Abstract
The aim of this study was to determine the methylation status of the p16INK4a, p14ARF and p15INK4b genes and the subsequent effect of hypermethylation on the expression of these genes in cervical cancer patients. Methylation-specific PCR (MSP) was performed to analyse the methylation status of p16INK4a, p14ARF and p15INK4b genes and was confirmed by sequencing. Reverse transcription PCR (RT-PCR) was carried out to determine changes in the expression of the genes due to hypermethylation. Hypermethylation of the p16INK4a, p14ARF and p15INK4b gene promoters was observed in 36, 8.8 and 11.2%, respectively, of 125 cervical cancer samples from a north Indian population. Methylation of p16INK4a was significantly (P<0.001) associated with the cervical cancer cases. Significant association of p16INK4a hypermethylation with passive smoking and oral contraceptive use was also observed. Methylation of p15INK4b was also found to be significant (P<0.05). Our findings did not reveal any correlation between the promoter methylation of p16INK4a, p14ARF and p15INK4b with factors, including age and human papillomavirus infection. mRNA expression was significantly reduced in patients with a methylated promoter (P<0.001) of p16INK4a compared to patients with an unmethylated promoter. In conclusion, this is the first study demonstrating significant hypermethylation of p15INK4b and p16INK4a genes among cervical cancer patients in a north Indian population, and a significant association of p16INK4a hypermethylation with passive smoking and oral contraceptive use.
Keywords: promoter hypermethylation, methylation-specific PCR, reverse transcription PCR, CpG island, human papillomavirus
Introduction
Cervical cancer is the third most common type of cancer in females, and the seventh most common type overall, with an estimated 529,000 new cases in 2008 (1,2). Epigenetic changes are as equally responsible as genetic changes in the development and progression of various types of cancer, including cervical cancer. These epigenetic changes include DNA methylation and histone deacetylation.
Aberrant promoter hypermethylation of tumor suppressor genes has been shown to be involved in human neoplasia (3). The genomic organization at 9p21 houses two members of the INK4 family of cyclin-dependent kinase inhibitors (CDKIs), p15 and p16, and an unrelated gene p14ARF. Notably, p14ARF utilizes two of the same exons as p16, but is translated in an alternative reading frame. p16 is the most commonly altered gene in human malignancies (4).
p15 is located 25 kb from the p16 gene and its protein shares significant areas of homologous amino acid sequences with p16 (5). p16 is a major target in carcinogenesis, rivaled in frequency only by the p53 tumor suppressor gene (6). The major mechanism of p15 gene inactivation in acute myelogenous leukemia (AML) is methylation of the 5′ promoter region of the gene, which leads to transcription silencing (7). The p15 gene is also aberrantly methylated in several human neoplasms, particularly among hematopoietic malignancies (8).
Hypermethylation of the p16 promoter region has been detected in various types of human cancer (9,10). In their study, Feng et al reported similar promoter methylation patterns in genes from exfoliated cell samples and corresponding biopsy specimens. Furthermore, the frequency of hypermethylation increased statistically significantly with the increasing severity of neoplasia present in the cervical biopsy (11). p16 hypermethylation is an early event in cervical carcinogenesis, but no study has examined its association with risk factors, including passive smoking, oral contraceptive (OC) use and age at first sexual intercourse (AFSI). Additionally, no study has examined the significant hypermethylation of p14 and p15 genes in cervical cancer.
Materials and methods
Sample collection
A total of 125 biopsy samples were collected with informed consent from patients diagnosed with cervical cancer. Ethics approval was provided by the Mohan Dai Oswal Cancer Treatment and Research Foundation, Ludhiana, India. One hundred blood samples from the healthy females (control) were collected in tubes with anticoagulant (heparin). Twenty-five biopsy samples, obtained from females in which hysterectomy had been carried out, but the cervix was normal, were used as controls.
