Abstract
microRNAs (miRNAs) are endogenous short RNAs that mediate vast networks of post-transcriptional gene regulation. Although computational searches and experimental profiling provide evidence for hundreds of functional targets for individual miRNAs, such data rarely provide clear insight into the phenotypic consequences of manipulating miRNAs in vivo. We describe a genome-wide collection of 165 Drosophila miRNA transgenes and find that a majority induced specific developmental defects, including phenocopies of mutants in myriad cell-signaling and patterning genes. Such connections allowed us to validate several likely targets for miRNA-induced phenotypes. Importantly, few of these phenotypes could be predicted from computationally predicted target lists, thus highlighting the value of whole-animal readouts of miRNA activities. Finally, we provide an example of the relevance of these data to miRNA loss-of-function conditions. Whereas misexpression of several K box miRNAs inhibited Notch pathway activity, reciprocal genetic interaction tests with miRNA sponges demonstrated endogenous roles of the K box miRNA family in restricting Notch signaling. In summary, we provide extensive evidence that misexpression of individual miRNAs often induces specific mutant phenotypes that can guide their functional study. By extension, these data suggest that the deregulation of individual miRNAs in other animals may frequently yield relatively specific phenotypes during disease conditions.
Keywords: Drosophila, Notch, Cell signaling, MicroRNA
INTRODUCTION
microRNAs (miRNAs) are ~22 nucleotide RNAs derived from transcripts bearing short inverted repeats, and are broadly distributed across eukaryotes (Axtell et al., 2011). Hundreds of miRNAs have been recorded in higher animals, and their biogenesis via canonical and alternative pathways has been extensively characterized (Yang and Lai, 2011). Computational strategies have been developed to identify target sites that are under evolutionary constraint, or that are species specific yet exhibit features of functional sites (Bartel, 2009; Garcia et al., 2011). These approaches yield predictions of hundreds to thousands of direct targets per animal miRNA. Evidence for such extensive target networks have been corroborated by systematic profiling of the transcriptome (Lim et al., 2005; Giraldez et al., 2006) and proteome (Baek et al., 2008; Selbach et al., 2008). Such quantitative studies indicate that most miRNA targets are influenced only subtly, even when measured using ectopic expression assays. This has been rationalized by the notion that animal miRNAs are often used for broad ‘fine-tuning’ of the transcriptome, or perhaps to silence spurious transcription.
A contrasting perspective emerges from genetic studies (Flynt and Lai, 2008; Smibert and Lai, 2010). In C. elegans, the first miRNAs and their targets emerged from the cloning of mutants that disrupted developmental progression and from knowledge of epistatic genetic interactions (Lee et al., 1993; Wightman et al., 1993; Reinhart et al., 2000). Contemporary studies in D. melanogaster demonstrated essential control of neural patterning by miRNA-binding sites within individual Notch target genes (Lai and Posakony, 1997; Lai et al., 1998; Lai and Posakony, 1998) and revealed the basic principle of seed-matched targeting (Lai, 2002). One may wonder if the first few miRNAs were somehow atypical, having been selected on the basis of visible morphological defects. A nearly genome-wide set of C. elegans miRNA deletions revealed surprisingly little in the way of obvious developmental or behavioral phenotypes (Miska et al., 2007), even when assayed as deletions of entire miRNA families (Alvarez-Saavedra and Horvitz, 2010). Similarly, most published deletions of Drosophila or mouse miRNAs are viable and of relatively normal exterior appearance (Smibert and Lai, 2008). Nevertheless, there is no shortage of compelling miRNA phenotypes in Drosophila (Dai et al., 2012), mouse (Xiao and Rajewsky, 2009; Small and Olson, 2011) and even C. elegans (Brenner et al., 2010), once the appropriate biological setting and the appropriate genetic background have been examined. However, such information is not easy to glean.
A complementary approach is the study of gain-of-function phenotypes. Although one must be cautious in divining the endogenous function of genes from elevating their activity, over- and mis-expression assays have long been crucial tools in the genetic arsenal. In several cases, miRNA gain-of-function phenotypes proved relevant to their loss-of-function phenotypes. One of the most overtly important Drosophila miRNAs, bantam, was isolated by gain-of-function assays that revealed its capacity to promote tissue growth and inhibit apoptosis (Brennecke et al., 2003). Bantam proved to mediate these functions as a direct transcriptional target of the Hippo and BMP signaling pathways, which have deeply conserved activities in controlling tissue patterning and organ size (Nolo et al., 2006; Thompson and Cohen, 2006; Oh and Irvine, 2011), and bantam loss severely impairs tissue growth and confers susceptibility to apoptosis (Hipfner et al., 2002; Brennecke et al., 2003; Jaklevic et al., 2008). As another example, deletion of Drosophila Hox locus mir-iab-4/8 exhibits spatial broadening of the Hox protein Ubx in the embryo (Bender, 2008), consistent with the striking Ubx phenocopies of haltere-to-wing transformation induced by ectopic miR-iab-4 and miR-iab-8 (Ronshaugen et al., 2005; Stark et al., 2008; Tyler et al., 2008).
We created a genome-wide resource for conditional expression of Drosophila miRNAs using the Gal4-UAS system. For maximal utility, we generated collections of random insertions using P transgenesis, as well as site-directed insertions using øC31 integrase. These miRNA transgenes collectively induced hundreds of dominant morphological phenotypes, many of which closely resemble specific alterations in core cell signaling pathways that mediate tissue patterning. The specificities of these phenotypes were not predictable from computational studies, highlighting the utility of in vivo phenotypic assays of miRNA function. We provide case illustrations of how these data can be used to determine relevant direct miRNA targets, and how they can inform the analysis of miRNA loss-of-function phenotypes in appropriately sensitized genetic backgrounds. Altogether, these transgenes comprise a genetic resource to study miRNA biology, and reveal unexpected capacity of different miRNAs to generate specific dominant phenotypes in the intact animal.
MATERIALS AND METHODS
Drosophila stocks
We obtained the following Gal4 driver stocks from the Bloomington Stock Center: sd-Gal4, bx-Gal4, ptc-Gal4, dpp-Gal4, GMR-Gal4 and da-Gal4. Stocks for generating mutant wings included UAS-neur (Lai and Rubin, 2001), UAS-E(spl)mgamma (Ligoxygakis et al., 1999), UAS-rolled[Sem] (Martin-Blanco, 1998), FRT40-Su(H)Δ47, P(B) (Morel and Schweisguth, 2000) and UAS-tkv[QD] (Gilboa and Lehmann, 2004). UAS-RNAi transgenes against dpp, hedgehog, Su(H), expanded and UAS-Dicer-2 for enhancing RNAi effects, were from the Vienna Drosophila RNAi Center. We also used the miRNA sensor tub-GFP-abrupt 3′UTR (Okamura et al., 2008).
