Abstract
Human embryonic stem cells have the capacity for self-renewal and pluripotency and thus are a primary candidate for tissue engineering and regenerative therapies. These cells also provide an opportunity to study the development of human tissues ex vivo. To date, numerous human embryonic stem cell lines have been derived and characterized. In this review, we will detail the strategies used to direct tissue-specific differentiation of embryonic stem cells. We also will discuss how these strategies have produced new sources of tissue-specific progenitor cells. Finally, we will describe the next generation of methods being developed to identify and select stem cell-derived tissue precursors for experimental study and clinical use.
Introduction
Stem cells have the ability to maintain long-term proliferation and self-renewal. Under specific conditions, stem cells can differentiate into a diverse population of mature and functionally specialized cell types. There are two main types of human stem cells classified according to their source and developmental potential: embryonic and adult, or tissue-specific, stem cells. Human embryonic stem cells (hESCs) are pluripotent cells that can differentiate into all types of somatic - and, in some cases, extraembryonic - tissues. Human adult stem cells are derived from non-embryonic tissues and are capable of generating specific cells from the organ or tissue of origin. Because of the unrestricted potential of hESCs, these cells have become a highly desirable experimental tool for understanding human development and are especially attractive for therapeutic applications. In addition, methods for inducing a pluripotent state in human somatic cells have created the opportunity to study and use patient-specific stem cells, or induced pluripotent stem cells (iPSCs), and their derivatives. For these reasons, methods for directing the differentiation of pluripotent stem cells and selecting these for analysis and clinical use have become an area of intense investigation.
Enrichment of tissue-specific precursors
Pluripotency is defined as the potential of a stem cell to differentiate into tissues originating from all of the three embryonic germ layers: endoderm (for example, gastrointestinal tract and lung), mesoderm (for example, heart, muscle, bone, blood, and urogenital), and ectoderm (for example, nervous system and epidermis). At the molecular level, highly regulated transcriptional circuitry as well as extrinsic factors control the pluripotent state [1,2]. In addition, chromatin remodeling and small non-coding RNAs have been implicated in the regulation of pluripotency [2,3]. Pluripotency can be tested by using in vivo and in vitro methods. A test of pluripotency in vitro involves determining the ability of hESCs and iPSCs to form human embryoid bodies (hEBs) when cultured in a non-adherent cell suspension in the absence of feeder cell layers. hEBs are spherical colonies of differentiating stem cells that contain cell types representative of all three embryonic germ layers [4]. The most commonly used in vivo method to test pluripotency involves the transplantation of undifferentiated stem cells into immunodeficient mice to induce the formation of teratomas [4-6]. Teratomas are benign tumors composed of disorganized tissue structures characteristic of the three embryonic germ layers. Analysis of embryonic tissues found in teratomas from engrafted stem cells can be used to test their differentiation potential (Figure 1).
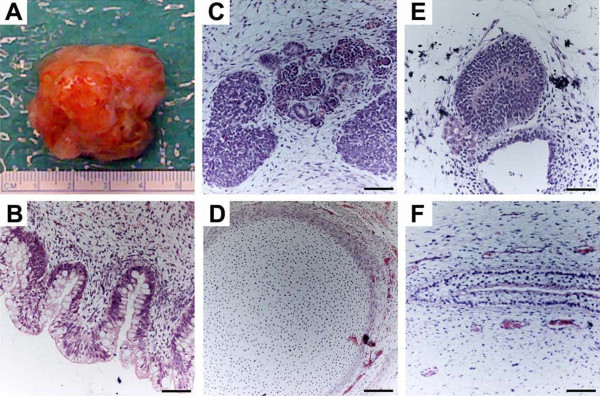
Figure 1.
Pluripotency assayed by teratoma formation. Proliferating cultures of human embryonic stem cells were used to form teratomas by renal capsule grafting by using established methods [5]. (a) An explanted teratoma is shown. (b-f) Teratomas were sectioned and stained with hematoxylin and eosin to identify embryonic tissues. Representative tissues from all three embryonic germ layers - endoderm (b), mesoderm (c, d), and ectoderm (e, f) - can be seen. (b) Glandular intestinal structure. (c) Nascent renal tubules and glomeruli within bed of primitive renal epithelium. (d) Cartilage surrounded by capsule of condensed mesenchyme. (e) Nascent neural tube. (f) Primitive squamous epithelium. Bar, 100 μm.
