Abstract
Background
Plant LIM domain proteins may act as transcriptional activators of lignin biosynthesis and/or as actin binding and bundling proteins. Plant LIM genes have evolved in phylogenetic subgroups differing in their expression profiles: in the whole plant or specifically in pollen. However, several poplar PtLIM genes belong to uncharacterized monophyletic subgroups and the expression patterns of the LIM gene family in a woody plant have not been studied.
Findings
In this work, the expression pattern of the twelve duplicated poplar PtLIM genes has been investigated by semi quantitative RT-PCR in different vegetative and reproductive tissues. As in other plant species, poplar PtLIM genes were widely expressed in the tree or in particular tissues. Especially, PtXLIM1a, PtXLIM1b and PtWLIM1b genes were preferentially expressed in the secondary xylem, suggesting a specific function in wood formation. Moreover, the expression of these genes and of the PtPLIM2a gene was increased in tension wood. Western-blot analysis confirmed the preferential expression of PtXLIM1a protein during xylem differentiation and tension wood formation. Genes classified within the pollen specific PLIM2 and PLIM2-like subgroups were all strongly expressed in pollen but also in cottony hairs. Interestingly, pairs of duplicated PtLIM genes exhibited different expression patterns indicating subfunctionalisations in specific tissues.
Conclusions
The strong expression of several LIM genes in cottony hairs and germinating pollen, as well as in xylem fibers suggests an involvement of plant LIM domain proteins in the control of cell expansion. Comparisons of expression profiles of poplar LIM genes with the published functions of closely related plant LIM genes suggest conserved functions in the areas of lignin biosynthesis, pollen tube growth and mechanical stress response. Based on these results, we propose a novel nomenclature of poplar LIM domain proteins.
Background
Plant LIM domain proteins are related to animal Cystein Rich Proteins (CRP), and contain two LIM domains characterized by the consensus sequence [C-X2-C-X17-H-X2-C]-X2-[C-X2-C-X17-C-X2-H] [1,2]. In animals, the CRP proteins mostly expressed in muscle tissues are involved in muscle differentiation, transcriptional regulation and actin organization [3,4]. In plant, the tobacco NtWLIM1 protein functions in the nucleus as a transcription factor regulating the expression of genes involved in lignin biosynthesis [5], and in the cytoplasm as an actin binding and bundling protein like the CRP1 protein in animal [6,7]. More recently, it has been shown that lily LlLIM1 and all Arabidopsis LIM domain proteins also regulate the actin cytoskeleton organization and dynamics [8,9].
Since the discovery of SF3/PLIM1, the first gene coding for a plant LIM domain protein [10,11], an increased number of LIM genes has been identified in a wide range of plant species including Arabidopsis, rice and poplar [1,2]. Whereas both Arabidopsis thaliana and Oryza sativa genomes contain six gene models, the Populus trichocarpa genome contains at least twelve PtLIM gene models resulting from the duplication of six ancestral genes [1]. These duplicated genes probably appeared during the "salicoid" whole-genome duplication event [1,12]. The plant LIM domain protein family has been divided into four groups: αLIM1, βLIM1, γLIM2, δLIM2 containing monophyletic subgroups differing according to their classification in plant taxonomic class or subclass and/or in their expression specificities [1]. In sunflower, tobacco and Arabidopsis, genes belonging to the WLIM1 and WLIM2 subgroups are widely expressed in plant, whereas genes belonging to the PLIM1 and PLIM2 subgroups are preferentially expressed in pollen [2,8,13]. However, the expression pattern of genes classified into the βLIM1 group and the XLIM1 and PLIM2-like subgroups remains unknown and there is no Arabidopsis and rice orthologs within these newly identified monophyletic groups [1].
Although EST distribution analysis in cDNA libraries from different poplar tissues may be indicative of their expression profile, the expression pattern of PtLIM genes has not been investigated. Using EST and microarray data, some poplar LIM genes (e.g. PtWLIM1a and b and PtXLIM1a and b) appear highly expressed in wood or in tension wood [1,14,15]. In trees, the formation of wood (or secondary xylem) is an important developmental process involving different steps: firstly cell division from the cambial meristem followed by xylem differentiation that include cell expansion, secondary wall synthesis, lignification and finally programmed cell death [16]. Produced in response to mechanical stresses, tension wood is a peculiar cellulose-rich and poorly lignified wood characterized by specific mechanical and structural properties that allow the tree to re-orientate its axis [17].
