Abstract
Saccharomyces cerevisiae has directly or indirectly contributed to the identification of arguably more mammalian genes that affect aging than any other model organism. Aging in yeast is assayed primarily by measuring replicative or chronological lifespan. Here, we review the genes and mechanisms implicated in these two aging model systems and key remaining issues that need to be addressed to optimize these assays. Because of its well-characterized genome and proteome that is remarkably amenable to genetic manipulation and high-throughput screening procedures, S. cerevisiae will continue to serve as a leading model organism to study pathways relevant to human aging and disease.
Introduction
Aging studies are becoming increasingly prominent in biomedical research. The reasons for this are obvious. The demographics of the world are rapidly changing, leaving a population with an increasing number of elders and a declining number of working age individuals to support them. Older people tend to have costly chronic diseases that negatively impact their quality of life and functional output. In fact, aging itself is the leading risk factor for an array of diseases that increasingly plague the world population. If researchers can understand aging and modify its rate, the consequences are likely to be a reduced incidence or progression of disease leading to increased healthspan, allowing older people to keep working and avoid high health care costs.
The potential of interventional approaches targeted at aging has yet to be realized in part because aging is a complicated multisystem process that has remained enigmatic in the face of research. However, findings in the last two decades have led to significant excitement. One of the most striking findings is that it is possible to administer a clinically approved drug, rapamycin, to mice at 20 months of age and extend both their lifespan and healthspan (Harrison et al., 2009). Surprisingly, much of the recent success of aging research can be traced back to one of its simplest model organisms: yeast. Two of the major pathways studied in the context of aging and age-related disease are the Sirtuin pathway and the TOR signaling pathway, and yeast was pivotal in their discovery.
There are two primary assays for yeast aging, replicative and chronological. Both will be discussed herein by four investigators with extensive experience in the field. The truth is that we all share the viewpoint that both yeast assays have and will continue to be strong models to understand aging. Regarding the particulars, we mostly agree but sometimes differ. Therefore, we have attempted to construct a review describing the consensus opinion but also not shying away from points of disagreement and from the technical issues that should be considered before studying aging in yeast. It is our hope that readers interested in aging will in one article be able to gain a strong understanding of the state of the field and that clearly articulated points that lack consensus will serve to stimulate further experiments leading to clarification.
The Replicative Life Span
Replicative life span (RLS) studies date back more than 50 years (Mortimer and Johnston, 1959). The assay is simple conceptually and takes advantage of the fact that yeast cells divide by asymmetric budding, with the daughter cell that is produced being smaller than the mother from which it is derived. The question Mortimer asked was: how many times can one cell divide. Daughter cells were isolated on a solid media substrate and, once they started dividing, all progeny were removed and tabulated. The key finding was that individual cells do not divide forever; instead they stop after a limited number of divisions (usually around 20–25) and enter a short post-replicative state followed by lysis.
After Mortimer’s initial work, studies of aging in yeast did not achieve widespread acceptance or notoriety until the early 1990’s when the power of this system was combined with newer and more sophisticated methodologies to begin identifying genes, pathways, and molecular mechanisms that modulate RLS. In recent years, attention has turned toward understanding which aspects of replicative aging in yeast are shared with multicellular eukaryotes, including mammals. These answers are beginning to be attained and in this section we will look at the pathways and mechanisms that modulate yeast RLS, the evidence that this assay is informative about aspects of the aging process in higher eukaryotes, and the likely future directions of these studies.
The Basic Methodology and New Variants
RLS analysis is typically performed by manual separation of daughter cells from mother cells using a standard tetrad dissection microscope equipped with a micromanipulator. Detailed written and video protocols have been published on how this assay is performed in the Kaeberlein and Kennedy labs, and we refer interested readers to these (Kaeberlein and Kennedy, 2005; Steffen et al., 2009). Recently, promising higher-throughput methods involving selective killing of daughter cells or microfluidic flow chambers have recently been described (discussed below). These methods have not become widely used thus far, however, and the manual dissection method remains the gold-standard life span assay for the field.
One issue surrounding studies of replicative aging in yeast involves the choice of growth conditions, which can have major effects on experimental outcome (see section on chronological life span below). For the vast majority of replicative aging studies, growth on rich medium in the presence of 2% glucose (YPD, Yeast Peptone Dextrose) is the method of choice. It is clear, however, that life span can be affected by these choices. For instance, lowering glucose concentration, a condition that is used to model dietary (or calorie) restriction in multicellular eukaryotes, causes life span extension in many, but not all, strain backgrounds. Alternate carbon sources have also been tested to a lesser extent. Budding yeast is a facultative anaerobe that generates most of its energy in the presence of ample glucose through fermentation, with only limited respiratory metabolism. Since most mammalian tissues rely primarily on respiration rather than fermentation, it has been argued that use of a respiratory carbon source such as glycerol may make for more relevant comparison to human aging (Botta et al., 2011). This assertion has yet to be rigorously evaluated. A relatively small number of studies have also examined RLS using synthetic defined (SD) medium, which is commonly used in the chronological life span assay (below), but a direct comparison of life span on SD versus YPD has not been performed. In summary, there is a need for a broader understanding for how different environmental factors, including the nutritional status of the growth medium, influence yeast RLS.
In addition to environmental considerations, there is growing recognition that genetic background plays a critical role in the outcome of replicative aging experiments. Several different laboratory strains have been used extensively in yeast replicative aging studies, and important differences have been uncovered. For example, the Jazwinski laboratory examined the effect of loss of mitochondrial DNA (rho0) on RLS in four different backgrounds, finding that in two cases life span was shortened, in one case there was no effect, and in the last case life span was extended (Kirchman et al., 1999). Likewise, it is now clear that the effects of dietary restriction by reducing glucose availability can vary from upwards of 30% in some strain backgrounds to no effect in others (Kaeberlein and Powers, 2007). The genetic variants that account for these differences have remained largely unexplored, but are likely to provide important insights into mechanisms of aging in yeast. Notably, the issue of strain differences and longevity is pertinent to all aging model organisms, including the CLS assay (see below).
Several approaches have been described to automate and enhance the throughput of the replicative aging assay. To generate large numbers of old cells, it is possible to biotin coat the surface of the yeast cell (Smeal et al., 1996). During cell division, the biotin is maintained specifically on the mother cell wall, since the cell wall of the daughter is newly synthesized. Following multiple divisions, the aged mothers can be separated from the unlabeled subsequent generations with avidin-coated magnetic beads. While not sufficient to measure life span, this method permits biochemical analysis of aged cells. More recently, a Mother Enrichment Program has been developed to monitor life span and isolate old cells from growing cultures (Lindstrom and Gottschling, 2009). This approach involves genetic manipulations to conditionally and specifically arrest cell division in daughter cells through excision of essential genes during their first cell division. While the benefits of this method have not been fully vetted, it has potential to significantly enhance yeast aging studies by generating old cells for analysis and measuring life span.
Genes and Pathways Modulating RLS
Nearly 100 yeast replicative aging genes have been identified where deletion results in enhanced longevity, and based on a partial screen of the deletion collection, it has been estimated that roughly 2% of the non-essential genes in the genome are likely to fall into this category (Kaeberlein et al., 2005c). Perhaps as many as 20% of the non-essential genes result in short lifespan when deleted; however, in the absence of further experiments, it is difficult to determine whether this represents accelerated aging or defects in processes that do not normally limit replicative potential. To a degree, it is possible to differentiate between these possibilities by determining whether known phenotypes associated with aging (e.g. ERC formation, sterility and reactive oxygen species formation) are also accelerated in short-lived mutants. Although it is not formally demonstrated, we suspect that many short-lived mutants are not experiencing accelerated aging, but rather have defects that limit their potential to divide. Thus, a majority of the focus has been on mutations that extend lifespan. The large number of yeast replicative aging genes is comparable to the situation in C. elegans, where hundreds of genes have been identified whose reduced expression leads to enhanced longevity. Although a detailed analysis of all of the known yeast aging genes is not possible here, we will highlight the best understood pathways (Figure 1) and their links to aging in the yeast chronological paradigm and/or other organisms.
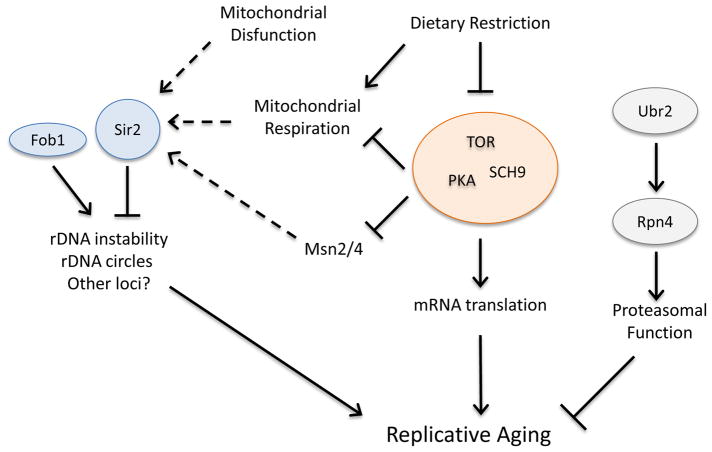
Figure 1.