UAS-miRNA transgenes
P element collection
We used these published UAS-DsRed-miRNA transgenes: mir-2 cluster, mir-4/5/286, mir-7 and mir-11 (Stark et al., 2003), as well as mir-6-1/6-2/6-3 and mir-79 (Lai et al., 2005). The remaining transgenes were prepared for this study. About 70 of these derived from our UAS-DsRed-miRNA plasmid collection (Silver et al., 2007). The rest were cloned using a similar strategy, placed into the 3′ UTR position of UAS-DsRed (Stark et al., 2003) using 5′ NotI and 3′ XbaI or 3′ XhoI sites. The miRNA inserts included ~200-250 nucleotides flanking each side of the pre-miRNA hairpin and were amplified from w[1118] or Canton S genomic DNA. In cases of clusters, we included a similar amount of genomic sequence flanking the 5′-most and 3′-most miRNA hairpins. When subdividing clusters so as to express only single miRNAs, we were not always able to include >200 nucleotide flanks; we maximized the endogenous flanking sequence as much as possible. These amplicons were either cloned directly into UAS-DsRed or were first cloned using TOPO-D-entr (Invitrogen) and then subcloned. In some cases, we used a version of UAS-DsRed in which we had inserted a Gateway compatible cassette for direct transferal of TOPO-D-entr inserts. The primers used for cloning are available upon request. P element lines were injected in house (with great help from Todd Laverty) or by BestGene (Chino Hills, CA, USA) using Δ2-3 helper transposase.
attP collection
To construct pWALIUM10-Luc, pWALIUM10-moe (http://www.flyrnai.org/TRiP-HOME.html) was digested with XbaI. Luciferase DNA fragment was amplified by PCR from pVALIUM10-Luciferase using specific primers (F, 5′-GGTCTAGAACCATGGAAGACGCCAAAAAC-3′; R, 5′-GGACTAGT TTACACGGCGATCTTTCCGC-3′). Luciferase fragment was digested with XbaI and SpeI, and was ligated into the linearized pWALIUM10-moe vector resulting in pWALIUM10-Luc. We amplified the miRNA inserts from the P element transgene collection using specific primers (XbaI-F, 5′-GCTCTAGAATCGTGGAGCAGTACGAGCG-3′; XbaI-R, 5′-GCTCTAGAGTAAGGTTCCTTCACAAAGATC-3′). PCR products were digested with XbaI and were cloned into the XbaI sites of the pWALIUM10-Luc vector. The sequence and the orientation of the individual miRNA fragments were confirmed by DNA sequencing. Landing site transformants were established at Genetic Services (GSI, http://www.geneticservices.com) by injecting miRNA constructs into flies carrying attP docking site, attP2, located on the third chromosome.
UAS-miRNA sponge transgenes
To generate the second generation miR-SP constructs used in this study, we modified the original miR-SP design (Loya et al., 2009) as follows: a silencing cassette of 20 repetitive microRNA complementary sequences separated by variable four-nucleotide linker sequences was introduced into the 3′UTR of mCherry. To avoid off-target effects, a sliding window of seven or eight nucleotides long between each linker and the adjacent sponge subunits was checked against every mature microRNA sequence in the Drosophila genome. The entire cassette was then cloned between NotI and XbaI sites into a modified pVALIUM10 vector (Ni et al., 2009) carrying the white+ selectable eye color marker instead of Vermilion (pWALIUM10-moe, http://www.flyrnai.org/TRiP-HOME.html) according to the manufacturer's protocols (BioBasic). To avoid epigenetic positional effects and obtain lines with equal expression levels, transgenic flies carrying one or two copies of the each miR-SP cassette were generated using phiC31 site-specific genomic integration (Genetic Services). The sequences of all K box miR-SP (miR-2bSP, miR-2cSP, miR-13aSP, miR-13bSP and miR-6SP), miR-7SP and SCRAMBLE-SP constructs are listed in supplementary material Table S3.
Immunostaining of imaginal discs
We used rabbit anti-GFP (Molecular Probes, 1:500), mouse anti-β-Gal (Developmental Studies Hybridoma Bank, 1:2000) and a previously described immunostaining protocol for imaginal disc histology (Lai and Rubin, 2001).
Luciferase sensor assays
3′ UTRs of predicted miRNA targets were cloned into the psiCHECK-2 vector (Promega) using the cold fusion cloning kit (System Biosciences). Sensors contained the entire annotated 3′ UTR, as well as a more than 150 nucleotide downstream sequence to ensure normal 3′ end formation; primers for cloning sensors are listed in supplementary material Table S3. Luciferase assays were performed as previously described (Okamura et al., 2007).
RESULTS
Genome-wide collections of conditionally activatable Drosophila miRNA transgenes Prior to the general recognition of miRNA genes in Drosophila, functional screening of large collections of randomly inserted P elements bearing UAS sites (i.e. ‘EP’ lines) identified certain loci that readily generated dominant phenotypes, but were not associated with obvious protein-coding genes (Abdelilah-Seyfried et al., 2000; Kraut et al., 2001; Hipfner et al., 2002). With the subsequent identification of scores of miRNA genes in flies (Lagos-Quintana et al., 2001; Aravin et al., 2003; Lai et al., 2003), some EP lines were recognized to hit miRNA loci, implying that miRNA genes could be conditionally regulated using the Gal4-UAS system.
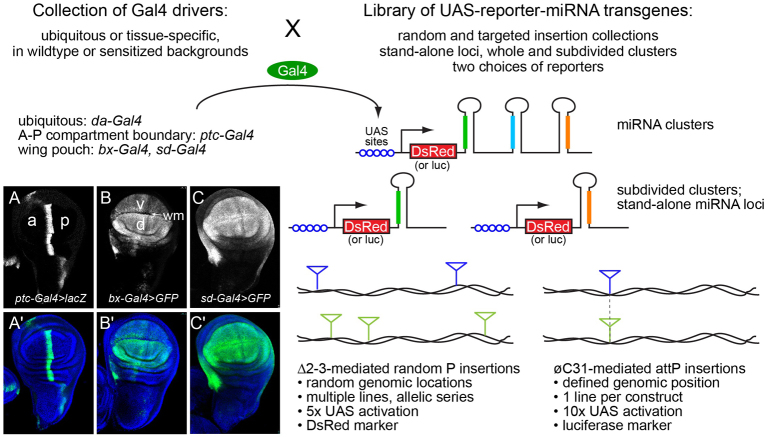
These observations prompted us to construct a large-scale set of UAS-miRNA transgenes. We initially created a dozen of these as straight UAS constructs bearing 400-500 bp fragments, including the miRNA hairpin. However, following the demonstration that UAS-DsRed-miRNA transgenes effectively produce active miRNAs and functional DsRed protein as a cell-autonomous marker (Stark et al., 2003), we switched to this format (Fig. 1). We remade the initial transgenes in the UAS-DsRed-miRNA layout and found that they induced identical phenotypes (supplementary material Table S1, S2). We then made transgenes covering the initially characterized miRNAs (Lagos-Quintana et al., 2001; Aravin et al., 2003; Lai et al., 2003), then updated the collection as additional canonical miRNAs (Ruby et al., 2007b; Berezikov et al., 2010) and non-canonical miRNAs (e.g. ‘mirtrons’) were annotated (Okamura et al., 2007; Ruby et al., 2007a). In many cases, miRNAs reside in local clusters that are co-expressed as operons from longer primary miRNA transcripts. Where practical, we generated transgenes of the entire cluster (usually for operons of less than 4 kb), as well as for the individual members of the cluster; occasionally, we constructed additional ‘sub-cluster’ transgenes where certain groups of miRNAs happened to be in close proximity. In total, this collection includes 665 lines, comprising 165 different transgenes that cover 149 distinct miRNA hairpins.
Fig. 1.
Use of the Gal4-UAS binary system for systematic in vivo screens of activated miRNA transgenes. We made two collections of UAS-miRNA transgenes, one consisting of random insertions in P-element backbones (165 different transgenes covering 149 different miRNA hairpins) and another consisting of defined insertions in attP landing sites (106 different transgenes covering 108 different miRNA hairpins), including stand-alone loci, miRNA clusters and subdivided clusters. The collections have complementary experimental strengths. We crossed these with a variety of ubiquitous and tissue-specific Gal4 drivers to assess systematically the consequences of ectopic miRNA activity. Examples of Gal4 driver patterns are shown on the left in wing imaginal discs (A-C) in which a lacZ or GFP reporter is driven by representative Gal4 insert; A′-C′ show reporter accumulation counterstained with DAPI. (A,A′) ptc-Gal4 is active at the border of the anterior (a) and posterior (p) compartments. (B,B′) bx-Gal4 is active throughout the wing pouch, but is elevated in the dorsal (d) relative to the ventral (v) compartment, and is not active along the presumptive wing margin (wm). (C,C′) sd-gal4 is active more uniformly throughout the wing pouch.