In this article, the expression of the twelve poplar PtLIM genes was investigated in different reproductive and vegetative tissues. Some PtLIM genes were ubiquitously expressed whereas others showed a preferential expression in pollen, cottony hairs or secondary xylem. Interestingly a subset of PtLIM genes was up-regulated in tension wood area and all duplicated genes exhibited distinct expression pattern suggesting subfunctionalisations. From this expression analysis and considering the previous phylogenetic classification we suggest a simplified nomenclature for the poplar LIM domain family.
Findings
Expression of poplar LIM genes in vegetative and reproductive organs
Previous EST distribution analyses indicated that PtXLIM1a and b and PtWLIM1a and b transcripts were mostly found in wood forming tissues whereas PtWLIM2a and b genes seemed to be ubiquitously expressed [1]. However, only a few PtLIM1a and b ESTs were found in undifferentiated cells and vegetative buds cDNA libraries. Moreover, PtPLIM2a, b, c and d transcripts were not or only poorly represented in poplar cDNA libraries preventing any inference on their expression profile. Therefore, the expression level of the twelve PtLIM genes was measured in different vegetative and reproductive tissues by semi quantitative RT-PCR.
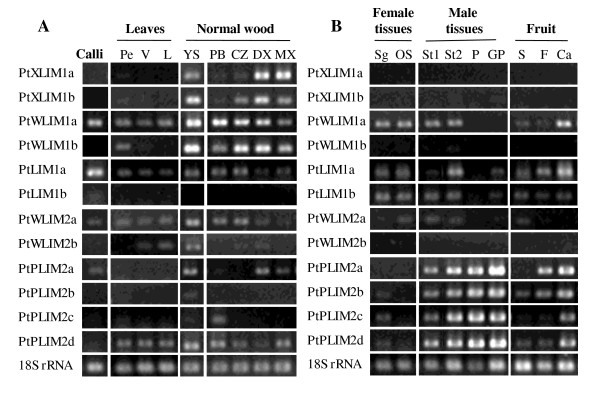
The PtXLIM1a and b and PtWLIM1a and b genes were strongly expressed in poplar stem particularly in wood, whatever the age of the tree (Figure 1 and Additional file 1). Moreover, they also showed a strong expression in the root differentiating xylem. These genes, except PtWLIM1a, were not or only weakly expressed in leaves, calli, and reproductive organs indicating a preferential expression in vascular tissues. PtXLIM1a expression appeared higher in developing and mature xylem (DX and MX) than in cambial zone (CZ) and phloem, suggesting specificity toward secondary xylem differentiation. Its duplicated gene PtXLIM1b as well as PtWLIM1b were more expressed in DX than in MX both in stem and root. Although the expression of PtWLIM1a gene was stronger in the stem, it seemed to be widely expressed in the whole tree except in pollen. From these results, genes from the XLIM1 and WLIM1 subgroups may play an important function during wood formation in poplar.
Figure 1.
Expression of PtLIM genes using semi quantitative RT-PCR analysis in poplar trees. A. Vegetative organs: Three straight young poplars (Populus tremula × P. alba) producing normal wood were cultivated in the greenhouse for three months. Phloem and bark (PB), cambial zone (CZ), developing xylem (DX) and mature xylem (XM) were scraped from the stem. To study primary growth, young green portions of stems (YS) were collected just below the apex. Leaf vascular tissues like petioles (Pe) and main vessels (V) were separated from limbs (L). Two-weeks-old dark-grown calli (C) were also harvested. B. Reproductive organs: female flowers were dissected from mature poplar trees (Populus nigra) growing in the nursery and the stigma (Sg) were separated from the ovary and style (OS). Two different developmental stages were analyzed on male flowers: partially-open flowers (St1) at an early developmental stage, and fully-open flowers at pre-anthesis stage (St2). Mature pollen (P) and germinated pollen (GP) were also analyzed, as well as seeds (S) separated from both the cottony fibers (F) and the capsule protecting seeds (Ca). 18S rRNA signals are indicative of the total RNA quantity in each sample
The duplicated PtLIM1a and b genes displayed distinct expression patterns. Whereas PtLIM1a gene was expressed in a wide range of tissues, the PtLIM1b gene appeared specifically expressed in reproductive organs including fruit, male and female flowers and to a lesser extent pollen (Figure 1). Interestingly, like PtWLIM1a, PtLIM1a expression is the highest in calli as well as in cottony hair (F). The expression pattern of the duplicated PtWLIM2a and b genes were strikingly different although their expression appeared rather weak in most poplar tissues. Indeed, PtWLIM2a was ubiquitously expressed in the plant when PtWLIM2b was mostly expressed in leaf and stem tissues. The PtWLIM1a, PtLIM1a and PtWLIM2a and b genes were also expressed in young roots collected from in vitro poplar plantlets, indicative of a preferential expression in tissues at primary growth stage (data not shown). Thus, PtLIM1a, PtWLIM1a and PtWLIM2a appeared widely expressed in poplar whereas the expression of their respective duplicated genes seemed more restricted to particular tissues.
