Abstract
Many incurable mitochondrial disorders result from mutant mitochondrial DNA (mtDNA) and impaired respiration. Leigh's syndrome (LS) is a fatal neurodegenerative disorder of infants, and Leber's hereditary optic neuropathy (LHON) causes blindness in young adults. Treatment of LHON and LS cells harboring G11778A and T8993G mutant mtDNA, respectively, by >90%, with healthy donor mtDNA complexed with recombinant human mitochondrial transcription factor A (rhTFAM), improved mitochondrial respiration by ∼1.2-fold in LHON cells and restored >50% ATP synthase function in LS cells. Mitochondrial replication, transcription, and translation of key respiratory genes and proteins were increased in the short term. Increased NRF1, TFAMB1, and TFAMA expression alluded to the activation of mitochondrial biogenesis as a mechanism for improving mitochondrial respiration. These results represent the development of a therapeutic approach for LHON and LS patients in the near future.
Iyer and colleagues use healthy mitochondrial DNA (mtDNA) complexed with a recombinant human mitochondrial transcription factor A protein as a platform for gene delivery to mitochondria. Using this approach, they are able to introduce healthy mtDNA into cytoplasmic hybrid (cybrid) cells containing platelets from a human subject with Leber's hereditary optic neuropathy and, subsequently, into primary skin fibroblasts from a human subject with Leigh's syndrome.
Introduction
Human mitochondrial DNA (mtDNA) disorders typically affect energy-intensive tissues and are clinically complex and often fatal. These disorders represent a large group of diseases with heterogeneous clinical and pathological expressions characterized by improper functions of and sometimes irreversible damage to specialized neuronal or cardiac cells or populations. The causes and mechanisms of neuronal or cardiac cell death and related defects in many of these disorders, although not fully understood, derive from mutations in mtDNA or decline in energy levels. Clinical severity can be influenced by the percentage of abnormal versus normal mtDNA genomes present in affected cells (heteroplasmy).
Mitochondrial disorders such as Leigh's syndrome (LS) and Leber's hereditary optic neuropathy (LHON) result from a high abundance of abnormal mtDNA leading to insufficient respiration (Leigh, 1951; Loeffen et al., 2000). Symptoms of LS usually include developmental, cardiac, respiratory, and muscle impairments and progress rapidly, because genetic mutations in mtDNA result in a chronic lack of energy in these cells (Loeffen et al., 2000). These mtDNA mutations influence the severity of the symptoms of the disease based on the replication and transcription of mutant mtDNA species (Loeffen et al., 2000). The 8993T>G mutation in the mtDNA-encoded ATP synthase 6 (ATPase 6) gene, when present in low abundance, results in NARP (neurogenic muscle weakness, ataxia, and retinitis pigmentosa) disease, and the individuals survive to adulthood. When present in very high abundance, LS results, leading to rapid lethality in infants (Holt et al., 1990; Shoffner et al., 1992; Tatuch et al., 1992).
LHON is a more common mitochondrial optic disorder that can be caused by mutations in mtDNA (Wallace et al., 1988). Symptoms of LHON usually include developmental, ocular, and respiratory impairments and progress because genetic mutations in mtDNA and/or nuclear DNA result in chronic lack of energy in these cells, leading to optic nerve degeneration (Yen et al., 2006). These mtDNA mutations influence the severity of the symptoms of the disease based on the abundance of mutant mtDNA transcripts (DiMauro and Schon, 2008). The most prevalent point mutation 11778 G>A occurs in the mtDNA-encoded ND4 gene (Wallace et al., 1988). When present in very high abundance, LHON results, leading to blindness in young adults (Yen et al., 2006). There is currently no specific treatment available for any of these disorders.
Although some of the basics of mtDNA replication, transcription, and transmission of the abnormal mtDNA are known, much remains to be discovered. Abnormalities of mtDNA replication and transcription (such as production of deleted species) or translation [due to mutations in tRNA or coding electron transfer system (ETS) genes] are responsible for illnesses that are present in childhood or adulthood. These “mitochondrial” diseases can display variable and overlapping phenotypes, and understanding their genotype–phenotype relationships remains a great challenge. Further insights into understanding how mitochondrial genomes replicate, transcribe, and transmit mtDNA to daughter cells will greatly aid in the development and refinement of current therapies for mitochondrial diseases. Current mitochondrial gene-therapy approaches used to date include ballistic introduction of mtDNA and cytosolic expression of mitochondrial-targeted restriction enzyme, an individual mitochondrial gene, or mitochondrial tRNA. These approaches are limited by technical challenges and poor efficiency of transformation (Vazquez-Memije et al., 1996; Chinnery et al., 1999; Manfredi et al., 2002).
In our earlier studies, we proposed the development of a novel approach that used a recombinant mitochondrial transcription factor A protein (rhTFAM) for external manipulation of the mitochondrial genome present inside cells (Khan and Bennett, 2004). In later studies, our results showed that rhTFAM improved respiration and mitochondrial gene expression of key genes by stimulating mitochondrial biogenesis in vitro and in vivo (Iyer et al., 2009b; Keeney et al., 2009; Thomas et al., 2011). In a more recent study, we also used this approach to introduce, replicate, and transcribe pathogenic mtDNA in human pluripotent stem cell–derived neural progenitor cells, while maintaining multipotency and successful differentiation into neuronal lineage in the short term (Iyer et al., 2011). Thus, we demonstrated and validated the applicability of the technology to facilitate a novel therapy for mitochondrial-derived dysfunction in the context of neuromitochondrial diseases (Iyer et al., 2011).
In this study, we used healthy mtDNA complexed with rhTFAM to transduce into the mitochondria two classic mitochondrial diseases, as cell models for proof-of-principle studies toward conducting mitochondrial gene therapy in the future. We introduced healthy mtDNA first into the cytoplasmic hybrid (cybrid) cells containing platelets from an LHON patient and, subsequently, into primary skin fibroblasts obtained from an LS patient. This study represents our first attempt at using healthy donor mtDNA circles complexed with rhTFAM to improve respiration and biogenesis in LS and LHON disease cell lines caused by different pathogenic mtDNA point mutations at near-homoplasmy.
Materials and Methods
Cell culture
SH-SY5Y cells carrying a G11778A mutation in the mtDNA Complex I ND4 gene that is responsible for the most common variant of LHON was used in the study. These cells were a kind gift from Dr. Russell Swerdlow and carried the mutation in high abundance. Fibroblasts from an 8-month-old Asian male LS patient carrying the 8993T>G mutation were obtained from Coriell Cell Repositories Camden, NJ) (GM13411-cell line identifier). All cells were passaged in culture using standard conditions described earlier (Iyer et al., 2009b).