Given the random nature of P insertions, occasional position effects can occur. We generally analyzed three to five P insert lines for each miRNA construct to verify that phenotypes were due to the transgene sequence, as opposed to genomic locale. The vast majority of phenotypes (described below) were qualitatively similar across each insertion panel, although their strength could vary to a certain degree. The ability to recover allelic series of transgene strengths is an advantage of random P insertions (Fig. 1). In addition, the fact that independent insertions are distributed throughout the genome facilitates downstream efforts to make recombinant stocks. Nevertheless, we recognized that it might be beneficial to have inducible miRNA transgenes inserted in a common genomic location, to provide uniformity of expression and to permit direct comparisons between different constructs. We therefore constructed a second set of 107 UAS-luciferase-miRNA inserts using an attP vector (Groth et al., 2004); these transgenes offer the ability to express miRNAs without accompanying DsRed (Fig. 1). These comprise the miRNAs that are well-conserved across the Drosophilids, as well as some less-conserved but reasonably expressed loci. We summarize the P and attP collections of miRNA transgenes in supplementary material Table S1.
A diversity of defects induced by miRNAs in the developing wing resembles alteration of known signaling and patterning genes
The binary Gal4-UAS system constitutes a flexible screening platform (Brand and Perrimon, 1993; Rorth et al., 1998), and the UAS-miRNA transgenes can be deployed with numerous Gal4 drivers towards diverse assessments of miRNA gain-of-function in vivo. Initial screening of the P lines against the ubiquitous driver da-Gal4 revealed that 108/165 miRNA transgenes were lethal at embryonic or larval stages (supplementary material Table S2). Although this reported on the adverse consequences of miRNA misexpression, it did not provide substantial phenotypic information. We therefore turned to the adult wing, as wing development requires the coordinate function of multiple signaling pathways (Molnar et al., 2011), and even minor alterations in wing development are visible under the dissecting microscope.
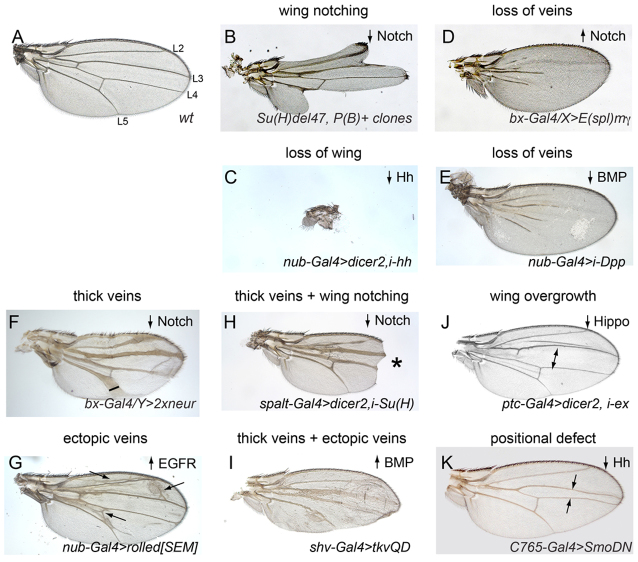
Fig. 2 illustrates how the patterning of the adult wing (Fig. 2A) is affected by gain- or loss-of-function of various signaling pathways and transcriptional regulators. For example, wing notching or wing loss is induced by loss of Notch signaling (Fig. 2B), Wnt signaling (not shown) or Hh signaling (Fig. 2C); loss of wing veins is observed upon loss of EGFR signaling (not shown), gain of Notch signaling (Fig. 2D) or loss of BMP signaling (Fig. 2E); vein thickening is caused by loss of Notch signaling (Fig. 2F) and ectopic veins are induced by gain of EGFR signaling (Fig. 2G). It is also possible to observe combinations of these phenotypes. For example, appropriate reduction of Notch signaling can simultaneously generate thick veins and wing notching (Fig. 2H), whereas appropriate activation of BMP signaling can generate both thick and ectopic wing veins (Fig. 2I). Alteration of many signaling pathways can affect wing growth and size. Inactivation of the Hippo pathway is particularly known for causing tissue overgrowth (Fig. 2J) and specific inactivation of the Hh pathway causes defects in anterior-posterior patterning of wing domains (Fig. 2K).
Fig. 2.
A diversity of patterning defects can be scored in the Drosophila wing. Shown are wild-type (A) and mutant (B-K) adult wings that illustrate major classes of mutant phenotypes. The directionality of pathway activity that yields these phenotypes is noted in the upper right corner of each panel. (A) The normal wing has a characteristic size and shape, and reproducible pattern elements such as the five longitudinal wing veins (L2-L5 are labeled) and sensory bristles that decorate the anterior margin of the wing. (B) Wing notching caused by mutant clones of the transcription factor in the Notch pathway, Su(H). (C) Loss of wing caused by knockdown of the Hedgehog ligand. (D,E) Examples of wing vein loss caused by misexpression of the Notch pathway target E(spl)mγ (D) or knockdown of the BMP ligand dpp (E). (F) Thick veins caused by high level expression of the Notch pathway component neuralized. (G) Expression of the activated MAP kinase rolled (Sevenmaker, Sem) causes ectopic wing veins. (H,I) Examples of wings that bear multiple mutant phenotypes. Asterisk indicates area of wing notching. (H) Knockdown of Su(H) induces thick veins and wing notching. (I) Misexpression of an activated BMP receptor tkv-QD causes both thick veins and ectopic veins. (J) Knockdown of the Hippo pathway component expanded in the central domain of the wing causes overgrowth (arrows). (K) Expression of a dominant-negative version of the Hedgehog receptor Smoothened (SmoDN) causes loss of the L3-L4 domain.
In order to capture a diversity of phenotypes, we screened the miRNA lines using wing drivers with broad as well as regionalized activity. We initially tested all insertions against bx-Gal4, which is active in the wing primordium, especially within the dorsal compartment. Systematic screening allowed us to categorize insertions with typical activity, as well as to identify weaker and stronger insertions. We subsequently recrossed two representative P lines to sd-Gal4, which is expressed more broadly in the wing primordium than bx-Gal4, as well as with ptc-Gal4, which is expressed in a restricted domain along the anterior-posterior compartment boundary (Fig. 1A-C). We also systematically analyzed crosses of all attP lines to sd-Gal4.