PtPLIM2a and b exhibited a very strong expression in stamens, mature pollen and especially in germinated pollen. Interestingly, the duplicated PtPLIM2c and d genes, closely related to PtPLIM2a and b, were also highly expressed in the same tissues. The rather weak expression of these four genes in stamens at an early developmental stage compared to older stamens, pollen and germinating pollen suggests an expression in microspores rather than in filament, anther wall, rachis or peduncle supporting stamens. Surprisingly, these four genes were also highly expressed in isolated cottony hairs and in the capsules bearing these fibers, indicative of a dual expression in both pollen and cotton fibers. The PtPLIM2a gene was also expressed, but to a lower extent, in DX and MX, and PtPLIM2d transcript was detected in all parts of leaves and stems. These results indicate that PtPLIM2a and b genes and their paralogs PtPLIM2c and d may have redundant functions during pollen tube growth as well as in cotton fiber development.
Comparison of poplar LIM gene expression between opposite and tension wood
Previously, PtXLIM1a ESTs were shown to be more abundant in tension wood DX than in opposite wood DX [1,15] and microarray analyses indicated a higher expression of PtXLIM1b in tension wood than in normal wood [14]. This prompted us to investigate more precisely the expression pattern of the PtLIM genes during tension wood differentiation on artificially tilted poplar trees. Tension wood, formed at the upper side of the tilted stem, is easily recognizable as a bright whitish crescent, and its presence was confirmed by the amplification of PopFLA1 gene (data not shown) a marker of tension wood formation [18].
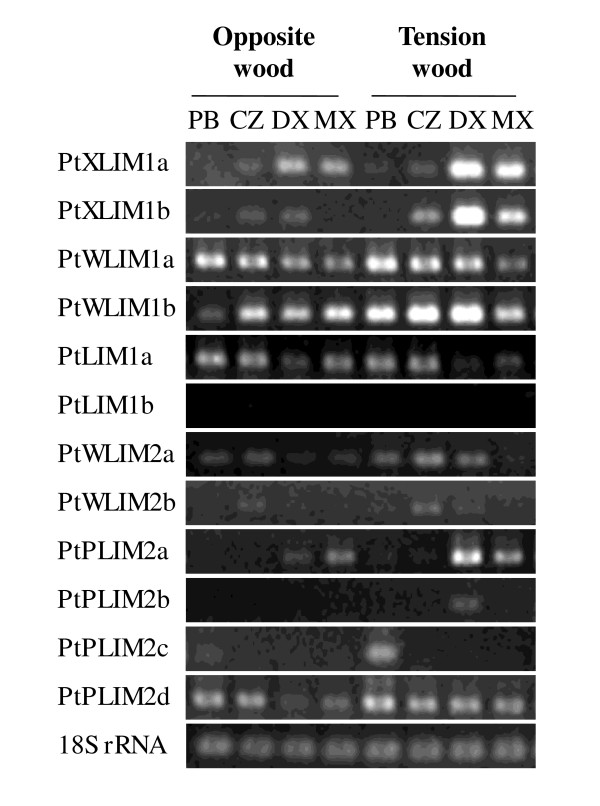
Unexpectedly, the semi quantitative RT-PCR results showed that a number of poplar PtLIM genes: PtXLIM1a and b, but also PtWLIM1b, PtWLIM2a, PtPLIM2a and PtPLIM2d exhibited a higher expression in tension wood compared to opposite wood (Figure 2). These results confirmed the increased accumulation of PtXLIM1a and b transcripts in DX from tension wood but also revealed a high expression level in tension wood MX. PtPLIM2a expression in tension wood appeared to be restricted to DX and MX. By contrast, the accumulation of PtWLIM1b transcript in tension wood was mainly localized in DX, CZ and phloem/bark. The accumulation of PtWLIM2a and PtPLIM2d in DX was only slightly higher in tension wood compared to opposite wood and the other PtLIM genes did not exhibit any tension wood preferential up-regulation. The induction of PtXLIM1a gene in tension wood was supported at the protein level by western blot analysis using purified anti-6His-PtXLIM1a antibodies. A 29 kDa polypeptide corresponding to the expected size of the PtXLIM1a protein was detected from the CZ to the MX but not in leaves and reproductive organs (Additional file 2, data not shown). The PtXLIM1a protein was particularly more expressed in DX from tension wood compared to opposite wood. In summary, PtXLIM1a and b as well as PtWLIM1b and PtPLIM2a are unambiguously tension wood-induced genes and their expression appears to fluctuate depending on xylem differentiation.