Restriction enzyme digestion analysis
A PCR product in the ATPase 6 gene spanning the SmaI site caused by the 8993T>G mutation was amplified from genomic DNA of LS cells from three independent treatment groups and digested with SmaI to check for the presence or absence of the mutation. The digested genomic DNA PCR products were analyzed using an automated electrophoresis system (Experion, Bio-Rad, Hercules, CA). Similarly, a PCR product in the ND4 gene spanning the G11778A mutation that removes a normally present SfaN1 restriction site was digested with SfaN1 and resolved the digestion products with a DNA chip-automated electrophoresis system.
Protein production
rhTFAM was obtained from Gencia Biotechnology (Charlottesville, VA) and produced as described earlier (Iyer et al., 2009b).
Human mtDNA preparation
All circular human mtDNA used in this study was obtained from Roche human genomic DNA and prepared and assayed by treating it with limiting amounts of ATP-dependent DNase as described earlier (Keeney et al., 2009) with slight modification, where it was further purified by RNase treatment to yield a clean circular ∼16-kb product when separated on a 0.8% agarose gel.
Mitochondrial protein transduction
LS and LHON cells were grown to ∼70% confluency, and 4 μg of freshly prepared human mtDNA circle was mixed with rhTFAM as described previously (Iyer et al., 2009b; Keeney et al., 2009). Cells were incubated with this protein–DNA mixture for ∼5 hr. The mtDNA+rhTFAM solution was removed; cells were placed in normal growth medium and cultured further prior to experimental analysis. Similarly, two independent T25 flasks were treated with the same amount of rhTFAM alone or PBS buffer as control (CTL) for LS cells, as had been conducted for LHON cells earlier (Iyer et al., 2009b).
Imaging of circular human mtDNA entry into LHON cybrid cells
Entry of circular human mtDNA into LHON cybrid cells was monitored using confocal microscopy. The LHON cells were propagated to ∼80% confluence on 35-mm glass-bottom dishes. The mitochondria in LHON cells were stained using MitoFluor Green (MitoSciences, Eugene, OR), whereas human mtDNA was labeled using Cy5 dye according to the manufacturer's instruction (Mirus, Madison, WI). The labeled circular human mtDNA was complexed with unlabeled TFAM, mixed in 2 ml of Dulbecco's modified Eagle's medium, and added to independent dishes of LHON cells with labeled mitochondria. Real-time single-plane time-lapse images were obtained every 5 min using an Olympus IX70 confocal microscope.
Mitochondrial respiration
Treated LS and LHON cells were harvested with trypsin; 2–5×106 cells/ml were assayed in culture medium using the OROBOROS Oxygraph-2k respirometer (Hütter et al., 2006). Respiratory oxygen flux was measured in real time and expressed as picomoles of O2 per second per 106 cells. The coupling control protocol started with ROUTINE respiration in the physiological coupling state R, as controlled by cellular energy demand, energy turnover, and the degree of coupling to phosphorylation of the intact cells. Then 2 μg/ml oligomycin was added to inhibit ATP synthase, followed by incremental additions to a maximum of 100 μM uncoupler (FCCP) and sequential inhibition of Complex I with 0.1 μM rotenone and of Complex III with 2.5 μM antimycin A/0.5 μM myxothiazole (Iyer et al., 2009b; Keeney et al., 2009). Aliquots of cells were saved for further analysis of mtDNA copy number, gene expression of mRNA, and respiratory protein levels. Analysis of variance was conducted between the three treatment groups. Paired Student's t tests were applied to the data, and p<0.05 was considered to be significantly different across samples.
Real-time PCR analysis
Total genomic DNA and RNA samples isolated from three different treatment groups from LS and LHON cells were analyzed by real-time quantitative PCR (qPCR) as described previously (Iyer et al., 2009b). In brief, 10 ng of genomic DNA from each treatment group was used for measuring the copy numbers of the mtDNA in LS and LHON cells. Individual copy numbers of ND2, COX3, and ND4 mitochondrial genes and gene expression of the cDNAs of the mitochondrial (ND2, COX3, and ND4) and nuclear (NRF-1, TFAMB1, PGC1-α, and TFAM-1a) transcripts were assayed in a multiplex qPCR assay (Iyer et al., 2009b). Combined levels of 18S, β-actin, and GAPDH were used as a normalized geometric mean. Data were obtained from three biological replicates from the three treatment groups over four time points for LHON disease and from two biological replicate samples at two time points from the three treatment groups for LS disease (due to the paucity of the fibroblast sample). Statistical comparisons were conducted where possible using paired Student's t test and one-way ANOVA to determine significant difference in gene expression values between treatments.
Western-blot analysis
Aliquots (100 μg) of total cell protein obtained from different treatment groups and different diseases were separated on 4–12% Bis-Tris Criterion precast gels, transferred, and immunoblotted for Complex I subunits or Complex I–V subunits as described previously (Iyer et al., 2009b). β-Actin was used as a loading control. The band intensity was quantified using the Odyssey infrared imaging system (LI-COR, Lincoln, NE).
Results
Entry of circular human mtDNA complexed with TFAM into cells
Human skin fibroblast cells were obtained from an infant who died of LS by 8 months and displayed poor respiration, neurodevelopmental delay, and sudden cardiac arrest (Holt et al., 1990; Shoffner et al., 1992). Quantitative analysis of SmaI-treated ATPase 6 PCR fragments from LS genomic DNA samples revealed >95% of 8993T>G mutant mtDNA in this cell line, with a slight decrease up to ∼94% over 2 weeks upon addition of healthy mtDNA, compared with mtDNA present in normal healthy fibroblast cells (data not shown). Similar quantitative analysis of SfaN1-treated ND4 PCR fragments of LHON genomic DNA samples revealed up to 98% of 11778G>T mutant mtDNA and was unable to detect any significant change in mutant DNA copies over time (data not shown). Although our methodology was sensitive to detecting the presence of wild-type or mutant mtDNA containing the SfaN1 site or SmaI site in LHON and LS cells, respectively, we were unable to find evidence of significant dilution by introduction of the donor mtDNA within the time course of these initial experiments.