DNA extraction
Cells obtained from tissue biopsies and blood samples were lysed in digestion buffer (10 mM Tris-HCl, pH 8.0, 10 mM EDTA, 150 mM NaCl and 2% SDS) containing proteinase K (0.2 mg/ml). DNA was then purified using the standard phenol-chloroform extraction and ethanol precipitation.
Human papillomavirus (HPV) infection and HPV-16 typing
The HPV consensus primers, MY09 and MY11, were used in the PCR assay for HPV infection (12). A total of100 ng of HPV-16 viral genome cloned into pBR322 was used as the positive control. The HPV-16 pBR322 plasmid DNA was a gift from E.M. DeVilliers of Deutsches Krebsforschungszentrum, Heidelberg, Germany. For typing of HPV-16, the primer sets and the methodology used was performed as previously described (13).
Methylation-specific PCR (MS-PCR)
DNA isolated from the biopsy and blood samples was modified with sodium bisulphite and MS-PCR was carried out using specific primers for methylation and unmethylation for the p16INK4a, p14ARF and p15INK4b genes (14–16). The amplified products were run on a 2% agarose gel.
Bisulfite sequencing
For sequencing, the MS-PCR product was purified using a gel purification kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. The sequencing was carried out by a 3100 ABI sequencer and the DNA sequence was then collected using a chromatogram.
Reverse transcription PCR (RT-PCR)
RNA was isolated from the biopsy and blood samples using TRIzol reagent. An equal amount of RNA was used to synthesize cDNA using the RevertAid first-strand cDNA synthesis kit (Fermentas, Glen Burnie, MD, USA). RT-PCR was carried out to determine the alteration in the level of mRNA expression due to promoter hypermethylation using specific primers (15–17). β-actin was used as the internal control.
Statistical analysis
The association between hypermethylation of the genes and risk of cervical cancer was estimated by computing odds ratios (ORs) and 95% confidence intervals (CI) using the Chi-square test, Fisher’s exact test and multivariate logistic regression analysis, which included several potential confounding variables. The reported OR may be interpreted as age-adjusted estimates of the relative risk of developing cervical cancer with the methylation of studied genes. The amount of mRNA expression was obtained by grading a ratio between the densitometry results of the gene and β-actin. Statistical analysis was performed using SPSS version 11.5 and Epi Info version 3.2. The γ-coefficient was calculated to correlate the hypermethylation of tumor suppressor genes with an increase in the stage of cervical cancer. P<0.05 was considered to indicate a statistically significant difference.
Results
Epidemiological characteristics
The cases and the controls were well-matched with respect to age, gender and residence. The mean age ± SD of the cases and controls was 48.33±10.23 and 46.71±11.85 years, respectively. A total of 50.4% of patients were in the ≤45 years age-group, while 49.6% of patients were in the >45 years age-group (data not shown).
HPV infection and HPV-16 typing
It was observed that 116 out of 125 (92.8%) cervical cancer patients were HPV-positive. Typing was carried out for HPV-16, which demonstrated that out of the HPV-infected patients, 69.8% were HPV-16 (Table I).
Table I.
HPV infection and HPV-16 typing in cervical cancer patients and controls.
| Patients (%) | Control (%) | |
|---|---|---|
| HPV-positive | ||
| (+) ve | 116 (92.8) | 5 (5) |
| (−) ve | 9 (7.2) | 95 (95) |
| HPV-16 typing | ||
| (+) ve | 81 (69.8) | 3 (60) |
| (−) ve | 35 (30.1) | 2 (40) |
HPV, human papillomavirus.
Status of the promoter hypermethylation of p14ARF and INK family
No significant difference in the methylation of p14ARF (P>0.05) between patients and controls was observed, but a statistically significant difference in the methylation of p15INK4b (P<0.05) between patients and controls was observed. Methylation of p15INK4b was found to marginally increase the risk of cervical cancer (OR=1.54; 95% CI=1.20–1.99). Hypermethylation of p16INK4a was observed in 36% of cervical cancer patients (Fig. 1). A statistically significant difference in the methylation of p16INK4a (P<0.001) between patients and controls was observed. The hypermethylation of p16INK4a in cervical cancer patients was associated with an approximately 2.19-fold increase in the risk of cervical cancer (OR=2.19; 95% CI=1.85–2.59) (Table II). No statistically significant correlation was observed between p14ARF, p15INK4b and p16INK4a methylation and HPV infection in cervical cancer patients (P>0.05) (data not shown).