Figure 2.
Expression of PtLIM genes using semi quantitative RT-PCR analysis in opposite and tension wood. Three young poplars (Populus tremula × P. alba) were cultivated in the greenhouse for two months, and tilted for one month to induce tension wood formation. Phloem and bark (PB), cambial zone (CZ), developing xylem (DX) and mature xylem (XM) samples were scraped from the upper side (tension wood) and the lower side (opposite wood) of tilted stems. 18S rRNA signals are indicative of the total RNA quantity in each sample
Discussion
Expression of LIM duplicated genes in poplar, toward a simplified nomenclature
The previous classification and nomenclature of plant LIM domain protein was complex because of the lack of expression data concerning genes belonging to newly identified XLIM1, PLIM2-like subgroups and βLIM1 group [1]. Furthermore, the expression of poplar genes from WLIM1, WLIM2 and PLIM2 subgroups was predicted from EST analysis and phylogenetic comparison. Thus, the expression pattern of PtLIM genes was analyzed on an exhaustive set of poplar tissues covering most of the tree developmental stages. From our results, the 12 PtLIM genes were all expressed in poplar indicating that pseudogenization events did not occur. Poplar PtLIM genes are organized in six pairs of duplicated genes, and the high sequence similarity between duplicated PtLIM genes, ranging from 85 to 95% of amino acid identity, suggests that they share similar functions [1]. However, a few pair of genes exhibit distinct expression patterns suggesting a subfunctionalisation in specific tissues or possibly a neofunctionalisation [19]. For example, the PtWLIM1a and PtLIM1a genes almost constitutively expressed may be involved in a general cellular function, whereas the corresponding duplicated genes probably evolved towards specific functions such as xylem development for PtWLIM1b and eventually reproductive processes for PtLIM1b. The finding that PtXLIM1a and b genes were preferentially expressed in xylem strengthens their classification into the XLIM1 monophyletic subgroup distinct from the WLIM1 subgroup (Additional file 3) [1]. XLIM1 genes probably result from an older duplication of the ancestral WLIM1 gene with a further specialization towards new vascular specific functions potentially important for tree physiology. From our results, we have now strong evidence supporting a pollen preferential expression for the PtPLIM2c and d genes classified into the PLIM2-like subgroup phylogenetically close to the pollen specific PLIM2 subgroup (Additional file 3) [1]. It is thus likely that orthologous genes from sunflower and tobacco classified within the PLIM2-like subgroup also share the same pollen preferential expression. According to their expression profile and phylogeny, we propose a new nomenclature for the poplar LIM domain protein family (Table 1). For simplification and clarity, the formerly named PtβLIM1a and b, PtδLIM2a and b and PtGLIM1a and b genes were renamed PtLIM1a and b, PtPLIM2c and d and PtXLIM1a and b respectively. This revised and more consistent gene nomenclature taking into consideration the expression pattern of poplar LIM gene could serve as a basis for gene naming in other plant species and will hopefully improve communication among scientists working on the LIM domain family.
Table 1.