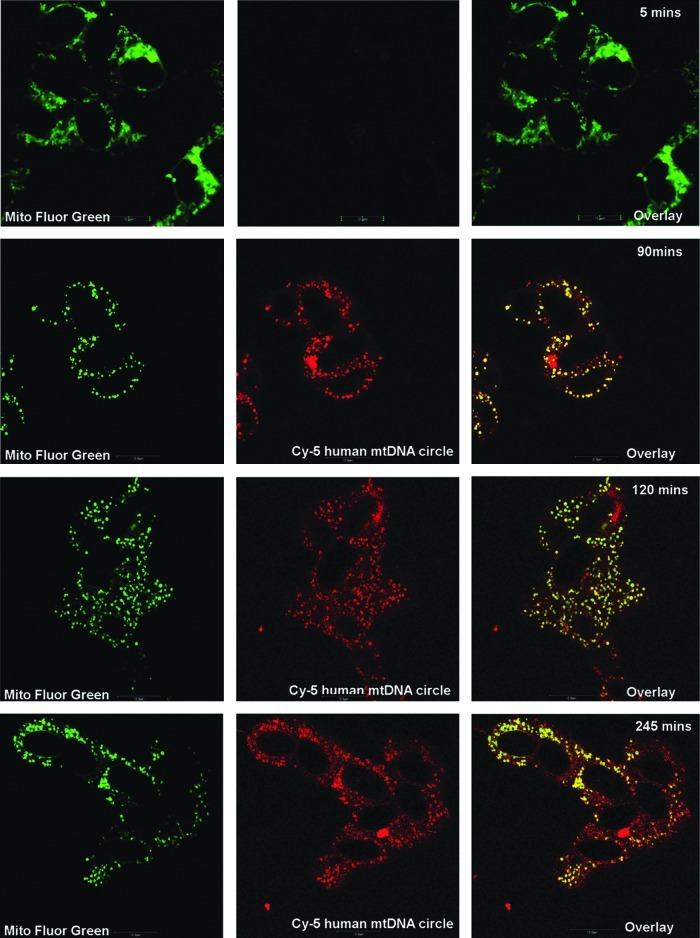
We next fluorescently labeled healthy mtDNA with Cy5 and complexed it with unlabeled TFAM. This mixture was added to LHON cybrid cells, as described in Materials and Methods and in earlier studies (Iyer et al., 2009b, 2011; Keeney et al., 2009). Live cell confocal images over several minutes up to 4.5 hr revealed a gradual entry of mtDNA in LHON cells. These results demonstrate successful entry of the donor mtDNA and are consistent with previous results of the formation of a larger DNA–protein complex, as 100% entry was achieved within half an hour and persisted up to 6 hr (Fig. 1).
FIG. 1.
Time-lapse confocal images to detect entry of donor mtDNA complexed with rhTFAM into LHON cybrid cells. Entry and colocalization (yellow overlay) of Cy5-human LHON mtDNA (red) complexed with rhTFAM was observed after 4 hr of incubation. Mitochondria within the cybrids have been stained with MitoFluor Green. Scale bar=10 μm.
We treated the LS and LHON cells with healthy donor mtDNA in complex with rhTFAM along with their respective CTLs, as described in our earlier studies (Iyer et al., 2009b; Keeney et al., 2009). Then we analyzed the effects of the treatment of two consecutive independent experiments on two passages after 3 weeks for LS disease and three consecutive independent experiments over four passages after 3 weeks for LHON disease cells. The reason for such an experimental design was due to the paucity of patient sample fibroblasts; therefore, the results obtained with LS fibroblast cells are more along the lines of a case study. In contrast, the ample availability of sample material enabled us to conduct extensive analysis using LHON cybrid samples.
Mitochondrial gene therapy improved respiration in LHON cells
The intact cells from the different treatment groups were analyzed using high-resolution respirometry with glucose as energy substrate in culture medium (Hütter et al., 2006; Iyer et al., 2009b). Approximately 8×106 cells were added to each of the two Oxygraph-2k chambers and tested for the effect of treatment on ROUTINE respiration (R; no additions), LEAK respiration (L; oligomycin, inhibiting ATP synthesis), and ETS capacity (E; maximum noncoupled respiration at optimum uncoupler concentration) in LHON cells (Gnaiger, 2008; Iyer et al., 2009b; Pesta and Gnaiger, 2012). The difference (R – L) represents net respiration coupled to ATP synthesis. Addition of rotenone blocks Complex I, and thus prevents formation of succinate (functional inhibition of Complex II). This is followed by inhibition of Complex III by antimycin A/myxothiazole, revealing residual oxygen consumption (ROX), which is largely nonmitochondrial respiration (typically 10–20% of ETS capacity).
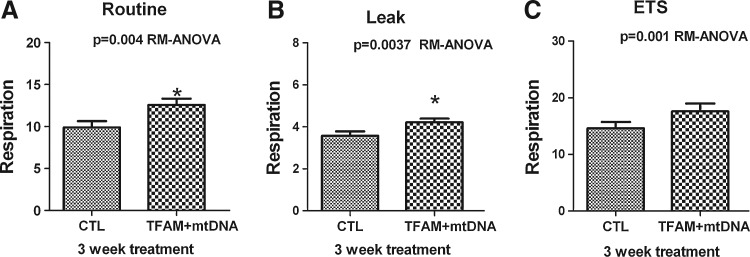
LHON cybrid cells treated with mtDNA in complex with TFAM showed increased ROUTINE respiration by ∼1.2-fold more than CTL (Fig. 2A, Table 1). Subsequently, LEAK respiration and ETS capacity were also increased over those of CTL cells (Fig. 2B and C, Table 1). The increases of R and L respiration were highly significant (p<0.05), and one-way ANOVA showed significant changes of R, L, and E with mtDNA+rhTFAM treatment (p=0.004, p=0.0037, p=0.001) over 3 weeks in LHON cells. Normalization of respiration to ETS capacity yields coupling control ratios that eliminate differences across experiments arising from cell numbers, cell sizes, or cellular mitochondrial densities. Table 1 shows that treatment with normal mtDNA+rhTFAM into diseased cybrid cells modestly increased normalized ROUTINE respiration (R/E) and LEAK respiration (L/E) due to some uncoupling, which was compensated by an increase in ROUTINE respiration levels, leading to a near constant (R – L)/E ratio over 3 weeks.
FIG. 2.
Mitochondrial gene therapy increases respiration in LHON cells. Three weeks after exposure to mtDNA complexed with rhTFAM, LHON cybrid cells showed increases in (A) ROUTINE by ∼1.2-fold, (B) LEAK by ∼1.3-fold, and noncoupled (C) ETS capacity by ∼1.2-fold compared with CTL cells (left column). Respiration is expressed as pmol of O2/(sec/106 cells).
Table 1.
Calculated Coupling Control Ratios on ROUTINE (R), LEAK (L), and Noncoupled Respiration (E)
| |
Respiration |
Coupling CTL ratios |
||||
|---|---|---|---|---|---|---|
| Cells | ROUTINE | LEAK | ETS | R/E | L/E | (R – L)/E |
| LHON CTL, 3 wk | 9.91 | 4.16 | 13.93 | 0.711 | 0.298 | 0.413 |
| LHON TFAM+mtDNA, 3 wk | 12.62 | 5.04 | 16.13 | 0.782 | 0.312 | 0.470 |
The quantitative effects of three independent treatments with mtDNA+TFAM of LHON samples (n=11) over 3 weeks are shown with respect to untreated CTL cells.
