Abstract
ETV6 (or TEL), a transcriptional repressor belonging to the ETS family, is frequently involved in chromosomal translocations linked with human cancers. It displays a DNA-binding mode distinct from other ETS proteins due to the presence of a self-associating PNT domain. In this study, we used NMR spectroscopy to dissect the structural and dynamic bases for the auto-inhibition of ETV6 DNA-binding by sequences C-terminal to its ETS domain. The CID (C-terminal inhibitory domain) contains two helices, H4 and H5, which sterically block the DNA-binding interface of the ETS domain. Importantly, these appended helices are only marginally stable as revealed by amide hydrogen exchange and 15N relaxation measurements. The CID is thus poised to undergo a facile conformational change as required for DNA-binding. The CID also dampens millisecond timescale motions of the ETS domain hypothesized to be critical for the recognition of specific ETS target sequences. This work illustrates the use of appended sequences on conserved structural domains to generate biological diversity, and complements previous studies of the allosteric mechanism of ETS1 auto-inhibition to reveal both common and divergent features underlying the regulation of DNA-binding by ETS transcription factors.
Keywords: ETS transcription factor, transcription repressor, protein structure and dynamics, NMR spectroscopy
Introduction
The human genome encodes 28 ETS transcription factors that play central roles in both the regulation of normal cellular growth and the development of malignancies.1, 2 A subset of the ETS genes are involved in human chromosome rearrangements that associate with specific cancers. ETV6 (or TEL) is best known for its involvement in a variety of hematopoietic malignancies (reviewed in 3). The altered ETV6 loci encode chimeric oncoproteins with the self-associating PNT (or SAM) domain of ETV6 fused to either a tyrosine kinase catalytic domain or the DNA-binding domain of another transcription factor. In less frequent cases, the ETV6 DNA-binding domain is linked to the transcription activation domains of MN1.4
Within its native context, ETV6 is a transcriptional repressor and putative tumor suppressor.5–8 The repressive activity of ETV6 results from its recruitment of several general co-repressors, including SMRT, mSin3A, and N-CoR, that are ultimately linked to histone deacetylases.9–12 In addition, the PNT domains of ETV6 and its Drosophila ortholog Yan form helical polymers, which are proposed to create extended repressive complexes at target DNA sites.13,14 In support of this proposal, mutations that restrict Yan to a monomeric state reduce its activity as a transcriptional repressor and impair its function during embryonic and retinal development.15
As a member of the ETS family, ETV6 contains a conserved ETS domain that mediates binding to ~9 bp DNA sequences with a core GGA(A/T) motif.16 Recently, we have demonstrated that ETV6 DNA-binding is strongly inhibited by residues C-terminal to its ETS domain (Fig. 1A).17 Intriguingly, the effect of this “CID” (C-terminal inhibitory domain) is partially reduced in larger ETV6 fragments due to dampening effects of sequences between the PNT and ETS domain. This currently undefined region is termed the “LID” (linker inhibitory damper). Furthermore, self-association of ETV6 via its PNT domain facilitates cooperative DNA-binding to tandem ETS sites, thereby compensating for the low affinity caused by auto-inhibition.14, 17 These target DNA sites can occur in either orientation and at variable spacing (up to 5 helical turns), suggesting that the linker region provides a high degree of conformational flexibility for promoter recognition.
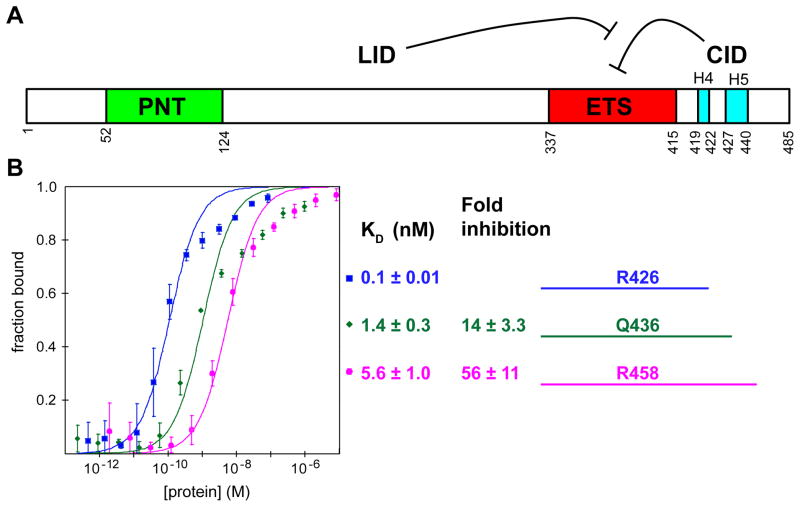
Fig. 1. ETV6 modular structure and DNA-binding affinity.
(A) Cartoon of murine ETV6 showing the PNT (green) and ETS (red) domains, along with the LID and the inhibitory CID composed of helices H4 and H5 (cyan) (adapted from 17). (B) The primary truncation fragments used in this study span from Arg335 to the indicated C-terminal residue. Affinities for a DNA duplex displaying a consensus ETS-binding site were determined by EMSA from three or two (in the case of ETV6R426) independent binding isotherms. The KD values and errors reflect the precision of a single curve fit of all data points for each fragment to the equation for a binary interaction. Fold inhibition represents decreased affinity relative to ETV6R426.
Auto-inhibition of DNA-binding has been reported for several ETS transcription factors, yet the phenomenon has been dissected structurally only in the cases of ETS1 and ETV6.2 An undocumented model of a human ETV6 fragment determined by the Riken Structural Genomics Initiative suggested that the CID forms two inhibitory helices, which could sterically block its DNA-binding interface. This stands in contrast to the allosteric mechanism of ETS1 auto-inhibition18–20 and reveals both common and divergent features underlying the regulation of DNA-binding by ETS transcription factors.
In this study, we investigated the mechanism of ETV6 auto-inhibition using NMR spectroscopy. The tertiary structures of two auto-inhibited fragments of murine ETV6 confirm that the CID forms two helices abutted against the DNA-binding interface of the winged helix-turn-helix ETS domain. In addition, using amide hydrogen exchange (HX) and 15N relaxation measurements, we show that the CID inhibitory helices are only marginally stable and thus poised to undergo a necessary conformational change to remove the steric blockage of the DNA-binding interface. Furthermore, the CID dampens msec timescale motions within the ETS domain that are likely required for both interacting with non-specific DNA sequences and forming high affinity complexes with specific ETS target sites.21,22 Collectively, these data lead to a model of ETV6 auto-inhibition in which the ETS domain exists in a conformational equilibrium between a rigid, sterically blocked state that is recalcitrant to DNA-binding and an unblocked flexible state that can associate with DNA. This model provides a structural and dynamic framework for understanding the regulation of ETV6 in normal cellular context and the aberrant activities of the chimeric oncoproteins resulting from its alteration by chromosomal translocations.
Results
DNA-binding affinity measurements
To investigate the mechanism of ETV6 auto-inhibition, we focused on three protein fragments that start at Arg335, span the ETS domain (337–415), and extend to Arg426, Gln436, or Arg458 (Fig. 1B; Supplemental Fig. S1). These fragments were based approximately on a previously characterized set of murine ETV6 variants, which uncovered the strong inhibitory function of residues C-terminal to the ETS domain.17 After an initial analysis of ETV6Q436, it became apparent that structured regions of the CID might extend beyond Gln436. Thus, we subsequently investigated ETV6R458, a fragment with 22 additional C-terminal residues that was identified by partial proteolysis as a stable species. In parallel, we also studied an uninhibited variant, ETV6R426, lacking most of the CID. An ETS domain-only fragment (ETV6T418) could not be characterized due to its poor expression and/or stability.
To confirm that these three ETV6 fragments indeed recapitulated auto-inhibition, equilibrium dissociation (KD) constants for DNA bearing an ETS consensus motif were determined by electrophoretic mobility shift assays (EMSAs; Fig. 1B). The presence of the additional C-terminal residues in ETV6Q436 and ETV6R458 led to 14- and 56-fold inhibition, respectively, relative to the higher affinity ETV6R426 fragment. Parenthetically, the KD values of ETV6R426 and ETV6R458 are both ~5-fold lower than those obtained under the current experimental conditions for previously characterized ETV6 fragments spanning residues 331–426 and 331–458.17 We speculate that this slightly higher affinity might reflect altered electrostatic interactions with DNA due to the deletion of residues 331Ile-Ala-Asp-Cys334 and/or the presence of the non-native N-terminal sequence Gly-Ser-His-Met remaining after thrombin cleavage of the His6-tag used for affinity purification of ETV6R426 and ETV6R458. Nevertheless, we deemed these fragments suitable for further analysis of the structural and mechanistic bases of ETV6 auto-inhibition.
Structural studies reveal a steric mechanism of ETV6 auto-inhibition
The well-dispersed 15N-HSQC spectra of the three ETV6 fragments confirmed that each adopted a similar, stable fold in isolation (Supplemental Fig. S2). Signals from the mainchain and sidechain 1H, 13C, and 15N nuclei of ETV6Q436 and ETV6R458 were assigned by standard heteronuclear NMR methods, whereas those from the amides of ETV6R426 were readily annotated by a direct comparison with the 15N-HSQC and 15N-relaxation spectra of ETV6Q436.