Phylogenetic classification and nomenclature of poplar PtLIM genes
| Group | Subgroup | Poplar | Arabidopsis | Rice | Sunflower | Expression profile |
|---|---|---|---|---|---|---|
| αLIM1 | XLIM1 (FLIM1) |
PtXLIM1a (PtGLIM1a) PtXLIM1b (PtGLIM1b) |
not found | not found | not found | secondary xylem |
| WLIM1 | PtWLIM1a PtWLIM1b |
AtWLIM1 | OsWLIM1 | HaWLIM1 | ubiquitous | |
| PLIM1 | not found | not found | not found | HaPLIM1a-SF3 HaPLIM1b |
pollen | |
| βLIM1 | βLIM1 | PtLIM1a (PtβLIM1a) PtLIM1b (PtβLIM1b) |
not found | not found | HaLIM1 (HaβLIM1) | ubiquitous |
| γLIM2 | WLIM2 | PtWLIM2a PtWLIM2b |
AtWLIM2a AtWLIM2b |
OsWLIM2 | HaWLIM2 | ubiquitous |
| δLIM2 | PLIM2 | PtPLIM2a PtPLIM2b |
AtPLIM2a AtPLIM2b AtPLIM2c |
OsPLIM2a OsPLIM2b OsPLIM2c |
HaPLIM2a | pollen |
| PLIM2-like (δLIM2) |
PtPLIM2c (PtδLIM2a) PtPLIM2d (PtδLIM2b) |
not found | not found | HaPLIM2b (HaδLIM2a) HaPLIM2c (HaδLIM2b) |
pollen | |
The old nomenclature of LIM gene and phylogenetic subgroups [1] is indicated under bracket
Expression pattern of LIM genes in differentiating secondary xylem and tension wood
According to the expression study, the PtXLIM1a and b and PtWLIM1b genes were preferentially expressed in differentiating secondary xylem both in root and shoot, and were up-regulated in tension wood (Figures 1 and Figure 2, Additional file 1). Wood formation involves cell expansion and deposition of a highly lignified secondary cell wall [15,16]. The tobacco NtWLIM1 ortholog probably acts as a transcription factor activating the expression of PAL, 4CL and CAD genes coding for enzymes of the lignin biosynthesis pathway [5]. Interestingly, this protein also acts on the stabilization of actin bundles [6]. Actin cables, longitudinally oriented in vessels and fibers, certainly play an essential role in the cytoplasmic streaming and the distribution of cellulose and lignin precursors throughout these very elongated cells [20]. Taken together, both expression patterns and potential functions suggest the involvement of PtXLIM1a and b and PtWLIM1b in the differentiation of xylem cells, during cell elongation and secondary cell wall formation. Although some variations do exist, the common up-regulation of several PtLIM genes in tension wood may be indicative of a functional redundancy during tension wood formation. We know that tension wood is enriched in cellulose and contains less lignin than opposite or normal wood [17], and in accordance, the expression of PAL, 4CL and CAD genes involved in lignin biosynthesis is lower in poplar tension wood [14]. However, this merely applies during the deposition of the cellulose rich gelatinous layer (G-layer), specific to tension wood fibers, that is possibly devoid of any lignin, whereas the sub layers (S1 and S2) deposited before the G-layer contains regular amount of lignin [21]. Therefore, we cannot rule out that poplar LIM domain proteins act as transcriptional activators of lignin biosynthetic genes as proposed for the NtWLIM1 protein [5]. Interestingly, it was demonstrated that muscle specific CRP2 and CRP3 proteins are involved in the stretch response and the regulation of cell contractile force by interacting with actin stress fibers [22,23]. The actin binding and bundling functions between plant LIM domain proteins and the related animal CRP proteins is highly conserved [6-9]. According to the high expression of several PtLIM genes in tension wood, it is tempting to speculate that poplar LIM proteins may also be implicated in the perception/transduction of mechanical stresses through interactions with actin cables just as proposed for CRP in muscle cells [3,22]. However, such an involvement of actin bundles in tensile or contractile strain still needs to be demonstrated in tree wood fibers.
Plant LIM domain proteins and anisotropic cell elongation
Poplar PLIM2 genes are preferentially expressed in mature and germinating pollen. These genes are potentially involved in pollen tube growth as already suggested for their orthologs in tobacco, sunflower and Arabidopsis. Indeed, NtPLIM1, NtPLIM2, HaPLIM2, AtPLIM2a, b and c genes are highly expressed in pollen and HaPLIM1-SF3 protein strongly accumulates in germinating pollen cones [2,8,13]. Poplar PLIM2 genes together with the PtWLIM1a and PtLIM1a genes were also strongly expressed in cottony fibers and capsules. Poplar cottony fibers resemble to cotton fibers from Gossypium sp. even though they originate from placenta and are borne by the capsule base [24]. Cotton fibers are unicellular trichomes composed of 95% cellulose that undergo considerable cell elongation with thick longitudinally oriented actin filaments [25,26]. Remarkably a high number of EST homologous to LIM domain protein has been detected in cotton fibers libraries [1]. Both our data and previous studies indicate that most of the plant LIM genes are expressed in cells with anisotropic elongation, such as xylem fibers, pollen tubes, and cottony fibers. This expression pattern is in agreement with the capacity of plant LIM proteins to bind and bundle actin filaments in order to favor the formation of thick actin cables [6,8,9]. Indeed longitudinally-oriented thick actin cables may be very important for effective tip growth and anisotropic cell expansion. Further genetics, biochemical and cell biological studies are required to characterize the function of plant LIM domain protein during cell elongation.