Mitochondrial gene therapy increased mitochondrial copy number and gene expression in LHON cells
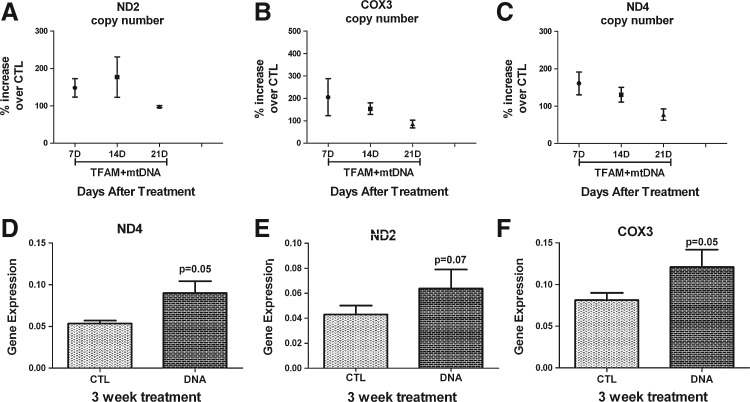
We used real-time PCR assays to determine alterations in mitochondrial gene copy numbers from genomic DNA obtained between treated (mtDNA complexed with TFAM) and untreated LS and LHON cells over 3 weeks (Iyer et al., 2009b). The mtDNA replication copy numbers for LHON cells showed a reversible change, with the peak increase around the first week and a gradual decline over 3 weeks, as shown in Fig. 3A–C. The values for each of the three mitochondrial genes (ND2, ND4, and COX3) over 3 weeks are shown as a percentage of increase over buffer CTL for LHON cells. Similarly, cDNA obtained by reverse transcription of mRNA of the three mitochondrial genes (ND2, ND4, and COX3) between treated and untreated LHON samples showed an increase (approximately twofold) in gene expression for ND4 (p=0.05), ND2 (p=0.07), and COX3 (p=0.05) over 3 weeks with mtDNA+rhTFAM treatment (Fig. 3D–F). The results (n=11) have been normalized to 18S rRNA for copy-number values and to the geometric mean of 18S, GAPDH, and β-actin for gene-expression studies.
FIG. 3.
Mitochondrial gene therapy increases mtDNA copy numbers and gene expression in LHON cells. (A–C) Real-time qPCR analysis of candidate mitochondrial genes (ND2, COX3, ND4) showed a reversible increase in mtDNA copy numbers over CTL on treatment with mtDNA complexed with rhTFAM over 3 weeks (n=11). These results were not statistically significant. (D–F) RNA from the different mitochondrial genes was reverse-transcribed to cDNA and assayed by real-time qPCR. Shown are normalized mean values of significant increases in gene expression for ND4 (p<0.05; an increase by ∼1.8-fold) and COX3 (p<0.05; an increase by ∼1.7-fold) and a not so significant increase in gene expression for ND2 (p<0.07; an increase by ∼1.5-fold) over CTL for each gene comparison.
Mitochondrial gene therapy increased nuclear respiratory protein levels in LHON cells
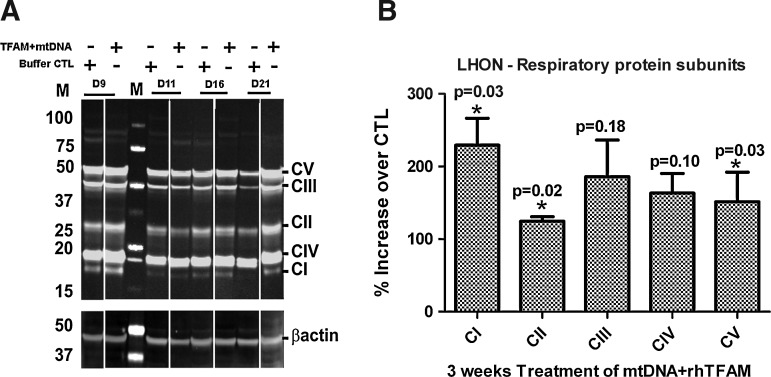
In the third experimental series, we also examined the treatment groups of LHON samples to assess the levels of multiple individual ETS proteins with western blots and assembly of ETS macrocomplexes with immunohistochemistry using antibodies directed against mtDNA-encoded catalytic subunits of Complexes I and IV, compared with that of an antibody against a nuclear genome-encoded component of Complex V (ATP synthase) as a marker for general mitochondrial distribution. Western blot experiments (Fig. 4A) revealed that the relative mitochondrial mass in cells, expressed as a ratio of the outer mitochondrial membrane protein mitofilin to that of cytosolic β-actin, increased by ∼1.5-fold in the cells exposed to mtDNA complexed with TFAM, indicative of an increase in the mitochondrial biogenesis program. The levels of multiple ETS proteins from several Complexes (I, II, and V) increased significantly (p<0.05; Fig. 4B) in the mtDNA+rhTFAM–treated cells by 3 weeks (Fig. 4B). Confocal microscopy did not reveal any effects of exposure of LHON cells complexed with mtDNA on the proportions of cells (95–100%) with intact ETS Complex I or Complex IV macro-assemblies (data not shown).
FIG. 4.
Mitochondrial gene therapy increases nuclear protein levels in LHON cells. Cells were passed, pellets were extracted, and total cellular protein was separated by SDS-PAGE. (A) The left panel is an immunoblot for members of ETS Complexes I–V and the outer membrane protein mitofilin using MitoSciences monoclonal antibodies. (B) The right panel shows ratios (n=11) of all protein bands normalized to β-actin loading control densities and expressed as percentage of buffer CTL cells for each time point. One-way ANOVA showed p<0.05 for Complexes I, II, and V and p>0.05 for Complexes III and IV for each comparison.