Figure 1.
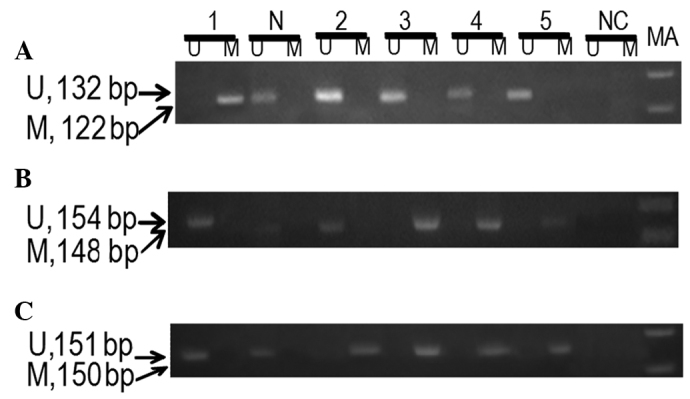
A representative agarose gel demonstrating the MS-PCR products of (A) p14ARF, (B) p15INK4b and (C) p16INK4a. N, sample from normal female (control); Lanes 1–5, samples from cervical cancer patients; MA, marker 100 bp; M, methylated primer; U, unmethylated primer; NC, negative control; MS-PCR, methylation-specific PCR.
Table II.
Frequency of methylation of p14ARF, p15INK4b and p16INK4a with relative risk in cervical cancer patients and controls.
| Methylation | Patient n=125 (%) | Control n=100 (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| p14ARF | 11 (8.8) | 2 (2) | 1.57 (1.21–2.05) | 0.059 |
| p15INK4b | 14 (11.2) | 3 (3) | 1.54 (1.20–1.99) | 0.03 |
| p16INK4a | 45 (36) | 1 (1) | 2.19 (1.85–2.59) | 0.000 |
Methylation of p16INK4a was found to be statistically significant in patients of the ≤45 and >45 years age groups (P<0.05). The risk of cervical cancer was increased 2.15-fold in the ≤45-year age group (OR=2.15; 95% CI=1.69–2.74), and 2.22-fold in the >45-year age group (OR=2.22; 95% CI=1.75–2.81). p16INK4a hypermethylation was found to be statistically significant in the passive smokers (P<0.05). The risk of cervical cancer increased 2.20-fold (OR=2.20; 95% CI=1.39–3.48) and 2.16-fold (OR=2.16; 95% CI=1.67–2.80) in association with p16INK4a methylation in the passive smokers and OC users, respectively (Tables III and IV).
Table III.
Frequency of methylation of p14ARF, p15INK4b and p16INK4a compared to passive smoking in cancer patients and controls.
| Methylation | Patient n=26 (%) | Control n=12 (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| p14ARF | 7 (26.9) | 2 (16.6) | 1.19 (0.77–1.84) | 0.68 |
| p15INK4b | 8 (30.7) | 1 (8.3) | 1.43 (0.99–2.07) | 0.22 |
| p16INK4a | 16 (61.5) | 0 (0) | 2.20 (1.39–3.48) | 0.001 |
Table IV.
Frequency of methylation of p14ARF, p15INK4b and p16INK4a compared to use of oral contraceptives in cancer patients and controls.
| Methylation | Patient n=62 (%) | Control n=43 (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| p14ARF | 4 (6.4) | 1 (2.3) | 1.38 (0.86–2.20) | 0.64 |
| p15INK4b | 9 (14.5) | 3 (6.9) | 1.32 (0.91–1.91) | 0.35 |
| p16INK4a | 28 (45.1) | 1 (2.3) | 2.16 (1.67–2.80) | 0.000 |
A significant association between the methylation of p16INK4a and risk of cervical cancer was observed in patients with AFSI ≤20 and >20 years (P<0.001). In addition, the hypermethylation of p16INK4a was associated with a 2.25-fold increased risk of cervical cancer in patients with AFSI ≤20 years and 2.11-fold in patients with AFSI >20 years.