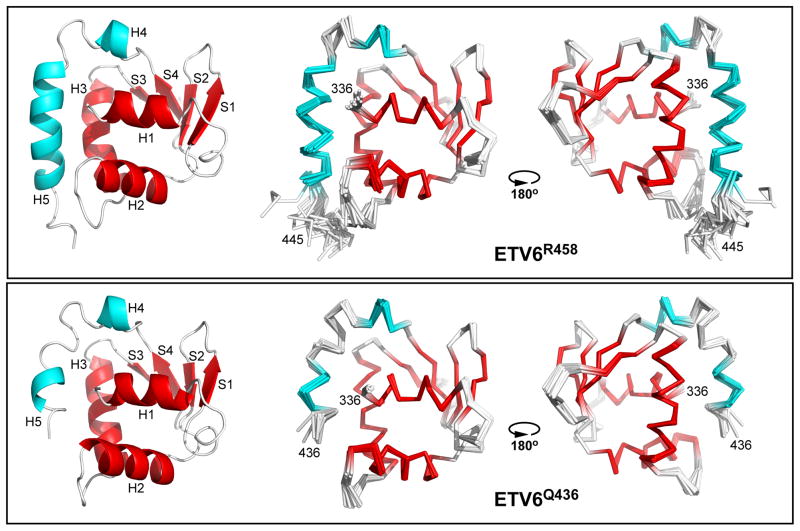
The structural ensembles of ETV6Q436 and ETV6R458 were determined with ARIA/CNS using NOE-derived distance and chemical shift-derived dihedral angle restraints (Table 1). As illustrated in Fig. 2, each protein adopts a very similar globular fold composed of five α-helices and four β-strands (ETV6R458 H1: residues 337–344; S1: 354–358; S2: 363–366; H2: 369–379; H3: 387–399; S3: 403–405; S4: 412–415; H4: 419–422; H5: 427–440). A small helical turn also links H1 and S1, but is not explicitly named to maintain consistency with the previous descriptions of other ETS factors.23 As discussed below, the primary difference between the two fragments is the shorter helix H5 (residues 430–433) in the smaller ETV6Q436 construct. The ETV6Q436 and ETV6R458 ensembles also resemble the undocumented NMR-derived structure of the ETS domain and partial CID of the human ETV6 ortholog deposited in the Protein Data Bank (2DAO.pdb; compared in Supplemental Fig. S3).
Table 1.
NMR Restraints and Statistics for the ETV6R458andETV6Q436 Ensembles
| ETV6R458 | ETV6Q436 | |
|---|---|---|
| Number of restraints | ||
| NOEsa | ||
| Intraresidue | 1302 (146) | 1182 (215) |
| Sequential | 450 (75) | 555 (167) |
| Medium Range (|i−j| = 2–4) | 316 (72) | 475 (190) |
| Long Range (|i−j| ≥ 5) | 485 (146) | 692 (311) |
| Total | 2553 (439) | 2904 (883) |
| Dihedral angles | ||
| (ψ,φ) | 95, 95 | 84,84 |
| Deviation from restraints | ||
| NOE (Å) | 0.0470 ± 0.0214 | 0.0390 ± 0.0018 |
| Dihedral angles (degrees) | 0.62 ± 0.026 | 0.64 ± 0.013 |
| Deviation from idealized geometry | ||
| Bonds (Å) | 0.0049 ± 0.0002 | 0.0058 ± 0.0002 |
| Angles (degrees) | 0.6238 ± 0.0261 | 0.6419 ± 0.0127 |
| Improper angles (degrees) | 1.570 ± 0.1051 | 1.588 ± 0.084 |
| Residues located within the generously allowed regions of the Ramachandran plot | 99.6 % | 99.7 % |
| rmsd from the average structures (Å) b | ||
| Backbone | 0.48 ± 0.07 | 0.40 ± 0.03 |
| Backbone and sidechains | 1.05 ± 0.09 | 1.07 ± 0.05 |
Number of unambiguous and, in parentheses,ambiguous restraints.
rmsd valuesfor the 20 member ensembles calculated for non-hydrogen atoms over all residues inthe range of Leu336 to Gln441 for ETV6R458 and Leu336 to Glu434 for ETV6Q436.
Fig. 2. Structural ensembles of ETV6Q436 and ETV6R458.
Shown are Cα-ribbon diagrams of the refined twenty member ensembles of ETV6R458 (upper) and ETV6Q436 (lower), along with a cartoon diagram of one low energy structure of each protein (left). The ribbon diagrams are rotated by ~180° about the vertical axis. The secondary structural elements of the ETS domain and CID are highlighted in red and cyan, respectively. The disordered N-terminal Gly-Ser-His-Met and C-terminal residues 446–458 have been removed for clarity.
As expected, residues 337–415 of both ETV6Q436 and ETV6R458 exhibited the well known architecture of an ETS domain with a winged helix-turn-helix motif (H2-turn-H3) affixed to a four-stranded anti-parallel β-sheet scaffold. Based on the structures of numerous ETS factor-oligonucleotide complexes, helix H3 is expected to bind in the major groove of target DNA sequences, with the invariant Arg392 and Arg395 hydrogen bonding to the guanines of the core GGA recognition motif. Additional direct and water-mediated contacts are likely provided by the “turn” between helices H2 and H3, the β-hairpin “wing” between strands S3 and S4, and the N-terminus of helix H1.23
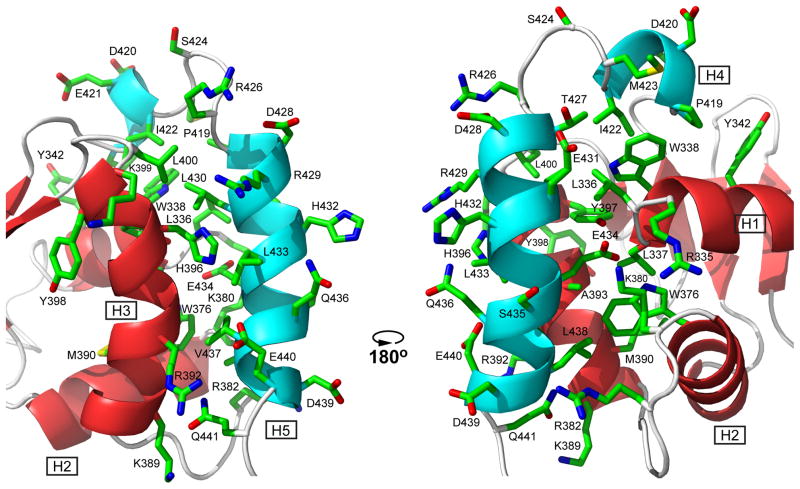
The conformational analysis of the inhibited ETV6 fragments reveals that the structured CID spans residues 419 through 440 and is composed of two amphipathic helices, H4 and H5. The interface between the CID and the ETS domain is illustrated in Fig. 3. The short helix H4 lies anti-parallel along a small N-terminal portion of helix H1, with the sidechains of Pro419, Ile422, and Met423 in van der Waals contact with those of Trp338, and to a lesser extent, Tyr342. More substantially, the longer helix H5 packs anti-parallel to the DNA-recognition helix H3, making extensive interactions with residues along this helix, as well as at the N-terminal end of helix H1 and the C-terminal end of helix H2. Key hydrophobic contacts involve Leu430, Leu433, Val437, and Leu438 from helix H5 with Leu336 and Trp338 of helix H1, Trp376 and Lys380 of helix H2 and the following “turn”, respectively, and Arg392, Ala393, His396, Tyr397, and Leu400 of helix H3. Many of these interactions were defined by unambiguous, manually assigned NOE restraints. In addition, the juxtaposition of complementarily charged sidechains indicates that the docking of the CID along the ETS domain is also established by electrostatic interactions. Most notably, the buried carboxylate of Glu434 in the middle of helix H5 is adjacent to the amino group of Lys380 at the end of helix H2 and within hydrogen bonding distance of the amide of Leu337 at the start of helix H1. This localizes the negatively-charged sidechains of Glu434, as well as Glu431, near the positively-charged end of the H1 helical dipole, which is predicted to contact the phosphodiester DNA backbone. Consistent with their key structural roles, mutation of either of these two residues to an alanine relieves inhibition.17 Asp439 and Glu440 in helix H5 are also near Arg382 and Arg392 in the “turn” and at the start of H3, respectively.
Fig. 3. Helices H4 and H5 of the CID dock along helices H1, H2, and H3 of the ETS domain via hydrophobic and electrostatic interactions.
Shown are the sidechains of all CID residues and selected ETS domain residues in one member of the ETV6R458 structural ensemble (carbon, green; oxygen, red; nitrogen, blue; hydrogens removed for clarity). In this position, the CID sterically blocks the DNA-binding interface of the ETS domain, which is centered around the recognition helix H3 (see Fig. 7) and includes the N-terminus of helix H1 and residues in the turn between helices H2 and H3. Cartoons of the helices and strands are colored as in Fig. 2. The two views are rotated by ~180° about the vertical axis.
Overall, the structural ensembles of ETV6Q436 and ETV6R458 are very similar (Fig. 2). However, helix H5 is shorter in the smaller construct due to the absence of Val437 through Glu440. These four residues complete the helix in the full length CID. Although it is somewhat surprising that a truncated H5 folds and is partially inhibitory, this likely reflects the importance of the above mentioned electrostatic and hydrophobic interactions involving residues from helices H1 and H3 with the partial H5, combined with the conformational dynamics of the CID, presented below. The different C-terminal sequences of ETV6Q436 and ETV6R458 also result in a few modest structural variations between the ETS domains of the two proteins. Most notably, the indole ring of Trp376 is flipped by ~180° in ETV6R458 relative to ETV6Q436 due to a hydrophobic interaction with Leu438 in helix H5 that is only possible in the larger fragment. Residues in the “turn,” including Asn384, also adopt different conformations due to the proximity of the longer helix H5 in ETV6R458.