Mitochondrial gene therapy triggered the expression of the biogenesis pathway over 3 weeks in LHON cells
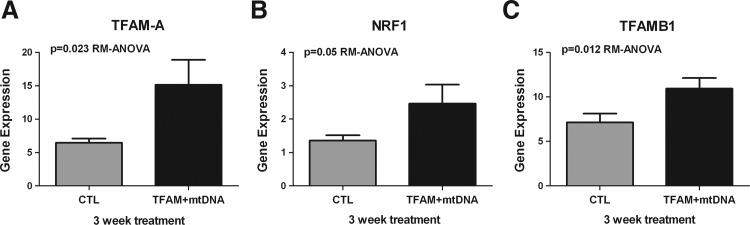
Our earlier studies demonstrated that mitochondrial gene therapy triggered an increase in the mitochondrial biogenesis pathway, especially of PGC-1α expression in cybrid cells (Keeney et al., 2009). Therefore, we hypothesized the possibility that mtDNA complexed with rhTFAM would increase downstream transcription targets of the mitochondrial biogenesis pathway. By real-time PCR analysis of the RNA samples from the individual treatments, we analyzed the expression of the cDNAs for nuclear respirator factor-1 (NRF1) gene, a master regulator of the nuclear genes responsible for the mitochondrial respiratory function, and TFAMB1 gene, a mitochondrial transcription factor that activated mitochondrial transcription in complex with TFAM1 (Scarpulla, 2008a,b). Treatment of LHON cells with mtDNA complexed with rhTFAM led to significant increases in gene expression of NRF1 (p=0.05, RM-ANOVA), TFAM1 (p=0.023, RM-ANOVA), and TFAMB1 (p=0.012, RM-ANOVA) after 3 weeks of treatment, as shown in Fig. 5 for LHON cells. We also observed an increase in PGC-1α expression over CTL, which was not significant in LHON cells (data not shown).
FIG. 5.
Mitochondrial gene therapy increases mitochondrial biogenesis in LHON cells. RNA from the different candidate biogenesis genes was reverse-transcribed to cDNA and assayed by real-time qPCR. Shown are geometric mean normalized cDNA values of mtDNA+rhTFAM treatment indicating significant increases in gene expression over CTL. (A) Increase in cDNA expression of TFAM1A by ∼1.5-fold over CTL. One-way ANOVA showed p<0.05. (B) Increase in cDNA expression of NRF1 by ∼1.6-fold over CTL. One-way ANOVA showed p=0.05. (C) Increase in cDNA expression of TFAMB1 by∼1.7 fold over CTL. One way ANOVA showed p<0.05. All CTLs are shown in gray and individual genes in black for each comparison.
Mitochondrial gene therapy improved respiration and restored partial ATP synthase capacity in LS cells
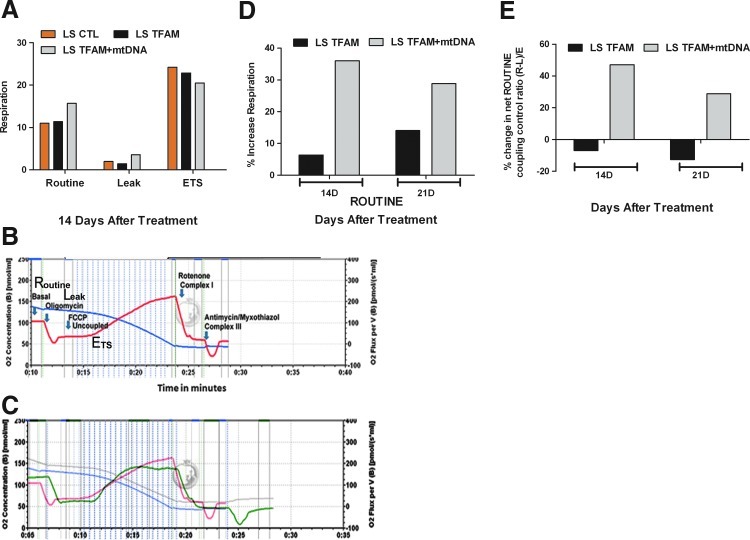
The positive response of mitochondrial gene-therapy treatment on LHON cybrid cells in this study provided the impetus to treat primary patient fibroblast cells from LS disease with healthy mtDNA complexed with rhTFAM. As the patient had died of respiratory and cardiac failure, we conducted our first experiment to analyze the effect of mitochondrial gene therapy on mitochondrial respiration. Approximately 8×106 intact cells were added to each of the two respiration chambers and tested for the effect of treatment on ROUTINE, LEAK, and ETS respiration in LS and cells over 3 weeks (Gnaiger, 2008; Iyer et al., 2009b; Pesta and Gnaiger, 2012). Treatment of LS fibroblasts with donor mtDNA+rhTFAM increased ROUTINE respiration compared with buffer CTL or treatment with rhTFAM alone (Fig. 6D and E, Table 2) over 14 days and 21 days. Treatment with mtDNA+rhTFAM increased (R – L)/E by ∼50% relative to CTL, the fraction of ROUTINE respiration sensitive to inhibition by oligomycin, and thus coupled to ATP synthase activity. This effect was not observed in LS cells treated with rhTFAM alone (Fig. 6E). An experimental tracing of overall respiration components and quantitative analysis are shown in Fig. 6A–C and Table 2 for LS fibroblast cells. Normalization of respiration to ETS capacity yielded coupling CTL ratios shown in Table 2. We observed that treatment with mtDNA+rhTFAM increased normalized ROUTINE respiration (R/E), LEAK respiration (L/E), and net coupled respiration [(R – L)/E] (Pesta and Gnaiger, 2012) for LS cells after 2 weeks.
FIG. 6.
Mitochondrial gene therapy increases respiration in LS cells. (A) Two weeks after exposure to mtDNA+rhTFAM (gray) or rhTFAM only (black), LS cells showed increased respiration, increased sensitivity to oligomycin, and lowered uncoupled respiration compared with untreated CTL (beige). Respiration is expressed as pmol of O2/(sec/106 cells). (B) A recording of the different aspects of the respiration profile of LS (CTL) cells after 2 weeks. (C) An overlay of the respiration recording of LS cells treated with mtDNA+rhTFAM (green) over CTL (red). (D) Two to 3 weeks after exposure to mtDNA+rhTFAM (gray), LS cells showed increased ROUTINE respiration compared with rhTFAM only (black). Respiration is expressed as pmol of O2/(sec/106 cells). (E) mtDNA+rhTFAM treatment but not rhTFAM alone increased oligomycin-sensitive LEAK respiration after 2 and 3 weeks.
Table 2.
Effects of Single Treatments with rhTFAM or rhTFAM+mtDNA
| |
Respiration |
Coupling CTL Ratios |
||||
|---|---|---|---|---|---|---|
| Cells | ROUTINE | LEAK | ETS | R/E | L/E | (R – L)/E |
| LS CTL, 2 wk | 11.01 | 1.97 | 24.20 | 0.45 | 0.08 | 0.37 |
| LS TFAM, 2 wk | 11.35 | 1.46 | 22.86 | 0.50 | 0.06 | 0.43 |
| LS TFAM+mtDNA, 2 wk | 15.70 | 3.60 | 20.51 | 0.77 | 0.18 | 0.59 |
The effects of single treatments with rhTFAM or rhTFAM+mtDNA on ROUTINE respiration (R), respiration with oligomycin (LEAK, L), and respiration with oligomycin and uncoupled with FCCP (ETS, E) were determined. The coupling CTL ratios were calculated as indicated.