Methods
Sampling of poplar tissues
Vegetative tissues have been harvested on Populus tremula x P. alba, INRA clone 717-1-B4, poplar trees. Coppiced poplar trees kept at 4°C during the winter were transferred mid-March in the greenhouse, potted in compost (3 L) and individually supplied with water and fertilizers by a drip system. Straight stems and leaves were collected on three individual 3 month-old young poplar trees. Differentiating xylem (DX) and mature xylem (MX) samples were sequentially scraped from the debarked stem with a scalpel (Additional file 4). Cambial zone (CZ) was first lightly scraped from the peeled bark, while the remaining tissues consisted of the phloem and bark (PB) samples. Young stem (YS) samples enriched in primary growth tissues were the green stem portions collected below the apex. Following the sampling procedure described above, tension and opposite wood samples were collected, respectively at the upper and lower sides of the stems of three trees tilted for one month. Petiole (Pe) and main vessels (V) were separated from the limb (L) of leaves with a scalpel.
A number of samples were also collected during the spring, when cambium was very active, on a four year-old mature poplar tree cultivated in the tree nursery. CZ samples were lightly scraped from the peeled bark of the trunk. Then, phloem (Ph) samples were peeled from bark (B) samples. DX samples were first gently scraped from the debarked stem, while MX was sampled as wood chips from the remaining stem. The different samples from the main root were harvested following the same procedure, except that the peeled cortex contains both secondary phloem and root epidermis (PE), and after DX removal, the root MX consisted of the entire central cylinder including the pith. Undifferentiated cells were collected from calli (INRA clone 717-1-B4) cultivated for 2 weeks at 24°C in the dark on MB5 agar plates [27].
Because Populus tremula x P. alba trees from INRA 717-1-B4 clone were female hybrid clones, female and male flowers have been collected on Populus nigra and P. nigra var. Italica poplar trees. Excised branches bearing immature male or female flower catkins from trees growing on the Loire riverside were harvested in early April 2006. Female and male branches were respectively transferred in compost and water. One month later, after anthesis, the rachis, peduncles and bracts were removed from female flowers with a forceps and a scalpel. The stigmas (Sg) were separated from the ovaries and styles (OS) (See Additional file 4). After removal of protecting calyx, stamens with peduncles and rachis were collected from male flowers at early (St1) or late (St2) developmental stages. St1 corresponded to young and small partially-open male flowers, bearing sessile stamens on the rachis, whereas at St2, just before anthesis, male flowers were fully expanded and bore elongated rachis and peduncles with spreader stamens. Mature pollen (P) was spread on modified MS/2 media [28,29] and germinated pollen (GP) with elongated pollen tubes was harvested 24 h after germination. In order to produce and collect seeds, female flowers were manually fecundated with a paintbrush; one month later, the cottony hairs or fibers (F) were separated from seeds (S) and capsules (Ca).
Samples were immediately frozen upon collection, ground in liquid nitrogen and stored at -80°C before use. Samples collected from leaves and stem of 3 different greenhouse-grown poplar trees were pooled after grinding and before RNA extraction.
RNA extraction and semi quantitative RT-PCR analyses
Total RNAs were prepared using the method described by Chang and colleague [30]. After overnight precipitation with LiCl, RNAs were further purified using QIAGEN RNEasy kit ® according to manufacturer's instructions. RNA concentration and quality were determined by OD measurement at 260 nm and 280 nm. Two micrograms of total RNA were reverse transcribed using 500 ng of oligo(dT)12-18 and 200 units of SuperScript™ II RT in a total volume of 20 μl following the manufacturer's instructions (Invitrogen™, Life technologies). After first-strand cDNA synthesis, RNA complementary to the cDNA was removed by incubating the reaction with 2 units of E. coli RNase H at 37°C for 20 min. Gene-specific PCR primers were designed to amplify PtLIM genes in Populus trichocarpa Nisqually genotype as well as in P. tremula × P. alba (INRA clone 717-1-B4) when cDNA sequences (PtXLIM1a and b, PtWLIM1a and b, PtWLIM2a and PtPLIM2a) were available [1]. Primers were located at both ends of gene coding sequences in order to amplify the four introns and therefore detect any genomic contamination. PCR amplification was done with 1/60 of the first-strand cDNA as template, 0,5 unit of rTaq DNA polymerase (Invitrogen™, Life technologies), 1,25 mM MgCl2 and 0,2 μM of gene specific primers (Additional file 5) in a total volume of 20 μl. For each primer pair, gene-specific amplification was optimized and verified on available PtLIM cDNA clones used as template. For PtLIM genes, the cycling conditions were 94°C for 5 min for one initial step followed by 94°C for 45 s, 65 or 60°C for 45 s and 72°C for 1 min, for 25 to 40 cycles (See Additional file 5). For 18S rRNA, the cycling conditions were 94°C for 5 min for one initial step followed by 94°C for 45 s, 55°C for 1 min and 72°C for 1 min 30, for 20 cycles [18]. The PCR was terminated with one extra step at 72°C for 10 min and hold at 4°C. The amplification was done using a GeneAmp® PCR System 9700 (Applied Biosystems). Ten microliters of each PCR product were electrophoresed on a 1% agarose gel that was scanned under UV light after ethidium bromide staining. Results presented are the representative illustration of at least three independent semi quantitative RT-PCR experiments. The identity of each PCR product was checked by sequencing.
