SUMMARY
Stem and progenitor cells maintain the tissue they reside in for life by regulating the balance between proliferation and differentiation. How this is done is not well understood. Here, we report that the human exosome maintains progenitor cell function. The expression of several subunits of the exosome were found to be enriched in epidermal progenitor cells, which were required to retain proliferative capacity and to prevent premature differentiation. Loss of PM/Scl-75 also known as EXOSC9, a key subunit of the exosome complex, resulted in loss of cells from the progenitor cell compartment, premature differentiation, and loss of epidermal tissue. EXOSC9 promotes self-renewal and prevents premature differentiation by maintaining transcript levels of a transcription factor necessary for epidermal differentiation, GRHL3, at low levels through mRNA degradation. These data demonstrate that control of differentiation specific transcription factors through mRNA degradation is required for progenitor cell maintenance in mammalian tissue.
Keywords: Stem Cell, Differentiation, Epidermis, Skin Differentiation, EXOSC9, PM/SCL-75, EXOSC7, hRrp42, EXOSC10, PM/SCL-100, GRHL3, Exosome, XRN1, mRNA Degradation, mRNA Decay
INTRODUCTION
The epidermis is an ideal system to address how stem and progenitor cells maintain tissue in homeostasis and regeneration. Mammalian epidermis is a stratified epithelium that resides above the dermis separated by the basement membrane zone. The self-renewal capacity of the epidermis is the result of stem and progenitor cells that reside in the basal layer of the epidermis(Sen, 2011). As the stem and progenitor cells differentiate, they withdraw from the cell cycle, detach from the basement membrane and migrate upwards to form the spinous layer and then the granular layer. Terminal differentiation results in the acquisition of the stratum corneum which is necessary for skin barrier function(Blanpain and Fuchs, 2006). Failure to properly regulate self-renewal and differentiation can result in a variety of skin disorders which afflict a large percentage of the population(Segre, 2006). Stem and progenitor cell self-renewal and differentiation can potentially be regulated by both transcriptional and post-transcriptional control of key genes. In the epidermis, p63 has been shown to be necessary for both stem cell self-renewal and differentiation while transcription factors such as ZNF750, KLF4, GRHL3, and CEBP alpha/beta are necessary for differentiation(Lopez et al., 2009; Segre et al., 1999; Sen et al., 2012; Senoo et al., 2007; Ting et al., 2005; Truong et al., 2006). We, and, others have also recently demonstrated that epigenetic factors such as DNMT1 and EZH2, can actively suppress differentiation gene expression in epidermal stem and progenitors cells to allow for self-renewal(Ezhkova et al., 2011; Ezhkova et al., 2009; Sen et al., 2010; Sen et al., 2008). Another potential mechanism for controlling stem and progenitor cell fate decisions is through the control of mRNA degradation by exonucleases
mRNA degradation in cells can occur through either the 5′-3′ or 3′-5′ mRNA decay pathways(Garneau et al., 2007). In the 5′-3′ decay pathway, the 5′ cap is removed by decapping enzymes which allows the mRNA body to be degraded by the 5′-3′ exoribonuclease, XRN1. Alternatively, transcripts can be degraded via the 3′-5′ pathway by the exosome complex(Houseley and Tollervey, 2009). The exosome complex is a large multimeric protein complex consisting of 11 subunits. The human exosome has a cap consisting of three S1/KH-domain subunits (hCsl4/EXOSC1, hRrp4/EXOSC2, and hRrp40/EXOSC3), which rests upon a ring of six RNase PH-domain subunits (hRrp41/EXOSC4, hRrp42/EXOSC7, OIP2/EXOSC8, PM/Scl-75/EXOSC9, hRrp46/EXOSC5, and hMtr3/EXOSC6)(Houseley et al., 2006). Two additional subunits Dis3 and PM/Scl-100/EXOSC10, which are RNase II/R and RNase D homologs respectively, contain the active exonuclease activity necessary for function. Both pathways have been implicated in the degradation of normal as well as abnormal (nonsense-mediated decay) transcripts(Belostotsky, 2009; Garneau et al., 2007). XRN1 has also been shown to degrade antisense non-coding RNAs while the exosome has been implicated in the destabilization of intergenic transcripts such as cryptic unstable transcripts and promoter upstream transcripts(Neil et al., 2009; Preker et al., 2008; van Dijk et al., 2011). In Arabidopsis, mutations in exosome subunits results in developmental defects unique to the subunit targeted(Belostotsky and Sieburth, 2009; Chekanova et al., 2007). In mammalian cells, exosome component, EXOSC10 has been implicated in X inactivation through the regulation of Xist RNA(Ciaudo et al., 2006). Despite these pathways being important for development and RNA regulation it is not known whether either of these pathways has any role in stem cell self-renewal and differentiation in mammals.
Here, we show that several subunits of the exosome complex are highly expressed in epidermal progenitor cells and downregulated upon differentiation consistent with a role in maintaining epidermal progenitor status. Depletion of a critical member of the exosome complex, EXOSC9, blocked self-renewal of progenitor cells residing in the basal layer of the epidermis, which lead to premature differentiation and eventual tissue loss in vivo. Loss of other key subunits, EXOSC7 and EXOSC10, of the exosome complex also resulted in the premature differentiation of progenitor cells. This is in contrast to XRN1 whose loss has no impact on self-renewal of epidermal tissue. Gene expression profiling reveals that EXOSC9 regulates 423 differentiation associated genes in progenitor cells. These effects are largely mediated through EXOSC9 destabilization of GRHL3, a transcription factor necessary for epidermal differentiation. Our results suggest that the exosome mediates progenitor self-renewal and inhibition of differentiation by destabilizing transcription factors such as GRHL3 that would otherwise promote differentiation.