Mitochondrial gene therapy showed an increased trend in mitochondrial copy number and gene expression in LS cells
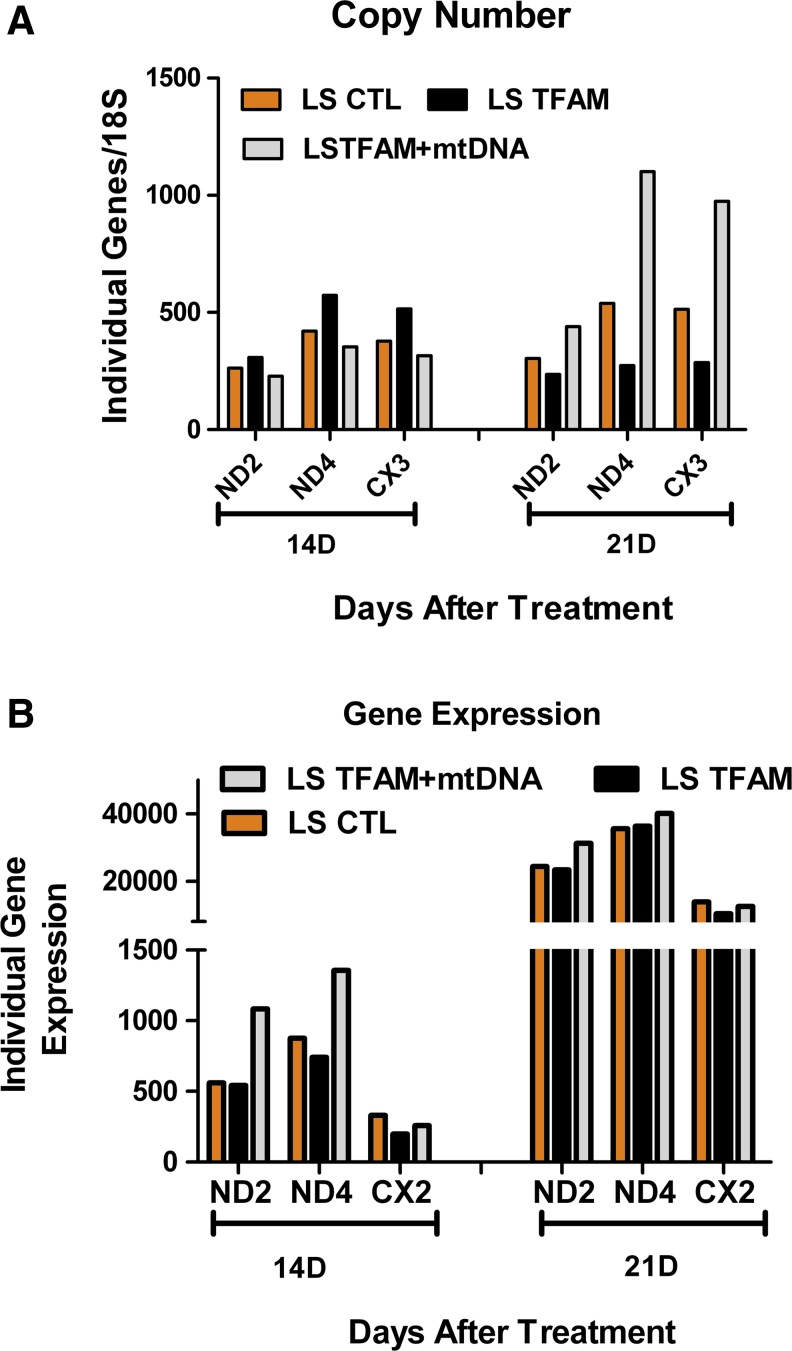
The second set of experiments tested the effects of mitochondrial gene therapy on mitochondrial replication and transcription of key genes in LS cells. Similar real-time PCR assays were conducted as described earlier for LHON cells in an earlier section. Genomic DNA and cDNA obtained from 14 and 21 days after treatment were analyzed for mtDNA copy number and expression in LS cells by qPCR analysis. Three weeks after a single dosage with mtDNA+rhTFAM, qPCR assays showed an increasing trend in replication and transcription over CTL in LS cells. The mtDNA gene copy numbers (ND2, ND4, and COX3) increased over CTL (Fig. 7A) by 21 days, and gene expression (transcription of the individual genes) also increased for the examined genes by 21 days (Fig. 7B).
FIG. 7.
Mitochondrial gene therapy increases mtDNA copy numbers in LS cells. (A) Increased trend of 18S rRNA normalized mtDNA gene copy numbers of ND2, ND4, and COX3 genes. (B) Increased trend of geometric mean values of 18S rRNA, GAPDH, and β-actin normalized cDNA gene expression of ND2, ND4, and COX3 genes. CTL, beige; TFAM, black; mtDNA+rhTFAM, gray. Color images available online at www.liebertonline.com/hum
Mitochondrial gene therapy increased nuclear respiratory protein levels in LS cells
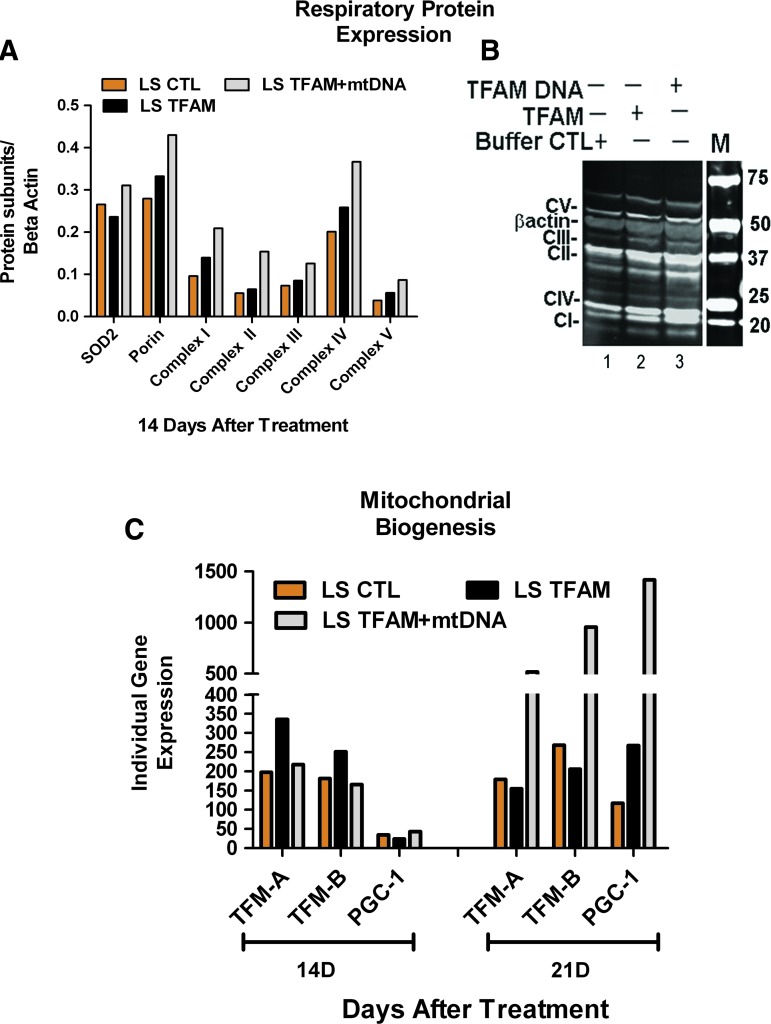
In the third experimental series, LS cells were analyzed for levels of multiple individual respiratory proteins and mitochondrial mass by western blots to assess if mitochondrial gene therapy triggered the biogenesis pathway (Iyer et al., 2009b). Two weeks after treatment with mtDNA+rhTFAM, western blots showed that mitochondrial mass and all assayed ETS proteins increased up to ∼1.8-fold relative to LS CTL (Iyer et al., 2009b) (Fig. 8B), indicative of a trigger in the mitochondrial biogenesis pathway. In all cases, the increases observed in respiratory proteins after treatment with mtDNA+rhTFAM were greater than those observed after treatment with rhTFAM alone. The ETS proteins assayed are all coded by nuclear genes, with the exception of the representative from Complex IV, which is coded by the mtDNA gene COX2 (Fig. 8A).