The docking of helix H5 against all three helices of the ETS domain provides an immediate explanation for the inhibitory effect of the CID. Specifically, helix H5 sterically blocks the DNA-binding interface of ETV6. This also requires that the CID be unfolded or otherwise displaced to allow the well-characterized interactions of the ETS domain with target DNA sequences. Given that the CID reduced the affinity of these ETV6 fragments for DNA by only ~10- to 50-fold, helices H5 and possibly H4 must be dynamic in order to undergo a facile conformational transition. We tested this prediction using amide HX and 15N relaxation experiments on the three isolated ETV6 variants.
Amide HX reveals a conformationally dynamic CID that stabilizes the ETS domain
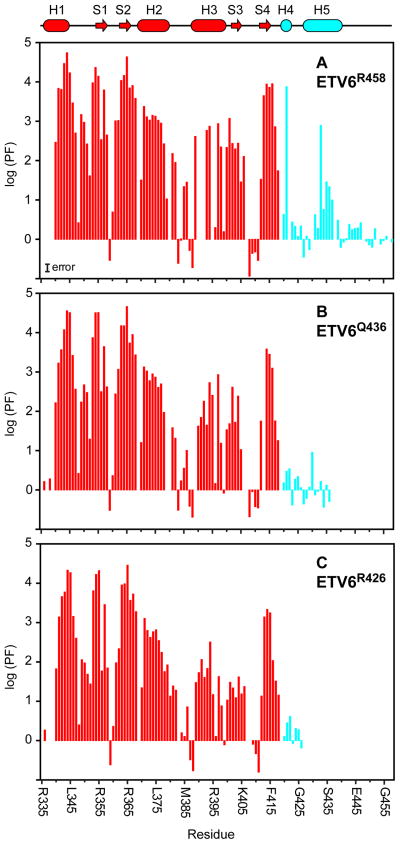
Using complementary proton-deuterium and proton-proton HX experiments measured with sample pH values ranging from 5.8 and 8.2, we determined the protection factors (PF) for most amides in ETV6R458, ETV6Q436, and ETV6R426 (Fig. 4). A PF is the ratio kpred/kex, where kex is the measured HX rate constant for a given amide and kpred is the predicated rate constant for a corresponding random coil polymer under the same conditions of pH, temperature, and solvent. According to the standard model of HX in the commonly observed pH-dependent EX2 regime, the PF is the inverse of an equilibrium constant for fluctuations between a closed state, where exchange is prevented by the structural (i.e., hydrogen bonding) and electrostatic environment of an amide, and a transient open exchange-competent state.24 Thus, PF’s provide a residue-specific measure of the free energy landscape (ΔG°HX = RTln(PF)) of a protein governing global and local conformational equilibria linked with exchange.
Fig. 4. Amide HX studies demonstrate that the CID is flexible and stabilizes the ETS domain.
HX protection factors (PF’s) for the ETS domain (red) and CID (cyan) of (A) ETV6R458, (B) ETV6Q436, and (C) ETV6R426 measured at 30 °C via either slow proton-deuterium or fast proton-proton exchange experiments. The relatively low PF’s for helix H5 demonstrate that the CID is only marginally stable and poised to unfold. Furthermore, inclusion of the partial (ETV6Q436) or full CID (ETV6R458) progressively stabilizes helix H3 against HX. Missing data points correspond to prolines, amides with overlapping signals, or amides for which HX rates could not be determined reliably by either exchange method (i.e., exchange lifetimes between several seconds and a few minutes). The indicated magnitude of the estimated PF error, on a logarithic scale, is based on assumed worse case errors of ± 20% in both measured and predicted exchange rates. The PF’s less than 1 reported for a few residues undergoing rapid HX are attributed to exchange outside the EX2 regime, inaccurate kpred values (particularly for ionizable sidechains), and errors associated with the CLEANEX-PM technique.77 The cartoon in the upper region of the plot defines the secondary structure regions of ETV6R458 with coloring as described in Fig. 2. Plots showing the relative PF’s of ETV6Q436 and ETV6R426 versus ETV6R458 are presented in Supplemental Fig. S3.
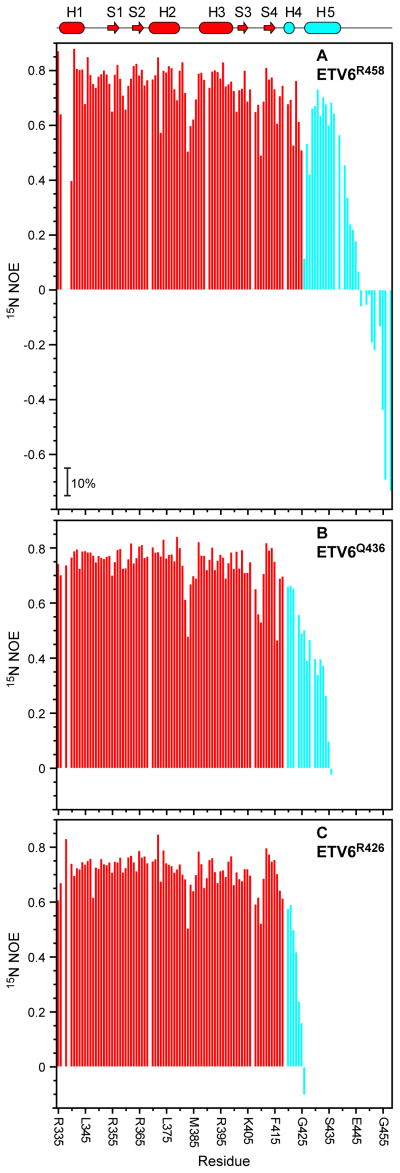
HX measurements for the ETV6 constructs reveal a wide range of dynamics. For each protein, the most slowly exchanging amides (PF ~ 2.5 × 104) are located in helix H1 and strands S1 and S2. These residues likely exchange via a global unfolding pathway and thus form the stable core of the ETS domain.25 The corresponding ΔG°HX ~ 6 kcal/mol also provides lower bounds on the unfolding free energy of the ETV6 constructs under native conditions. In contrast, amides in loop regions, including the “turn” between helices H2 and H3 and the “wing” between strands S3 and S4, show relatively low PF’s and hence must exchange via more localized conformational fluctuations. This is consistent with their generally higher solvent accessibilities and elevated rms deviations in the structural ensembles of ETV6R458 and ETV6Q436 (Fig. 2). Finally, residues Gln441 through Arg458 in the C-terminal tail of ETV6R458 show random coil behavior with PF ~ 1, indicating that they are disordered.
HX measurements clearly demonstrate that the CID region is marginally stable relative to the ETS domain core. In the case of ETV6R458, the PF’s for residues in helix H5 are ~ 2–30 with Leu430 exhibiting the highest value of only 780 (Fig. 4A). These correspond to an average ΔG°HX ~ 1.3 kcal/mol for fluctuations leading to an open, exchange competent state. Thus, in the absence of DNA, helix H5 is poised for a facile conformational change as required to remove steric blockage and allow DNA-binding by ETS domain. Helix H4 also shows low PF’s, indicative of limited stability. However, the first three amide nitrogens of a regular α-helix are generally not involved in intrahelical hydrogen bonding and are not expected to show protection from HX. Hence, it is difficult to draw conclusions on the dynamic properties of this short helix in ETV6R458 from the exchange behavior of a limited number of residues. Although helices H4 and H5 are well defined in the structural ensemble of ETV6Q436, the entire truncated CID of this intermediate length fragment shows PF’s less than 10 (Fig. 4B). This demonstrates that, while still partially inhibitory, the residues forming these helices are in equilibrium with a substantially populated unfolded state. Finally, in the case of ETV6R426, Asp420 through Arg426 show essentially random coil exchange kinetics (Fig. 4C). Therefore, in the absence of further NMR data, it is uncertain if helix H4 is formed at a measurable level in this uninhibited ETV6R426 fragment.
In addition to demonstrating the conformational dynamics of the CID, HX experiments provide a second key insight into the mechanism of ETV6 auto-inhibition. As summarized in Fig. 4A, the PF’s of helices H2 and H3 in ETV6R458 are ~ 10 - 100 fold lower than those of the stable core of the ETS domain. This indicates that the HX of these helices results from sub-global fluctuations (ΔG°HX ~ 3–4 kcal/mol) and that the DNA-binding interface of ETV6 is conformationally flexible. Similar exchange behavior has been reported for the ETS domains of FLI1,26 SPI1,27 and ETS1,19 and is consistent with the general theme that a dynamic interface is required for sequence specific DNA-binding.21 More strikingly, the PF’s of helix H3, and to a lesser extent, helix H2 and strand S4, decrease progressively with truncation of the CID to form ETV6Q436 and then ETV6R426 (Figs. 4B,C and Supplemental Fig. S4). As a result, amides throughout helix H3 exchange only ~ 100 fold slower in ETV6R426 than expected for a random coil polymer. This is indicative of substantial conformational fluctuations in the uninhibited ETV6 fragment, and demonstrates that the full CID in ETV6R458 stabilizes the DNA-binding interface of the ETS domain. We therefore hypothesize that the CID reduces the affinity of ETV6 for DNA via both steric effects and by dampening necessary flexibility of the ETS domain. This idea is bolstered by 15N relaxation measurements.