RESULTS
EXOSC9 expression is enriched in progenitor cells
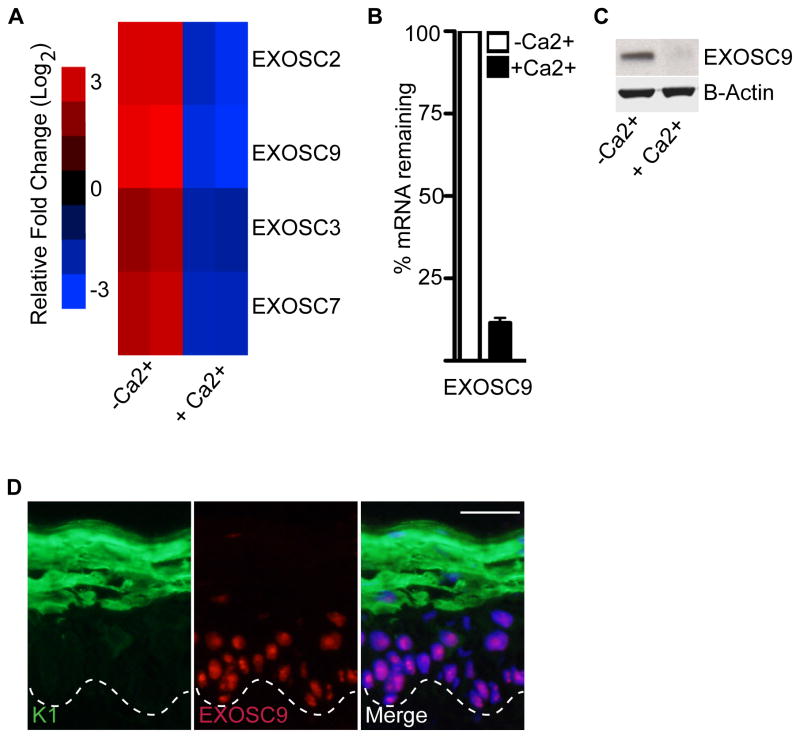
To determine a potential role for the exosome and XRN1 in epidermal self-renewal and differentiation, their expression patterns were analyzed. Consistent with a role for the exosome complex in maintaining the undifferentiated state of progenitor cells, several subunits of the exosome complex including EXOSC2, EXOSC9, EXOSC3, and EXOSC7 are highly enriched in progenitor cells and subsequently downregulated upon calcium induced differentiation (Figure 1A). Upon calcium induced differentiation both transcript and protein levels of exosome subunit, EXOSC9 is downregulated (Figure 1A–C). In adult human epidermis, EXOSC9 expression is enriched in the undifferentiated, basal layer suggesting a potential role in regulating self-renewal of the tissue (Figure 1D). In contrast, there is no change in the transcript levels of XRN1 during epidermal differentiation (Figure S1A).
Figure 1. Exosome component, EXOSC9 is downregulated during epidermal differentiation.
(A) Microarray analysis showing several components of the exosome being downregulated during calcium-induced differentiation (genes repressed, blue; genes induced, red; log2 scale). Heat map with (-Ca2+) denote undifferentiated and (+Ca2+) denote differentiated cells. (B–C) Downregulation of EXOSC9 during differentiation on the mRNA level and protein level. (-Ca2+) denote undifferentiated and (+Ca2+) denote differentiated cells. Error bars=mean with SEM for QRT-PCR data in (B). (D) EXOSC9 staining in adult human epidermis. EXOSC9 staining is shown in red and differentiation protein keratin 1(K1) is shown in green. Scale bar=50μm; dashed lines denote basement membrane zone.