Three genetically distinct pathways modulating replicative life span. Based on evidence from many labs, it now seems clear that at least 3 genetically distinct pathways modulate RLS. Sir2 and Fob1 influence RLS largely through their role in the rDNA, although there is evidence for aging-related functions for Sir2 at other loci, including telomeres, as well as by modulating asymmetric inheritance of cytoplasmic damage. DR is mediated at least in part through reduced TOR/PKA/Sch9 signaling. Ubr2 and Rpn4 appear to influence RLS by modulating proteasome activity. The relationship of the retrograde response and mitochondrial dysfunction to these pathways remains unclear, although there is some evidence for altered Sir2 activity in one mitochondrial translation factor mutant. Dotted lines represent potential points of cross-talk between pathways that have been proposed.
One of the most famous yeast aging genes is SIR2, which encodes the founding member of the sirtuin family of NAD-dependent protein deacetylases. Sirtuins have been extensively studied in recent years for their potential role as conserved modulators of aging in a variety of organisms, including mammals (Finkel et al., 2009). The first data directly linking these proteins to aging comes from yeast, where overexpression of Sir2 was shown to extend RLS (Kaeberlein et al., 1999). Whereas Sir2-orthologs have many targets in multicellular eukaryotes and non-histone targets have been identified in yeast (Lin et al., 2009), yeast Sir2 is thought to be primarily a histone deacetylase (Imai et al., 2000). One mechanism by which Sir2 activity promotes replicative longevity is by suppressing homologous recombination in the rDNA that can cause the formation of extrachromosomal rDNA circles (ERCs). These circular DNA molecules are self-replicating and asymmetrically segregated to the mother cell during cell division (Sinclair and Guarente, 1997). It is believed that accumulation of ERCs can limit the RLS of mother cells, at least in certain strain backgrounds, although this idea has been challenged recently by the suggestion that rDNA instability in general, rather than ERCs, is the primary defect causing senescence (Lindstrom et al., 2011). Providing strong evidence for the role of the rDNA in yeast aging, deletion of FOB1, which reduces rDNA recombination and ERC formation, can increase life span in some strain backgrounds and also suppresses the short the lifespan of sir2Δ mother cells (Kaeberlein et al., 1999). This finding is in sharp contrast to most other longevity mutations, which have no effect in the sir2Δ background, despite the fact that they extend wild type life span (Delaney et al., 2011b). The probable reason for this is that multiple types of damage can limit the life span of wild type cells, but in the absence of Sir2, rDNA instability becomes the predominant factor driving senescence. Only mutations that suppress this instability (such as deletion of FOB1) are able to overcome the defect limiting the replicative capacity of sir2Δ mother cells.
While ERCs and rDNA instability are part of the story, recent studies have suggested other roles for SIR2 in yeast aging. For example, loss of Sir2 results in a defect in asymmetric retention of oxidatively damaged cytoplasmic proteins in the mother cell (Aguilaniu et al., 2003; Erjavec et al., 2007). This defect causes sir2Δ daughter cells to inherit higher levels of oxidative damage, which may also contribute to the short RLS of these cells, as evidenced by the observation that overexpression of Hsp104 can extend the life span of cells lacking Sir2 (Erjavec et al., 2007; Erjavec and Nystrom, 2007).
Interestingly, Sir2 protein levels decline with age, perhaps explaining why overexpression or Sir2 can extend lifespan (Dang et al., 2009). Also changing with aging are epigenetic modifications to histones. For instance, H4K16 acetylation near telomeres increases concomitantly with a decline in Sir2 levels (Dang et al., 2009) and mutations that increase expression of histones or alter their deposition at sites near telomeres can extend RLS (Dang et al., 2009; Feser et al., 2010). These findings suggest that Sir2 function at telomeres, a location where Sir2 is known to promote gene silencing, may also influence replicative aging.
Overexpression of SIR2 orthologs in worms and flies has been reported to extend lifespan, and several studies have suggested that activation of the mammalian Sir2-ortholog, SirT1, can enhance healthspan in mice (Finkel et al., 2009). Although the data in worms and flies has recently been called into question (Burnett et al., 2011; Rizki et al., 2011; Viswanathan and Guarente, 2011), there seems to be general consensus that SirT1 interacts with important aging-related pathways in mammals, if not necessarily directly modulating the aging process itself. Thus, understanding the molecular mechanisms by which Sir2 influences RLS in yeast, and its relationship to other longevity pathways, is imperative in order to meaningfully interpret the complex and controversial data emerging from studies in other model organisms.
Dietary restriction, a reduction in nutrient availability without malnutrition, is known to extend lifespan in a wide range of organisms from yeast (replicative and chronological) to primates (Anderson and Weindruch, 2012; Katewa and Kapahi, 2010). Extensive effort has been devoted to understanding the pathways that mediate the benefits of dietary restriction, since interventions that target these pathways may be effective in humans against the diseases of aging. In the yeast replicative assay, dietary restriction is usually evoked by reducing glucose concentration from 2% to 0.5–0.05% (Lin et al., 2000), although restriction for amino acids has also been reported to extend lifespan (Jiang et al., 2000). Much of the research on dietary restriction with respect to replicative aging initially focused on the role of Sir2 downstream of DR. It was initially proposed that DR increased RLS by activation of Sir2; however, subsequent studies have called this model into question. Although it is still debated as to whether part of the effect of DR on RLS is mediated by Sir2, there is now general consensus that DR can also extend life span via Sir2-independent mechanisms. In addition to Sir2, yeast has four other sirtuin proteins. It has been proposed that Hst1 and Hst2 (a homologue of the human sirtuin histone deacetylase SirT2) can compensate for the loss of Sir2 under glucose-limited conditions. This model also remains controversial, however, and we (MK and BKK) have reported that triple-mutant cells lacking SIR2, HST1, and HST2 still show a robust RLS extension from DR so long as ERCs are kept low (Easlon et al., 2007; Kaeberlein et al., 2006a; Lamming et al., 2005a; Tsuchiya et al., 2006). Loss of both HST3 and HST4 leads to a dramatic reduction in RLS, possibly through increased genome instability (Hachinohe et al., 2011; Tsuchiya et al., 2006). The role, if any, of Sirtuins in RLS extension from DR has been discussed extensively elsewhere and we refer the reader to published reviews on this topic for additional discussion (Kaeberlein, 2010; Kaeberlein and Powers, 2007; Lu and Lin, 2010).
More recently, evidence has accumulated that life span extension from DR is mediated largely via reduced signaling through overlapping nutrient-responsive Ras-PKA and TOR/Sch9 pathways. Both pathways play a concerted role in appropriately regulating growth, metabolism, and stress resistance in response to nutrient availability, and signaling through these pathways is reduced by DR. Mutations that impair Ras-PKA or TOR/Sch9 signaling are sufficient to extend RLS even when nutrients are plentiful, and combining such mutations with DR fails to result in further RLS extension (Fabrizio et al., 2004c; Kaeberlein et al., 2005c). Importantly, both of these pathways play a similar role in modulating yeast chronological life span, as well as longevity in worms, flies, and mice, providing strong evidence for their conserved effects on aging throughout eukaryotes (Fontana et al., 2010).
A recent report also suggests that Sch9 activity can be regulated independently of DR to influence RLS through acetylation of the Snf1 complex component, Sip2 (Lu et al., 2011). Sip2 acetylation decreased with replicative age, and enhancing Sip2 acetylation extends life span. Interestingly, Snf1 is the yeast AMP-activated protein kinase, and there is abundant evidence that activation of AMP kinase can extend life span in C. elegans (Apfeld et al., 2004).
Currently, major efforts are being directed at understanding the downstream mediators of the Ras-PKA and TOR/Sch9 pathways that are important for replicative aging. At least one of those involves reduced mRNA translation, which is apparent from polysome profiles of sch9Δ cells or cells treated with the TOR inhibitor rapamycin (Steffen et al., 2008). Several mutations that decrease mRNA translation initiation increase RLS, including deletion of genes encoding ribosomal proteins and mRNA translation initiation factors. Again, it is interesting to note that several of these replicative aging genes that promote mRNA translation have also been shown to play a similar role modulating longevity in C. elegans (Smith et al., 2008). This has led to the idea that modulation of mRNA translation via reduced TOR signaling may be conserved mechanism for linking nutrient response to aging in evolutionarily diverse species. Studies in flies show, however, that while reduced protein translation does occur in response to DR, that the phenotype is genetically separable from life span extension (Kabil et al., 2011). These findings suggest that altered translation for specific mRNAs might underlie the longevity benefits of DR. Reduced signaling through these pathways also has a range of other effects in addition to decreasing mRNA translation that plays a role in yeast replicative and chronological aging, as well as aging in other species. These include activation of stress-responsive transcription factors such as Gcn4 and Msn2/4, increased autophagy, and altered mitochondrial metabolism.