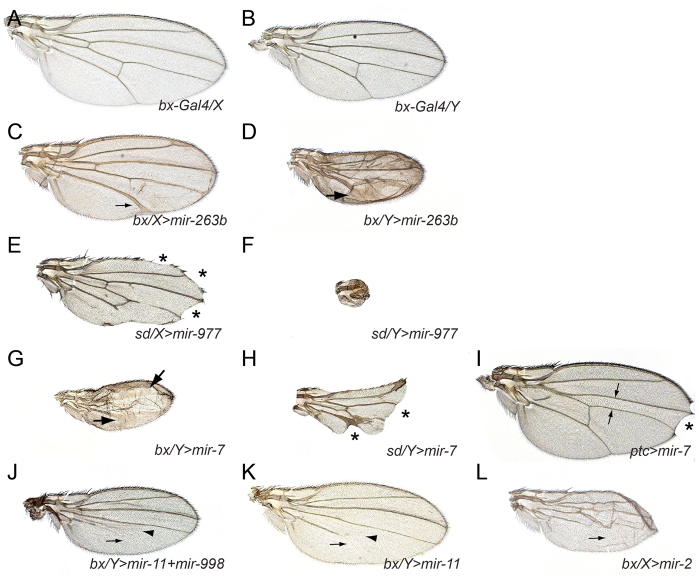
Several general properties of the UAS-miRNA phenotypes bear mention. First, the phenotypes induced were collectively very diverse. This is likely to reflect the consequences of miRNA-guided target regulation, as opposed to saturating the miRNA biogenesis pathway, as has been observed in other experimental systems (Grimm et al., 2006); such an outcome would have been manifest as non-specific phenotypes that were common to most miRNA transgenes. Second, the defects induced by miRNA transgenes were, without exception, dose sensitive. This was easily observed with X-linked drivers such as bx-Gal4 and sd-Gal4. When virgin Gal4 females were mated to UAS-miRNA males, the sons always exhibited stronger phenotypes than the daughters. For example, mir-263b induced thicker wing veins and mir-977 induced stronger loss of wing tissue, in bx-Gal4 males compared with females (Fig. 3A-F). However, in these and almost all other examples, the nature of the phenotype was usually qualitatively similar between the sexes. Exceptions included the occasional cases where Gal4 females did not induce a miRNA phenotype, whereas one was evident in males, or, conversely, where Gal4 females exhibited a phenotype, whereas the corresponding phenotype in males was fatal (supplementary material Table S2).
Fig. 3.
Selected examples of miRNA-induced wing phenotypes illustrate general properties of the UAS-DsRed-miRNA transgenes. (A,B) Wings of a heterozygous bx-Gal4 (‘bx’) female (A) and hemizygous male (B); males are smaller than females, which accounts for size difference. (C-F) Examples of dose effects. Vein thickening (arrows) induced by mir-263b is weaker in bx-Gal4 females (C) than males (D). Wing notching (asterisks) induced by mir-977 is weaker in sd-Gal4 (sd) females (E) than males (F). (G-I) Different miRNA misexpression phenotypes are evident in different Gal4 backgrounds. (G) bx/Y>mir-7 exhibits massive vein thickening (arrows), but the margin is continuous. (H) sd/Y>mir-7 exhibits massive wing notching (asterisks), and only mild vein thickening. (I) ptc-Gal4>mir-7 exhibits distal wing notching and reduction in the L3-L4 domain (arrows). (J,K) Dissection of a miRNA cluster. (J) Activation of the mir-11/mir-998 operon induces vein (arrow) and crossvein (arrowhead) loss; these phenotypes are recapitulated by ectopic mir-11 (K). (L) Similarity of seed families; mir-2 is in the same family as mir-11 and also induces vein loss (arrows).
Third, we observed that the same miRNA could induce different phenotypes when activated by different Gal4 lines. For example, depending on the driver, ectopic mir-7 caused wing notching, thickened veins or a decrease in the L3-L4 intervein domain (Fig. 3G-I) (Stark et al., 2003; Lai et al., 2005). As is the case with gain-of-function screens for protein-coding genes, then, profiling multiple drivers maximizes knowledge of their functional capacity (Rorth et al., 1998). Fourth, in many cases, miRNAs that generated phenotypes when expressed alone produced similar phenotypes when expressed as part of clusters. For example, misexpression of the mir-11/mir-998 cluster with bx-Gal4 induced loss of the L5 wing vein and the crossveins (Fig. 3J), effects recapitulated by misexpression of mir-11 alone (Fig. 3K). Exceptions included cases where the aggregate effects of multiple co-expressed miRNAs were strong enough to obscure some individual phenotypes, the clearest example being when miRNA clusters induced synthetic lethality. Fifth, we frequently observed that members of miRNA seed families often induced similar phenotypes. For example, misexpression of mir-2, which shares a ‘K box’ seed with mir-11, also induced L5 vein loss (Fig. 3L). Phenotypic overlap among miRNA seed families provided further evidence that the observed effects were driven by misregulation of specific targets, as opposed to non-specific titration of miRNA pathway components. The K box miRNAs also illustrate that distinct phenotypes are observed with different drivers, as mir-2 and mir-11, along with other family members mir-6 and mir-13, can all induce wing notching (supplementary material Fig. S1).
Of the 165 miRNA transgenes, 96 induced fully penetrant phenotypes with bx-Gal4, 98 with sd-Gal4 and 73 with ptc-Gal4 (supplementary material Table S2). Interestingly, although many of these could be grouped into qualitatively similar cohorts, the majority of miRNAs that generated similar phenotypes were not related in sequence. This was not a trivial outcome, given that different miRNA seed families typically exhibit little overlap in predicted target sets. Some of the prominent phenotypic classes that we observed included loss of wing veins (Fig. 4F-J), thickened wing veins (Fig. 4K-T), notched wings (Fig. 4U-AA), ectopic wing veins (Fig. 4BB-DD), specific loss of wing margin bristles (Fig. 4EE-OO), wing overgrowth (Fig. 4PP-UU), wing undergrowth/loss of L3-L4 domain (Fig. 4VV-ZZ), wing blistering (Fig. 4AAA-EEE), compression of the anterior-posterior axis (Fig. 4FFF) and shortening of the proximal-distal axis (Fig. 4GGG).
Fig. 4.
Summary of wing phenotypes caused by misexpression of different Drosophila miRNAs. Shown are adult female wings, except as noted for X-linked Gal4 drivers (/X= female; /Yμmale). (A-E) Except for the wild-type [w1118] wing (A), all other flies contain a single copy of Gal4 and UAS-DsRed-miRNA transgene. (B) bx-Gal4/X heterozygous females exhibit a normal wing, as do sd-Gal4/X females (not shown). (C) Gal4 activity in sd-Gal4/Y males results in a minor loss of posterior wing margin, especially near the wing hinge (boxed regions in A and C are enlarged in D and E, respectively). (F-J) Examples of vein loss induced by different miRNAs. (K-T) Examples of vein thickening induced by different miRNAs. (U-AA) Examples of wing notching induced by different miRNAs. Note that P-S and U-X highlight miRNAs that induce vein thickening or margin loss, respectively, depending on the driver. Other combinations of phenotypes are evident by inspection. (BB-DD) Examples of ectopic wing veins (arrows) induced by different miRNAs. (EE-OO) Examples of miRNAs that have a selective effect on wing margin bristles; close-ups of the anterior margin are shown in JJ-OO. (PP) ptc-Gal4 heterozygous female; the ptc+ domain includes the L3-L4 region marked by the double arrow. (QQ-UU) Examples of miRNAs that induced overgrowth of the L3-L4 domain. (VV-ZZ) Examples of miRNAs that induced undergrowth or loss of the L3-L4 domain. (AAA-EEE) Examples of miRNAs that induced wing blisters. (FFF) miRNA that induces a potential defect along the anterior-posterior axis. (GGG) miRNA that induces a potential proximal-distal defect. (HHH-OOO) Examples of other severe wing deformities or wing loss induced by different miRNAs.
We also recorded a number of examples of curly wing, small crumpled wings or wing vestiges (Fig. 4HHH-OOO), the developmental bases of which were less obviously attributable to any specific process. Nevertheless, these phenotypes were a reminder of the detrimental consequences of miRNA deregulation. Altogether, the gain-of-function of different miRNAs collectively yields a surprising variety of characteristic phenotypes. A comprehensive compilation of phenotypes is presented in supplementary material Fig. S1, and we catalog these line by line using a controlled vocabulary in supplementary material Table S2. These resources permit visual browsing as well as text searching for phenotypes of interest. Reassuringly, systematic tests of the phenotypes induced using the attP insertion lines when crossed to sd-Gal4 showed that they were qualitatively similar to the companion UAS-miRNA P insertion lines. There was not a consistent trend for the attP lines to be stronger or weaker, highlighting the advantage of the allelic diversity possible with random insertions.