EXOSC9 is necessary to sustain proliferation and prevent premature differentiation of progenitor cells in the basal layer of the epidermis
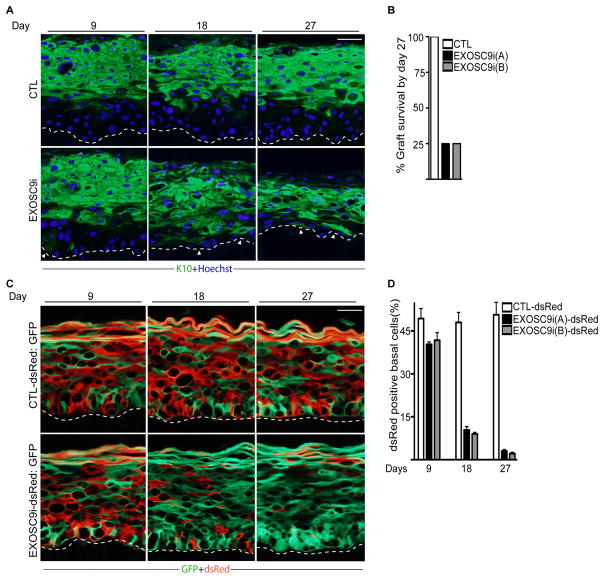
To study whether the 5′-3 or the 3′-5′ mRNA decay pathways have any role in epidermal progenitor function in vivo, XRN1 and EXOSC9 expression were depleted using short hairpin RNA constructs (XRN1i and EXOSC9i) delivered through retroviruses(Figure S1B and Figure S2A–B). EXOSC9i, XRN1i and control knockdown cells were used to regenerate human epidermis on immune-deficient mice, which recapitulates features of intact tissue such as progenitor cell self-renewal and differentiation(Khavari, 2006; Sen et al., 2010). By 18 days post-grafting, EXOSC9 knockdown tissue was hypoplastic with differentiation protein, keratin 10 (K10) expression in the normally undifferentiated basal layer(Figure 2A). By 27 days, only 25% of the EXOSC9i tissue generated had survived as compared to 100% of the controls (Figure 2B). This loss was not due to increased apoptosis but a result of ectopic differentiation of the basal layer (Figure 2A and Figure S2C). The hypoplastic tissue resulting from loss of EXOSC9 expression suggests that the progenitor cells had prematurely differentiated and exited the basal compartment. Consistent with this, EXOSC9 depletion dramatically diminished cell proliferation in the basal layer to less than 5% of control by 27 days post grafting (Figure S2D–E). Cell cycle analysis of EXOSC9i cells also demonstrated a loss of proliferation as the cells withdrew from the S phase and accumulated in G0/G1 (Figure S2F). In contrast, XRN1i tissue had no effects on differentiation, thickness of the tissue, or survivability of the grafts (Figure S1B–D). These results suggest that EXOSC9 but not XRN1 is necessary for promoting self-renewal of basal layer cells.
Figure 2. EXOSC9 loss results in premature differentiation and loss of self-renewal in human epidermal tissue.
(A) Keratinocytes expressing shRNAs for EXOSC9 (EXOSC9i) or control shRNA (CTL) were used to regenerate human epidermis on immune deficient mice. Experiments were performed using two separate shRNAs targeting different regions of EXOSC9i [EXOSC9i(A) and EXOSC9i(B)]. Tissues were harvested at 9, 18, and 27 days post grafting on mice. Keratin 10 (K10) staining shown in green marks differentiated epidermal layers. Hoechst staining in blue marks the nuclei. White arrowheads denote areas of ectopic basal layer differentiation. The dashed lines denote basement membrane zone (Scale bar=40μm; n=4 grafted mice per shRNA construct per timepoint). (B) Graft survival. Knockdown grafts that survived on mice by day 27. (C) Human epidermal progenitor competition assay. Keratinocytes were first transduced with a retrovirus to constitutively express dsRed. dsRed expressing cells were then knocked down for EXOSC9 (EXOSC9i) or control (CTL) and mixed at a 1:1 ratio with GFP expressing keratinocytes. The mixed cells were used to regenerate human epidermis on immune deficient mice. GFP expressing cells are shown in green while dsRed expressing cells are shown in red. Scale bar=40μ n=4 grafted mice per shRNA construct per timepoint. (D) Quantification of dsRed cells in the basal layer. Error bars=mean with SEM. See also Figure S1 and S2.
EXOSC9 maintains self-renewal of progenitor cells through cell autonomous mechanisms
To determine whether EXOSC9 is acting through cell or non-cell autonomous mechanisms we used a modified version of the progenitor cell competition assay we previously developed(Sen et al., 2010). Cells were first transduced with retroviral vectors encoding for dsRed and then knocked down for control or EXOSC9. These cells were mixed with control cells expressing GFP at a 1:1 ratio and used to regenerate human epidermis on immune compromised mice. By 18 days postgrafting, only ~8–10% of EXOSC9i cells expressing dsRed were present in the basal layer (Figure 2C–D). By 27 days, less than 4% of the cells in the basal layer were expressing dsRed with a vast majority of the dsRed positive-EXOSC9i cells in the upper differentiated layers of the epidermis (Figure 2C–D). When control knockdown cells expressing dsRed were mixed with control GFP cells, the ratio of GFP to dsRed expressing cells remained equivalent throughout the entire time course in both the basal layer and the differentiated layers of the epidermis(Figure 2C–D). To rule out off-target RNAi effects, competition assays were performed expressing full length EXOSC9 in EXOSC9i-dsRed cells. Expression of full length EXOSC9 but not LacZ control rescued the loss of EXOSC9i-dsRed cells from the basal layer as well as the rest of the epidermis (Figure S2G–H). This suggests that EXOSC9 maintains self-renewal in a cell autonomous manner.