15N relaxation measurements reveal fast dynamics of the ETS domain and CID
To characterize the global hydrodynamic and local dynamic properties of the three ETV6 fragments, amide 15N T1, T2 and heteronuclear NOE relaxation data were also collected for each protein at 30 °C using a 600 MHz NMR spectrometer. Based on the resulting T1/T2 ratios for well ordered amides, the fit isotropic tumbling correlation times τc for ETV6R458, ETV6Q436, and ETV6R426 were 8.02 ± 0.1 ns, 6.75 ± 0.04 ns, and 6.17 ± 0.09 ns, respectively. These global τc values are consistent with the molecular masses of the three proteins, and confirm that each is indeed monomeric under these experimental conditions.28
Heteronuclear {1H}-15N NOE values decrease from approximately +0.8 to −4 with increasing mobility of the amide 15N-1HN bond vector on the sub-nsec timescale and thus provide a sensitive measure of the fast internal backbone dynamics of a protein.29 Focusing first on the ETS domain, the heteronuclear NOE values of residues in helices and sheets are uniformly high for all three ETV6 constructs (ETV6R458 average 0.78 ± 0.05), demonstrating that they are well ordered on this fast timescale (Fig. 5). The lower PF’s for helices H2 and H3 relative to helix H1 and strands S1 and S2 thus result from flexibility on a slower timescale, not detectable by this 15N relaxation experiment. In contrast, loop regions, including the “turn” and “wing” show NOE values as low as ~ 0.5, indicative of enhanced fast backbone dynamics. This behavior is expected for these solvent exposed residues of the ETS domain, which also exhibit limited HX protection (Fig. 4) and elevated rms deviations in the structural ensembles of ETV6R458 and ETV6Q436 (Fig. 2).
Fig. 5. Fast nsec-psec timescale mobility of amides in ETS domain and CID are revealed by reduced steady-state heteronuclear {1H}-15N NOE values.
Shown are the relaxation data for (A) ETV6R458, (B) ETV6Q436, and (C) ETV6R426. Decreasing NOE values correspond to increasing mobility of the 15N-1HN bond on a sub-nsec timescale. Missing data points correspond to prolines and amides with weak or overlapping signals. Average errors are ± 5% (magnitude indicated in panel A). The cartoon representation of the secondary structure of ETV6R458 is also given with coloring as in Fig. 2.
Residues forming helices H4 and H5 in the CID of ETV6R458 have average NOE values of 0.64 ± 0.1, indicating that they are generally well ordered, yet exhibit a degree of enhanced sub-nsec mobility relative the ETS domain (Fig. 5A). This provides further support for the hypothesis that flexibility of the CID is a key mechanistic feature of the auto-inhibition of ETV6 DNA-binding. In sharp contrast, residues following helix H5 have significantly reduced NOE values, as well as elevated T2 lifetimes (Supplemental Fig. S5), random coil chemical shifts,17 PF’s ~ 1 (Fig. 4A), and high rms deviations in the structure ensemble of ETV6R458 (Fig. 2), thus confirming that they are conformationally disordered. These data establish Glu440 as the C-terminal boundary of the structured CID, and indeed the 15N-HSQC spectrum of ETV6D446′, with residues Glu447-Arg458 deleted, closely resembles that of ETV6R458 (not shown). Parenthetically, the C-terminus of human ETV6 corresponds to residue 448 in the murine ortholog, suggesting that the non-conserved extension (Thr449 to Glu485) of the latter may be dispensable. Truncation of the CID leads to greater fast-time flexibility of the partial helix H5 in ETV6Q436, as evidenced by reduced amide NOE values (average 0.32 ± 0.17) and longer T2 lifetimes (Fig. 5B and not shown). Helix H4 retains the same backbone order detected with ETV6R458. The increased flexibility of the truncated CID in ETV6Q436 relative to the full CID in ETV6R458 is consistent with the relative levels of auto-inhibition exhibited by these two proteins (Fig. 1B).
15N relaxation dispersion measurements detect conformational fluctuations of the uninhibited ETS domain
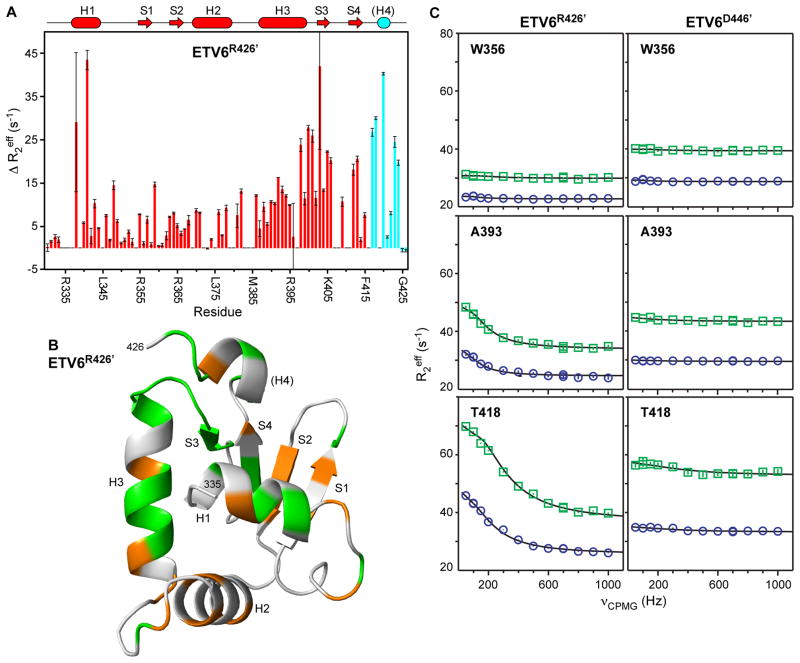
Amide 15N T2 relaxation measurements also reveal msec-μsec timescale motions in the ETS domain that are dampened by the CID. A comparison of the relaxation profiles presented in Supplemental Fig. S5 shows a pattern of reduced T2 lifetimes for residues spanning the C-terminal end of helix H3 to strand S3 in uninhibited ETV6R426, but not inhibited ETV6R458. Such behavior is indicative of conformational dynamics that lead to NMR signal broadening.29 This suggests that part of the DNA-binding interface of the ETV6 ETS domain is also flexible on the msec-μsec timescale, and that the CID dampens this mobility. To examine this further, we undertook relaxation dispersion experiments as a more robust approach for detection of the contributions of conformational exchange on this timescale to the effective decay of the transverse 15N signal.30
Numerous residues in the ETS domain of ETV6R426′ show exchange broadening, as reflected by differences in effective relaxation rates, ΔR2eff, with slow versus fast relaxation dispersion CPMG refocusing pulses. This confirms that the uninhibited ETS domain indeed undergoes conformational fluctuations between states with different amide chemical shifts. As summarized in Figure 6A, the effect is most pronounced for amides in helix H1, the “turn”, and from the end of helix H3 through strand S3. When mapped onto a model of the predicted ETV6R426′ structure (Fig. 6B), amides withΔR2eff > 5 s−1 are distributed broadly throughout the ETS domain with some bias towards residues closest to the DNA-interface. Global fitting the relaxation dispersion profiles for 32 amides in ETV6R426′ to a two-state model yielded an exchange rate constant kex = 820 ± 10 s−1 and a population of 4.0 ± 0.1% for the minor state involved in this conformational equilibrium (Fig. 6C). Importantly, the same residues did not exhibit relaxation dispersion behavior in the inhibited ETV6D446′ (Fig. 6C and Supplemental Fig. S6) or ETV6R458 (not shown). Thus, amide T2 relaxation and relaxation dispersion, as well as HX measurements, demonstrate that the ETV6 ETS domain is dynamic, and that the CID dampens its mobility.
Fig. 6. Amide 15N relaxation dispersion experiments reveal msec-μsec conformational exchange in the ETS domain that is dampened by the CID.
(A) ΔR2eff values for ETV6R426′ from relaxation dispersion experiments with νCPMG of 50 Hz versus 1000 Hz, recorded using an 850 MHz NMR spectrometer. The cartoon representation of the secondary structure of ETV6R426 is also given with coloring as in Figure 2; however, it is unknown if helix H4 remains stably folded in this variant. (B) When mapped on the predicted structure of ETV6R426′, residues with an arbitrary cut-off of ΔR2eff > 5 s−1 (orange) or ΔR2eff > 10 s−1 (green) are broadly distributed through the ETS domain with some bias towards to the DNA-binding interface (centered around helix H3; see Fig. 7). In contrast, such exchange broadening is not seen for ETV6D446′ (Supplemental Fig. S6) or ETV6R458 (not shown) with an intact CID. (C) Complete relaxation dispersion profiles recorded with 500 MHz (blue boxes) and 850 MHz (green circles) spectrometers for uninhibited ETV6R426′ (left) and inhibited ETV6D446′ (right). Both Ala393 (helix H3) and Thr418 (between strand S4 and helix H4) show exchange broadening in ETV6R426′ but not ETV6D446′. Also presented is a control amide in Trp356 (strand S1) showing no dispersion in either protein. Error bars are smaller than the symbols, and the lines are best fits to a two-state exchange model.