The ability of hESCs and iPSCs to mimic in vitro and in vivo the events occurring during human development makes them not only valuable tools for understanding the mechanisms involved in developmental processes but also stepping stones toward the generation of desired cell types suitable for cell therapies. Recent studies have shown that it is possible to generate lineage-restricted progenitors that are capable of differentiating into specialized post-mitotic cell types such as cardiomyocytes, pancreatic islet cells, chondrocytes, hematopoietic cells, endothelial cells, and neurons. Furthermore, the ability of pluripotent stem cells to divide indefinitely makes these a potential large-scale source of specific progenitors. In the following sections, we will provide examples of how stem cell differentiation can be directed toward specific cell/tissue types.
Developmental programming
Some of the most effective strategies for directing the differentiation of pluripotent stem cells into specific cell types have taken advantage of our understanding of human development (Table 1). Endodermal derivatives include cells that populate the lung, liver, and pancreas. Directing the differentiation of hESCs and iPSCs toward definitive endoderm would help generate specific cell types, such as islet cells or hepatocytes, which could be used in the treatment of diseases such as diabetes or liver disease, respectively. D'Amour and colleagues [7] showed that selective induction of endoderm could be achieved through the addition of high concentrations of Activin A, under low serum conditions, and in a stage-specific manner. Activin A mimics the action of Nodal, a ligand that activates transforming growth factor-beta (TGFβ) signaling, which in turn leads to the induction of endoderm differentiation. The effect of Activin A in inducing definitive endoderm is enhanced when additional factors such as Wnt3a [8] and Noggin [9] are present or when coupled with the suppression of the phosphoinositide 3-kinase pathway [10].
Table 1.
Examples of directed differentiation of human embryonic stem cells into specific cell types
| Embryonic germ layers | Differentiation factors or culture conditions or both | Example of differentiated cells |
|---|---|---|
| Endoderm | FGF, BMP4, hepatocyte growth factor, oncostatin M, dexamethasone | Hepatocytes [12,13] |
| Activin A, Wnt3A, keratinocyte growth factor/FGF7, retinoic acid, cyclopamine, noggin | Pancreatic islet progenitors [11] | |
| Mesoderm | Serum-free conditions; BMP4 | Dendritic cells [16] |
| Micromass of dissociated embryoid bodies; BMP2 | Chondrocytes [100] | |
| High-density culture of dissociated embryoid bodies; ascorbic acid, dexamethasone | Chondrocytes [101] | |
| Co-culture with primary chondrocytes; poly-D, L-lactide scaffold | Chondrocytes [102] | |
| Serum-free conditions | Blood cells [15] | |
| Dense monolayer of hESCs; Activin A, BMP4 | Cardiomyocytes [25] | |
| BMP4, BMP4/bFGF/Activin A, VEGF/DKK1, VEGF/DKK1/bFGF | Cardiomyocytes [26] | |
| Ectoderm | FGF8, SHH | Dopaminergic neurons [29,103] |
| Ciliary neutrotrophic factor, neuregulin 1β, dbcAMP | Schwann cells [34] | |
| Retinoic acid, SHH | Motor neurons [33] | |
| Withdrawal of FGF2, BDNF; addition of GDNF, NGF, dbcAMP | Peripheral sympathetic and sensory neurons [34] | |
| Serum-free conditions; Activin A, nicotinamide | Retinal pigment epithelium [104] | |
| B27, thyroid hormone, retinoic acid, FGF2, EGF, insulin | Oligodendrocytes [35] | |
| BMP4, ascorbic acid | Basal keratinocytes [105] |
BDNF, brain-derived neurotrophic factor; bFGF, basic fibroblast growth factor; BMP, bone morphogenetic protein; dbcAMP, dibutyryl cyclic adenosine monophosphate; DKK1, Dickkopf-related protein; EGF, epidermal growth factor; FGF, fibroblast growth factor; GDNF, glial cell-derived neurotrophic factor; hESC, human embryonic stem cell; NGF, nerve growth factor; SHH, sonic hedgehog; VEGF, vascular endothelial growth factor.
Induction of definitive endoderm can lead to the generation of specific progenitor populations after the addition of other factors. Among the most successful examples to date is the generation of pancreatic islet progenitors, devised by Kroon and colleagues [11] and accomplished through the sequential exposure of hESCs to Activin A and Wnt3A, followed by the addition of keratinocyte growth factor or fibroblast growth factor 7 to induce the formation of the primitive gut tube. Subsequently, retinoic acid, cyclopamine, and Noggin are added to inhibit hedgehog and TGFβ signaling and thus induce the differentiation of posterior foregut cells, the source of pancreatic cell progenitors. These are cultured further to generate pancreatic endoderm cells. When engrafted in immunodeficient mice, these cells display the histological and structural characteristics of pancreatic islet cells and are able to sustain insulin production for at least 100 days [11].