EXOSC9 controls a gene expression program that maintains proliferation and inhibits differentiation
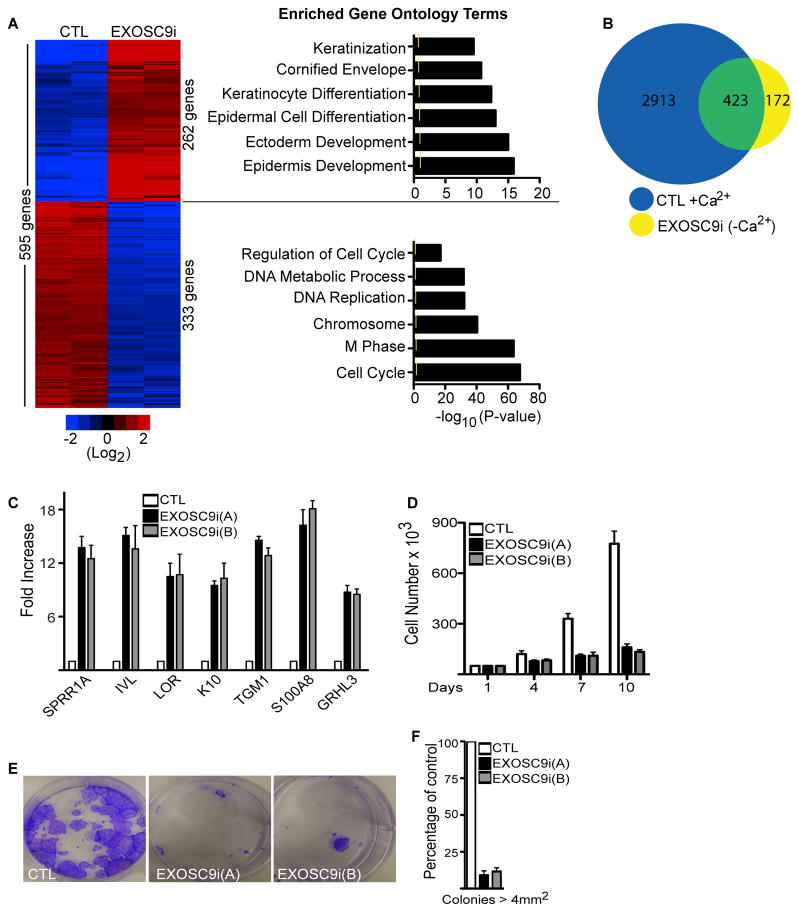
To understand the mechanism by which EXOSC9 maintains progenitor status, global gene expression profiles of EXOSC9 depleted epidermal cells cultured in growth conditions were compared to that of control cells. 595 genes changed significantly in EXOSC9i cells with 262 genes having increased and 333 genes having decreased expression (Figure 3A and Table S1). Genes with increased expression were enriched for gene ontology terms such as keratinization, cornified envelope, and epidermal cell differentiation whereas genes with decreased expression were enriched for proliferation related gene ontology terms (Figure 3A). To determine the extent to which EXOSC9 regulates differentially expressed epidermal differentiation genes, the EXOSC9i regulated genes were compared to our previously generated set of 3,336 genes that change significantly (increase or decrease) during epidermal differentiation(Sen et al., 2010). 423 or 71% of EXOSC9 regulated genes are differentiation regulated (Figure 3B and Table S2). Loss of EXOSC9 resulted in increased expression of structural and enzymatic differentiation genes such as IVL, LOR, and TGM1 (Figure 3C). Loss of EXOSC9 also impaired the proliferation of primary human epidermal cells(Figure 3D). In clonogenic assays, EXOSC9i cells failed to proliferate into larger colonies and only produced small colonies (Figure 3E–F). To determine if other subunits of the exosome are necessary to maintain progenitor function, EXOSC7 and EXOSC10 were also knocked down (Figure S3A–B). EXOSC7 and EXOSC10 loss resulted in an increase in epidermal differentiation gene expression and inhibited proliferation (Figure S3C–E). This suggests that exosome complex proteins are necessary to prevent premature differentiation and sustain proliferation in epidermal progenitor cells.
Figure 3. EXOSC9 is necessary to repress differentiation gene expression and maintain proliferation.
(A) Heat map (left panel) of the 595 genes that change significantly upon EXOSC9 knockdown. Heat map is shown in red (induced genes) and blue (repressed genes) on a log2-based scale. Gene ontology analysis (right panel) of the genes with increased or decreased values upon EXOSC9 knockdown. Yellow mark in bar graphs demark p value=0.5. (B) Overlap of differentiation regulated gene set with EXOSC9i genes. 3,336 genes (blue; CTL+Ca2+) change significantly during epidermal differentiation. These 3,336 genes that change during differentiation were overlapped with the 595 genes that change when EXOSC9 is knocked down in growth conditions (EXOSC9i (–Ca2+)). Shown in the overlap (423 genes:green) are EXOSC9 regulated genes that are also differentiation regulated. (C) QRT-PCR verification of microarray data. Error bars=mean with SEM. (D) Cell proliferation assay. Cells were knocked down for EXOSC9 or control. 50,000 cells were seeded and counted over 10 days (n=3). Error bars=mean with SEM. (E) Clonogenic assay of control shRNA (CTL) and EXOSC9i keratinocytes plated at limiting dilution. (F) Quantification of colonies > 4 mm2 (n=3 per group), error bars=mean with SEM. See also Figure S3, Table S1 and Table S2.