Interestingly, reduced TOR signaling also leads to decreased rDNA recombination and ERC formation, suggesting that the Sir2 and TOR pathways may, at least in part converge on similar downstream events relevant to life span. The mechanisms for this are not entirely clear. For instance, reduced TOR signaling mediated by rapamycin leads to elevated Pnc1 levels and increased Sir2 rDNA association (Ha and Huh, 2011; Medvedik et al., 2007b). However, both tor1Δ and sch9Δ strains retain their ability to extend life span in the absence of SIR2 and FOB1 (Kaeberlein et al., 2005b), indicating that other pathways are sufficient to mediate the longevity effects. Another possibility is that reduced rDNA recombination occurs independently of Sir2 regulation (Prusty and Keil, 2004; Riesen and Morgan, 2009), perhaps as a consequence of decreased rRNA transcription, although it is clear that reduced rDNA recombination is separable from life span extension in some mutants (Delaney et al., 2011b). Unraveling these connections, as well as their relevance to rDNA recombination and RLS will require more study.
Mitochondrial function plays a critical, but poorly understood, role in RLS determination. This was first clearly shown by studies from the Jazwinski lab demonstrating that induction of the retrograde response pathway, which transmits signals of mitochondrial stress to the nucleus, can increase RLS in certain genetic backgrounds and that this retrograde response is involved in life span extension through the Ras-PKA pathway (Borghouts et al., 2004; Kirchman et al., 1999). The effects of mitochondrial perturbation on life span are strongly influenced by the nuclear genome, however, since, as mentioned above, loss of mitochondrial DNA is associated with reduced, unaltered, or enhanced lifespan depending on strain background. It has also been proposed that DR activates Sir2 by altering the NAD/NADH ratio through up-regulation of mitochondrial respiration (Lin et al., 2002); however, other studies have reported that neither mitochondrial DNA nor a functional electron transport chain are required for RLS extension from DR (Kaeberlein et al., 2005a). In addition, a number of strains lacking nuclear encoded mitochondrial genes, including those in the TCA cycle, are associated with lifespan extension. For example, a recent study reported that deletion of the nuclear gene encoding SOV1, a mitochondrial translation factor, leads to increased RLS by a mechanism that involves activation of Sir2 (Caballero et al., 2011). Clearly, much more work is needed to clarify the important role that mitochondria play in yeast replicative aging, particularly the interplay between mitochondrial and nuclear genomes.
Recently, it was reported that enhanced proteasome activity is sufficient to increase RLS by a mechanism that is genetically distinct from both DR and Sir2 (Kruegel et al., 2011). This finding is consistent with a growing body of work linking aging with loss of normal proteostasis in a variety of organisms, further supporting the idea that multiple yeast longevity pathways are conserved (Taylor and Dillin, 2011). Like ERCs, damaged proteins are asymmetrically segregated to the mother cell during cell division, and it may be the case that elevated proteasome activity extends life span by reducing the age-related increase in this type of damage. Longevity-associated mutations in the Ras-PKA and TOR/Sch9 pathways are also associated with resistance to oxidative stress, leading to the possibility that oxidative damage is a primary driver of yeast replicative aging. Despite these findings, however, it is been difficult in yeast, as with multicellular eukaryotes, to link oxidative damage and aging in a causal fashion. For instance, overexpression of superoxide dismutases shorten RLS rather than extending it (Fabrizio et al., 2004c). Interestingly a recent finding supports a model whereby DR extends lifespan at least partially by countering age-associated oxidation and inactivation of the peroxiredoxin, Tsa1 (Molin et al., 2011). Further studies will be needed to clarify the role of oxidative damage in yeast replicative aging, as well as aging in other species.
Strengths/ Limitations of the Replicative aging model
How reasonable is it that the replicative potential of a yeast mother cell can be related to aging in multicellular eukaryotes? This has been extensively debated, with one hypothesis being that replicative aging may be linked to aging of mitotic or stem cell populations in a complex organism, whereby chronological aging may more closely resemble aging of non-dividing cells. One recent study tested links between replicative aging and that of C. elegans, a post-mitotic organism (except for germline) upon adulthood (Smith et al., 2008), finding that a yeast ortholog of a worm aging gene was five times more likely to modulate yeast aging that a yeast ortholog of a randomly chosen worm gene. This study establishes quantitatively that yeast RLS and worm aging share significant overlap, but also points to the existence of aging pathways that are unique to both organisms. Interestingly, both S6 kinase (Sch9) and components of the TOR pathway, including the TOR kinase, multiple ribosomal proteins, and mRNA translation initiation factors, were among the homolog-pairs that modulate longevity in both yeast and C. elegans.
One difficulty with interpreting what replicative aging means is that the dividing cell is the organism. It is generally accepted that there is often a trade-off between reproduction and life span, and a majority of mutations that extend life span reduce reproduction and organismal fitness. In this regard, it is important to note that cell division rate, not the number of daughter cells produced, is the appropriate measure of fecundity and the primary factor determining fitness. A recent study illustrated this point by examining the relative fitness of more than 30 long-lived yeast mutants in direct competition assays involving co-culture with wild type cells (Delaney et al., 2011a). As predicted, a majority of the mutants showed substantial fitness defects, which could often be directly attributed to a reduced maximal growth rate and a G1-specific cell cycle delay. Whether environmental conditions exist in the wild that select for reproductive capacity rather than maximal growth rate remain to be determined.
In order for an organism to be evolutionarily successful, maintaining an untarnished germline is essential. In part, aging yeast do this by maintaining damaged molecules in mother cells, in an attempt to protect their daughters from aging. Meiosis, however, is likely the principal manner by which yeast recombine their genome in the wild. A recent study suggests that mechanisms exist to preserve the integrity of spores of aging diploid cells induced to sporulate (Unal et al., 2011). By mechanisms that are currently unknown, aging diploid cells that are induced to sporulate appear to remove age-associated damage to a point that is no longer detectable. Further studies will be needed to address the mechanism of this surprising finding, but it suggests that maintenance of the germline is of paramount importance in the face of aging, even in single-celled yeast.
The Chronological Life Span
Chronological life span (CLS) is the length of time that a non-dividing yeast cell survives. CLS is typically measured by growing a culture of yeast cells into the post-diauxic state, following which most cells exit the cell cycle. The post-diauxic phase is the period that begins approximately 24 hours after initial inoculation when cells deplete extracellular glucose, dramatically reduce growth and switch to a mitochondrial respiratory mode of metabolism dependent on the ethanol generated during fermentation (Werner-Washburne et al., 1996). Stationary phase begins at the end of the post-diauxic phase between day 2 and 7, depending on the medium used in the experiment, and is characterized by lower metabolic rates and up-regulation of stress-resistance pathways. The yeast CLS assay was developed to complement the RLS assay, by providing the ability to model aging of non-dividing cells of higher organisms (Fabrizio and Longo, 2007; Longo, 1997; Longo et al., 1996). Below we will summarize some of the major findings to date with regard to pathways and mechanisms of aging delineated using this system as well as describe the salient features and caveats of the established methods currently used to measure yeast CLS.
Genes and Pathways Modulating CLS
Although many mutations and conditions affect CLS, the purpose of this review is to focus on those that have been confirmed by multiple studies or laboratories to extend longevity in higher eukaryotes including mammals. The major genes and pathways regulating the yeast CLS show remarkable similarities to those in worms, flies, and mammals. We propose that there are two major pro-chronological aging pathways in S. cerevisiae, both of which sense nutrient availability and control their utilization. These are the Tor/S6K pathway (Fabrizio et al., 2001), which is activated by amino acids and other nutrients, and the Ras/adenylate cyclase (AC)/PKA pathway (Longo et al., 1999, Fabrizio et al., 2003), which is principally activated by glucose, but also affected by other nutrients (Fig. 3)(De Virgilio, 2011; De Virgilio and Loewith, 2006; Dechant and Peter, 2008; Fontana et al., 2010; Smets et al., 2010). These two signaling pathways have partially overlapping, yet distinct pro-aging effects (Wei et al., 2008; Wei et al., 2009). A primary mode of action of these pathways is their convergence on the stress-resistance regulon that includes Rim15 and transcription factors Msn2/Msn4, and Gis1 (Fig. 3)(Cameroni et al., 2004; Fabrizio et al., 2004c; Fabrizio et al., 2001; Pedruzzi et al., 2000; Pedruzzi et al., 2003; Reinders et al., 1998; Wei et al., 2008). In addition to stress, these factors regulate metabolism and the accumulation and utilization of intracellular and extracellular carbon sources (Bonawitz et al., 2007; Wei et al., 2008).