Discussion
ETV6 auto-inhibition: steric blockage and reduced flexibility of the ETS domain
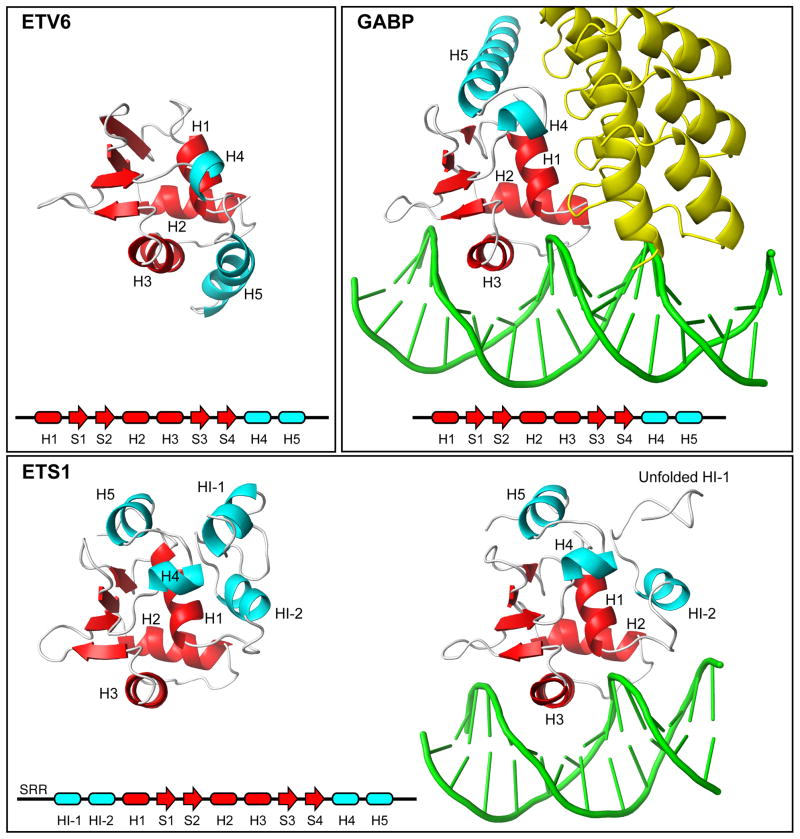
In this study, we dissected the mechanism of ETV6 auto-inhibition by using NMR spectroscopy to characterize the structural and dynamic properties of three ETV6 fragments that recapitulated this phenomenon. The structural ensembles of the inhibited ETV6R458 and ETV6Q436 fragments revealed that the CID contains two helices, H4 and H5, interfaced closely with the ETS domain of this transcriptional repressor. Most notably, helix H5 contacts all three helices of the ETS domain and, thereby, sterically blocks its DNA-binding interface. In contrast to the well characterized structures of the ETS proteins ETS1 and GABP in complex with DNA (Fig. 7), the CID would prevent both base-specific major groove interactions mediated by the recognition helix H3, as well as key phosphodiester contacts involving residues in the H2-H3 “turn” and at the N-terminus of helix H1.23,31
Fig. 7. Appended helices provide specificity to ETS factors.
Shown are the ETS domains (red) and appended helices (cyan) present in free ETV6, the complex of GABPA with GABPB (yellow) and DNA (1AWC.pdb), and ETS1 in its free (1R36.pdb) and DNA-bound (1MDM.pdb) forms. A comparison of these structures shows that the CID helix H5 sterically blocks the DNA-binding interface of ETV6. In contrast, ETS1 contains a distal inhibitory module formed by helices flanking the ETS domain, and helix HI-1 unfolds in an allosteric response to DNA-binding. Multi-site phosphorylation of an unstructured Serine Rich Region (SRR) stabilizes the inhibitory module and thereby increases auto-inhibition.18 In the case of GABP, helix H5 contributes to an interface with the ankyrin repeats of GABPB, and the presence of GABPB indirectly enhances the affinity of GABPA for DNA.42
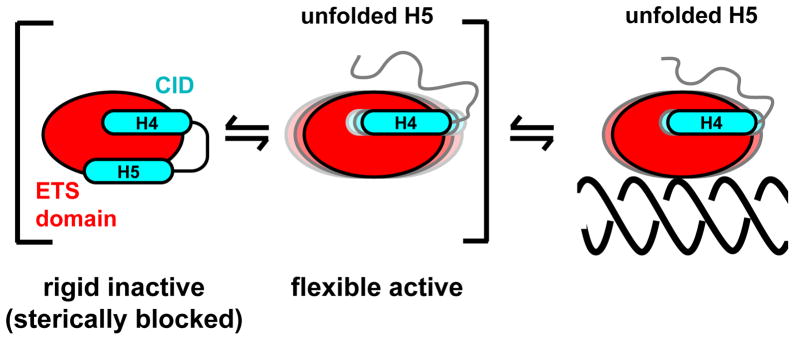
Based on these structural data, DNA-binding by ETV6 must require displacement of helix H5 from its inhibitory position on the ETS domain. Although this could result from a shift of the intact CID to an alternative tertiary conformation, the simplest mechanism would involve the transient unfolding of helix H5, and possibly H4, to expose the ETV6 DNA-binding interface (Fig. 8). Strong support evidence for the latter scenario is provided by two complementary experimental approaches that detect protein dynamics. First, amides within the CID undergo facile amide HX, with PF’s ranging from 2 to 780 for ETV6R458 and < 10 for ETV6Q436. Thus, the inhibitory helices are marginally stable relative to the ETS domain and readily undergo local conformational fluctuations to unfolded, exchange-competent states. Second, although well defined in the structural ensembles of ETV6R458, the CID helices have lower average heteronuclear 15N NOE values than do the secondary structural elements of the ETS domain. This is indicative of enhanced backbone mobility on the nsec-psec timescale. These NOE values are reduced further for the truncated, yet still inhibitory, CID in ETV6Q436. However, msec-μsec timescale conformational exchange was not detected for the CID of ETV6R458 by relaxation dispersion methods, and thus the interconversion rates to the unfolded, exchange-competent states were not determined. The increased flexibility and decreased stability of the truncated helix H5 in the CID of ETV6Q436 detected by 15N relaxation and HX measurements, respectively, are consistent with the intermediate level of auto-inhibition of this fragment relative to ETV6R458. These NMR-based data are also consistent with previous limited proteolysis studies that revealed a similar inhibited ETV6 fragment (residues Cys334 - Gln436) is susceptible to cleavage at Arg426 17. Furthermore, alanine substitution of Glu431 and Glu434 in helix H5 relieved inhibition and increased the protease sensitivity of this site. Arg426 is located at the N-terminus of helix H5 and, thus, a structural transition such as helical unfolding would likely be required for access to the active site of the enzyme. Collectively, these dynamic measurements indicate that unbound ETV6 exists in a conformational equilibrium between an inhibited state with the folded CID sterically blocking the ETS domain and an uninhibited state with the CID transiently unfolded.
Fig. 8. A dynamic model of ETV6 auto-inhibition.
The CID both sterically blocks the DNA-binding interface of the ETS domain and shifts a conformational equilibrium towards a more rigid, inactive state. Helix H5 (and possibly H4) is either displaced or more likely unfolded (as shown) upon DNA binding. Not included is the LID region, which attenuates the inhibitory effect of the CID by an unknown mechanism, as depicted in Fig. 1A.17
Dynamic studies revealed that, in addition to sterically blocking ETV6, the CID also stabilizes the ETS domain and reduces its flexibility. In the absence of the intact CID, relaxation dispersion measurements uncovered conformational fluctuations for amides throughout the ETS domain of ETV6R426′. The fluctuations occur with kex = 820 ± 10 s−1 and involve a minor, higher energy state of the protein with a population of 4.0 ± 0.1%. Complementary amide HX experiments also demonstrated that the recognition helix H3 in this uninhibited fragment has PF’s of only ~ 100 and thus undergoes substantial local unfolding. These results are in accord with a general observation of functionally relevant flexibility in the DNA-binding domains of transcription factors.21,22,32 In contrast, ETV6R458 did not exhibit detectable exchange broadening and showed increased protection of helix H3 against HX.
Based on our investigation of the structural and dynamic properties of the ETV6 fragments, we propose a mechanistic model of auto-inhibition (Fig. 8). Specifically, ETV6 exists in a conformational equilibrium between a rigid state that is sterically blocked by the folded CID and a more flexible state with the CID unfolded. The latter state has both an exposed DNA-binding interface and the necessary mobility to scan non-specific DNA and to form a high affinity complex with specific target sequences. Aspects of this model will be tested by investigating the structure and dynamics of the ETV6 fragments bound to DNA bearing either ETS consensus motifs or non-specific sequences.
Diverse mechanisms of ETS factor auto-inhibition
Auto-inhibition of DNA-binding is a general phenomenon for ETS family members, yet arises from a range of mechanisms with both common and distinct properties.2 Best characterized in vitro and in vivo is ETS1, for which a dynamic inhibitory module is formed by helices both N-terminal (HI-1 and HI-2) and C-terminal (H4 and H5) to the ETS domain (Fig. 7). In contrast to ETV6, this flanking inhibitory module is distal to the DNA-binding interface, and auto-inhibition results from the allosteric coupling of DNA-binding with a conformational change highlighted by the unfolding of HI-1.19,33,34 Furthermore, a predominantly unstructured serine rich region (SRR) preceding helix HI-1 transiently interacts with the inhibitory module to reinforce auto-inhibition. This reinforcement increases in a graded fashion with progressive multi-site phosphorylation of the SRR by CaMKII, thereby providing “rheostatic” control of ETS1 at the level of promoter recognition.18,20
Although ETV6 and ETS1 differ structurally in terms of having steric versus allosteric inhibitory elements, they also share common underlying dynamic features. In both cases, the inhibitory helices are marginally stable, as evidenced by HX, NMR relaxation, and proteolysis measurements, and thus poised to undergo a facile conformational change.18,19 By thermodynamic linkage, the energetic penalty of this conformational equilibrium reduces the net affinity for DNA. Furthermore, relaxation dispersion and HX measurements revealed that the DNA-binding interfaces of ETV6 and ETS1 are also flexible, and this flexibility is dampened by the inhibitory helices.18, 20 In the case of ETS1, this provides a route for regulation via alternative splicing, post-translational modifications, and protein partnerships.2 As discussed below, we speculate that such regulatory mechanisms may also exist for ETV6.