Production of 6His-PtXLIM1a protein, protein extraction and Western blot
See Additional file 6 for detailed description of the procedure.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DA participated in the conception and design of the study, carried out semi-quantitative RT-PCR experiments as well as the production and purification of recombinant protein, and wrote the manuscript. AD and J-CL participated in the design of the study, manuscript revising and provided helpful discussions. M-CL-D performed sequencing of PCR products. MV participated in the collection of reproductive samples. NB carried out protein extraction, antibodies purification and western blot. HB coordinated and contributed to production and purification of recombinant protein. GP coordinated the study, participated in its conception and design as well as revisions to the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
PtLIM genes expression in wood tissues of an adult poplar tree. Expression of PtLIM genes using semi quantitative RT-PCR analysis in the trunk and main root of a four-year old poplar tree.
Expression of PtXLIM1a protein in opposite and tension wood. Immunodetection of PtXLIM1a protein in total protein extracts collected from tilted trunk of four-year old poplar trees.
Phylogenetic tree of plant LIM domain proteins. Phylogenetic tree of poplar, Arabidopsis, rice, tobacco and sunflower LIM domain proteins.
Sampling of wood tissues and reproductive organs. Illustration of samples collected from wood forming tissues on stems and description of male and female flowers used in the study.
Primers used in semi-quantitative RT-PCR. Forward and reverse primer sequences (5' to 3') for all PtLIM genes analyzed and PCR conditions used for semi-quantitative RT-PCR.
Supplemental methods for protein extraction and western blot. Method used for protein extraction, western-blot analysis, production and purification of 6His-PtXLIM1a recombinant protein.
Contributor Information
Dominique Arnaud, Email: arnaud@evry.inra.fr.
Annabelle Déjardin, Email: Annabelle.Dejardin@orleans.inra.fr.
Jean-Charles Leplé, Email: Jean-Charles.Leple@orleans.inra.fr.
Marie-Claude Lesage-Descauses, Email: Marie-Claude.Lesage@orleans.inra.fr.
Nathalie Boizot, Email: Nathalie.Noel@orleans.inra.fr.
Marc Villar, Email: Marc.Villar@orleans.inra.fr.
Hélène Bénédetti, Email: helene.benedetti@cnrs-orleans.fr.
Gilles Pilate, Email: pilate@orleans.inra.fr.
Acknowledgements
DA was supported by a PhD grant from the 'Région Centre'. The authors acknowledge Alain Roussel and Franck Coste (CNRS-CBM Orléans) for their help to produce the recombinant PtXLIM1a protein.