EXOSC9 directly regulates the stability of GRHL3 transcript levels
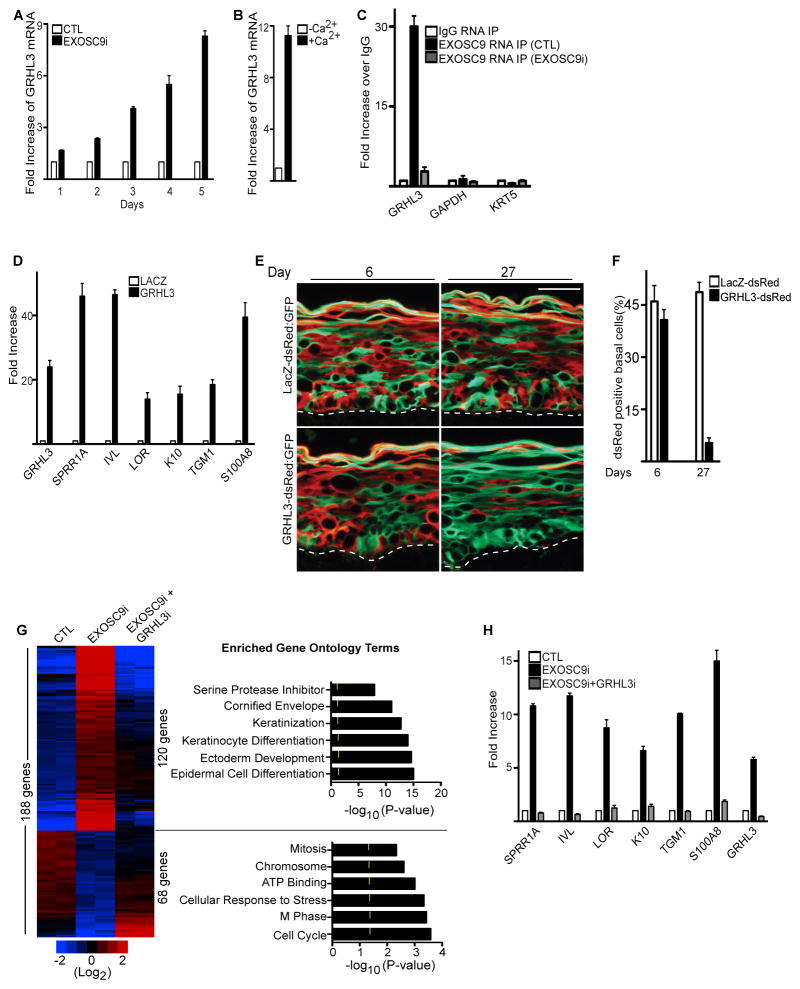
There are 262 genes that increase in expression upon EXOSC9 depletion and 333 genes that decrease. All of these genes are unlikely to be direct targets of the exosome especially the transcripts that decrease in expression. Epidermal differentiation specific structural genes with increased expression upon EXOSC9 knockdown are also unlikely to be direct targets of the exosome complex since these genes are kept silent through repressive epigenetic marks such as H3K27me3 and DNA methylation in progenitor cells(Ezhkova et al., 2011; Ezhkova et al., 2009; Sen et al., 2010; Sen et al., 2008). This suggests that the exosome complex may be targeting genes that have major impacts on proliferation and differentiation such as epigenetic or transcription factors. One possible candidate is GRHL3 whose transcript levels increase upon knockdown of EXOSC9(Figure 3C and Table S1). GRHL3 is a transcription factor whose expression increases during epidermal differentiation and has been shown to be necessary for differentiation(Ting et al., 2005). Although GRHL3 is expressed at low levels in progenitor cells, its chromatin marks with both H3K4me3 and RNA polymerase II binding suggests an actively transcribed gene(Figure S4A–B). To test whether the exosome is necessary to destabilize GRHL3, EXOSC9 was knocked down and the kinetics of GRHL3 stabilization was examined. Within 48 hours post EXOSC9 depletion, GRHL3 mRNA levels double and by 96 hours the levels have increased by ~6 fold similar to levels seen during epidermal differentiation without increased RNA polymerase II binding to the GRHL3 promoter(Figure 4A–B and Figure S4C). To determine if EXOSC9 directly degrades GRHL3, cells were treated with actinomycin D to determine the half-life of GRHL3 in control and EXOSC9i cells. Loss of EXOSC9 increased the half-life of GRHL3 transcript from 92.6 minutes to 170minutes(Figure S4D). During differentiation EXOSC9 is downregulated and the half-life of GRHL3 transcript increases from 92.6 to 152minutes suggesting that the increase of GRHL3 during differentiation is partly due to the increased stability of the transcript (Figure S4D). Notably, knockdown of XRN1 had no impacts on the stability of GRHL3 suggesting that the degradation is specific to EXOSC9 (Figure S1E). To explore if EXOSC9 directly binds GRHL3 transcripts, RNA immunoprecipitations (RIP) were performed using an EXOSC9 antibody on extracts isolated from epidermal progenitor cells. Binding of EXOSC9 to GRHL3 transcripts could be detected in control but not EXOSC9i cells (Figure 4C). Furthermore, GRHL3 bound specifically to EXOSC9 as no binding could be detected for GAPDH or KRT5 which are transcripts whose levels are not effected by EXOSC9 knockdown(Figure 4C). GRHL3 transcripts could also be detected in the immunoprecipitates of EXOSC7 and EXOSC10 suggesting these factors are part of the same complex(Figure S3F–G). These data indicate that GRHL3 mRNA is actively transcribed in progenitor cells but is directly destabilized by exosome components.
Figure 4. EXOSC9 prevents premature differentiation and maintains self-renewal in progenitor cells by regulating GRHL3 transcript levels.