DNA-binding auto-inhibition has been reported for other ETS factors. For example, members of the PEA3 subfamily have inhibitory sequences flanking their ETS domains.35 However, these appended regulatory elements do not appear to be helical as judged by proline-scanning mutagenesis.36 In the cases of the ESE and TCF subfamilies, inhibition of DNA-binding is mediated in complex manners by sequences distant from their ETS domains.1,37,38 Although the detailed auto-inhibitory mechanisms remain to be elucidated for these ETS factors, we hypothesize each shares a common underlying feature of dynamic inhibitory elements quenching motions of the ETS domain that are necessary for specific DNA-binding.
Appended helices provide biological specificity
All ETS family members are characterized by a conserved ETS domain that mediates binding to very similar target DNA sequences with a core GGA(A/T) motif. This exemplifies a conundrum, often seen with transcription factors, for understanding how such similar proteins can still exhibit diverse biological roles. As reviewed recently, specificity can arise from numerous avenues including structural variations, post-translational modifications, and protein partnerships.2 For example, it has been proposed that the preference of ETV6 for DNA sequences with an adenine following the GGA core arises from the unusual presence of a histidine (His396), rather than a highly conserved tyrosine, along its recognition helix H3.16
One intriguing source for ETS factor specificity is the occurrence of appended dynamic helices on the highly conserved ETS and PNT domains.2 For example, unlike many characterized ETS family members (including FLI1, SPI1, SPDEF, ELF3, ELF5, and ELK1), ETV6 and GAPBA have two C-terminal helices affixed to their ETS domains (Fig. 7). Furthermore, ETS1 has two N-terminal helices and two C-terminal helices that fold as a helical bundle closely interfaced with intervening ETS domain. As discussed above, the appended helices of ETV6 and ETS1 serve to auto-inhibit DNA-binding, yet by distinct steric and allosteric mechanisms, respectively. In contrast to ETV6, helices H4 and H5 of ETS1 are not directly inhibitory, but rather form a scaffold upon which the allosterically-coupled helices HI-1 and HI-2 fold. In the absence of these N-terminal inhibitory helices, ETS1 binds DNA with high affinity and structural studies confirm that helices H4 and H5 remain intact.39 However, deletion of the latter impairs the expression and/or solubility of both ETV6 and ETS1 fragments,40,41 indicating that the C-terminal helices also contribute to the overall folding and stability of their ETS domains. In the case of GAPBA, helices H4 and H5 have not been reported to have an inhibitory role, but rather serve as a docking site for the ankryin repeats of GABPB.42 Unlike other ETS factors, GABP is a heterotetramer with the α-subunit providing the ETS domain and the β-subunit contributing the transactivation domain. GABPB also indirectly slows the dissociation of GABP from DNA by stabilizing GABPA in a high affinity conformation.
In accord with their functional differences, the appended C-terminal helices adopt very different tertiary positions on the ETS domain of ETV6 relative to those of ETS1 and GABPA (Fig. 7). Isolated GABPA has not been characterized structurally and thus it is plausible that helices H4 and H5 are positioned primarily because of their extensive intermolecular interactions with the ankyrin repeats of GABPB.42 However, this is not the case for ETS1, as deletion of helices HI-1 and HI-2 does not perturb the helices H4 and H5, despite their many intramolecular interactions.34, 39 Based on sequence alignments (Supplemental Fig. S1) and modeling studies (Supplemental Fig. S7), we hypothesize that the conformational variation between the three ETS factors arise from several non-conserved amino acid differences in both their ETS domains and appended C-terminal regions. For example, the packing of helix H5 against helices H1, H2, and H3 of ETV6 to sterically block DNA-binding involves Leu336 (preceding H1), Leu400 (following H3), and Leu438 (end of H5). The corresponding residues in ETS1 (Gln336, Lys399, and Pro437) and GABPA (Gln320, Gly384, and Glu422) would likely disfavor such a hydrophobic interface involving a long helix. Also, the buried Glu434 from helix H5 appears to play an important electrostatic or hydrogen bonding role in positioning the CID against the N-terminus of helix H1 in ETV6. The corresponding Leu433 of ETS1 and Val418 of GABPA could not serve such a function. Conversely, the sequence of ETV6 would not likely adopt the fold of ETS1 or GABPA for several reasons. In the case of ETS1, Trp361 (between strands S1 and S2) and Leu433 (following H5) play important roles in forming the appended helical bundle. The corresponding charged residues from ETV6, Lys362 and Glu434, would most certainly disfavor the packing of helix H5 in any comparable position. Similarly, in the case of GABPA, Val418 and Glu422 help dock helix H5 against the ETS domain. The corresponding Glu434 and Leu438 of ETV6 have the reversed charge and hydrophobic properties, and again would disfavor helix H5 from adopting a similar position. In summary, although sharing many conserved ETS domain features, sequence variations between ETV6, ETS1, and GABPA can account for the structural and functional differences between their appended helices, and thus help establish biological diversity.
Auto-inhibition and the regulation of ETV6
Auto-inhibition provides several potential routes for the control of ETV6 function in vivo. Previously, we demonstrated that dimerization of ETV6 via its PNT domain enables cooperative binding to tandem consensus DNA sites with variable spacing and orientation.17 This compensates for the low affinity of the auto-inhibited ETS domain towards a single site and should direct ETV6 to promoters with multiple ETS target sequences, such as those found for the stromelysin-143 and BcL-XL genes.8 We also discovered that the sequences linking the PNT and ETS domains of ETV6 partially relieve the auto-inhibitory effect of the CID (Fig. 1)17. The mechanism by which these presumably flexible LID (“linker inhibitory damper”) sequences mediate this effect remains to be established through further DNA-binding and NMR spectroscopic studies of systematic ETV6 deletion fragments. However, the structural ensembles of ETV6Q436 and ETV6R458 revealed that the N-terminus of helix H1 abuts the inhibitory helix H5. Thus, its is plausible that the LID is spatially adjacent to the CID and could alter its structure, stability, or dynamics, and thereby shift the conformational equilibrium of ETV6 towards a less inhibited state.
By analogy with ETS1, we speculate that the LID and CID integrate regulatory pathways for ETV6. For example, the repressive activity of human ETV6 is reduced in response to cellular stress because phosphorylation of Ser257 by the p38 MAP kinase leads to its nuclear export.44 However, it has also been reported that phosphorylation of Ser213 and Ser257 by the ERK MAP kinase reduces the affinity of ETV6 for DNA.45 Although the mechanism underlying this effect is currently unknown, it is plausible that the modification of these potential LID residues impairs their dampening effect on the CID to enhance auto-inhibition. Adding layers of complexity, sumoylation of Lys11 has also been reported to inhibit the repressive function of ETV6, possibly by impeding DNA-binding.46 Consistent with this observation, an ETV6 isoform formed using Met43 as the initiation codon is strongly repressive. Alternatively, the interactions of ETV6 with several co-repressor complexes including mSin3A, SMRT,9 N-CoR,10,12 and Tip6047,48 have been mapped to the residues between the PNT and ETS domains. These partnerships could conceivably relieve or enhance auto-inhibition by modulating the dampening effect of the LID. Precedence for this regulation of auto-inhibition is seen by the relief of ETS1 auto-inhibition through cooperative interactions with transcription factors including RUNX149 and PAX550 and re-enforcement auto-inhibition by calcium-dependent phosphorylation.18 The structural and dynamic analyses of ETV6 presented herein provide a foundation for investigating these and other potential regulatory pathways mediated through the auto-inhibition of DNA-binding.
Materials and Methods
Protein expression and purification
All murine ETV6 fragments were expressed with an N-terminal His6-affinity tag and thrombin proteolysis site from pET28b+ vectors in E. coli BL21 (λDE3) cells. The samples were purified by conventional Ni+2-affinity chromatography, followed by thrombin cleavage of the affinity tag, and gel filtration chromatography. Gel filtration chromatography was used as a polishing step to remove thrombin and the cleaved histidine tag, and to buffer exchange the NMR sample. Proteins used in EMSA were subjected additionally to anion and cation exchange chromatography prior to gel filtration to ensure purity necessary for accurate protein concentration determination. Detailed procedures are provided as Supplementary Methods. The final proteins are denoted as ETV6R426 (residues Arg335-Arg426), ETV6Q436 (Arg335-Gln436), and ETV6R458 (Arg335-Arg458). At the end of this study, two additional constructs were generated for relaxation dispersion measurements, namely ETV6R426′ (Gly329-Arg426) and ETV6D446′ (Gly329-Asp446) with six additional N-terminal residues included before the start of helix H1. Both have the sole cysteine (Cys334) mutated to serine, and the latter also lacks the disordered C-terminal residues after helix H5. All proteins contain four non-native N-terminal residues (Gly-Ser-His-Met) remaining from the cleavage site.
Electrophoretic mobility shift assays
The equilibrium dissociation constants (KD) of the ETV6 fragments for a 23-bp double stranded duplex containing a consensus ETS binding site (underlined), 5′-TCGACGGCCAAGCCGGAAGTGAGTGCC-3′ (top strand), were measured with EMSAs, as described previously17 except for following details. Binding equilibria and gel electrophoresis were performed at 4 °C and the use of a Biorad mini-gel system enabled loading and running to be executed in 45 min. All protein concentrations were determined by use of calculated molar absorptivities.