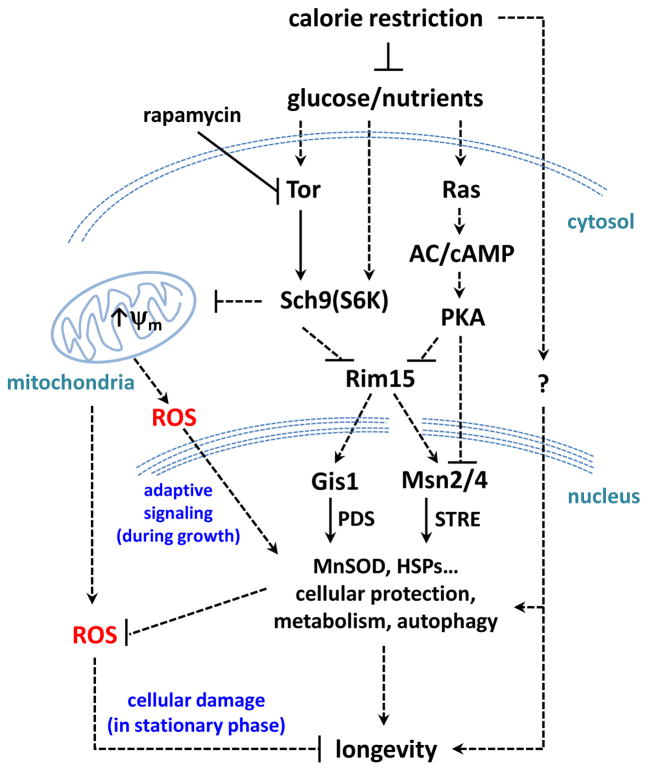
Figure 3.
Yeast Chronological Life Span Major Regulatory Pathways. The nutrient-sensing pathways controlled by Sch9, Tor, and Ras converge on the protein kinase Rim15. A major portion of the effect of CR on longevity appears to be mediated by the down-regulation of the Ras/AC/PKA and Tor/Sch9 pathways and consequent activation of the Rim15-controlled Msn2/4 and Gis1 stress-responsive transcription factors. Reduced Tor/Sch9 signaling (genetic mutations or rapamycin) also increases coupled mitochondrial respiration and membrane potential (Δψ m) during growth phase, which leads to an adaptive mitochondrial ROS signal. During stationary phase (i.e. chronological aging), Tor and Sch9 deficiencies and adaptive mitochondrial ROS signaling decrease ROS production and enhance cellular stress responses, culminating in life span extension.
Interestingly, Sir2 is also connected to Msn2/4 activity and regulation of both RLS and CLS (Medvedik et al., 2007a)(Fabrizio et al., 2005; Smith et al., 2007). Whereas Sir2 is intimately linked to RLS regulation (see RLS section), similar genetic manipulations of Sir2 do not appear to affect CLS in standard medium and SIR2 deletion actually extends CLS under CR conditions (Fabrizio et al., 2005; Smith et al., 2007). Also, sir2Δ mutants, like the long-lived sch9Δ and ras2Δ mutants, cause premature depletion of extracellular ethanol, which contributes to CLS extension (Fabrizio et al., 2005). Notably, the deacetylation of histone H3, mediated not by Sir2 but by other deacetylases in response to spermidine, is associated with reduced oxidative stress and CLS extension (Eisenberg et al., 2009). These results indicate that the effects of Sir2 on CLS are complex and raise the interesting possibility that Sir2 could both interfere with and promote the beneficial effects of CR (Longo and Kennedy, 2006).
Since chronological aging culminates in a form of cellular death with features of mammalian apoptosis, the CLS system has also been pivotal for the mechanistic elucidation of apoptotic and necrotic pathways including Endonuclease G (Buttner et al., 2007), BH3-only proteins (Buttner et al., 2011), cathepsin D (Carmona-Gutierrez et al., 2011) and cdc48/VCP (Heo et al., 2010; Madeo et al., 1997).
Mechanisms of CLS regulation: longevity reprogramming
Numerous studies utilizing genetic, nutritional and pharmacological manipulation have addressed the mechanism of chronological aging in budding yeast and have greatly expanded our knowledge of this process (Chen et al., 2005; Eisenberg et al., 2009; Fabrizio and Longo, 2003; Madia et al., 2007). The systems biology-based studies indicate that many pathways with a wide variety of functions affect CLS. Thus CLS extension observed by inhibiting the two major pro-chronological aging pathways, Tor/S6K or Ras/AC/PKA, no doubt requires large-scale and coordinated changes in gene expression and perhaps epigenetic reprogramming (Cheng et al., 2007; Ge et al., 2010). One major downstream effects of such a reprogramming program is clearly protection from the macromolecular damage and cellular stress that limit CLS, as the PKA and Tor/S6K pathways converge on transcription factors that mediate expression of stress-responsive genes (Fig. 3) (Cameroni et al., 2004; Cheng et al., 2007; Fabrizio et al., 2004c; Gorner et al., 1998; Gorner et al., 2002; Pedruzzi et al., 2000; Pedruzzi et al., 2003; Wei et al., 2008). Furthermore, results from many studies highlight the multifactorial nature of yeast chronological aging, and point to several damage and stress pro-senescence pathways including oxidative stress (Chen et al., 2005; Fabrizio and Longo, 2003; Fabrizio et al., 2001; Longo et al., 1996), mitochondrial dysfunction and reactive oxygen species (Aerts et al., 2009; Bonawitz et al., 2007; Bonawitz et al., 2006; Goldberg et al., 2010; Heo et al., 2010; Herker et al., 2004; Longo et al., 1999), reduced autophagy (Eisenberg et al., 2009; Fabrizio et al., 2010; Yorimitsu et al., 2007), nuclear DNA damage, mutagenesis and replication stress (Fabrizio et al., 2005; Maclean et al., 2003; Madia et al., 2008; Madia et al., 2009; Qin et al., 2008; Weinberger et al., 2007; Weinberger et al., 2010), metabolic alterations (Goldberg et al., 2009; Wei et al., 2009), extrinsic stress (Burtner et al., 2009; Burtner et al., 2011; Fabrizio et al., 2004a; Fabrizio et al., 2005), and other factors (Alvers et al., 2009; Fabrizio et al., 2010; Goldberg et al., 2009). Such a diversity of processes and pathways so far implicated in CLS studies has limited the development of a consensus regarding mechanisms of CLS regulation, particularly downstream of the major stress resistance transcription factors. This should be a primary goal of the field moving forward. However, taking stock at this point, we suggest that convergence of nutrient-sensing signaling pathways, mitochondrial respiratory capacity and stress responses in the control of CLS is one provocative network hypothesis, which is discussed in detail below.
Nutrient signaling and oxidative stress
The diversion of available resources away from anabolic activities and toward stress-resistance pathways is apparently a key mechanism through which reduced Tor/Sch9 signaling extends yeast CLS. However, reduced Tor (TORC1) signaling also increases mitochondrial translation, OXPHOS density and coupled respiration during log-phase growth leading to increased mitochondrial ROS (mROS) production (Bonawitz et al., 2007; Pan and Shadel, 2009). Growth-phase production of mROS is sufficient to promote significant CLS extension; therefore, a key component of the effects of reduced Tor signaling on yeast chronological aging appears to be a mitochondrial adaptive (or mitohormesis) stress response (Fig. 3) (Pan et al., 2011). The ability of pre-growth on a respiratory carbon source to extend yeast CLS (Piper et al., 2006), the observation that CLS extension can be achieved by overexpression of the Hap4 transcription factor, which induces mitochondrial respiration during log phase, (Piper et al., 2006) and the observation that caloric restriction elevates respiration (Lin et al., 2002) and log-phase ROS levels (Goldberg et al., 2009), is consistent with an adaptive mitochondrial longevity signal (Mesquita et al., 2010), as are studies in C. elegans that implicate early increases in mitochondrial ROS but late decreases in ROS as pro-longevity cues (Fabrizio et al., 2003; Fabrizio et al., 2001; Schulz et al., 2007; Yang and Hekimi, 2010). In fact, elevated SOD2 has been detected in a variety of long-lived mutants in yeast (Fabrizio et al., 2003) and higher eukaryotes (Brown-Borg et al., 2002; Honda and Honda, 1999), and overexpression of both SOD1 and SOD2 extend CLS and life span in yeast (Fabrizio et al., 2003) and Drosophila (Orr and Sohal, 1993; Sohal et al., 1995; Sun et al., 2002; Sun et al., 2004; Sun and Tower, 1999). The impact of signaling ROS on aging by such a complex mechanism will require more evolved versions of the mitochondrial and free radical theories of aging, particularly because overexpression of antioxidant enzymes has such a small effect on CLS compared to that caused by lack of the Tor/S6K and Ras/AC/PKA pathways (Bonawitz et al., 2006; Fabrizio et al., 2003) (Table 1). In addition, mitochondrial (or other) hormetic effects on stress-resistance pathways may also help explain why some aging studies do not document a positive correlation between ROS levels and macromolecular damage (Ristow and Zarse, 2010). The most likely interpretation of all the studies above is that coordinated regulation of a variety of protective systems (e.g. antioxidant enzymes) by stress (hormesis), dietary restriction or mutations in nutrient signaling pathways promotes longevity extension. Importantly, repair and replacement systems, as well as epigenetic changes, which are only beginning to be implicated in yeast longevity regulation, are also likely to play a key role in age-dependent cellular damage and mortality.
