a hotspot for recombination between CRF22_01A1 and other subtypes/CRFs. The exact prevalence of the CRF22_01A1 intersubtype recombinants is still unclear. However, according to the current study, given that the CRF22_01A1-containing recombinants were possibly introduced to the population as early as 2000, CRF22_01A1 could be more broadly distributed in Cameroon even outside this country (Figure 1B). Although the CRF22_01A1 strain was prevalent over the 10-year period, the numbers of reported CRF22_01A1 cases were limited since reference strains of CRF22_01A1 were not well classified until now. The complexity of CRF22_01A1 strains in Cameroon seemed to increase during the 2002–2010 period. The CRF22_01A1 strain has also been identified in Saudi Arabia24, 25, Equatorial Guinea26, 27, Central African patients28 and USA29 indicating that the CRF22 strain was spreading to different geographic regions.
It has been well known that HIV-1 CRFs behave like a pure subtype of HIV-1 and are able to recombine with other subtypes or CRFs during HIV-1 viral reverse transcription when co-infection or super-infection occurs. We document here that CRF22_01A1 is circulating in Cameroon and is involved in complex intersubtype recombination events with subtypes (D or F), subsubtypes (A1 or F2) and CRFs (CRF01_AE or CRF02_AG). CRF22_01A1 recombinants seem to co-circulate at higher proportions than its pure prototype. Our findings further demonstrate the dynamic evolution of emerging variants in Cameroon which could potentially impact the phlyogenetic nature of the epidemic in this region in the future.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge Drs. Mingjie Zhang, Krishnakumar Devadas and Robin Biswas for review of the manuscript. The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
Source of Funding; Supported by The National Heart, Lung, and Blood Institute for funding part of this work through an IAA – National Heart, Lung and Blood InterAgency Agreement BY1-HB-5026-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to disclose.
SUPPLEMENTAL CONTENT
Supplemental Content1.docx
Table. Phylogenetic analysis of CRF22_01A1 containing recombinant segments
REFERENCES
- 1.Gao F, Bailes E, Robertson DL, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999 Feb 4;397(6718):436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 2.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000 Jan 28;287(5453):607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 3.Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006 Jul 28;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr JK, Torimiro JN, Wolfe ND, et al. The AG recombinant IbNG and novel strains of group M HIV-1 are common in Cameroon. Virology. 2001 Jul 20;286(1):168–181. doi: 10.1006/viro.2001.0976. [DOI] [PubMed] [Google Scholar]
- 5.Plantier JC, Leoz M, Dickerson JE, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009 Aug 2; doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 6.Powell RL, Zhao J, Konings FA, et al. Circulating Recombinant Form (CRF) 37_cpx: An Old Strain in Cameroon Composed of Diverse, Genetically Distant Lineages of Subtypes A and G. AIDS Res Hum Retroviruses. 2007 Jul;23(7):923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 7.Powell RL, Zhao J, Konings FA, et al. Identification of a Novel Circulating Recombinant Form (CRF) 36_cpx in Cameroon That Combines Two CRFs (01_AE and 02_AG) with Ancestral Lineages of Subtypes A and G. AIDS Res Hum Retroviruses. 2007 Aug;23(8):1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- 8.Vergne L, Bourgeois A, Mpoudi-Ngole E, et al. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology. 2003 Jun 5;310(2):254–266. doi: 10.1016/s0042-6822(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 9.Wilbe K, Casper C, Albert J, Leitner T. Identification of two CRF11-cpx genomes and two preliminary representatives of a new circulating recombinant form (CRF13-cpx) of HIV type 1 in Cameroon. AIDS Res Hum Retroviruses. 2002 Aug 10;18(12):849–856. doi: 10.1089/08892220260190326. [DOI] [PubMed] [Google Scholar]