NMR experiments
Unless stated otherwise, NMR experiments were performed using Varian Unity 500 MHz and Inova 600 MHz spectrometers and proteins were 0.4–0.6 mM in 90% sample buffer (50 mM phosphate, 200 mM NaCl, pH 5.8) with 10% D2O lock solvent at 30 °C. Spectra were processed and analyzed using NMRPipe51 and Sparky.52 Initial HSQC spectra, 15N relaxation measurements, and hydrogen exchange (HX) experiments were recorded using uniformly 15N labeled samples. Standard multidimensional NMR spectra53 for resonance assignments and NOE distance restraints were obtained using uniformly 13C/15N labeled samples. A long range 15N-HSQC experiment was used to determine the chemical shifts and protonation/tautomeric states of the histidine residues.54 The signals from aromatic nuclei were assigned with 2D 1H-13C correlation experiments.55,56 Simultaneous 3D 1H-15N/13C-1H NOESY-HSQC (aliphatic/aromatic; τmix = 100 ms), and constant time methyl-methyl and amide-methyl 15N/13C-13C-1H NOESY spectra (τmix = 140 ms) were collected for NOE distance restraints.57,58 The chemical shift assignments of ETV6Q436 and ETV6R458 have been deposited in the BioMagResBank under accession codes 17741 and 17742, respectively.
Structure calculations
The structural ensembles of ETV6Q436 and ETV6R458 were determined using the automated iterative ARIA v2.3 assignment protocol59 with the program CNS60 with chemical shift assignments, dihedral angle restraints, and NOESY crosspeaks and distance restraints as input data (Table 1). The dihedral angles were predicted from the 13Cα, 13Cβ, 13C′, 1Hα, and 1HN chemical shifts with TALOS+.61 Interproton NOESY crosspeaks were picked manually and initially assigned either manually or automatically. Additional manual assignments were added throughout the refinement stages. Automated assignments were obtained with chemical shift tolerances set as Proton1 0.03 ppm, Hetero1 0.04 ppm, Proton2 0.03 ppm, and Hetero2 0.04 ppm. For iterations it0–it3 and i4-it8, 60 and 30 structures, respectively, were calculated per cycle, with the 20 lowest energy structures carried over to each subsequent iteration, as well as to a final water refinement step. Default ARIA parameters were used from it0 to it5. The tolerance threshold, violation threshold, and maximum contributions were then adjusted to tighten the criteria for including an assigned peak in the calculations. The parameters were lowered from it6 to it8 as follows: violation tolerance 0.09, 0.07, 0.05 Å; violation threshold 0.5, 0.4, 0.3 Å; maximum contributions 15, 10, 5.62 The refinement steps involved manual inspection of distance violations over 0.5 Å, restraints not used in the calculations, and ambiguous restraints with three or more contributions. Secondary structure boundaries were determined using Promotif63 and Procheck-NMR,64 and figures rendered with PyMol.65 The final structural ensembles for ETV6Q436 and ETV6R458 have been deposited in the Protein Data Bank under accession codes 2LF7.pdb and 2LF8.pdb, respectively.
Amide hydrogen exchange
Slow (minutes-days timescale) amide proton-deuterium exchange rates for ETV6R426, ETV6Q436, and ETV6R458 were measured at 30 °C from a series of sensitivity-enhanced 15N-HSQC spectra66 recorded following rapid transfer of the 15N-labeled proteins through a Sephadex G-25 spin column equilibrated with sample buffer (uncorrected pH* 5.8) prepared in 99% D2O.67 After a deadtime of ~ 8 min, six 15N-HSQC experiments were collected in succession with acquisition times of ~ 8 min per spectra. Subsequent spectra were recorded over the period of 8 days with increasing numbers of transients per t1 increment for improved signal-to-noise ratios. Pseudo-first order rate constants for exchange, kex, were obtained by non-linear least squares fitting (Matlab) of peak intensity, It (scaled according to the number of transients), versus the mid-point time of each spectrum to the equation It = Ioexp(−kext) + I∞, where I∞ accounts for residual protons from the initial H2O buffer.
Rapid (seconds timescale) amide proton-proton exchange rates for 15N-labeled ETV6R426, ETV6Q436, and ETV6R458 in sample buffer adjusted pH 6.4, 7.4 or 8.2 were measured at 30 °C by the CLEANEX-PM method.68,69 For each sample, a series of six spectra with transfer periods ranging from 10 to 60 msec were recorded in 4 hr per spectrum using recycle delays of 1.5 s. Reference spectra were recorded in 2.5 hr using a recycle delay of 12.0 s to ensure complete water relaxation. Pseudo-first order rate constants for chemical exchange, kex, were obtained by non-linear least squares fitting of peak intensities versus transfer time to using a Matlab module provided by W.-Y. Choy (University of Western Ontario). A scaling factor of 0.7 was applied to account for the steady-state water magnetization.
The protection factor (PF) for each amide was determined as the ratio kpred/kex where kpred is the predicted exchange rate constant for an unstructured polypeptide with the sequence of the ETV6 fragment. The kpred values were calculated with the program Sphere70 using poly-DL-alanine reference data corrected for amino acid type, temperature, pH, and isotope effects.71,72 The reported PF of a given amide corresponds to the kex measured most reliably by the proton-deuterium or proton-proton exchange protocols. The comparison of PF values thus assumes an EX2 mechanism with first-order dependence of exchange rate constants on sample pH over the conditions utilized for these studies.24
Amide 15N relaxation
Amide 15N T1, T2, and steady-state heteronuclear NOE experiments were collected for 15N-labeled ETV6R426, ETV6Q436, and ETV6R458 at 30 °C on a Varian Inova 600 MHz spectrometer73. Spectra for the T1 (10, 30, 50, 100, 151, 301, 452, 602, 853 and 1254 msec) and T2 (17, 34, 51, 68, 84, 101, 118, 135, 152 and 169 msec) time series were recorded in a random order. Each set of peak intensities was fit to a single exponential decay with associated errors using the “relaxation peak heights” function of Sparky with the “heights at assigned peak positions only” option.52 The heteronuclear {1H}-15N NOE values were determined by taking the ratios of corresponding peak heights, acquired with and without 3 sec of 1H saturation and a total recycle delay of 5 sec. Global correlation times τc were extracted from these relaxation data using Tensor2.74
Amide 15N relaxation dispersion
Amide relaxation-compensated 15N-CPMG-HSQC spectra were recorded with Bruker Avance III 500 and 850 MHz spectrometers for 15N-labeled ETV6R426′ and ETV6D446′ in 20 mM phosphate, 50 mM NaCl, pH 6.5 at 25 °C.75 Interleaved spectra corresponding to the effective B1 fields, νCPMG (50, 100, 150, 200, 300, 400, 500, 600, 700, 800, 900, and 1000 Hz with repeat points at 100 and 700 Hz for error analysis), were collected in random order with a constant time delay T = 40 msec. The effective field νCPMG = 1/(4τCPMG) with 2τCPMG being the time between the centers of successive refocusing pulses. The spectra were analyzed using the autoFit.tcl script of NMRpipe to obtain values of R2eff = (−1/T)ln(ICPMG/Io) as a function of νCPMG, where ICPMG and Io are peak intensities with and without the CPMG pulse train, respectively. The resulting dispersion curves for 32 residues (Met(−1), Tyr340, Gln343, Ser346, Ser348, Glu357, Ile363, Phe364, Arg365, Asn370, Gly371, Trp376, Asn378, Arg382, Thr386, Lys389, Met390, Ser391, Arg392, Ala393, Arg395, Tyr398, Lys405, Glu406, Gln409, Leu412, Phe413, Lys417, Thr418, Ile422, Met423 and Ser424) were fit globally to obtain populations and the conformational exchange rate constant kex using the Matlab program GUARDD.76
Supplementary Material
Highlights.
Conserved DNA-binding ETS domain of ETV6 is auto-inhibited
Two C-terminal inhibitory helices sterically block the DNA-binding interface
Inhibitory helices are dynamic, marginally stable, and poised to unfold
Inhibitory helices also dampen ETS domain motions
Protein dynamics control DNA-binding and differentiate ETV6 from other ETS factors
Acknowledgments
We thank Desmond Lau for help with DNA-binding assays, Shaheen Shojania for advice with NMRpipe/Sparky, Eric Escobar for assistance with ARIA, Lewis Kay for NMR pulse sequences, and Suzanna Perry of the UBC Proteomic Core Facility for help with MALDI-TOF mass spectrometry. HJC is grateful to I. Bertini, L. Banci and S. Cioffi-Bafoni for early training. This study was funded by the Canadian Cancer Society Research Institute (CCSRI 017308 and 2011–700772 to L.P.M.) and the National Institutes of Health (R01GM38663 to B.J.G. and P50CA42014 to the Huntsman Cancer Institute for use of core facilities). Funding from the Huntsman Cancer Institute/Huntsman Cancer Foundation is also acknowledged. NMR instrument support was provided by the Canadian Institutes for Health Research (CIHR), the Canadian Foundation for Innovation (CFI), the British Columbia Knowledge Development Fund (BCKDF), the UBC Blusson Fund, and the Michael Smith Foundation for Health Research (MSFHR).