Table I.
Chronological life span1.
| Mean CLS in 2% glucose (SDC) | Mean CLS in water | |||
|---|---|---|---|---|
|
| ||||
| days | % of wild type CLS | days | % of wild type CLS | |
| WT (DBY746) | 6.5 | 100 | 12.7 | 100 |
| sch9Δ | 15.3 | 235 | 30.0 | 236 |
| tor1Δ | 8.6 | 132 | 15.0 | 118 |
| ras2Δ | 18.8 | 290 | 38.9 | 306 |
| ras2Δ sch9Δ | 35.4 | 545 | 63.0 | 496 |
| ras2Δ sch9Δ rim15Δ | 16.8 | 258 | 49.6 | 390 |
Adapted from Wei et al 2009 (see supplementary material for full tables).
With regard to yeast CLS it remains to be determined how a mitochondrial ROS signal is sensed and transduced to the nucleus, and what key aspects of cellular homeostasis are affected to extend life span. One possibility is that ultimately redox-sensitive transcriptional and epigenetic responses stabilize the nuclear genome and promote or maintain overall cellular or population homeostasis (e.g. by modulating DNA-damage responses, protein homeostasis, autophagy, apoptosis, etc). While other scenarios can certainly be postulated to underlie CLS regulation, the central role of mitochondria in metabolism, ROS production, and apoptosis makes this organelle an attractive central player in aging that, through added effects from mROS and stress signaling, can link nutrient-sensing and stress-response pathways to other key processes that affect longevity such as nuclear genome maintenance Observations of elevated respiration and ROS during the early stages of caloric restricted growth in yeast further supports mitochondrial stress as a potential centerpiece of a network theory that could explain key aspects of how chronological life span is regulated in yeast and perhaps other organisms.
While we have outlined only one such network theory of CLS above, we fully acknowledge that this is currently only a strong hypothesis and other scenarios need to be simultaneously considered. For the CLS field, many outstanding issues remain. These include determining which factors act in parallel and downstream of transcription factors Msn2/4 and Gis1, what signals precisely trigger TOR/S6K and RAS/AC/PKA pathway signaling, how these two primary pathways crosstalk with each other (and possibly other pathways), to what extent mitochondrial ROS signaling and damage contribute to CLS, and finally what are the key downstream effects and effectors of disruption of these pathways with age (e.g. apoptosis, nuclear genome instability, cellular damage/senescence, protein aggregation, epigenetic changes etc).
CLS Methods
Because changes in the medium, conditions or techniques used can have large effects on chronological survival, it is important to be aware of the major methods to measure it and of the potential artifacts that can affect the interpretation of CLS studies. There are at least three established methods to measure CLS: 1) monitoring survival of cells grown in 2% glucose (SDC) medium and maintained in the medium modified by the cells during the growth, and post-diauxic phases, 2) monitoring survival of cells grown and maintained in the medium described above but switched to water during the post-diauxic phase, and 3) monitoring survival on 2% glucose agar plates containing all the nutrients except tryptophan (TRP-drop out, or trp-, SDC plates). The rationale and potential caveats of these CLS assays will be described in the following sections.
CLS in 2% glucose SDC
This method is initiated with the growth of the yeast cells in liquid 2% glucose SDC medium in flasks (Fabrizio and Longo, 2007; Longo et al., 1996). After the growth phase, the cells are maintained in the glucose-depleted medium, which is composed principally of ethanol as the carbon source. Slight variations of this method are used by different laboratories (e.g. different types of caps used and media/flask volume ratios), which can cause differences in culture aeration, which can effect ethanol accumulation, redox status and other variables and cause quantitative differences in CLS (see supplementary material). However, by and large, similar results are obtained with regard to genes and pathways that have been observed to affect CLS (Table I). This method has been adopted so frequently for CLS studies by the Longo lab, that it is often assumed that it is the only way to assess chronological survival. However, chronological aging refers to the senescence of non-dividing cells under a variety of commonly encountered conditions including liquid ethanol medium, water and solid complete SDC media. Thus, to identify genes and pathways likely to be relevant for mammalian aging, it is important to confirm the CLS results with additional methods such as those described below.
CLS in Water and Under Calorie Restriction
The monitoring of chronological survival of cells grown in SDC medium, described above, but switched to water on day 3 has been carried out in a number of studies for three reasons: 1) To determine the effects of a particular mutation on life span independently of the nutritional environment (ethanol, acetic acid, etc) or of the acidification of the medium (Table I), 2) To promote starvation (extreme calorie restriction conditions) in addition to the standard CR method (growth in 0.05–0.5%-glucose instead of 2% glucose medium). The switch to water or reduced glucose medium causes ~2-fold or higher survival extension (Table I), which is consistent with the fact that both calorie restriction (CR) and complete nutrient deprivation are well-known to extend life span in different organisms (Kaeberlein et al., 2006c; Lee et al., 2006), 3) To rule out regrowth (gasping) of the stationary population (see “Potential CLS Artifacts section)(Fabrizio et al., 2004a). To minimize further the potential for regrowth, some experiments in water are accompanied by three washes with sterile water every 48 hours, with great care not to introduce contamination, which are common during long studies.
CLS on plates
A recently developed method to study chronological aging is the monitoring of survival on plates with solid agar containing all the nutrients contained in the standard 2% glucose SD medium but lacking tryptophan (SD – trp). In this method, which is only appropriate for cells lacking tryptophan biosynthetic capacity, approximately 200 cells from the same day 1 liquid culture are plated onto approximately 10 different plates and every two days tryptophan is added to one of the 10 plates to monitor viability (Madia et al., 2007; Wei et al., 2009). This -trp solid medium yields CLS results similar to those obtained with the standard liquid SDC medium, but in principle, this approach could be amenable to starvation for other amino acids on plates. It would be interesting to determine whether survival using this method is differentially affected by starvation for different amino acids.
CLS in buffered media
Buffering the pH of cells grown in 2% glucose SDC medium to 6.0 or neutralizing the pH with NaOH has been shown to be sufficient to extend CLS to an extent comparable to DR (>2-fold) in several different laboratory yeast strains, as well as a vineyard strain RM11 (Burtner et al., 2009; Fabrizio et al., 2004a; Fabrizio et al., 2005; Murakami et al., 2011; Pan et al., 2011). This has led to concerns in the CLS field that cell death due to acidification of the culture medium may be a primary determinant the survival of yeast cells in 2% glucose SDC. Interpretation of the importance these findings remains controversial (see Supplementary Materials), however, and the central aging regulatory pathways that influence CLS under unbuffered conditions appear to play a similar role when acidification is no longer limiting survival (Table I and (Wei et al., 2008; Wei et al., 2009)). In order to resolve the uncertainty regarding the importance of media acidification in CLS, more direct comparisons of strains and genetic backgrounds in buffered and unbuffered media, systematically as a function of pH, are needed.
A recommendation for multiple approaches
Given the complexities of the CLS assay, the authors recommend that multiple methods be used to confirm results. One approach used in the Longo laboratory is to perform the great majority of the experiments using the acidic ethanol/acetic acid containing medium derived from growth in 2% glucose SDC medium supplemented with a four-fold excess of the amino acids/bases whose biosynthesis has been compromised by mutations that make the cells amenable to genetic manipulations. Results are then confirmed with the water and solid agar plate methods described above. For at least some long-lived mutants, including sch9Δ, tor1Δ, ras2Δ, and cyr1::Tn, the extension of CLS achieved in SDC medium is also observed in the water as well as in the plate assays, although each assay causes a shift in the survival of both wild type cells and long-lived mutants (Table 1 and Table S1, S2 supplementary material). Survival in trp- plates is slightly shorter than that in SDC medium, and survival in water is approximately twice as long as that in SDC (Table I) (Madia et al., 2007; Wei et al., 2009). An alternative approach used in the Kaeberlein and Kennedy labs is to measure CLS in both unbuffered 2% glucose SDC medium and in the same medium buffered to pH 6.0 (Burtner et al., 2009; Burtner et al., 2011; Murakami et al., 2011). This allows for a direct comparison of how medium acidification impacts CLS in response to different environmental and genetic interventions. It is important to note that when using any of the liquid based CLS methods gasping/regrowth of cells must be carefully monitored (see Supplemental Materials).
The consistent CLS extending role of these mutants using the various methods strongly suggest that the effect of these genes on aging is largely independent of the environment but also underlines the importance of confirming the effect of mutants on CLS in the various media, if the goal is to identify genes with conserved pro-longevity effects.