- 10.Zhong P, Burda S, Urbanski M, et al. HIV type 1 group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J Acquir Immune Defic Syndr. 2002 Dec 15;31(5):495–505. doi: 10.1097/00126334-200212150-00007. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi J, Bodelle P, Vallari AS, et al. HIV infections in northwestern Cameroon: identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res Hum Retroviruses. 2004 Sep;20(9):944–957. doi: 10.1089/aid.2004.20.944. [DOI] [PubMed] [Google Scholar]
- 12.Machuca A, Tang S, Hu J, et al. Increased genetic diversity and intersubtype recombinants of HIV-1 in blood donors from urban Cameroon. J Acquir Immune Defic Syndr. 2007 Jul 1;45(3):361–363. doi: 10.1097/QAI.0b013e318053754c. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Tang S, Ragupathy V, et al. Identification and genetic characterization of a novel CRF22_01A1 recombinant form of HIV type 1 in Cameroon. AIDS Res Hum Retroviruses. 2010 Sep;26(9):1033–1045. doi: 10.1089/aid.2009.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan C, Bodelle P, Coffey R, et al. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr. 2008 Dec 1;49(4):432–439. doi: 10.1097/QAI.0b013e31818a6561. 2008. [DOI] [PubMed] [Google Scholar]
- 15.Ragupathy V, Zhao J, Wood O, et al. Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virology Journal. 2011;8:185. doi: 10.1186/1743-422X-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Circulating Recombinant Forms (CRFs) Los Alamos National Laboratory (online) 2011 Oct 25;12:26. http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html. Last modified: Tue. [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997 Dec 15;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999 Jan;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kijak GH, Sanders-Buell E, Wolfe ND, et al. Development and application of a high-throughput HIV type 1 genotyping assay to identify CRF02_AG in West/West Central Africa. AIDS Res Hum Retroviruses. 2004 May;20(5):521–530. doi: 10.1089/088922204323087778. [DOI] [PubMed] [Google Scholar]
- 21.McCutchan FE. Understanding the genetic diversity of HIV-1. Aids. 2000;14(Suppl 3):S31–S44. [PubMed] [Google Scholar]
- 22.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000 Apr 7;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 23.Swanson P, de Mendoza C, Joshi Y, et al. Impact of Human Immunodeficiency Virus Type 1 (HIV-1) Genetic Diversity on Performance of Four Commercial Viral Load Assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J Clin Microbiol. 2005 Aug;43(8):3860–3868. doi: 10.1128/JCM.43.8.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi J, Badreddine S, Swanson P, Bodelle P, Devare SG, Brennan CA. Identification of new CRF43_02G and CRF25_cpx in Saudi Arabia based on full genome sequence analysis of six HIV type 1 isolates. AIDS Res Hum Retroviruses. 2008 Oct;24(10):1327–1335. doi: 10.1089/aid.2008.0101. [DOI] [PubMed] [Google Scholar]
- 25.Badreddine S, Smith K, van Zyl H, et al. Identification and characterization of HIV type 1 subtypes present in the Kingdom of Saudi Arabia: high level of genetic diversity found. AIDS Res Hum Retroviruses. 2007 May;23(5):667–674. doi: 10.1089/aid.2007.0185. [DOI] [PubMed] [Google Scholar]
- 26.Yebra G, Rivas P, Herrero MD, et al. Clinical differences and viral diversity between newly HIV type 1-diagnosed African and non-African patients in Spain (2005–2007) AIDS Res Hum Retroviruses. 2009 Jan;25(1):37–44. doi: 10.1089/aid.2008.0134. [DOI] [PubMed] [Google Scholar]
- 27.Djoko CF, Wolfe ND, Vidal N, et al. HIV type 1 pol gene diversity and genotypic antiretroviral drug resistance mutations in Malabo, Equatorial Guinea. AIDS Res Hum Retroviruses. 2010 Sep;26(9):1027–1031. doi: 10.1089/aid.2010.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moussa S, Pinson P, Pelembi P, et al. First Data on HIV-1 Resistance Mutations to Antiretroviral Drugs in Central African Republic. AIDS Res Hum Retroviruses. 2010 Oct 12;26(11):1247–1248. doi: 10.1089/aid.2010.0091. [DOI] [PubMed] [Google Scholar]
- 29.Pyne MT, Holzmayer V, John Hackett J, Hillyard1 DR. Analysis of HIV Subtype Prevalence and Geographic Distribution in a Large U.S. Population. 16th Conference on Retroviruses & Opportunistic Infections; February 8–11, 2009; Montreal, Canada. 2009. Poster #292. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.