Bisulfite sequencing
The bisulfite sequencing of samples hypermethylated in the promoter region of the three studied genes revealed a conversion of unmethylated cytosine, but not methylated cytosine (Fig. 2).
Figure 2.
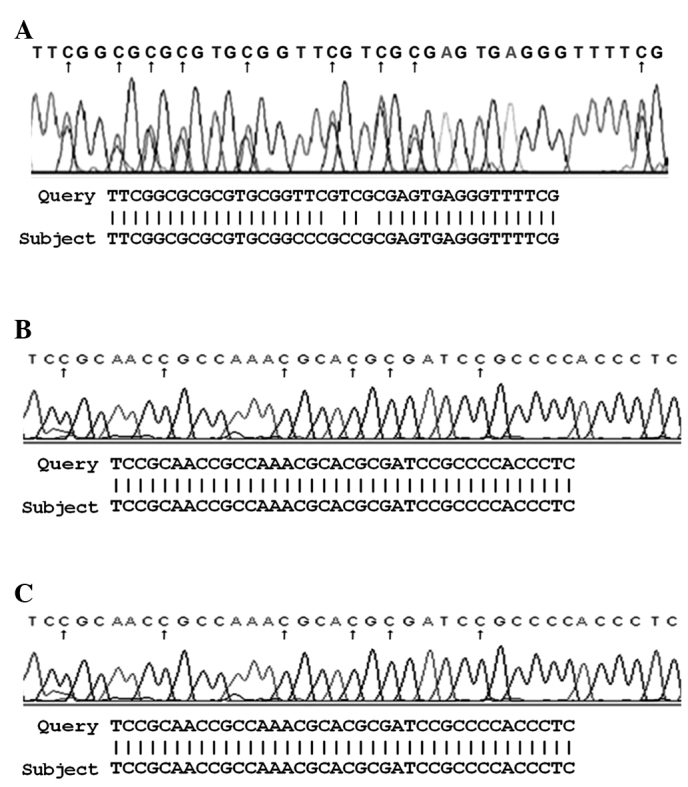
Bisulfite sequence of (A) p14ARF, (B) p15INK4b and (C) p16INK4a genes.
Correlation of methylation status with stage of cervical cancer
There was no increase in the percentage of methylation of p14ARF and p15INK4b with stage (data not shown). By contrast, an increase in the percentage of methylation of p16INK4a was observed with the increase in stage of cervical cancer. The methylation of p16INK4a was found to be higher in the advanced stages. However, to calculate the significance of methylation with the stage of disease more samples should be studied. The γ-coefficient was also not found to be significant for the three genes; p16 (γ=0.200, P=0.115), p14 (γ=0.314, P=0.166) and p15 (0.348, P=0.079).
Semi-quantitative RT-PCR
The mRNA expression of p14ARF and p15INK4b was significantly reduced in patients with a hypermethylated promoter compared to the patients with an unmethylated promoter (Fig. 3). The mRNA expression was significantly reduced in patients with a methylated promoter (P<0.001) of p16INK4a compared to patients with an unmethylated promoter. Downregulation at the transcriptional level was approximately 85.3% due to the hypermethylation of p16INK4a (Table V).
Figure 3.