(A) Loss of EXOSC9 results in increased levels of GRHL3. QRT-PCR on GRHL3 mRNA levels on days post EXOSC9 knockdown. (B) GRHL3 expression is increased during epidermal differentiation. QRT-PCR measurements on levels of GRHL3 increase during epidermal differentiation. Error bars=mean with SEM. (C) EXOSC9 binds to GRHL3 but not GAPDH or KRT5 transcripts. RNA immunoprecipitations (RIP) were performed using an antibody against EXOSC9 or mouse IgG on paraformaldehyde crosslinked nuclear extracts isolated from control (CTL) or EXOSC9i cells. QRTPCR was used to determine the levels of binding. Error bars=mean with SEM. (D) Forced expression of GRHL3 in epidermal progenitor cells results in premature differentiation. 4 days post-transduction of cells with a GRHL3 or LacZ encoding retrovirus, cells were harvested and assayed using QRT-PCR. Error bars=mean with SEM. (E) Epidermal progenitor competition assay. dsRed expressing epidermal cells were transduced with a retroviral construct encoding GRHL3 or LacZ. These cells were mixed at a 1:1 ratio with GFP expressing cells and used to regenerate human epidermis. GFP cells =green; dsRed cells= red. The dashed lines denote basement membrane zone (Scale bar=40μm; n=3 grafted mice per construct per timepoint). (F) Quantification of dsRed positive cells in the basal layer. (G) Knockdown of GRHL3 in EXOSC9i cells rescues the differentiation and growth arrest phenotypes of EXOSC9i cells. Heat map (left panel) of control (CTL), EXOSC9i, and EXOSC9i+GRHL3i cells. Red(induced) and blue (repressed), log2-based scale. Gene ontology analysis (right panel) of the genes rescued by double knockdown. Yellow mark in bar graphs demark p value=0.5. (H) GRHL3 is necessary for the differentiation effects of EXOSC9i cells. QRT-PCR verification of array data. Error bars=mean with SEM. See also Figure S4 and Table S3.
EXOSC9 prevents premature differentiation and maintains self-renewal in progenitor cells through the regulation of GRHL3
To determine the impacts of increased levels of GRHL3 on progenitor self-renewal and differentiation, GRHL3 was overexpressed in epidermal progenitor cells delivered by retroviruses. Increased expression of GRHL3 resulted in induction of epidermal differentiation genes similar to levels that was observed with EXOSC9 knockdown (Figure 4D and Figure 3C). Progenitor cell competition assays with GRHL3 overexpression also demonstrate that increased levels of GRHL3 negatively impacts progenitor cell self-renewal and maintenance in the basal layer. By 27 days post-grafting, GRHL3 expressing cells co-transduced with dsRED were depleted from the basal layer of the epidermis with only control GFP expressing cells remaining (Figure 4E–F). In contrast, control LacZ expressing cells co-transduced with dsRED showed equivalent percentages of cells as control GFP expressing cells at all timepoints tested (Figure 4E–F). This suggests that the levels of GRHL3 are critical in determining whether epidermal progenitor cells self-renew or differentiate. To determine if the impacts of EXOSC9 loss on proliferation and differentiation is mediated primarily through increased GRHL3 transcript levels, GRHL3 and EXOSC9 were simultaneously knocked down and compared to EXOSC9i and control cells. Global gene expression analysis show that 31.5% (188/595) of the genes that were increased or decreased in EXOSC9i cells were reverted to control levels upon simultaneous knockdown of GRHL3 and EXOSC9 (Figure 4G and Table S3). 120 of the genes with increased expression upon EXOSC9 knockdown were restored to control levels upon co-knockdown with GRHL3 (Figure 4G and Table S3). These genes were enriched for gene ontology terms such as keratinocyte differentiation, cornified envelope, and included genes such as SPRR1A, IVL, and TGM1(Figure 4G–H). 68 genes with decreased values upon EXOSC9 knockdown were restored to control levels upon co-knockdown with GRHL3 and were enriched for gene ontology terms involved in cell cycle and proliferation (Figure 4G and Table S3). Notably, simultaneous knockdown of GRHL3 with either EXOSC7 or EXOSC10 rescued the premature differentiation phenotype of EXOSC7 or EXOSC10 knockdown cells suggesting that these exosome components are also acting through GRHL3 (Figure S3C–D). To determine if increased levels of GRHL3 were the cause of premature differentiation in the basal layer of EXOSC9 knockdown epidermis, GRHL3 and EXOSC9 were simultaneously knocked down in tissue. Loss of EXOSC9 resulted in premature differentiation of the basal layer as evidenced by the hypoplastic epidermis and expression of K10 in the basal layer (Figure S4E, middle panel). Double knockdown of EXOSC9 and GRHL3 prevented premature differentiation of the basal layer similar to control tissue as seen by the absence of K10 expression in the basal layer (Figure S4E, right panel). The proliferative capacity of the epidermis was also restored upon double EXOSC9 and GRHL3 knockdown as evidenced by the more than 8 fold increase in the number of cells staining for ki67 as compared to EXOSC9 knockdown (Figure S4E–F). This suggests that EXOSC9 sustains proliferation and inhibits differentiation in progenitor cells by controlling the mRNA levels of GRHL3.