Association of miRNA gain-of-function phenotypes with relevant target genes
Perhaps the simplest explanation for the ability of sequence-unrelated miRNAs to induce similar phenotypes is that they may target common pathways. Indeed, the phenotypic similarity of groups of mutants is the fundamental basis of using genetics to assemble molecular pathways. The sundry miRNA-induced phenotypes were strikingly reminiscent of phenotypes caused by dysfunction of cell signaling components and transcription factors that control wing development (Fig. 2). Although it cannot be ruled out that these miRNA-induced phenotypes were the outcome of coordinately mild reduction in the activity of tens or hundreds of targets, it seems more parsimonious to infer that they might be due to the suppression of specific wing patterning genes. The similarity of miRNA gain-of-function phenotypes to known mutants provides a way to focus target searches from among lists of hundreds of conserved targets.
For example, miR-8 was reported as an inhibitor of Wnt signaling, and shown to directly repress wntless and CG32767 (Kennell et al., 2008). These findings are consistent with the observation that ectopic miR-8 could generate wing notching (Fig. 2B), but did not induce vein thickening (which might have been consistent with a reduction of Notch signaling). The Wnt pathway ligand Wingless (Wg) is required for wing formation, and its 3′ UTR bears a highly conserved 8-mer target site for miR-8 (supplementary material Fig. S2B). The wg 3′ UTR was responsive to ectopic miR-8 in luciferase sensor assays in S2 cells (supplementary material Fig. S2C). Therefore, targeting of wg may be relevant to miR-8 activity.
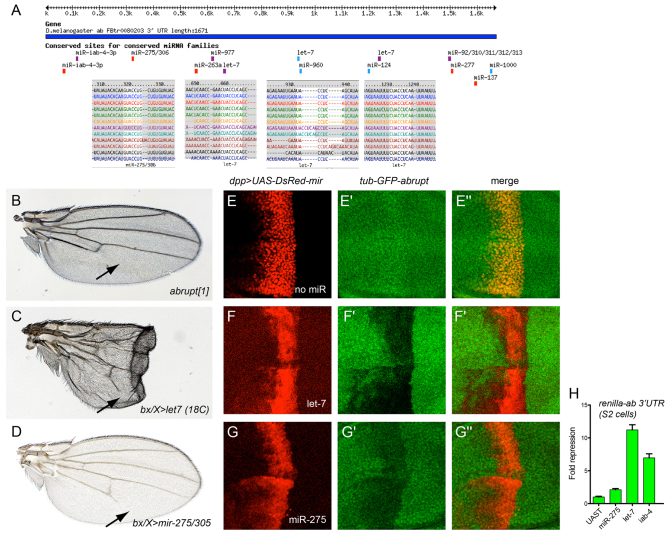
Studies of let-7 deletion indicated that the transcription factor encoded by abrupt is a key direct target (Fig. 5A), as Abrupt is misexpressed in let-7 clones and abrupt heterozygosity can suppress certain let-7 mutant phenotypes (Caygill and Johnston, 2008). Although ectopic let-7 strongly perturbed wing development, these wings also exhibited shortening of L5 wing vein, corresponding to the phenotype of viable abrupt mutants (Diaz-Benjumea and Garcia-Bellido, 1990) (Fig. 5B,C). We used a genetic assay to confirm that the 3′ UTR of abrupt can be directly repressed by let-7. We stained for the expression of a ubiquitously active tub-GFP-abrupt 3′ UTR sensor transgene (Okamura et al., 2008) in the presence of spatially restricted let-7 driven along the AP compartment boundary using dpp-Gal4. We observed strong cell-autonomous reduction of the abrupt GFP sensor in the dpp+ domain (Fig. 5E,F). Browsing our collection of wing phenotypes, we observed that ectopic expression of mir-275 was among the miRNAs that also led to loss of wing veins, particularly of L5 (Fig. 5D). We observed the abrupt 3′ UTR contains a highly conserved seed match for miR-275 (Fig. 5A), and that ectopic mir-275 suppressed the abrupt sensor in vivo (Fig. 5G). The stronger repression of the abrupt sensor by let-7 compared with miR-275 was recapitulated in luciferase assays in S2 cells (Fig. 5H). Together with our previous observation that mir-iab-4 is another strong in vivo repressor of abrupt (Okamura et al., 2008) (Fig. 5H), it appears that abrupt is substantially targeted by multiple miRNAs in Drosophila.
Fig. 5.
Validation of miRNAs that directly target abrupt. (A) Targetscan predictions of conserved miRNA-binding sites in the abrupt 3′ UTR. (B-D) Adult female wings. (B) The viable abrupt[1] mutant exhibits loss of distal L5 wing vein (arrow). (C) Misexpression of let-7 induces wing deformity even when cultured at low temperature to limit Gal4 activity; in addition, loss of the distal region of L5 is seen (arrow). (D) Misexpression of mir-275/mir-305 also induces loss of L5. (E-G″) Transgenic sensor assays in wing imaginal discs that carry tub-GFP-abrupt 3′ UTR, dpp-Gal4 and UAS-DsRed (linked to a miRNA in F-G″); the central domain of the wing pouch is shown. (E-E″) Control staining shows that expression of DsRed does not repress the abrupt sensor. (F-F″) Ectopic let-7 strongly represses the abrupt sensor. (G-G″) Ectopic miR-275 mildly represses the abrupt sensor. (H) Renilla-abrupt 3′ UTR sensor assays in S2 cells. Consistent with the in vivo results, mir-275 weakly repressed the abrupt sensor, while let-7 strongly repressed it; mir-iab-4 has previously been validated to repress the abrupt 3′ UTR (Okamura et al., 2008). Data are mean±s.e.m.
We suggest that the forward phenotypic screening is likely to serve as a rich resource for pairing many miRNAs to biological pathways. Strikingly, the in vivo work revealed many cases of miRNAs whose misexpression phenocopied strong or nearly null situations of known wing patterning genes and pathways (Figs 2, 3 and 4), for which perusal of available computational predictions of miRNA binding sites (e.g. http://www.targetscan.org/) did not reveal any candidate hits. It is also worth stating the obvious that the mere presence of a conserved miRNA binding site in a given gene does not guarantee that misexpression of the corresponding miRNA will necessarily induce a corresponding mutant phenotype.
The connections of a given miRNA to a specific pathway were bolstered in some instances by independent phenotypes. For example, ectopic miR-7 can induce both vein thickening and wing notching, both of which are associated with Notch pathway loss-of-function (Fig. 2H). We observed that mir-980, mir-12, mir-190 and mir-982 similarly induced both strong vein thickening (Fig. 4P-S) and strong notching (Fig. 4U-X), suggesting their potential connection to Notch signaling. As another example, among Drosophila miRNAs that regulate neurogenesis, mir-9a mutants exhibit ectopic notum sensory bristles (Li et al., 2006; Bejarano et al., 2010), while mir-279 mutants exhibit ectopic CO2-sensing neurons (Cayirlioglu et al., 2008). These phenotypes broadly place these miRNAs as having anti-neural properties. Consistent with this, we observed that ectopic expression of miR-279 and members of the miR-9a/b/c family specifically disrupted wing margin sensory bristles while retaining relatively normal patterning of the wing proper (Fig. 4EE-HH,KK-NN). These observations are consistent with anti-neural activity of these miRNAs. Interestingly, mir-252 similarly induced strong loss of wing margin bristles with relatively little effect on the wing margin (Fig. 4II,OO), suggesting it may also have a role in regulating neurogenesis.
Endogenous functional relevance of gain-of-function phenotypes: K box miRNAs
Our systematic phenotypic profiling tests provide the first in vivo evidence of the biological activities of the majority of Drosophila miRNAs. Certainly, many of these may be misexpression effects caused by introduction of the miRNA into an ectopic setting. Such phenotypes still provide useful insights on the in vivo functional capacities of deregulated miRNAs, and offer insights into the possible roles of these miRNAs in their normal locations. Nevertheless, it is pertinent to consider whether any of these phenotypes are relevant to the endogenous function of the miRNA.