FIG. 8.
Mitochondrial gene therapy increases mitochondrial biogenesis in LS cells. (A) Representative western blot of ETS complex proteins in LS fibroblast cells. (B) β-Actin-normalized levels of mitochondrial proteins. (C) Increased trends of normalized geometric mean of cDNA gene expressions of TFAM1A, TFAMB1, and PGC1-α genes. CTL, beige; TFAM, black; mtDNA+rhTFAM, gray. Color images available online at www.liebertonline.com/hum
Mitochondrial gene therapy triggered an increased trend in the expression of the biogenesis pathway in LS cells
The last experimental set with LS cells showed a similar increase in trend upon treatment with mtDNA+rhTFAM by 21 days in RNA expression of regulators involved in triggering the mitochondrial biogenesis pathway, as described earlier (Fig 7A and B).
Shown in Fig. 8C are the results of qPCR analysis of increases in gene expression of PGC-1α, TFAM, and TFAMB1 14 and 21 days after treatment with mtDNA complexed with rhTFAM. The results for PGC-1α expression alone were markedly low around day 14 and very robust by day 21. At this time, we are unsure of the reason for this trend. For purely graphical representation, the data have been multiplied by 100 for both individual replicates for the gene PGC-1α, as shown in Fig. 8C.
Discussion
The goal of this study was to determine the effects of donor human mtDNA in complex with rhTFAM on mitochondrial physiology in an LHON cybrid line and in an LS fibroblast disease cell line carrying a high mutant mtDNA load and inefficient respiration. Our first important observation was that mtDNA complexed with TFAM entered the mitochondria by half an hour and persisted for several hours, consistent with other studies in human cells using the same technology (Iyer et al., 2011). Mitochondrial gene therapy (mtDNA+rhTFAM) significantly increased mitochondrial respiration as well as increased gene expression and copy number of mitochondrial genes in LHON and LS cells by 3 weeks. In LS fibroblasts, not only was routine cell respiration increased, we also observed an improvement in the ATP synthase activity by 3 weeks, which was not observed by treatment with only rhTFAM.
These respiratory improvements occurred with mild changes in the very high baseline abundance of the 8993T>G ATPase 6 mutation. It is possible that expression of normal ATPase 6 was increased by treatment with healthy mtDNA by 2 weeks. However, analysis conducted at 4 weeks did not show any detectable difference between untreated and treated samples. It is important to note that samples obtained for analysis were from senescing cells and could have influenced the continued presence of normal mtDNA. Our future studies are focused on determining the stability of the introduced mtDNA into patient fibroblasts and factors that might influence their transmission and maintenance. Our inability to find evidence of alteration of the high abundance of mtDNA carrying the LHON G11778A mutation was disappointing, but may reflect a low mtDNA turnover rate in the background cybrid cells that do not have any evidence for mtDNA depletion. Therefore, appropriate studies to understand such mechanistic shifts in the mutational mtDNA burden (homoplasmy levels) over time are currently under way in human pluripotent stem cells. Our results have shown that the introduced mtDNA in complex with rhTFAM selectively amplified and transcribed in human pluripotent stem cells devoid of their own mtDNA, without altering their “stemness” or capacity to differentiate (Iyer et al., 2009a, 2011). Given the scarcity of animal models, we believe that these cell models will serve as good tools for understanding the genetic transmission and threshold effect of abnormal mtDNA in complex mitochondrial diseases during development. Further, the use of this technology in vivo to alter mtDNA composition and expression in mitochondrial diseases will be strengthened by answering these fundamental questions in these model systems.
The present study also provides strong support for the use of mtDNA complexed with rhTFAM for stimulating mitochondrial biogenesis based on increases in mitofilin, porin, and multiple key respiratory proteins coded by the nuclear and mitochondrial genomes and statistically significant increases in expression in NRF1, TFAM1, and TFAMB1 over 3 weeks in LHON cells. These striking results strengthen our previous observation of increased PGC-1α expression as a marker for mitochondrial biogenesis following treatment with mtDNA in complex with recombinant TFAM (Iyer et al., 2009b; Keeney et al., 2009; Thomas et al., 2011). Similar increasing trends were also observed in LS cells along with increased expression of PGC-1α, TFAM1, and TFAMB1 over 21 days, which strengthened our overall hypothesis that the mechanism for increases in mitochondrial respiration was by triggering the biogenesis pathway in vitro and in vivo (Iyer et al., 2009b; Keeney et al., 2009; Thomas et al., 2011).
Although our findings are encouraging for the use of mitochondrial gene therapy to improve respiration and biogenesis in LHON and LS, we are unclear concerning how and why the addition of healthy mtDNA circles in the mitochondrial compartment would trigger such an important nuclear event related to mitochondrial biogenesis. Our findings point us toward an important adaptive response that clearly links external stimuli of introducing exogenous donor mtDNA to the regulation of mitochondrial biogenesis and respiration. Previous studies have shown that triggering the biogenesis of mitochondria and the maintenance of mtDNA is a complex biological process and requires communication from mitochondria to the nucleus to influence gene expression and coordinate many cellular functions (Rebelo et al., 2011). In favor of this possibility, it is conceivable that the introduction of healthy mtDNA complexed with rhTFAM triggers crosstalk between mitochondria and the nucleus to increase transcription and biogenesis in the cells under normal or diseased conditions.