DISCUSSION
Here, we report that of the two major mRNA decay pathways, only the 3′-5′ decay pathway is necessary for maintaining the undifferentiated state. Depletion of EXOSC9 resulted in premature differentiation and loss of progenitor cells from the basal layer of the epidermis. Several other subunits of the exosome complex (EXOSC7 and EXOSC10) are also essential for preventing premature differentiation of progenitor cells. The exosome maintains the progenitor state by directly binding to and degrading GRHL3 transcripts. During differentiation, several subunits of the exosome are downregulated, which results in the stabilization of GRHL3 transcripts allowing for differentiation gene expression. Furthermore, preventing the increase in GRHL3 levels in EXOSC9 knockdown cells can rescue EXOSC9 regulated genes and tissue phenotype suggesting that maintaining GRHL3 at low levels is critical for preventing differentiation. This suggests that EXOSC9 serves to keep differentiation specific transcription factors at low levels to promote self-renewal in progenitor cells.
The exosome can function in both the cytoplasm and nucleus. The cytoplasmic exosome can interact with RNA binding proteins and degrade transcripts from the nonsense mediated decay pathway as well as AU rich transcripts in the cytoplasm(Houseley et al., 2006). GRHL3 does not have canonical AU rich sites in its 3′UTR making it unlikely to be targeted to the exosome through AU rich binding proteins. The nuclear exosome has been shown in S. cerevisiae to be necessary for the degradation of cryptic unstable transcripts(Neil et al., 2009). In Drosophila, the nuclear exosome associates with chromatin at sites of active transcription to degrade pre-mRNAs as soon as they are transcribed(Andrulis et al., 2002). Our data suggests that the nuclear exosome is responsible for the degradation of GRHL3 since EXOSC9 is localized to the nucleus in the epidermis.
Although our data suggests that the nuclear exosome is targeting GRHL3 transcripts for degradation, it is unclear how the exosome specifically recognizes its target. In Arabidopsis, it has been shown that mutations in different subunits of the exosome give rise to unique developmental defects. The different subunits also regulated distinct but also overlapping RNAs(Belostotsky and Sieburth, 2009; Chekanova et al., 2007). In Drosophila, microarray analysis of cells depleted for different subunits of the exosome also showed that each subunit regulated their own distinct set of RNAs(Kiss and Andrulis, 2010). This type of substrate specificity may potentially occur in the context of the entire exosome or in the context of smaller subcomplexes. These data suggest that the function of certain subunits of the exosome is to potentially recognize distinct RNA substrates which then allow EXOSC10 and DIS3, which contain the exonuclease activity, to degrade the target RNA. Our data is not inconsistent with this model. EXOSC9 and EXOSC7 may directly target GRHL3 transcripts whether through the structure of the mRNA or through sequence recognition and thus allow degradation of the transcript through EXOSC10. In line with this, EXOSC9 and EXOSC7 expression is downregulated during differentiation, which would thus prevent further targeting of GRHL3 by the exosome and lead to an increase in its expression. A majority of the exosome subunits are not downregulated during differentiation. Notably, EXOSC10 and DIS3 which contain the exoribonuclease activity are not downregulated during differentiation which may allow other functions of the exosome such as processing of sn/snoRNAs and pre-rRNAs to be maintained(Callahan and Butler, 2008; Schilders et al., 2007; Tomecki et al., 2010). However, we cannot exclude the possibility that processing of certain sn/snoRNAs that may be specifically targeted by differentially expressed exosome components such as EXOSC9 will be lost upon differentiation.
The exosome may also recognize GRHL3 transcripts through interactions with adaptor proteins. In yeast, Mtr4p is necessary to activate the function of the nuclear exosome while Rrp47p binds to RNA with certain structures and Mpp6p binds to poly(U) RNA(de la Cruz et al., 1998; Milligan et al., 2008; Mitchell et al., 2003). Furthermore, Sen1p, Nrd1p, and Nab3p along with RNA polymerase II associate with the nuclear exosome to degrade snoRNAs and cryptic unstable transcripts(Vasiljeva and Buratowski, 2006). Human homologues of these yeast adaptor proteins have also been identified which include hMTR4(Mtr4p), MPP6(Mpp6p), and C1D(Rrp47p)(Schilders et al., 2005; Schilders et al., 2007). In addition to these factors, ZCCHC8 and RBM7 were recently identified as part of the human MTR4 complex that target the nuclear exosome to promoter upstream transcripts(Lubas et al., 2011). It will be interesting to determine whether any of these proteins are involved in recruiting the exosome to GRHL3 to promote self-renewal in progenitor cells.
In conclusion, we describe a mechanism for maintaining cells undifferentiated through the rapid degradation of differentiation specific transcription factors by the exosome.
EXPERIMENTAL PROCEDURES
The Supplemental Experimental Procedures section which includes a list of retroviral constructs, siRNAs, antibodies, and primer sequences is available online.
Tissue culture
Primary human keratinocytes were derived from fresh foreskin. Cells were grown in KSF-M (GIBCO-BRL) supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE). Cells were induced to differentiate by the addition of 1.2 mM calcium for 3 days in full confluence. Amphotrophic phoenix cells were maintained in DMEM and 10% fetal bovine serum.
Gene transfer
Amphotrophic phoenix cells were transfected with 3 ug of each retroviral construct to either knockdown or overexpress genes. Transfections were done in 6 well plates using Fugene 6 (Roche). Viral supernatants were collected 48 hours post transfection and polybrene added (5ug/ml). These supernatants were placed on primary human keratinocytes and centrifuged for 1 hour at 1000rpm. Cells were transduced a total of 2 times and selected using puromycin (2ug/ml) after the last transduction.