We previously showed that multiple members of the K box family (sharing the UGUGAU seed) directly inhibit Notch target genes, and induce phenotypes reminiscent of Notch loss of function when misexpressed (Lai et al., 1998a; Lai, 2002; Lai et al., 2005). For example, miR-2 and miR-6 induce wing notching and loss of wing margin when ectopically expressed, and we extend these activities to other members of the family, including miR-11 and miR-13 (supplementary material Fig. S1). The pervasive capacity of different K box miRNAs to suppress Notch signaling motivated us to test whether their loss of function could promote Notch signaling.
Recently, we showed that transgenes bearing multimers of bulged target sites (‘sponges’) could induce miRNA loss-of-function phenotypes in Drosophila (Loya et al., 2009). We generated sponges for several K box miRNAs, including miR-2b, miR-2c, miR-13a and miR-13b. These did not induce obvious dominant phenotypes, even in animals carrying two copies of the sponge transgene (Fig. 6B and data not shown). However, several Notch pathway components lack substantial phenotypes in certain settings unless the genetic background is sensitized (Schrons et al., 1992; Zeng et al., 1998; Duan et al., 2011). We therefore analyzed females heterozygous for Notch, which normally exhibit notching of the distal wing (Fig. 6A). Two-thirds of N/+ flies expressing control sponges exhibited notching. However, four independent K box miRNA sponges (miR-13a, miR-13b, miR-2b and miR-2c) rescued the haploinsufficient Notch phenotype (Fig. 6C,E-G), indicating that one or more miRNAs of this family has an endogenous function to limit Notch signaling during wing margin specification. Fig. 6I quantifies the degree of rescue provided by these sponges. The miR-13b sponge nearly completely suppressed wing notching, whereas the others reduced notching to only 10% of flies with either miR-2b/13a sponges and to 20% of flies in the case of miR-2c sponge. We note miR-2c is slightly offset in its seed region from canonical K box miRNAs, providing a rationale for its sponge being less effective than the other K box sponges in suppressing wing notching.
Fig. 6.
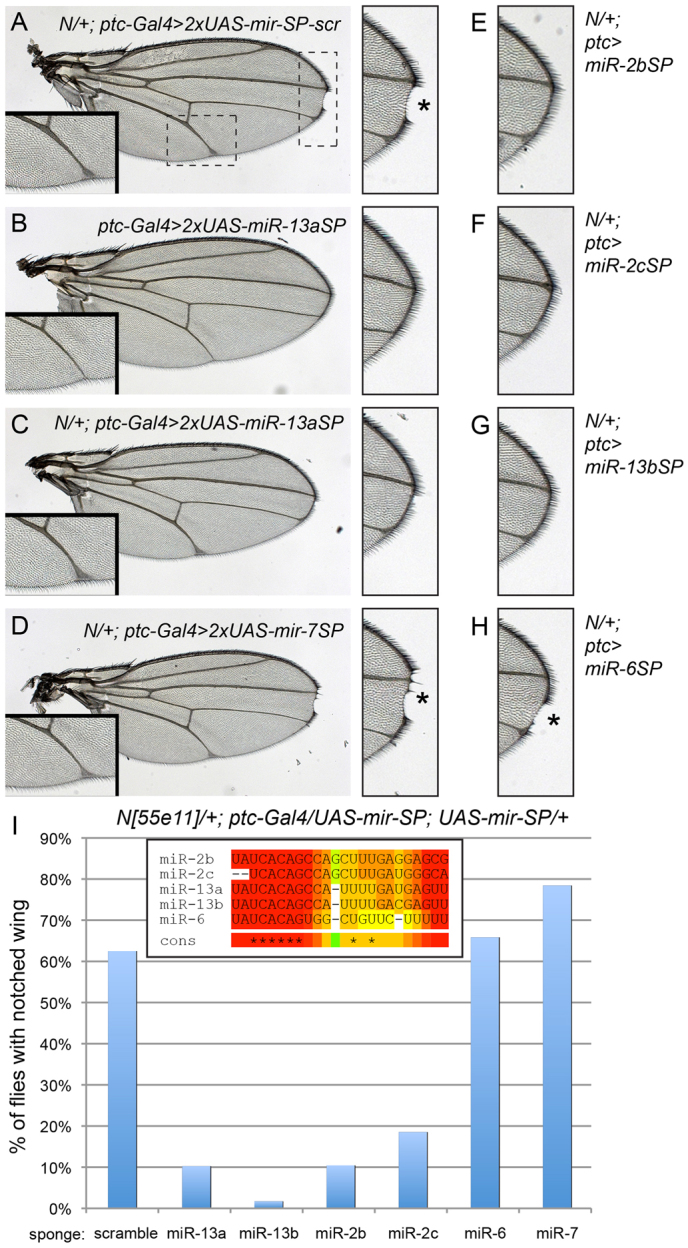
K box miRNA sponges enhance Notch signaling during wing development. (A,C-H) Notch[55e11]/+ (N/+) heterozygous females that carry ptc-Gal4 and two copies of the indicated miRNA sponges (SP); scr, scrambled sponge control. (A) N/+ females expressing control sponges exhibit a notch (asterisk) at the distal tip and mild vein thickening; the regions outlined are magnified to highlight these phenotypes. (B) Misexpression of any of the sponges used in this figure did not alter wing development; ptc-Gal4>2xmir-13aSP is shown as an example. The magnified insets exhibit normal vein thickness and can be used to judge N/+ haploinsufficiency. (C) Misexpression of the miR-13aSP rescued N/+ notching, but not vein thickening. (D) Misexpression of miR-7SP did not rescue either N/+ phenotype. Asterisk indicates area of wing notching. (E-H) Magnifications of the distal wing tips to highlight the status of wing notching in other sponge backgrounds. Asterisk indicates area of wing notching. (E) miR-2bSP, (F) miR-2cSP and (G) miR-13bSP all rescued N/+ notching, but (H) miR-6SP could not (asterisk). (I) Quantification of rescue of wing notching in various genotypes. N/+ in various ptc-Gal4>UAS-mir-SP backgrounds exhibit notching in about two-thirds of animals, this is reduced to less than 20% in the presence of miR-2cSP, to 10% in miR-2bSP and miR-13aSP, and to less than 2% in miR-13bSP. Inset shows the sequence relationship of these K box miRNAs.
Expression of K box sponges using ptc-Gal4 only rescued notching and did not suppress vein thickening in N/+ (Fig. 6A-C, inset panels), providing evidence for the specificity of their activity. As a further check, we assayed sponges for the K box family member miR-6 and the GY box family member miR-7 (Lai, 2002). Both of these miRNAs induce wing notching when misexpressed (Stark et al., 2003; Lai et al., 2005), but neither of their sponges suppressed N/+ (Fig. 6D,H). This can be rationalized by the fact that these miRNAs are not endogenously expressed in the developing wing margin: miR-6 is a member of a miRNA cluster that is exclusively expressed in the early embryo (Aboobaker et al., 2005; Bushati et al., 2008), whereas miR-7 is specifically expressed in the eye and in proneural domains of imaginal discs (Li and Carthew, 2005; Li et al., 2009).