We observed significant positive responses to treatment in LHON and LS cells in this study. However, our data regarding potential clinical utility in treating LS or LHON disease should be viewed with cautious optimism at this time. Our experiments were carried out with a single fibroblast line from an LS patient and a single cybrid line from an LHON patient. As a first step, our study provided us the opportunity to test the effect of a single dosage of mtDNA+rhTFAM treatment to the cells on mitochondrial replication, transcription, translation, and biogenesis events. The partial responses observed were based on the effect of this single dose in the short term. Unfortunately, given the fact that it was a primary fibroblast line, we were unable to conduct long-term studies to see if full rescue could be achieved, given that the cells senesced by 3.5 weeks. In future studies, we plan on conducting experiments at multiple doses in additional LS and LHON cell lines. Nonetheless, these experimental outcomes are important first steps toward development of mitochondrial gene therapy, which provides hope, as a restorative treatment for patients suffering from mitochondrial disorders characterized by energy failure.
Acknowledgments
We would like to thank members of the Genetics and CSBC department for stimulating discussions and Dr. Joann Bodurtha for critical reading of the manuscript. We would also like to thank Charles Arthur, Lisa Dunham, and R.R. Thomas for assistance with the experiments. The rhTFAM used in this work was obtained through an MTA with Gencia Corp. (Charlottesville, VA). This work was supported in part by funds from the National Institutes of Health (J.P.B.) and from the Parkinson's Disease Foundation and American Parkinson's Disease Association (S.I.). This work has been dedicated to the author's father (Ram Iyer), who passed away during this study.
Author Disclosure Statement
No authors have any financial interests in the use of rhTFAM or in Gencia Corp. Dr. Erich Gnaiger is founder and director of OROBOROS INSTRUMENTS Corp., the company responsible for development and distribution of the Oxygraph-2k for high-resolution respirometry. No competing financial interests exist for other authors of this article.
References
- Chinnery P.F. Taylor R.W. Diekert K., et al. Peptide nucleic acid delivery to human mitochondria. Gene Ther. 1999;6:1919–1928. doi: 10.1038/sj.gt.3301061. [DOI] [PubMed] [Google Scholar]
- DiMauro S. Schon E.A. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Gnaiger E. Polarographic oxygen sensors, the oxygraph and high-resolution respirometry to assess mitochondrial function. In: Dykens J.A., editor; Will Y., editor. Mitochondrial Dysfunction in Drug-Induced Toxicity. John Wiley & Sons, Inc.; Hoboken, NJ: 2008. pp. 327–352. [Google Scholar]
- Holt I.J. Harding A.E. Petty R.K. Morgan-Hughes J.A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- Hütter E. Unterluggauer H. Garedew A., et al. High-resolution respirometry—a modern tool in aging research. Exp. Gerontol. 2006;41:103–109. doi: 10.1016/j.exger.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Iyer S. Alsayegh K. Abraham S. Rao R.R. Stem cell-based models and therapies for neurodegenerative diseases. Crit. Rev. Biomed. Eng. 2009a;37:321–353. doi: 10.1615/critrevbiomedeng.v37.i4-5.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. Thomas R.R. Portell F.R., et al. Recombinant mitochondrial transcription factor A with N-terminal mitochondrial transduction domain increases respiration and mitochondrial gene expression. Mitochondrion. 2009b;9:196–203. doi: 10.1016/j.mito.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. Xiao E. Alsayegh K., et al. Mitochondrial gene replacement in human pluripotent stem cell derived neural progenitors. Gene Ther. 2011 doi: 10.1038/gt.2011.134. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney P.M. Quigley C.K. Dunham L.D., et al. Mitochondrial gene therapy augments mitochondrial physiology in a Parkinson's disease cell model. Hum. Gene Ther. 2009;20:897–907. doi: 10.1089/hum.2009.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.M. Bennett J.P., Jr. Development of mitochondrial gene replacement therapy. J. Bioenerg. Biomembr. 2004;36:387–393. doi: 10.1023/B:JOBB.0000041773.20072.9e. [DOI] [PubMed] [Google Scholar]
- Leigh D. Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatry. 1951;14:216–221. doi: 10.1136/jnnp.14.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen J.L. Smeitink J.A. Trijbels J.M., et al. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum. Mutat. 2000;15:123–134. doi: 10.1002/(SICI)1098-1004(200002)15:2<123::AID-HUMU1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Manfredi G. Fu J. Ojaimi J., et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- Pesta D. Gnaiger E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibres from small biopsies of human muscle. Methods Mol. Biol. 2012;810:25–58. doi: 10.1007/978-1-61779-382-0_3. [DOI] [PubMed] [Google Scholar]
- Rebelo A.P. Dillon L.M. Moraes C.T. Mitochondrial DNA transcription regulation and nucleoid organization. J. Inherit. Metab. Dis. 2011;34:941–951. doi: 10.1007/s10545-011-9330-8. [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N.Y. Acad. Sci. 2008a;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008b;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Shoffner J.M. Fernhoff P.M. Krawiecki N.S., et al. Subacute necrotizing encephalopathy: oxidative phosphorylation defects and the ATPase 6 point mutation. Neurology. 1992;42:2168–2174. doi: 10.1212/wnl.42.11.2168. [DOI] [PubMed] [Google Scholar]
- Tatuch Y. Christodoulou J. Feigenbaum , et al. Heteroplasmic mtDNA mutation (T—-G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am. J. Hum. Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- Thomas R.R. Khan S.M. Portell F.R., et al. Recombinant human mitochondrial transcription factor A stimulates mitochondrial biogenesis and ATP synthesis, improves motor function after MPTP, reduces oxidative stress and increases survival after endotoxin. Mitochondrion. 2011;11:108–118. doi: 10.1016/j.mito.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Memije M.E. Shanske S. Santorelli F.M., et al. Comparative biochemical studies in fibroblasts from patients with different forms of Leigh syndrome. J. Inherit. Metab. Dis. 1996;19:43–50. doi: 10.1007/BF01799347. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. Singh G. Lott M.T., et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Yen M.Y. Wang A.G. Wei Y.H. Leber's hereditary optic neuropathy: a multifactorial disease. Prog. Retin. Eye Res. 2006;25:381–396. doi: 10.1016/j.preteyeres.2006.05.002. [DOI] [PubMed] [Google Scholar]