The controversial role of acetic acid in CLS
Media acidification limits CLS (Burtner et al., 2009; Fabrizio et al., 2004a; Fabrizio et al., 2005) and is accompanied by ROS production and mitochondrial changes that are associated with aging in yeast and other organisms (Burhans and Weinberger, 2009; Pan et al., 2011). One of our laboratories (VDL) showed that ethanol and acidification reduce the CLS and proposes that the effect of ethanol removal on life span extension is in agreement with the caloric contribution of this alcohol, since ethanol is the primary carbon source being metabolized by yeast during chronological aging (Fabrizio et al., 2004a; Fabrizio et al., 2005). Acidification can also be important since it contributes to preventing growth even though there are high levels of carbon sources in the medium (Fabrizio et al., 2004a; Fabrizio et al., 2005). However, why acidification causes an acceleration of chronological aging is not known. In contrast, Burtner et al propose that it is acetic acid, and not ethanol, that increases mortality and is the primary cause of chronological aging in yeast (Burtner et al., 2009; Kaeberlein, 2010). In addition, they proposed that the effects of DR can be explained by reduced acidification of the medium, based on the observations that the pH of 0.5% glucose medium does not change significantly during a CLS experiment, while the pH of 0.05% glucose medium actually becomes more basic. This finding has been recently been extended to a variety of alternative carbon sources with the observation that culture pH at day 2 or day 4 of the experiment shows a significant inverse correlation with CLS (Murakami et al., 2011) in agreement with earlier results (Fabrizio et al., 2004a; Fabrizio et al., 2005). They also suggested that CLS extension resulting from mutations in the Ras/AC/PKA and Tor/S6K pathway might be explained in part by their enhanced resistance to cell death induced by very high levels of acetic acid accumulation in the medium. Finally, bolstering their original conclusions, Burtner et al., using a modified high-throughput CLS method, performed a screen of 550 deletion strains and concluded that gene deletions that increase CLS (as measured by the 2% glucose SDC method) often result in reduced medium acidification and do not overlap well with known genes that affect life span in C. elegans when their function is reduced (Burtner et al., 2011). Below, the authors’ alternative viewpoints on the role of acetic acid in CLS are detailed further.
Viewpoint 1
We (VDL, GSS) disagree with many aspects and the primary interpretations of the two Burtner et al. studies for the following reasons:
-
The yeast CLS assay has been remarkably successful in identifying genes and pathways confirmed to play pro- and anti-aging roles in mammals. Both targeted and unbiased approaches based on CLS analysis in the acidic pH have provided the first evidence that Ras, adenylate cyclase, and PKA promote chronological aging and that SOD2 is required for the life span extension effect of mutations in these pathways (Fabrizio et al., 2003; Longo, 1997; Longo et al., 1996; Wei et al., 2008; Wei et al., 2009). Since then, Ras, adenylate cyclase and PKA have all been shown to promote aging and diseases in mice and mechanisms similar to those described for the regulation of CLS in both acidic and neutral pH conditions (Table I, S1, S2) have been proposed (Borras et al., 2011; Enns et al., 2009a; Enns et al., 2010; Yan et al., 2007), which involve stress resistance transcription factors and SOD2. More recently, reduced expression of Ras and PKA, which reduces age-dependent DNA damage in the CLS assay, was found to be associated with IGF-I deficiency and increased cellular protection in a study of a growth hormone receptor deficient population, shown to be protected against diabetes and cancer (Guevara-Aguirre et al., 2011). The CLS system also led to the initial identification of the role of the Tor/S6k pathway in stress resistance and aging (Fabrizio et al., 2001) and of the mechanisms involved (Bonawitz et al., 2007; Fabrizio et al., 2003; Pan et al., 2011; Powers et al., 2006), which was later confirmed in worms, flies, and mice (Fontana et al., 2010).
Studies of yeast CLS also provided some of the first conclusive evidence for mechanisms involved in the effect of calorie restriction on longevity. For example, the down-regulation of the Ras/AC/PKA and Tor/Sch9 pathways and up-regulation of stress resistance transcription factors Msn2, Msn4, and Gis1 are required for a major portion of the starvation/CR-dependent CLS extension (Wei et al., 2008). In addition, yeast CLS studies were the first to reveal the role of spermidine in longevity (Eisenberg et al., 2009), which also extends life span in worms and flies. Finally, yeast CLS studies were among the first to show that the Tor inhibitor rapamycin extends organismal life span, which was confirmed recently in mice (Harrison et al., 2009; Powers et al., 2006).
-
Burtner et al. showed that wild type BY4743 cells generate 2–8 mM acetic acid, yet in the experiments used to demonstrate the toxicity of acetic acid they use up to 100 times that concentration (200 mM) (Burtner et al., 2009). Furthermore, they used a CLS method that promotes depletion of ethanol, perhaps due to their use of plastic caps that promote high oxygenation versus the standard method in the Longo lab using aluminum foil caps (Fig. 2), which reduces oxygen levels and better recapitulates the conditions encountered by yeast cells in a colony. Although Burtner et al. propose that acetic acid is an extrinsic toxic factor, at the physiological levels generated and released during chronological aging, it is instead a non-toxic carbon source which can be used by yeast for growth (Brown et al., 1975; Gilvarg and Bloch, 1951; Weinhouse and Millington, 1947). Together with the well-established effect of ethanol in increasing metabolic rates and the role of carbon sources including acetic acid in activating pro-aging pathways, we (VDL, GSS) believe that the studies of Fabrizio et al. and Burtner et al. most likely demonstrate that at physiological levels ethanol and acetic acid act as carbon sources that prevent entry of cells into a calorie restriction-like mode.
One of the findings we all agree with is that acidification accelerates aging (Burtner et al., 2009; Fabrizio et al., 2004a). However, because extracellular acidification promotes intracellular acidification, which in turn can cause an increase in Ras signaling (Colombo et al., 1998) and ROS (Burhans and Weinberger, 2009), we believe that low pH simply accelerates aging through the activation of nutrients signaling pathways and an increase in oxidative damage (i.e. by exacerbating known pathways involved in yeast chronological aging)..
The proposal that the effects of mutations in the Ras/AC/PKA and Tor/S6K pathway on CLS are due to increased resistance to the toxic acetic acid does not explain their effect on CLS, as these mutations extend CLS in a range of media in which acetic acid is absent or present at very low levels (i.e. at least an order of magnitude below toxic levels). These include CLS experiments in water (Table 1 and Table S1, S2) and trp- plates (Fabrizio et al., 2005; Wei et al., 2008; Wei et al., 2009). In addition experiments in which the medium of wild type and long-lived cells was swapped after the accumulation of acetic acid had commenced had no effect on the CLS of either strain (Pan et al., 2011) and the effect of spermidine in extending CLS was shown to be independent of pH (Eisenberg et al., 2009).
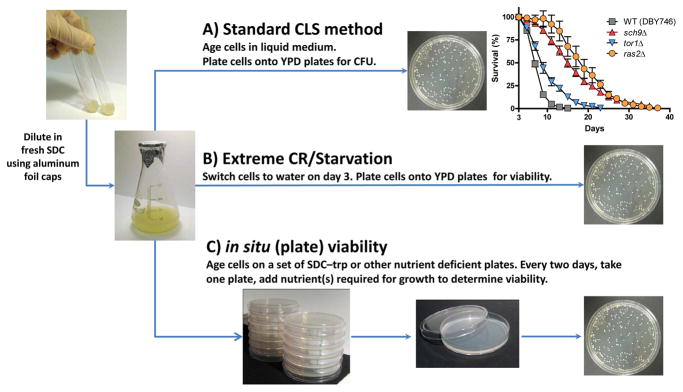
Figure 2. Yeast chronological life span (CLS).
(A) The standard 2% glucose (SDC) CLS assay. Overnight cultures are diluted (1:200) into 10 ml of fresh SDC medium (with flask to culture volume of 5:1) at 30°C with shaking (200 rpm) to ensure equal aeration of all cells. Cells grow logarithmically until they reach the mostly non-dividing high metabolism post-diauxic phase within 24 hours. Notably, aluminum foil, which reduces the level of oxygen in the flask, is used to cap the flask. The inoculation time point is time 0. Every two days, aliquots from the culture are diluted according to the estimated survival and plated on to YPD (Yeast Peptone Dextrose) plates. The YPD plates are incubated at 30°C for 2–3 days and viability in the flask is estimated by Colony Forming Unit (CFUs) counts. Viability at day 3, when the great majority of the cells stop dividing, is considered to be the initial survival (100%). Representative results of chronological survival of the wild type (DBY746), sch9Δ, tor1Δ and ras2Δ are shown.
(B) For extreme CR/starvation, cells from three-day-old SDC culture are washed three times with sterile distilled water, and resuspended in water. Cells are incubated in water at 30°C with shaking. Every 2–4 days, cells from the water cultures are washed to remove nutrients released from dead cells. For CR modeled by glucose reduction, overnight SDC cultures are diluted (1:200) into fresh SC medium supplemented with 0.5% instead of 2% glucose.