In a similar manner, hepatocytes can be obtained after differentiation of hESCs into definitive endoderm [12,13]. A robust population of functional hepatocytes was generated with the sequential addition of low serum medium, collagen I matrix, and hepatic differentiation factors that include fibroblast growth factor (FGF), bone morphogenetic protein 4 (BMP4), hepatocyte growth factor, oncostatin M, and dexamethasone [13]. These cells expressed known markers of mature hepatic cells, exhibited appropriate function, and were able to integrate and differentiate into mature liver cells when injected into mice with liver injury [13].
Directing the differentiation of stem cells into mesoderm requires activation of the TGFβ signaling pathway and can be accomplished through the stepwise and dosage-dependent addition of Activin A, BMP4, vascular endothelial growth factor (VEGF), and basic fibroblast growth factor (bFGF) [14]. Mesodermal derivatives have also been successfully obtained by spontaneous differentiation of hESCs through hEB formation without first directing them toward mesoderm. Robust differentiation of hESCs into hematopoietic lineage cells, which give rise to all blood cell types and components of the immune system, has been achieved under serum-free conditions through spin hEB formation [15]. Specific hematopoietic cells, such as functional dendritic cells, have been successfully differentiated from hESCs through spontaneous hEB formation under serum-free conditions with the addition of BMP4 at specific time points [16]. Hematopoietic progenitor cells that give rise to functional T and natural killer cells capable of targeting human tumor cells both in vitro and in vivo have also been derived from hESCs co-cultured with stromal cells [17]. Thus, the ability to differentiate hESCs into hematopoietic lineage cells promises to be useful in improving existing therapies that require blood cell transplantation and in immune therapies that require induction of the immune response in an antigen-specific manner [18].
Cardiomyocytes, which represent another therapeutically important derivative of mesoderm, have been successfully generated from pluripotent stem cells by using several methods [19]. Spontaneous differentiation of stem cells under appropriate culture conditions can produce cardiomyocytes that exhibit morphological, molecular, and electrophysiological properties similar to those of adult cardiomyocytes [20] and display quantifiable responses to physiological stimuli reminiscent of atrial, ventricular, and pacemaker/conduction tissue [21-24]. Cardiomyocytes have also been generated by directed differentiation with Activin A and BMP4 of a dense monolayer of stem cells; these cells successfully form specific cardiac lineages when transplanted in vivo [25]. Another study used additional medium supplements that included VEGF and the Wnt inhibitor, DKK1, followed by the addition of bFGF to promote cardiomyocyte differentiation in culture [26]. The success of these studies was measured by the expression of proteins specific for mature cardiac cells, such as cardiac troponin T, atrial myosin light chain 2, and the cardiac transcription factors Tbx5 and Tbx20.
The dominant differentiation pathway in hESC cultures leads to the formation of ectoderm, which makes up cells of the nervous system and the epidermis. hESC-derived neural progenitor cells are characterized by rosette-like neural structures that form in the presence of growth factors FGF2 or EGF either through spontaneous differentiation from an overgrowth of hESCs or after hEBs are plated onto adherent substrates [27,28]. These neural rosettes have become the signature of hESC-derived neural progenitors, capable of differentiation into a broad range of neural cells in response to appropriate developmental signals. Thus, many studies are exploring ways to enhance the formation of neural rosettes in order to generate an enriched population of specific neural cell types. One example is the use of specific stromal cell lines [29]. With this method, stromal cells provide ectodermal signaling factors required for neural induction, as determined in animal model studies, and therefore promote the formation of neural rosettes [30,31].
The withdrawal of FGF2 and EGF and the addition of specific compounds can lead to the differentiation of neural rosettes into specific neural subtypes. For example, hESC-derived neural progenitors treated with FGF8 and sonic hedgehog give rise to dopaminergic neurons [32], whereas treatment with sonic hedgehog and retinoic acid induces motor neuron differentiation [33]. Neural crest stem cells derived from neural rosettes can differentiate into peripheral sympathetic and sensory neurons by withdrawing FGF2/EGF and adding brain-derived neurotrophic factor (BDNF), glial cell-derived neurotrophic factor (GDNF), nerve growth factor (NGF), and dibutyryl cyclic adenosine monophosphate (dbcAMP) or into Schwann cells in the presence of ciliary neurotrophic factor (CNTF), neuregulin 1β, and dbcAMP [34]. Neuroglial cells, such as oligodendrocytes, are generated with B27, thyroid hormone, retinoic acid, FGF2, epidermal growth factor, and insulin [35].