References
- Arnaud D, Déjardin A, Leplé J-C, Lesage-Descauses M-C, Pilate G. Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 2007;14:103–116. doi: 10.1093/dnares/dsm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson Å, Gass N, Mundel C, Baltz R, Kräuter R, Evrard J-L, Steinmetz A. Molecular and expression analysis of a LIM protein gene family from flowering plants. Molecular and General Genetics. 2000;264:257–267. doi: 10.1007/s004380000312. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Günther K. The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays. 2003;25:152–162. doi: 10.1002/bies.10226. [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H. Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 2000;22:289–301. doi: 10.1046/j.1365-313x.2000.00737.x. [DOI] [PubMed] [Google Scholar]
- Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, Steinmetz A. Tobacco WLIM1 is a novel F-Actin binding protein involved in actin cytoskeleton remodeling. Plant Cell. 2006;18:2194–2206. doi: 10.1105/tpc.106.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T, Singleton C, Fraley T, Greenwood J. Cysteine-rich protein 1 (CRP1) regulates actin filament bundling. BMC Cell Biol. 2005;6:45. doi: 10.1186/1471-2121-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A, Thomas C. Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell. 2010;22:3034–3052. doi: 10.1105/tpc.110.075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-J, Wan A-R, Jauh G-Y. An actin-binding protein, lllim1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 2008;147:1619–1636. doi: 10.1104/pp.108.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R, Domon C, Pillay D, Steinmetz A. Characterization of a pollen-specific cDNA from sunflower encoding a zinc finger protein. Plant J. 1992;2:713–721. [PubMed] [Google Scholar]
- Baltz R, Evrard JL, Domon C, Steinmetz A. A LIM motif is present in a pollen-specific protein. Plant Cell. 1992;4:1465–1466. doi: 10.1105/tpc.4.12.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Baltz R, Schmit A-C, Kohnen M, Hentges F, Steinmetz A. Differential localization of the LIM domain protein PLIM-1 in microspores and mature pollen grains from sunflower. Sexual Plant Reproduction. 1999;12:60–65. doi: 10.1007/s004970050172. [DOI] [Google Scholar]
- Andersson-Gunneras S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B. Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 2006;45:144–165. doi: 10.1111/j.1365-313X.2005.02584.x. [DOI] [PubMed] [Google Scholar]
- Déjardin A, Leplé J-C, Lesage-Descauses M-C, Costa G, Pilate G. Expressed sequence tags from poplar wood tissues - a comparative analysis from multiple libraries. Plant Biol. 2004;6:55–64. doi: 10.1055/s-2003-44744. [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol. 2001;47:239–274. doi: 10.1023/A:1010699919325. [DOI] [PubMed] [Google Scholar]
- Timell TE. The chemical composition of tension wood. Svensk Papperstidning. 1969;72:173–181. [Google Scholar]
- Lafarguette F, Leplé J-C, Déjardin A, Laurans F, Costa G, Lesage-Descauses M-C, Pilate G. Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytologist. 2004;164:107–121. doi: 10.1111/j.1469-8137.2004.01175.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Chaffey N, Barlow P, Sundberg B. Understanding the role of the cytoskeleton in wood formation in angiosperm trees: hybrid aspen (Populus tremula × P. tremuloides) as the model species. Tree Physiol. 2002;22:239–249. doi: 10.1093/treephys/22.4.239. [DOI] [PubMed] [Google Scholar]
- Pilate G, Chabbert B, Cathala B, Yoshinaga A, Leplé J-C, Laurans F, Lapierre C, Ruel K. Lignification and tension wood. Comptes Rendus Biologies. 2004;327:889–901. doi: 10.1016/j.crvi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kim-Kaneyama Jr, Suzuki W, Ichikawa K, Ohki T, Kohno Y, Sata M, Nose K, Shibanuma M. Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vivo. J Cell Sci. 2005;118:937–949. doi: 10.1242/jcs.01683. [DOI] [PubMed] [Google Scholar]
- Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang M-L, Hayashi T, Shiga N, Yasukawa H, Schaper W. et al. The cardiac mechanical stretch sensor machinery involves a z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/S0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- Boes TK, Strauss SH. Floral phenology and morphology of black cottonwood, Populus trichocarpa (Salicaceae) Am J Bot. 1994;81:562–567. doi: 10.2307/2445730. [DOI] [Google Scholar]
- Li X-B, Fan X-P, Wang X-L, Cai L, Yang W-C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell. 2005;17:859–875. doi: 10.1105/tpc.104.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Functional Plant Biol. 2007;34:1–10. doi: 10.1071/FP06234. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Leplé JC, Brasileiro ACM, Michel MF, Delmotte F. Transgenic poplars; expression of chimeric genes using four different constructs. Plant Cell Rep. 1992;11:137–141. doi: 10.1007/BF00232166. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Chang SJ, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PtLIM genes expression in wood tissues of an adult poplar tree. Expression of PtLIM genes using semi quantitative RT-PCR analysis in the trunk and main root of a four-year old poplar tree.
Expression of PtXLIM1a protein in opposite and tension wood. Immunodetection of PtXLIM1a protein in total protein extracts collected from tilted trunk of four-year old poplar trees.
Phylogenetic tree of plant LIM domain proteins. Phylogenetic tree of poplar, Arabidopsis, rice, tobacco and sunflower LIM domain proteins.
Sampling of wood tissues and reproductive organs. Illustration of samples collected from wood forming tissues on stems and description of male and female flowers used in the study.
Primers used in semi-quantitative RT-PCR. Forward and reverse primer sequences (5' to 3') for all PtLIM genes analyzed and PCR conditions used for semi-quantitative RT-PCR.
Supplemental methods for protein extraction and western blot. Method used for protein extraction, western-blot analysis, production and purification of 6His-PtXLIM1a recombinant protein.