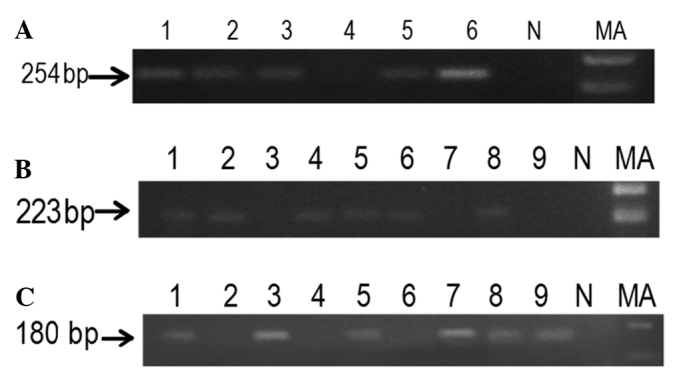
A representative agarose gel demonstrating the RT-PCR products of p14ARF, p15INK4b and p16INK4a. (A) Lanes 1–6 and (B and C) Lanes 1–9, RT-PCR products of methylated and unmethylated samples. MA, marker 100 bp. NC, negative control; RT-PCR, reverse-transcription PCR.
Table V.
mRNA expression in methylated and unmethylated p14ARF, p15INK4b and p16INK4a in cervical cancer patients.
| Gene | Mean (±SE)
|
Downregulated expression of genes (%) | P-value | |
|---|---|---|---|---|
| Unmethylated | Methylated | |||
| p14ARF | 0.858 (±0.02) | 0.594 (±0.03) | 31.0 | 0.00 |
| p15INK4b | 0.795 (±0.02) | 0.634 (±0.01) | 20.2 | 0.03 |
| p16INK4a | 0.940 (±0.03) | 0.118 (±0.03) | 85.3 | 0.000 |
Discussion
Cervical cancer is one of the most common types of cancer that affects female reproductive organs. It is the seventh most common type of cancer overall and the third most common among females (1). In developing countries, cervical cancer is often the most common cancer in females and constitutes up to 25% of all female cancers (2). The risk factors of cervical cancer include HPV infection and cofactors including age, smoking, oral contraceptives, low age at first sexual intercourse, deficient diet and a family history of cervical cancer.
HPV infection is the cause of almost all cases of cervical cancer (18). HPV 16 is the most commonly occurring subtype in cervical neoplasia, however, HPV 18 is associated with more advanced cervical neoplasia.
The incidence of cervical cancer has been reported to increase with age. The results of several case-control and cross-sectional studies indicate that females married to smokers experience a higher risk of cervical neoplasia than females married to non-smokers (19).
Several studies have reported that prolonged use of oral contraceptives increases the risk of cervical cancer. Early AFSI has been associated with an increased risk of high-risk HPV infection, a sexually transmitted infection, that in susceptible females is responsible for virtually all cases of invasive cervical cancer (ICC) (20).
The majority of recent studies have focused on the study of epigenetic changes resulting in many types of neoplasia (21). DNA methylation was the first epigenetic alteration to be observed in cancer cells (22). Recent studies have found that certain genes are hypermethylated in preinvasive lesions, raising the possibility that testing for methylation of these genes may prove to be a useful screening tool (23), particularly in cervical cancer as it evolves through a series of well-defined stages.
Methylation status of the p14ARF gene promoter has been found to be a useful biomarker for pathological and clinical outcome and prognosis of patients with colon, oral squamous cell carcinoma (24) and non-small cell lung cancer. However, of a panel of 16 genes selected for a study in cervical cancer, p14ARF did not demonstrate promoter methylation (23).
Hypermethylation of p14ARF was present in 8.8% of patients, and in the present population it was not found to be significant. Additionally, the risk factors did not have any significant impact on the methylation pattern of p14ARF.
In this study, due to a low frequency of p14ARF and no significant correlation with risk factors, p14ARF does not appear to be a significant biomarker for the diagnosis of cervical cancer.
p15 promoter methylation was approximately 11.1% (5/45) in non-small cell lung cancer (25). However, hypermethylation of p15INK4b has not yet been reported in cervical cancer.
In this study, p15INK4b hypermethylation was observed in 11.2% of patients. p15INK4b methylation was found to be significant in cervical cancer patients in the north Indian population, and its methylation was found to marginally increase the risk of cervical cancer (P<0.05). This is the first study demonstrating significant hypermethylation of this gene in cervical cancer.