Chemical enrichment
In some cases, the manipulation of key pathways during germ layer development has allowed significant enrichment of specific cell types without the complete recapitulation of embryonic development. Treatment with 5-azacytidine at days 6 to 8 of hESC differentiation significantly increases cardiac alpha-myosin heavy chain (αMHC) expression and enhances cardiomyocyte differentiation, suggesting that DNA demethylation is a key factor in directing tissue-specific differentiation [36]. Similarly, exposure to SB203580, a small-molecule inhibitor of p38MAPK, has been shown to significantly improve cardiomyocyte differentiation of hESCs grown in medium conditioned by mouse END2 cells, supporting a role for p38MAPK signaling in regulating human cardiomyocyte differentiation [37]. SB203580-treated hEBs display an increase in expression of both early mesoderm markers (Brachyury T, Tbx6, and Mesp1) and cardiac αMHC as well as increased cardiomyocyte numbers. Gaur and colleagues [38] subsequently showed that p38MAPK inhibition occurs in a dose- and stage-dependent manner, that it also causes the accelerated differentiation of hESC-derived cardiomyocytes by using the standard hEB formation method, and that it appears to act at the ectoderm/mesoendoderm branch point during hESC differentiation.
In the original study with SB203580, cells were subjected to an adapted differentiation system in which hESCs were differentiated in a suspension culture by using serum-free medium conditioned by the mouse END2 cell line [39]. END2-conditioned medium alone exhibits cardiomyocyte-inducing activity during hESC differentiation [37], and biochemical as well as microarray analysis of END2-conditioned medium and END2 cells, respectively, identified PGI2, a product of prostaglandin synthase enzymes, as an inducing factor in hESC cardiac differentiation [40]. Two key enzymes involved in PGI2 synthesis are upregulated in END2 cells compared with control MES1 cells [41], which lack cardiogenic activity. PGI2 levels are between 6- and 10-fold higher in END2-conditioned medium compared with control conditioned medium from MES1 cells. Moreover, insulin, a common supplement in media formulations, was discovered to be an inhibitor of hESC cardiac differentiation. END2-conditioned medium supplemented with increasing concentrations of insulin results in a dramatic decrease in hESC cardiomyocyte differentiation. Thus, addition of PGI2 in combination with insulin-free, unconditioned medium yields effective cardiac induction similar to that produced by END2-conditioned medium. Cardiac differentiation is further augmented in the presence of SB203580. Taken together, these three components provide a basic synthetic recipe for directing cardiomyocyte differentiation of hESCs.
Mechanical factors that influence differentiation
Since cardiac muscle is one of the few tissues that develop under the effects of dynamic force, it is not surprising that conditions generated by the force of fluids in motion can enhance cardiomyocyte differentiation as well. Supplying a constant rotary orbital motion for 7 days to suspension cultures of differentiating mouse EBs (mEBs) results in a significantly increased number of beating mEBs compared with mEBs cultured in static suspension [42]. Analysis of gene expression shows higher levels of mesodermal and cardiac proteins (Brachyury, GATA4, Nkx2-5, MEF2c, αMHC, and MLC2v) in rotary mEBs than in static mEBs. In addition, a greater proportion of rotary mEBs stain positive for α-sarcomeric actin compared with static EBs. The enhanced cardiomyocyte differentiation is independent of rotary speed ranging from 25 to 55 revolutions per minute as determined by the expression of cardiomyogenic genes [43].
Domian and colleagues [44] have examined the effects of surface tension on cardiomyogenic differentiation of murine cardiac progenitors. Progenitors derived from embryos and mouse ESCs (mESCs) are cultured on either fibronectin-coated slides or micropatterns of fibronectin alternating with a surfactant that blocks cell adhesion. When grown on these micropatterned surfaces, a population of cells form longitudinally aligned myocardial fibers. In addition, culturing this population on micropatterned surfaces results in a statistically significant increase in the proportion of cardiomyocytes, supporting a role for microenvironmental forces in cardiac muscle differentiation.