Gene knockdown and overexpression
ShRNA retroviral constructs were generated by cloning oligos into the pSuper Retro vector. Retroviral constructs expressing GFP or dsRed (LZRS-GFP or LZRS-dsRED) were a generous gift from the Khavari laboratory. The full-length open reading frame of GRHL3 was cloned into the LZRS retroviral vector.
Western blotting and immunofluorescence
50 ug of the cell lysates were used for immunoblotting. For immunofluorescence experiments, 7 μm thick epidermal sections from adult human skin or xenografts were used.
Grafts of regenerated human skin onto immune deficient mice
For xenografts, 1 million genetically modified keratinocytes were seeded onto devitalized human dermis for 48 hours and then grafted onto female 10–12 week old CB17 scid/scid mice for up to 27 days as previously described(Sen et al., 2010; Sen et al., 2008).
In vivo epidermal progenitor cell tracking system
Marked epidermal cells were generated by transducing the cells with a dsRed expressing retroviral construct. Cells were then transduced with retroviral constructs to knockdown EXOSC9, overexpress GRHL3 or control. These modified cells were mixed at a 1:1 ratio with GFP expressing cells and grafted onto immune deficient mice to regenerate human epidermis. The number of dsRed expressing cells in the basal layer was counted and divided by the total number of cells in the basal layer to generate the percentage of dsRed positive basal layer cells.
Quantitative reverse transcriptase-PCR analysis
Total RNA from cells was extracted using the RNeasy mini kit. One ug of total RNA was reverse transcribed and quantitative PCR was performed. Samples were run in triplicate and normalized to GAPDH.
Gene expression profiling
For gene expression profiling, cultured primary human keratinocytes were knocked down for EXOSC9, EXOSC9 and GRHL3, or control. RNA was harvested from the cells 5 days after knockdown. Microarray analysis using Affymetrix HG-U133 2.0 plus arrays was performed on duplicate samples. Significantly changed genes were identified as previously described(Sen et al., 2010).
Apoptosis assays
Epidermal tissue was stained for apoptosis using the In Situ Cell Death Detection kit (Roche: 12156792910) according to manufacturers protocol.
Clonogenic assays
For clonogenic assays with primary human keratinocytes, 100 cells were plated onto mitomycin C treated (15ug/ml) 3T3 feeder cells in a 30 mm Petri dish. Colonies were stained with crystal violet 3 weeks after plating.
Chromatin Immunoprecipitation
ChIP was performed as previously described(Sen et al., 2008). 5 million cells were used for ChIP for each antibody used. 3ug of antibody was used for each pulldown experiment. Experiments were performed in triplicates. Results were represented as a percent of input DNA.
RNA Immunoprecipitation
5 million cells were used for RNA IP for each antibody used. 3ug of antibody was used for each pulldown experiment. RNA IP was performed using the RNA IP kit (53024) from Active Motif according to manufacturer’s protocol.
Measurement of the stability and half-life of GRHL3 mRNA
Cells were treated with actinomycin D (10ug/ml) for 0, 0.5, 1.5, 2.5, and 4 hours to determine the half-life of GRHL3 mRNA. RNA was isolated from the samples and QRT-PCR was used to determine the levels of GRHL3. Half-life(Leclerc et al., 2002) was calculated using the formula T1/2= 0.3t/log(D1/D2).
Cell cycle analysis
Cell cycle analysis was performed by analyzing DNA content using flow cytometry. Cells were fixed in ice-cold 70% ethanol and then stained with PI solution (50ug/ml propidium iodide, 3.8mM NaCitrate, 150ug/ml RNase A in PBS) for 30 minutes at room temperature. Cells were then analyzed using a flow cytometer.
Supplementary Material
Highlights.
EXOSC9 expression is found in progenitor cells and decreases upon differentiation.
EXOSC9 is necessary to sustain proliferation in progenitor cells.
EXOSC9 prevents premature differentiation of progenitor cells in the basal layer.
EXOSC9 controls a gene expression program that maintains the progenitor state.
EXOSC9 does this by controlling the mRNA stability of GRHL3.
EXOSC7 and EXOSC10 are also necessary for maintaining progenitor status.
Acknowledgments
We thank Y. Wang for pre-submission review and helpful discussions. We thank D. Webster and P. Khavari for the LZRS GFP and LZRS dsRED phoenix cells. This work is supported by a K01AR057828–04 and Ray Thomas Edwards Award to G.L.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Belostotsky D. Exosome complex and pervasive transcription in eukaryotic genomes. Curr Opin Cell Biol. 2009;21:352–358. doi: 10.1016/j.ceb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol. 2009;12:96–102. doi: 10.1016/j.pbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2006;2:e94. doi: 10.1371/journal.pgen.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Khavari PA. Modelling cancer in human skin tissue. Nat Rev Cancer. 2006;6:270–280. doi: 10.1038/nrc1838. [DOI] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED. Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA. 2010;16:781–791. doi: 10.1261/rna.1906710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2:1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, Kurz E, Paramio JM, Nerlov C. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Schilders G, Raijmakers R, Raats JM, Pruijn GJ. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen GL. Remembering one’s identity: the epigenetic basis of stem cell fate decisions. FASEB J. 2011;25:2123–2128. doi: 10.1096/fj.11-182774. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.