Footnotes
Supplementary material related to this article will be provided on-line.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 2.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Ann Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15:162–74. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Buijs A, van Rompaey L, Molijn AC, Davis JN, Vertegaal AC, Potter MD, Adams C, van Baal S, Zwarthoff EC, Roussel MF, Grosveld GC. The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity. Mol Cell Biol. 2000;20:9281–9293. doi: 10.1128/mcb.20.24.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel H, Oury C, Carron C, Duprez E, Laabi Y, Tsapis A, Romana SP, Mauchauffe M, Le Coniat M, Berger R, Ghysdael J, Bernard OA. The TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene. 1997;14:349–357. doi: 10.1038/sj.onc.1200829. [DOI] [PubMed] [Google Scholar]
- 6.Lopez RG, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. J Biol Chem. 1999;274:30132–30138. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 7.Fenrick R, Wang L, Nip J, Amann JM, Rooney RJ, Walker-Daniels J, Crawford HC, Hulboy DL, Kinch MS, Matrisian LM, Hiebert SW. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol Cell Biol. 2000;20:5828–5839. doi: 10.1128/mcb.20.16.5828-5839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irvin BJ, Wood LD, Wang L, Fenrick R, Sansam CG, Packham G, Kinch M, Yang E, Hiebert SW. TEL, a putative tumor suppressor, induces apoptosis and represses transcription of Bcl-XL. J Biol Chem. 2003;278:46378–46386. doi: 10.1074/jbc.M305189200. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti SR, Nucifora G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem Biophys Res Comm. 1999;264:871–877. doi: 10.1006/bbrc.1999.1605. [DOI] [PubMed] [Google Scholar]
- 10.Guidez F, Petrie K, Ford AM, Lu H, Bennett CA, MacGregor A, Hannemann J, Ito Y, Ghysdael J, Greaves M, Wiedemann LM, Zelent A. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood. 2000;96:2557–2561. [PubMed] [Google Scholar]
- 11.Mavrothalassitis G, Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Hiebert SW. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20:3716–3725. doi: 10.1038/sj.onc.1204479. [DOI] [PubMed] [Google Scholar]
- 13.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao F, Song HY, Kim CA, Sawaya MR, Hunter JB, Gingery M, Rebay I, Courey AJ, Bowie JU. Derepression by depolymerization: Structural insights into the regulation of Yan by Mae. Cell. 2004;118:163–173. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Graham TG, Vivekanand P, Cote L, Cetera M, Rebay I. Sterile alpha motif domain-mediated self-association plays an essential role in modulating the activity of the Drosophila ETS family transcriptional repressor Yan. Mol Cell Biol. 2010;30:1158–1170. doi: 10.1128/MCB.01225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green SM, Coyne HJ, III, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem. 2010;285:18496–18504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 19.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 20.Lee GM, Pufall MA, Meeker CA, Kang HS, Graves BJ, McIntosh LP. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J Mol Biol. 2008;382:1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 22.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 23.Grishin AV, Alexeevsky AV, Spirin SA, Karyagina AS. Conserved structural features of ETS domain-DNA complexes. Mol Biol. 2009;43:612–619. [PubMed] [Google Scholar]
- 24.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleica cids. Quart Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 25.Li R, Woodward C. The hydrogen exchange core and protein folding. Protein Sci. 1999;8:1571–1590. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang H, Mao X, Olejniczak ET, Nettesheim DG, Yu L, Meadows RP, Thompson CB, Fesik SW. Solution structure of the Ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994;1:871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- 27.Jia X, Lee LK, Light J, Palmer AG, 3rd, Assa-Munt N. Backbone dynamics of a short PU. 1 ETS domain. J Mol Biol. 1999;292:1083–1093. doi: 10.1006/jmbi.1999.3123. [DOI] [PubMed] [Google Scholar]
- 28.Daragan VA, Mayo KH. Motional model analyses of protein and peptide dynamics using C-13 and N-15 NMR relaxation. Prog Nucl Mag Res Spect. 1997;31:63–105. [Google Scholar]
- 29.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 30.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, McIntosh LP, Graves BJ. Inhibitory module of Ets-1 allosterically regulates DNA binding through a dipole-facilitated phosphate contact. J Biol Chem. 2002;277:2225–2233. doi: 10.1074/jbc.M109430200. [DOI] [PubMed] [Google Scholar]
- 32.Milon A, Campagne S, Gervais V. Nuclear magnetic resonance analysis of protein-DNA interactions. J Royal Soc Interface. 2011;8:1065–1078. doi: 10.1098/rsif.2010.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen JM, Skalicky JJ, Donaldson LW, McIntosh LP, Alber T, Graves BJ. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 34.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem. 2002;277:45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 35.Bojovic BB, Hassell JA. The PEA3 ETS transcription factor comprises multiple domains that regulate transactivation and DNA binding. J Biol Chem. 2001;276:4509–4521. doi: 10.1074/jbc.M005509200. [DOI] [PubMed] [Google Scholar]
- 36.Greenall A, Willingham N, Cheung E, Boam DS, Sharrocks AD. DNA binding by the ETS-domain transcription factor PEA3 is regulated by intramolecular and intermolecular protein. protein interactions. J Biol Chem. 2001;276:16207–16215. doi: 10.1074/jbc.M011582200. [DOI] [PubMed] [Google Scholar]
- 37.Stinson J, Inoue T, Yates P, Clancy A, Norton JD, Sharrocks AD. Regulation of TCF ETS-domain transcription factors by helix-loop-helix motifs. Nucleic Acids Res. 2003;31:4717–4728. doi: 10.1093/nar/gkg689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopp JL, Wilder PJ, Desler M, Kinarsky L, Rizzino A. Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J Biol Chem. 2007;282:3027–3041. doi: 10.1074/jbc.M609907200. [DOI] [PubMed] [Google Scholar]
- 39.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 40.Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994;33:13509–13516. doi: 10.1021/bi00250a001. [DOI] [PubMed] [Google Scholar]
- 41.Werner MH, Clore GM, Fisher CL, Fisher RJ, Trinh L, Shiloach J, Gronenborn AM. Correction of the NMR structure of the ETS1/DNA complex. J Biomol NMR. 1997;10:317–328. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 42.Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABP alpha/beta: An ETS domain ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 43.Wood LD, Irvin BJ, Nucifora G, Luce KS, Hiebert SW. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc Natl Acad Sci USA. 2003;100:3257–3262. doi: 10.1073/pnas.0637114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson CA, Wood LD, Hiebert SW. Cellular stress triggers TEL nuclear export via two genetically separable pathways. J Cell Biochem. 2008;104:488–498. doi: 10.1002/jcb.21637. [DOI] [PubMed] [Google Scholar]
- 45.Maki K, Arai H, Waga K, Sasaki K, Nakamura F, Imai Y, Kurokawa M, Hirai H, Mitani K. Leukemia-related transcription factor TEL is negatively regulated through extracellular signal-regulated kinase-induced phosphorylation. Mol Cell Biol. 2004;24:3227–3237. doi: 10.1128/MCB.24.8.3227-3237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roukens MG, Alloul-Ramdhani M, Vertegaal AC, Anvarian Z, Balog CI, Deelder AM, Hensbergen PJ, Baker DA. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol Cell Biol. 2008;28:2342–2357. doi: 10.1128/MCB.01159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordentoft I, Jorgensen P. The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor. Biochem J. 2003;374:165–173. doi: 10.1042/BJ20030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putnik J, Zhang CD, Archangelo LF, Tizazu B, Bartels S, Kickstein M, Greif PA, Bohlander SK. The interaction of ETV6 (TEL) and TIP60 requires a functional histone acetyltransferase domain in TIP60. Biochim Biophys Acta. 2007;1772:1211–1224. doi: 10.1016/j.bbadis.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol Cell Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitzsimmons D, Lukin K, Lutz R, Garvie CW, Wolberger C, Hagman J. Highly cooperative recruitment of Ets-1 and release of autoinhibition by Pax5. J Mol Biol. 2009;392:452–64. doi: 10.1016/j.jmb.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.Goddard TD, Kneeler DG. Sparky. 3 1999. [Google Scholar]
- 53.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Spect. 1999;34:93–158. [Google Scholar]
- 54.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamazaki T, Formankay JD, Kay LE. 2-Dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- 56.Löhr F, Hansel R, Rogov VV, Dötsch V. Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J Biomol NMR. 2007;37:205–224. doi: 10.1007/s10858-006-9128-4. [DOI] [PubMed] [Google Scholar]
- 57.Pascal SM, Muhandiram DR, Yamazaki T, Formankay JD, Kay LE. Simultaneous acquisition of 15N-Edited and 13C-Edited NOE spectra of proteins dissolved in H2O. J Magn Reson Ser B. 1994;103:197–201. [Google Scholar]
- 58.Zwahlen C, Gardner KH, Sarma SP, Horita DA, Byrd RA, Kay LE. An NMR experiment for measuring methyl-methyl NOEs in 13C-labeled proteins with high resolution. J Am Chem Soc. 1998;120:7617–7625. [Google Scholar]
- 59.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 60.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D Biol Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 62.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 63.Hutchinson EG, Thornton JM. PROMOTIF-a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 65.DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstracts of Papers of the American Chemical Society. 2004;228:U313–U314. [Google Scholar]
- 66.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 67.Connelly GP, McIntosh LP. Characterization of a buried neutral histidine in Bacillus circulans xylanase: internal dynamics and interaction with a bound water molecule. Biochemistry. 1998;37:1810–1818. doi: 10.1021/bi972085v. [DOI] [PubMed] [Google Scholar]
- 68.Hwang TL, Shaka AJ. Multiple-pulse mixing sequences that selectively enhance chemical exchange or cross-relaxation peaks in high-resolution NMR spectra. J Magn Reson. 1998;135:280–287. doi: 10.1006/jmre.1998.1598. [DOI] [PubMed] [Google Scholar]
- 69.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–6. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y-Z. PhD Thesis, Structural Biology and Molecular Biophysics. University of Pennsylvania; PA, USA: 1995. Protein and peptide structure and interactions studied by hydrogen exchange and NMR. [Google Scholar]
- 71.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 73.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 74.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 75.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (μs-ms) time scale dynamics in protein side chains by 15N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 76.Kleckner IR, Foster MP. GUARDD: user-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J Biomol NMR. 2012;52:11–22. doi: 10.1007/s10858-011-9589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR. 15N H/D-SOLEXSY experiment for accurate measurement of amide solvent exchange rates: application to denatured drkN SH3. J Biomol NMR. 2010;46:227–244. doi: 10.1007/s10858-010-9398-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.