Studies of substrate stiffness and elasticity during stem cell differentiation have also demonstrated effects on skeletal muscle and bone development. Myosin/actin striations occurred only when myoblasts were cultured on gels with stiffness typical of normal muscle [45]. In addition, muscle stem cells cultured on hydrogel substrates that mimic the elasticity of muscle self-renew and fuse to existing myofibers with greater efficiency than cells grown on rigid plastic [46]. Substrate stiffness has also been shown to favor mesodermal formation from ESCs, specifically osteogenic differentiation [47].
These observations, as well as an evolving understanding of the tissue environment in which stem cells differentiate in vivo, underlie the recent application of biomaterials to promote the differentiation and retention of stem cells in a number of tissues [48,49]. For example, optimal sizing of pancreatic β-cell clusters derived from hESCs is essential to preventing nutrient and oxygen deprivation in β-cell transplant therapy for diabetes. As shown by Van Hoof and colleagues [50], the size of β-cell aggregates can be controlled by using micropatterned laminin. Polymer scaffolds have also been studied to facilitate transplantation of retinal progenitor cells for the treatment of macular degeneration. Steedman and colleagues [51] demonstrated that biodegradeable thin films of polycaprolactone could be topographically fabricated to enhance the attachment and organization of progenitor cells and induce their differentiation to photoreceptor cells.
Epigenetic reprogramming
MicroRNAs (miRNAs) are small non-coding RNAs thought to regulate the expression of 30% of protein-coding genes [52]. The biological importance of these RNAs in stem cell biology is underscored by recent studies demonstrating that mESCs lacking the miRNA-processing enzyme Dicer display differentiation and proliferation defects [53-56]. miR-1 and miR-133 specifically are expressed in the mouse heart [57,58]. Targeted deletion or knockdown of these miRNAs results in dysregulation of cardiac morphogenesis, electrical conduction, cell-cycle, and cardiac hypertrophy [57-60]. Recently, Ivey and colleagues [61] showed that miR-1 and miR-133 regulate the differentiation of mESCs and hESCs into the cardiac lineage. Both miRNAs are enriched in mESC-derived cardiomyocytes. Lentiviral introduction of either miR-1 or miR-133 into mESCs enhances early mesoderm differentiation as evidenced by increased expression of Brachyury. miR-1 and miR-133 also reinforce mesoderm lineage decisions by repressing endoderm and neuroectoderm differentiation. When stimulated to differentiate into either endoderm or neuroectoderm lineages, mEBs expressing either miR-1 or miR-133 express lower levels of endodermal and neural markers compared with control mEBs. However, further differentiation revealed opposing roles of miR-1 and miR-133. miR-1 promotes differentiation of mesoderm into the cardiac and skeletal muscle lineages as determined by enhanced Nkx2-5 and myogenin expression, respectively, whereas miR-133 blocks induction of both markers. Importantly, the differentiation of hESCs in the presence of miR-1 behaves comparably to mESC differentiation. Overexpression of miR-1 in hESCs increases Nkx2-5 expression and yields more than a threefold higher number of beating hEBs compared with wild-type controls.
Whereas miRNAs direct cell lineage determination by controlling protein dosage, epigenetic regulation through chromatin remodeling has been shown to control cell fate as well. Takeuchi and Bruneau [62] identified a minimal set of factors necessary to execute the cardiac transcriptional program. Baf60c, a cardiac-enriched subunit of the Swi/Snf-like BAF chromatin remodeling complex, in combination with cardiac transcription factors GATA4 and Tbx5, is able to induce cardiac differentiation in mouse embryos when ectopically expressed. With this combination, 90% of the transfected embryos display expression of the early cardiac marker Actc1 and 50% of the transfected embryos exhibit beating tissue. GATA4, together with Baf60c, is essential in initiating the cardiac gene program as assessed by expression of Actc1. Neither of the other transcription factors tested alone (Tbx5 and Nkx2-5) or in concert with Baf60c is able to induce Actc1 expression. GATA4/Baf60c, however, is not sufficient for generating spontaneously contracting embryonic tissue: Tbx5 is required to achieve beating cardiomyocytes.
Selection of tissue-specific precursors
As discussed earlier in this review, enrichment of specific cell types can be achieved by using molecules introduced at specific time points during culture. However, many of these methods yield only moderate enrichment that is not yet scalable for clinical application. In addition, it may be desirable to enrich first for partially differentiated, proliferative stem cell intermediates with specific fates. These could then be expanded before further differentiation into cells for therapy. In the following sections, we will discuss the variety of methods used to select tissuespecific precursors and their derivatives.