K box miRNAs comprise the largest family in Drosophila (Lai et al., 2003), and both miR-2 and miR-13 families are well-expressed in imaginal discs (Ruby et al., 2007b). As genetic 3′ UTR sensors bearing K boxes that exhibit only seed pairing to any family member direct potent suppression in imaginal discs (Lai et al., 1998) and are sensitive to multiple K box miRNAs (Lai et al., 2005), multiple members of this family probably contribute to restricting Notch signaling. Given such likely functional overlap, single K box miRNA mutants may not suffice to reveal these effects. These tests therefore provide a proof of principle of how miRNA gain-of-function phenotypes can be used to direct experimental approaches that reveal the endogenous contribution of miRNAs to tissue patterning, especially in cases that may require appropriate genetic sensitization.
DISCUSSION
A plethora of specific phenotypes induced by miRNA gain-of-function in vivo
There is abundant evidence from cell-based studies that animal miRNAs directly but mildly repress hundreds of targets (Lim et al., 2005; Hendrickson et al., 2009; Guo et al., 2010), even when measured in contexts of ectopic activity. Given this, the in vivo consequences of miRNA deregulation might reasonably have been supposed to often be subtle (with the view that few targets can be sufficiently suppressed to reveal loss-of-function phenotypes), or might often compromise general cell viability (with the view that the coordinate downregulation of hundreds of targets might cause cells to simply become unhealthy).
We describe a systematic in vivo examination of the consequences of targeted miRNA misexpression within the intact animal. In contrast to the aforementioned possibilities, we find that the majority of miRNAs tested generated diverse and relatively distinct mutant phenotypes, most of which could not be anticipated from target predictions or from the general fine-tuning model of miRNA function. Although a number of miRNAs have profoundly adverse consequences that might be due to cellular toxicity, many miRNA-induced phenotypes closely resemble those exhibited by mutants of genes in signaling/proliferation/apoptosis pathways that are crucial to tissue development and patterning. The present studies extend our earlier functional screens in cultured cells that linked miR-315 to activation of the Wnt pathway (Silver et al., 2007), and now permit diverse functional screening in the animal. Indeed, many miRNAs of unrelated sequences generated similar phenotypes in vivo, which might be explained if they hit different nodal points in the same pathways.
Many, if not most, genes contain conserved binding sites for multiple miRNAs. But it is clear that the simple presence of conserved miRNA-binding sites does not guarantee responsiveness in directed sensor assays. It is even more so the case that presence of cognate binding sites does not render a miRNA likely to be able to induce a corresponding loss-of-function phenotype in the animal, even when misexpressed (Silver et al., 2007). Even when using artificial shRNA constructs designed to have perfect complementarity for maximal effect, it is typical for them to elicit only partial knockdown or sometimes to not work at all. The known dose sensitivity of the core cell signaling pathways and patterning genes provides a genetic rationale for why they may be especially prone to be affected by miRNAs in a way that translates into overt mutant phenotypes (Hagen and Lai, 2008; Smibert and Lai, 2010). Such genetic connections can guide functional studies and point to likely target pathways, even when knowledge of relevant computationally predicted targets is lacking.
Beyond understanding the underlying genetic circuitry of insects, our studies highlight that ectopic miRNAs can generate specific developmental phenotypes, often as a result of altering tissue patterning, proliferation of apoptosis. This has substantial consequences for interpreting the etiology of disease and cancer. For example, the overexpression of a growing number of mammalian miRNAs can generate cell specification or metabolic defects, and miRNAs such as mir-21 (Medina et al., 2010) and mir-17-92 (He et al., 2005) are overt oncogenes. Our systematic screening in Drosophila strongly suggests that scores of vertebrate miRNAs may prove to induce relatively specific phenotypes in the animal, but that these may only rarely be predicted on the basis of computationally derived target associations.
A genetic resource for miRNA screening in vivo
A great deal of effort has been devoted to expanding collections of conditionally activated transgene insertions in Drosophila (Brand and Perrimon, 1993; Rorth et al., 1998). Over the past 15 years, these have been of tremendous use in revealing the biological activity and function of protein-coding genes. Here, we describe genome-wide collections of miRNA transgenes, and demonstrate their collectively diverse activities during wing development. These collections include both P insertion and attP insertion lines, providing a great deal of flexibility for their subsequent screening. The latter permits the activity of different miRNAs to be compared directly, whereas the former provides in many cases allelic series of transgene strengths. The availability of these lines permits a wide variety of screens using tissue- or cell-specific drivers, to evaluate the consequences of miRNA deregulation on development, as well as adult roles in physiology or behavior. Knowledge of their functional capacities can then inform the study of different miRNAs within their endogenous expression domains (Aboobaker et al., 2005; Berezikov et al., 2011).
While this work was under review, Cohen and colleagues described a smaller set of UAS-miRNA transgenes and their application towards searching for modifiers of a bristle phenotype of the cell cycle regulator minus (Szuplewski et al., 2012). Beyond a limited set of bristle modifiers, however, their analysis primarily revealed lethality as the outcome of miRNA expression (Szuplewski et al., 2012). We find over 100 of our miRNA transgenes induced lethality when broadly expressed with da-Gal4. However, our detailed analysis using a panel of wing drivers revealed a cornucopia of distinct phenotypes, many of which phenocopy the modulation of fundamental signaling pathways and patterning factors (Figs 2, 3, 4, 5 and 6). Our UAS-miRNA collections complement and substantially extend their transgenes, and together they constitute a formidable resource for in vivo analysis of miRNA activity. Many miRNAs have subtle if not undetectable loss-of-function phenotypes (Miska et al., 2007; Alvarez-Saavedra and Horvitz, 2010), but it is also the case that many miRNA mutants produce synthetic phenotypes in combination with other genetic insults (Brenner et al., 2010). Data such as ours provide a genetic basis for pursuing more than 100 demonstrable miRNA activities and many tens of compelling miRNA-target/pathway linkages, and can inform more complex interaction studies with miRNA sponges (Loya et al., 2009). Indeed, we provide proof of principle for how K box miRNA gain of function, which inhibited Notch signaling, informed sensitized genetic assays that revealed the endogenous activity of a likely highly redundant set of endogenous K box miRNAs in restricting Notch signaling during wing development. Our extensive assays provide compelling evidence of the usefulness of these genome-wide collections of conditionally activatable miRNA transgenes, and suggest that these may be well complemented by similar collections of miRNA sponge transgenes.
Supplementary Material
Acknowledgements
This work was initiated in the laboratory of Gerald Rubin and we are grateful for his support. We thank Todd Laverty for performing many of the embryo injections, and Stephen Cohen and Julius Brennecke for reagents. Kieran Harvey, Carole Poon, Ethan Bier and Jose de Celis provided enlightening discussion and kindly provided some images of mutant wings.