A significant trend towards an increase in the risk of cervical cancer was not observed with methylation of the p15INK4b gene in association with risk factors in the patients compared to the healthy controls.
The methylation profile and alteration in transcription suggests that the downregulated expression of p15INK4b by aberrant promoter methylation is an inactivating event in cervical cancer. However, this study requires extension to a larger population size in order to verify whether p15 can be used as a significant or reliable marker in cervical cancer in the north Indian population.
p16 is the most commonly altered gene in human malignancies (4). Hypermethylation of the p16 tumor suppressor gene and its effect on transcriptional downregulation or silencing is one of the major mechanisms of p16INK4a gene inactivation in various types of cancer, including cervical carcinoma (26).
p16INK4a methylation was found to be significant in cervical cancer patients in the north Indian population and its methylation was found to increase the risk of cervical cancer (P<0.001). The present result is consistent with other reports on cervical cancer (23,26) on other population groups.
Previous studies have not identified an association between p16 alterations, including mutation or promoter hypermethylation, and HPV infection (27). In this study, a significant trend towards an increased risk of cervical cancer in association with HPV infection was not observed with methylation of the p16INK4a gene in patient samples compared to healthy controls.
Findings of a recent study showed hypermethylation of the p16INK4a gene promoter to be unchanged according to the patient age (28). Similar observations were also obtained in the present study, suggesting that age is not associated with the increase in the risk of cervical cancer with respect to p16 hypermethylation.
Aberrant p16 methylation has been reported to be associated with active tobacco use in patients with squamous cell carcinoma and high-grade dysplasia (29). Methylation of p16INK4a was found to be statistically significant even in the case of passive smokers, which increased the risk of cervical cancer. It also significantly increased the risk of cervical cancer among OC users (P<0.001). AFSI did not have a significant impact on the methylation of p16INK4a in the present study.
Results of a previous study showed that high-stage cancers exhibited an increased promoter methylation frequency for p16 (22). In the present study, an increasing trend of methylation of p16 was observed with increasing pathological change, confirming p16 as a biomarker.
The results from the present study have shown that downregulation of the mRNA expression of p16INK4a by its aberrant promoter methylation is a significant inactivating event in cervical cancer. This gene may be used as a significant and reliable biomarker in cervical cancer in the north Indian population. Hypermethylation of p16 was also found to be significantly associated with passive smoking and OC use. Hypermethylation of p15 was also observed to be significant although at a low frequency.
Although the present findings require extension to a larger series, this study suggests that the pattern of aberrant methylation in females with or without dysplasia may help identify subgroups at an increased risk of histological progression or cancer development.
Acknowledgements
We acknowledge the financial assistance provided by the Council of Scientific and Industrial Research (CSIR), India, to AKJ.
References
- 1.Globocan. 2008 http://wwwdep.iarc.fr/globocan/database.htm.
- 2.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clinical Cancer Res. 2001;7:584–589. [PubMed] [Google Scholar]
- 4.Hirama T, Koeffer HP. Role of the cyclin dependent kinase inhibitors in the development of cancer. Blood. 1995;86:841–854. [PubMed] [Google Scholar]
- 5.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 6.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 7.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SJ, Endo S, Ichikawa T, Washiyama K, Kumanishi T. Frequent deletion and 5 CpG island methylation of the p16 gene in primary malignant lymphoma of the brain. Cancer Res. 1998;58:1231–1237. [PubMed] [Google Scholar]