(C) Chronological survival in the presence of various carbon sources using the in situ viability assay. Day one SDC cultures are diluted and plated onto at least 10 agar plates (extreme calorie restriction) or tryptohpan drop-out (SC-TRP) plates (only trp auxotrophic cells). Plates are incubated at 30°C for the duration of the assay. Every two days, one plate from each set is retrieved and either tryptophan or the required nutrients are added to allow the growth and colony formation by the surviving cells.
Viewpoint 2
We (MK and BK) stand behind both the data and interpretation of our studies. We proposed a model that acetic acid was the toxic component of the acidified culture medium, based on multiple experimental data points, including the observations that (1) acetic acid accumulates in the culture medium during aging, (2) among several organic acids tested, only acetic acid is sufficient to induce death, and (3) buffering the medium to pH 6.0 is sufficient to extend CLS to an extent comparable to the most potent reported interventions, including dietary restriction or deletion of SCH9. We agree that other components of the culture medium, including ethanol, may also contribute to cell death, particularly under different experimental conditions. The key point here is that, even under these conditions, preventing acidification of the medium is sufficient to extend CLS. Thus, regardless of whether ethanol or acetic acid is the primary cause, acidification is intimately linked to cell death when cells are grown in SD 2% glucose medium, the standard conditions employed by a majority of laboratories using this system. The other methods described in this review, such as buffering the medium, transfer to water, and use of trp- plates provide useful alternatives where acidification no longer limits CLS.
Potential CLS artifacts
Although the CLS methods appear to be relatively simple, there are a number of potential artifacts that can affect the results obtained and their interpretation:
The most common artifact in CLS experiments is regrowth or “gasping” of aging cells, which occurs when a small subset of the population escapes from quiescence and re-enters the cell cycle (Fabrizio et al., 2004b). Assuming that a sufficient number of age-points are determined, gasping is easily detected when in occurs late in the CLS experiment because it yields an apparent increase in survival, relative to the prior age-point. More problematic are cases where regrowth or continuous slow growth occurs early in the CLS experiment (e.g. days 3–6). The alternative CLS assays involving transfer to water or trp- plates have the advantage that this potential artifact is eliminated.
Interpretation of CLS experiments performed in rich YPD medium are complicated by the extended post-diauxic phase (up to one week) during which cells continue to grow slowly (Werner-Washburne et al., 1996). For this reason, we do not recommend performing CLS assays in YPD in the absence of a direct comparison to the same mutation/intervention using SDC as the culture medium.
The use of prototrophic strains that can biosynthesize all of the amino acids or the supplementation with amino acids or carbon sources after day 1 should generally be avoided since it promotes growth. However, survival of prototrophic strains can be studied in water with frequent washes.
High throughput screens or systematic analyses of mutants (transposon, YKO, etc) using only one method to identify long-lived mutants is useful for an initial identification of candidate genes but appropriate replication and validation steps are necessary to rule out false positive effects. Confirmation with at least 3 samples is required, and confirmation with additional methods as well as in strains of different genetic backgrounds is desirable. These confirmations are important to rule out the identification of mutants that appear to be long-lived because they undergo delayed growth or regrowth during stationary phase and also to remove false positives due to other causes.
Although, we all agree that medium acidification caused by ethanol and/or acetic acid accelerates aging and death in cells grown in complete glucose medium (SDC) one disagreement between the authors (see previous section) is whether acetic acid: 1) accumulates to biologically relevant levels in the medium during CLS experiments, 2) is a toxin or simply a carbon source and 3) has a significant effect on CLS in general. One way to counteract this acidification and extend CLS is to alkalinize the medium (see above). However, as for standard SDC experiments, the possibility of regrowth must be ruled out when using alkalinization of the medium (from the standard <4 pH) in liquid culture (see #1 above).
CLS vs RLS: the effect of the shared aging regulatory genes is conserved in higher eukaryotes
One important fact about aging research in yeast is that the genes and components of pathways that affect both RLS and CLS, including Tor/Sch9, and Ras/AC/PKA as well as many downstream factors affecting stress-dependent transcription and translation have conserved orthologs or analogs in higher eukaryotes. As discussed earlier the Tor/Sch9 pathway has now been shown to promote aging in all the major model organisms for aging and mutations that reduce Ras, AC, and PKA activity all extend the life span of mice (Borras et al., 2011; Enns et al., 2009b; Yan et al., 2007). They may also represent two of the major pathways responsible for the effects of nutrients and calories on aging. Because the S. cerevisiae aging process that will shed light on the mechanisms of mammalian aging is likely to be identifiable by both the RLS and CLS methods, it is important to continue to focus on the common denominators responsible for life span extension in both methods.
Conclusions
Both yeast assays, replicative and chronological, were designed to provide amenable systems to develop hypotheses about mammalian aging and, at least at the level of identifying conserved genes, they appear to have succeeded in their goals. The question remains: will the mechanisms also be conserved. Surprisingly, while aging gene identification has been successful, linking the genes to mechanisms driving aging has proven more difficult. We believe this derives from the complex nature of aging in eukaryotes and, as stated in the CLS section, the time is ripe in both assays for a systems biology approach designed to generate aging networks. Elaborate processes likely require elaborate explanations.
What is clear is that both yeast assays have been at the forefront of discovery in the aging field, leading or playing a prominent in the discovery of Sir2, TOR, RAS, adenylate cyclase, PKA and S6 kinase as conserved modulators of longevity. Moreover, evidence to date indicates that the downstream pathways discovered to date, such as regulation of stress responsive transcription factors (Msn2/4, Gis1 and Gcn4), reduced translation, enhanced autophagy, control of oxygen radicals and the protective responses they invoke have conserved effects on aging in multicellular eukaryotes. Time will tell if one yeast aging assay ultimately proves more informative than the other and to what extent they are measuring related outcomes, but the data to date clearly indicate that both assays are powerful and should continue to be exploited as we attempt to understand human aging and develop therapeutic approaches to mitigate the diseases that it enables.
Supplementary Material
Acknowledgments
We thank Dr. Min Wei for the careful reading of the manuscript and helpful comments. This study was funded, in part, by NIH/NIA grants AG20642, AG025135 and AG034906 to V.D.L, AG039390 to MK, AG033373 to B.K.K., AG025549 to M.K. and B.K.K., by grants from the U.S. Army Research Office and The Glenn Foundation to G.S.S. MK is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerts AM, Zabrocki P, Govaert G, Mathys J, Carmona-Gutierrez D, Madeo F, Winderickx J, Cammue BP, Thevissen K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009;583:113–117. doi: 10.1016/j.febslet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Weindruch R. The caloric restriction paradigm: Implications for healthy human aging. American journal of human biology : the official journal of the Human Biology Council. 2012;24:101–106. doi: 10.1002/ajhb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes & development. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell metabolism. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol Cell Biol. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1093/genetics/166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras C, Monleon D, Lopez-Grueso R, Gambini J, Orlando L, Pallardo FV, Santos E, Vina J, Font de Mora J. RasGrf1 deficiency delays aging in mice. Aging (Albany NY) 2011;3:262–276. doi: 10.18632/aging.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta G, Turn CS, Quintyne NJ, Kirchman PA. Increased iron supplied through Fet3p results in replicative life span extension of Saccharomyces cerevisiae under conditions requiring respiratory metabolism. Exp Gerontol. 2011;46:827–832. doi: 10.1016/j.exger.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG, Romanick MA, Kennedy MA. Effects of growth hormone and insulin-like growth factor-1 on hepatocyte antioxidative enzymes. Exp Biol Med (Maywood) 2002;227:94–104. doi: 10.1177/153537020222700203. [DOI] [PubMed] [Google Scholar]
- Brown HD, Satyanarayana T, Umbarger HE. Biosynthesis of branched-chain amino acids in yeast: effect of carbon source on leucine biosynthetic enzymes. J Bacteriol. 1975;121:959–969. doi: 10.1128/jb.121.3.959-969.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans WC, Weinberger M. Acetic acid effects on aging in budding yeast: are they relevant to aging in higher eukaryotes? Cell Cycle. 2009;8:2300–2302. doi: 10.4161/cc.8.14.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle. 2011;10:1385–1396. doi: 10.4161/cc.10.9.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, et al. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Buttner S, Ruli D, Vogtle FN, Galluzzi L, Moitzi B, Eisenberg T, Kepp O, Habernig L, Carmona-Gutierrez D, Rockenfeller P, et al. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011;30:2779–2792. doi: 10.1038/emboj.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Ugidos A, Liu B, Oling D, Kvint K, Hao X, Mignat C, Nachin L, Molin M, Nystrom T. Absence of mitochondrial translation control proteins extends life span by activating sirtuin-dependent silencing. Mol Cell. 2011;42:390–400. doi: 10.1016/j.molcel.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Cameroni E, Hulo N, Roosen J, Winderickx J, De Virgilio C. The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle. 2004;3:462–468. [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Bauer MA, Ring J, Knauer H, Eisenberg T, Buttner S, Ruckenstuhl C, Reisenbichler A, Magnes C, Rechberger GN, et al. The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis. 2011;2:e161. doi: 10.1038/cddis.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Ding Q, Keller JN. The stationary phase model of aging in yeast for the study of oxidative stress and age-related neurodegeneration. Biogerontology. 2005;6:1–13. doi: 10.1007/s10522-004-7379-6. [DOI] [PubMed] [Google Scholar]