Footnotes
Funding
Q.D. was supported by the Swedish Research Council. Work in N.P.'s laboratory was supported by the Howard Hughes Medical Institute, Starr Cancer Consortium and National Institutes of Health (NIH) [R01-GM084947]. T.A.F. and D.V.V. were supported by the NIH [R01-NS069695]. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund [1004721], the Starr Cancer Consortium [I3-A139] and the NIH [R01-GM083300 and U01-HG004261]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.079939/-/DC1
References
- Abdelilah-Seyfried S., Chan Y. M., Zeng C., Justice N. J., Younger-Shepherd S., Sharp L. E., Barbel S., Meadows S. A., Jan L. Y., Jan Y. N. (2000). A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155, 733-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboobaker A. A., Tomancak P., Patel N., Rubin G. M., Lai E. C. (2005). Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. USA 102, 18017-18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R. (2010). Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337-350 [DOI] [PubMed] [Google Scholar]
- Axtell M. J., Westholm J. O., Lai E. C. (2011). Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 12, 221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F., Smibert P., Lai E. C. (2010). miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 338, 63-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. (2008). MicroRNAs in the Drosophila bithorax complex. Genes Dev. 22, 14-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Liu N., Flynt A. S., Hodges E., Rooks M., Hannon G. J., Lai E. C. (2010). Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat. Genet. 42, 6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Robine N., Samsonova A., Westholm J. O., Naqvi A., Hung J. H., Okamura K., Dai Q., Bortolamiol-Becet D., Martin R., et al. (2011). Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 21, 203-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003). bantam Encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25-36 [DOI] [PubMed] [Google Scholar]
- Brenner J. L., Jasiewicz K. L., Fahley A. F., Kemp B. J., Abbott A. L. (2010). Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 20, 1321-1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501-506 [DOI] [PubMed] [Google Scholar]
- Caygill E. E., Johnston L. A. (2008). Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 18, 943-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Kadow I. G., Zhan X., Okamura K., Suh G. S., Gunning D., Lai E. C., Zipursky S. L. (2008). Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319, 1256-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Smibert P., Lai E. C. (2012). Exploiting Drosophila genetics to understand microRNA function and regulation. Curr. Top. Genes Dev. 99, 201-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Benjumea F. J., Garcia-Bellido A. (1990). Genetic analysis of the wing vein pattern of Drosophila. Roux's Arch. Dev. Biol. 198, 336-354 [DOI] [PubMed] [Google Scholar]
- Duan H., Dai Q., Kavaler J., Bejarano F., Medranda G., Negre N., Lai E. C. (2011). Insensitive is a novel corepressor of Suppressor of Hairless and regulates Notch-mediated cell fate decisions in the peripheral nervous system. EMBO J. 30, 3120-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A. S., Lai E. C. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9, 831-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., Bartel D. P. (2011). Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18, 1139-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L., Lehmann R. (2004). Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 14, 981-986 [DOI] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J., Schier A. F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75-79 [DOI] [PubMed] [Google Scholar]
- Grimm D., Streetz K. L., Jopling C. L., Storm T. A., Pandey K., Davis C. R., Marion P., Salazar F., Kay M. A. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537-541 [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775-1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835-840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen J. W., Lai E. C. (2008). microRNA control of cell-cell signaling during development and disease. Cell Cycle 7, 2327-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828-833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson D. G., Hogan D. J., McCullough H. L., Myers J. W., Herschlag D., Ferrell J. E., Brown P. O. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7, e1000238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner D. R., Weigmann K., Cohen S. M. (2002). The bantam gene regulates Drosophila growth. Genetics 161, 1527-1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklevic B., Uyetake L., Wichmann A., Bilak A., English C. N., Su T. T. (2008). Modulation of ionizing radiation-induced apoptosis by bantam microRNA in Drosophila. Dev. Biol. 320, 122-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell J. A., Gerin I., MacDougald O. A., Cadigan K. M. (2008). The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. USA 105, 15417-15422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R., Menon K., Zinn K. (2001). A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11, 417-430 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853-858 [DOI] [PubMed] [Google Scholar]
- Lai E. C. (2002). microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30, 363-364 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Posakony J. W. (1997). The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 124, 4847-4856 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Posakony J. W. (1998). Regulation of Drosophila neurogenesis by RNA:RNA duplexes? Cell 93, 1103-1104 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Rubin G. M. (2001). neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev. Biol. 231, 217-233 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Burks C., Posakony J. W. (1998). The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development 125, 4077-4088 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Tomancak P., Williams R. W., Rubin G. M. (2003). Computational identification of Drosophila microRNA genes. Genome Biol. 4, R42.1-R42.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., Tam B., Rubin G. M. (2005). Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 19, 1067-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854 [DOI] [PubMed] [Google Scholar]
- Li X., Carthew R. W. (2005). A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123, 1267-77 [DOI] [PubMed] [Google Scholar]
- Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W. (2009). A microRNA imparts robustness against environmental fluctuation during development. Cell 137, 273-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J. A., Gao F. B. (2006). MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20, 2793-2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P., Bray S. J., Apidianakis Y., Delidakis C. (1999). Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for notch activity in wing margin establishment in Drosophila. Development 126, 2205-2214 [DOI] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769-773 [DOI] [PubMed] [Google Scholar]
- Loya C. M., Lu C. S., Van Vactor D., Fulga T. A. (2009). Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E. (1998). Regulatory control of signal transduction during morphogenesis in Drosophila. Int. J. Dev. Biol. 42, 363-368 [PubMed] [Google Scholar]
- Medina P. P., Nolde M., Slack F. J. (2010). OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86-90 [DOI] [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., McGonagle S. M., Bartel D. P., Ambros V. R., Horvitz H. R. (2007). Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3, e215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar C., Resnik-Docampo M., Organista M., Martin M., Hevia C. F., de Celis J. F. (2011). Signalling pathways in development and human disease: a Drosophila wing perspective. In Human Genetic Diseases (ed. Plaseska-Karanfilska Dijana.). Croatia: InTech; [Google Scholar]
- Morel V., Schweisguth F. (2000). Repression by Suppressor of Hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 14, 377-388 [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Liu L. P., Binari R., Hardy R., Shim H. S., Cavallaro A., Booker M., Pfeiffer B. D., Markstein M., Wang H., et al. (2009). A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182, 1089-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R., Morrison C. M., Tao C., Zhang X., Halder G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895-1904 [DOI] [PubMed] [Google Scholar]
- Oh H., Irvine K. D. (2011). Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell 20, 109-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Phillips M. D., Tyler D. M., Duan H., Chou Y. T., Lai E. C. (2008). The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 15, 354-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F., Basson M., Pasquinelli A., Bettinger J., Rougvie A., Horvitz H. R., Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901-906 [DOI] [PubMed] [Google Scholar]
- Ronshaugen M., Biemar F., Piel J., Levine M., Lai E. C. (2005). The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 19, 2947-2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., Szabo K., Bailey A., Laverty T., Rehm J., Rubin G. M., Weigmann K., Milan M., Benes V., Ansorge W., et al. (1998). Systematic gain-of-function genetics in Drosophila. Development 125, 1049-1057 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P. (2007a). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., Lai E. C. (2007b). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrons H., Knust E., Campos-Ortega J. A. (1992). The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics 132, 481-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58-63 [DOI] [PubMed] [Google Scholar]
- Silver S. J., Hagen J. W., Okamura K., Perrimon N., Lai E. C. (2007). Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc. Natl. Acad. Sci. USA 104, 18151-18156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. M., Olson E. N. (2011). Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Lai E. C. (2008). Lessons from microRNA mutants in worms, flies and mice. Cell Cycle 7, 2500-2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Lai E. C. (2010). A view from Drosophila: multiple biological functions for individual microRNAs. Semin. Cell Dev. Biol. 21, 745-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R. B., Cohen S. M. (2003). Identification of Drosophila microRNA targets. PLoS Biol. 1, E60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Bushati N., Jan C. H., Kheradpour P., Hodges E., Brennecke J., Bartel D. P., Cohen S. M., Kellis M. (2008). A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22, 8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuplewski S., Kugler J. M., Lim S. F., Verma P., Chen Y. W., Cohen S. M. (2012). MicroRNA transgene overexpression complements deficiency-based modifier screens in Drosophila. Genetics 190, 617-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Cohen S. M. (2006). The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767-774 [DOI] [PubMed] [Google Scholar]
- Tyler D. M., Okamura K., Chung W. J., Hagen J. W., Berezikov E., Hannon G. J., Lai E. C. (2008). Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 22, 26-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855-862 [DOI] [PubMed] [Google Scholar]
- Xiao C., Rajewsky K. (2009). MicroRNA control in the immune system: basic principles. Cell 136, 26-36 [DOI] [PubMed] [Google Scholar]
- Yang J. S., Lai E. C. (2011). Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 43, 892-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Younger-Shepherd S., Jan L. Y., Jan Y. N. (1998). Delta and Serrate are redundant Notch ligands required for asymmetric cell divisions within the Drosophila sensory organ lineage. Genes Dev. 12, 1086-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.