- 9.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 10.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 12.D’Costa J, Saranath D, Dedhia P, Sanghvi V, Mehta RA. Detection of HPV-16 genome in human oral cancers and potentially malignant lesions from India. Oral Oncol. 1998;34:413–420. doi: 10.1016/s1368-8375(98)00028-1. [DOI] [PubMed] [Google Scholar]
- 13.Park JS, Dong SM, Kim HS, Lee JY, Jong Um S, Park IS, Kim SJ, Namkoong SE. Detection of p16 gene alteration in cervical cancer using tissue microdissection and LOH study. Cancer Lett. 1999;136:101–108. doi: 10.1016/s0304-3835(98)00366-8. [DOI] [PubMed] [Google Scholar]
- 14.Yeh KT, Chang GA, Lin TH, Wang YF, Tien N, Chang JY, Chen CJ, Shih MC. Epigenetics changes of tumor suppressor genes, p15, p16, VHIL and p53 in oral cancer. Oncol Rep. 2003;10:659–663. [PubMed] [Google Scholar]
- 15.Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14ARF is independent of p16INK4a methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 16.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation associated inactivation indicates a tumor suppressor role for p15INK4b. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 17.Perrone F, Tabano S, Colombo F, Dagrada G, Birindelli S, Gronchi A, Colecchia M, Pierotti MA, Pilotti S. p15INK4b, p14ARF and p16INK4a inactivation in sporadic and neurofibromatosis type 1-related malignant peripheral nerve sheath tumors. Clinical Cancer Research. 2003;9:4132–4138. [PubMed] [Google Scholar]
- 18.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 19.Tay SK, Tay KJ. Passive cigarette smoking is a risk factor for cervical neoplasia. Gynecol Oncol. 2004;93:116–120. doi: 10.1016/j.ygyno.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 20.Louie KS, de Sanjose S, Diaz M, Castellsagué X, Herrero R, Meijer CJ, Shah K, Franceschi S, Muñoz N, Bosch FX International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Early age at first sexual intercourse and early pregnancy are risk factors for cervical cancer in developing countries. Br J Cancer. 2009;100:1191–1197. doi: 10.1038/sj.bjc.6604974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jha AK, Nikbakht M, Parashar G, Shrivastava A, Capalash N, Kaur J. Reversal of hypermethylation and reactivation of the RARβ2 gene by natural compounds in cervical cancer lines. Folia Biol (Praha) 2010;56:195–200. [PubMed] [Google Scholar]
- 22.Jha AK, Kumar S, Nikbakht M, Sharma V, Kaur J. Epigenetics and its role in ageing and cancer. J Med Med Sci. 2011;2:696–713. [Google Scholar]
- 23.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV. Frequent promoter methylation of CDH1, DAPK, RARβ and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishida E, Nakamura M, Ikuta M, Shimada K, Matsuyoshi S, Kirita T, Konishi N. Promotor hypermethylation of p14ARF is a key alteration for progression of oral squamous cell carcinoma. Oral Oncol. 2005;41:614–622. doi: 10.1016/j.oraloncology.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Kurakawa E, Shimamoto T, Utsumi K. Hypermethylation of p16INK4a and p15INK4b gene in non-small cell lung cancer. Int J Oncol. 2001;19:277–281. [PubMed] [Google Scholar]
- 26.Jeong DH, Youm MY, Kim NY, Lee KB, Sung MS, Yoon HK, Kim KT. Promoter methylation of p16, DAPK, CDH1 and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer. 2006;16:1234–1240. doi: 10.1111/j.1525-1438.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi A, Banerjee S, Roy A, Roychowdhury S, Panda CK. Alterations of the p16 gene in uterine cervical carcinoma from Indian patients. Int J Gynecol Cancer. 2003;13:472–479. doi: 10.1046/j.1525-1438.2003.13330.x. [DOI] [PubMed] [Google Scholar]
- 28.Attaleb M, El hamadani W, Khyatti M, Benbacer L, Benchekroun N, Benider A, Amrani M, El Mzibri M. Status of p16INK4a and E-cadherin gene promoter methylation in Moroccan patients with cervical carcinoma. Oncol Res. 2009;184:185–192. doi: 10.3727/096504009790217416. [DOI] [PubMed] [Google Scholar]
- 29.Lea JS, Coleman R, Kurien A, Schorge JO, Miller SD, Minna JD, Muller YC. Aberrant p16 methylation is a biomarker for tobacco exposure in cervical squamous cell carcinogenesis. Am J Obstet Gynecol. 2004;190:674–679. doi: 10.1016/j.ajog.2003.09.036. [DOI] [PubMed] [Google Scholar]