- Cheng C, Fabrizio P, Ge H, Longo VD, Li LM. Inference of transcription modification in long-live yeast strains from their expression profiles. BMC Genomics. 2007;8:219. doi: 10.1186/1471-2164-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde JH, Gorwa MF, Colavizza D, et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C. The essence of yeast quiescence. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2011.00287.x. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Dechant R, Peter M. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Delaney JR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle. 2011a;10:156–165. doi: 10.4161/cc.10.1.14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Sutphin GL, Dulken B, Sim S, Kim JR, Robison B, Schleit J, Murakami CJ, Carr D, An EH, et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011b doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Dilova I, Wang C, Lu SP, Skinner C, Lin SJ. The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J Biol Chem. 2007;282:6161–6171. doi: 10.1074/jbc.M607661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona- Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Enns LC, Morton JF, Treuting PR, Emond MJ, Wolf NS, Dai DF, McKnight GS, Rabinovitch PS, Ladiges WC. Disruption of protein kinase A in mice enhances healthy aging. PloS one. 2009a;4:e5963. doi: 10.1371/journal.pone.0005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns LC, Morton JF, Treuting PR, Emond MJ, Wolf NS, McKnight GS, Rabinovitch PS, Ladiges WC. Disruption of protein kinase A in mice enhances healthy aging. PLoS One. 2009b;4:e5963. doi: 10.1371/journal.pone.0005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns LC, Pettan-Brewer C, Ladiges W. Protein kinase A is a target for aging and the aging heart. Aging (Albany NY) 2010;2:238–243. doi: 10.18632/aging.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes & development. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. The Journal of cell biology. 2004a;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004b;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, Valentine JS, Gralla EB, Longo VD. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS letters. 2004c;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Wei M, Fabrizio P, Hu J, Cheng C, Longo VD, Li LM. Comparative analyses of time-course gene expression profiles of the long-lived sch9Delta mutant. Nucleic Acids Res. 2010;38:143–158. doi: 10.1093/nar/gkp849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilvarg C, Bloch K. The utilization of acetic acid for amino acid synthesis in yeast. J Biol Chem. 1951;193:339–346. [PubMed] [Google Scholar]
- Goldberg AA, Bourque SD, Kyryakov P, Gregg C, Boukh-Viner T, Beach A, Burstein MT, Machkalyan G, Richard V, Rampersad S, et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp Gerontol. 2009;44:555–571. doi: 10.1016/j.exger.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Goldberg AA, Richard VR, Kyryakov P, Bourque SD, Beach A, Burstein MT, Glebov A, Koupaki O, Boukh-Viner T, Gregg C, et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging (Albany NY) 2010;2:393–414. doi: 10.18632/aging.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachinohe M, Hanaoka F, Masumoto H. Hst3 and Hst4 histone deacetylases regulate replicative lifespan by preventing genome instability in Saccharomyces cerevisiae. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16:467–477. doi: 10.1111/j.1365-2443.2011.01493.x. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Kabil H, Kabil O, Banerjee R, Harshman LG, Pletcher SD. Increased transsulfuration mediates longevity and dietary restriction in Drosophila. Proc Natl Acad Sci U S A. 2011;108:16831–16836. doi: 10.1073/pnas.1102008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS genetics. 2005a;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kennedy BK. Large-scale identification in yeast of conserved aging genes. Mechanisms of ageing and development. 2005;126:17–21. doi: 10.1016/j.mad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life-span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing research reviews. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005c;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006a;312:1312b. doi: 10.1126/science.1124608. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006b;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006c;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Katewa SD, Kapahi P. Dietary restriction and aging, 2009. Aging Cell. 2010;9:105–112. doi: 10.1111/j.1474-9726.2010.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, et al. Elevated Proteasome Capacity Extends Replicative Lifespan in Saccharomyces cerevisiae. PLoS genetics. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005a;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005b;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez P, Culotta VC, Fink G, Guarente L. Calorie restriction extends Saccharomyces cerevisiae life span by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. 411SI–413SI. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002015. doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD. The Pro-senescence Role of Ras2 in the Chronological Life Span of Yeast. Los Angeles: University of California Los Angeles; 1997. pp. 112–153. [Google Scholar]
- Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Longo VD, Liou LL, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- Lu JY, Lin YY, Sheu JC, Wu JT, Lee FJ, Chen Y, Lin MI, Chiang FT, Tai TY, Berger SL, et al. Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction. Cell. 2011;146:969–979. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SP, Lin SJ. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim Biophys Acta. 2010;1804:1567–1575. doi: 10.1016/j.bbapap.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean MJ, Aamodt R, Harris N, Alseth I, Seeberg E, Bjoras M, Piper PW. Base excision repair activities required for yeast to attain a full chronological life span. Aging Cell. 2003;2:93–104. doi: 10.1046/j.1474-9728.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Fabrizio P, Longo VD. A simple model system for age-dependent DNA damage and cancer. Mech Ageing Dev. 2007;128:45–49. doi: 10.1016/j.mad.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Wei M, Fabrizio P, Burhans WC, Weinberger M, Galbani A, Smith JR, Nguyen C, Huey S, et al. Longevity mutation in SCH9 prevents recombination errors and premature genomic instability in a Werner/Bloom model system. J Cell Biol. 2008;180:67–81. doi: 10.1083/jcb.200707154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Wei M, Yuan V, Hu J, Gattazzo C, Pham P, Goodman MF, Longo VD. Oncogene homologue Sch9 promotes age-dependent mutations by a superoxide and Rev1/Polzeta-dependent mechanism. J Cell Biol. 2009;186:509–523. doi: 10.1083/jcb.200906011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007a;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to Sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007b;5:230–241. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leao C, Costa V, Rodrigues F, Burhans WC, Ludovico P. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci U S A. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M, Yang J, Hanzen S, Toledano MB, Labarre J, Nystrom T. Life Span Extension and H(2)O(2) Resistance Elicited by Caloric Restriction Require the Peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell. 2011;43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Murakami CJ, Wall V, Basisty N, Kaeberlein M. Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PloS one. 2011;6:e24530. doi: 10.1371/journal.pone.0024530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Effects of Cu-Zn superoxide dismutase overexpression of life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1993;301:34–40. doi: 10.1006/abbi.1993.1111. [DOI] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell metabolism. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY) 2009;1:131–145. doi: 10.18632/aging.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Burckert N, Egger P, De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000;19:2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mechanisms of ageing and development. 2006;127:733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty R, Keil RL. SCH9, a putative protein kinase from Saccharomyces cerevisiae, affects HOT1-stimulated recombination. Mol Genet Genomics. 2004;272:264–274. doi: 10.1007/s00438-004-1049-x. [DOI] [PubMed] [Google Scholar]
- Qin H, Lu M, Goldfarb DS. Genomic instability is associated with natural life span variation in Saccharomyces cerevisiae. PloS one. 2008;3:e2670. doi: 10.1371/journal.pone.0002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Boller T, Wiemken A, De Virgilio C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998;12:2943–2955. doi: 10.1101/gad.12.18.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesen M, Morgan A. Calorie restriction reduces rDNA recombination independently of rDNA silencing. Aging Cell. 2009;8:624–632. doi: 10.1111/j.1474-9726.2009.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS. The evolutionarily conserved longevity determinants HCF-1 and SIR- 2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet. 2010;56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome research. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Agarwal A, Agarwal S, Orr WC. Simultaneous overexpression of copper- and zinc-containing superoxide dismutase and catalase retards age-related oxidative damage and increases metabolic potential in Drosophila melanogaster. J Biol Chem. 1995;270:15671–15674. doi: 10.1074/jbc.270.26.15671. [DOI] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009 doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Molitor J, Tower J. Effects of simultaneous over-expression of Cu/ZnSOD and MnSOD on Drosophila melanogaster life span. Mech Ageing Dev. 2004;125:341–349. doi: 10.1016/j.mad.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- Unal E, Kinde B, Amon A. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science. 2011;332:1554–1557. doi: 10.1126/science.1204349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, Longo VD. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Feng L, Paul A, Smith DL, Jr, Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. DNA replication stress is a determinant of chronological lifespan in budding yeast. PloS one. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Mesquita A, Caroll T, Marks L, Yang H, Zhang Z, Ludovico P, Burhans WC. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2010;2:709–726. doi: 10.18632/aging.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S, Millington RH. Acetate metabolism in yeast, studied with isotopic carbon. J Am Chem Soc. 1947;69:3089–3093. doi: 10.1021/ja01204a048. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun EL, Crawford ME, Peck VM. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.