Use of cell surface markers
Even before the tools of genetic engineering were employed to manipulate stem cells, proteins expressed on the cell surface were used to identify distinct populations of stem cells and their ontogeny. Perhaps the best-known application of this approach is to the intermediates of hematopoiesis [63]. Over the past 10 years, similar attempts to create lineage maps of other tissues have focused on the identification of lineage-specific cell surface markers (Table 2). For example, pluripotent stem cells can be identified by the expression of stage-specific embryonic antigen (SSEA) 3 and 4 and the embryonal carcinoma marker Tra-1-60 on their surface [4]. The expression of the cell surface antigen CD133 on proliferating hESCs identifies cells predestined toward a neuroectodermal fate [4]. Hemangioblasts that ultimately give rise to hematopoietic stem cells, smooth muscle progenitors, and endothelial progenitors can be selected on the basis of the surface expression of CD143, E-cadherin, VE-cadherin, platelet endothelial cell adhesion molecule 1 (PECAM-1), and vascular endothelial growth factor receptor 2 (VEGFR2) [64]. Mesenchymal stem/stromal cells that differentiate into muscle, fat, cartilage, and bone cells can be identified by the expression of the cell surface proteins CD105/endoglin, CD73, and CD90/Thy-1 in the absence of surface expression of CD45, CD34, CD14 or CD11b, CD79α or CD19, and HLA-DR [65]. Recently, cardiomyocytes that have, until now, eluded detection by specific surface protein expression were shown to express signal-regulatory protein alpha (SIRPA) [66].
Table 2.
Examples of surface markers expressed on stem, progenitor, and differentiated cells
| Stem cell | Progenitor cell | Differentiated cell | |
|---|---|---|---|
| Pluripotent stem cell | SSEA-3/4, TRA-1-60 | ||
| Ectoderm | Neural stem cell CD133, ABCG2 [4,106,107] |
Neuron-restricted progenitor NCAM [108] |
Neuronal cell PSD-95, Synaptophysin [109] |
| Glial-restricted progenitor FGFR [110-112] |
Oligodendrocyte MOG, CD140a [113,114] |
||
| Type 2 astrocyte | |||
| Type 1 astrocyt FGFR3 [110,111] |
|||
| Motor neuron progenitor | Motor neuron | ||
| Surface ectoderm | Epidermal stem cell α6-integrinhigh, CD71low [115] |
Epithelial cell CEACAM-1, EpCAM [116,117] |
|
| Mesoderm | Hemangioblast | Hematopoietic stem cell | |
| CD143, E-cadherin, VE-cadherin, PECAM-1, VEGFR2 [64] | CD34, CD44, CD150, VEGFR2, Sca-1 [63] | Myeloid/lymphoid lineages [118] | |
| Smooth muscle progenitor VE-cadherin [119] |
Smooth muscle cell VE-cadherin [119] |
||
| Endothelial progenitor CD31, CD105, CD144, CD146, VEGFR2, VWF [120] |
Endothelial cell CD31 [120] |
||
| Mesenchymal stem cell CD105/endoglin, CD73, CD90/Thy-1 [65] |
Myogenic precursor α7-integrin, M-cadherin [121] |
Cardiac muscle SIRPA [66] |
|
| Skeletal muscle α7β1-integrin [122] |
|||
| Pre-adipocyte | Adipocyte Glut4 [123] |
||
| Chondrocyte precursor | Chondrocyte CD44 [124] |
||
| Osteoblast | Osteocyte | ||
| Endoderm | Primitive endoderm | ||
| Definitive endoderm | Hepatic progenitor c-Met, CD49f [125] |
Hepatocyte Il-6 [126] |
|
| Pancreatic progenitor | α cell | ||
| β cell Glut2 [127] |
|||
| PPγ cell | |||
| δ cell | |||
| ε cell |
ABCG2, ATP-binding cassette sub-family G member 2; CEACAM-1, carcinoembryonic antigen-related cell adhesion molecule 1; EpCAM, epithelial cell adhesion molecule; FGFR, fibroblast growth factor receptor; Glut, glucose transporter; MOG, myelin oligodendrocyte glycoprotein; NCAM, neural cell adhesion molecule; PECAM-1, platelet endothelial cell adhesion molecule 1; PSD-95, post-synaptic density protein 95; SIRPA, signal-regulatory protein alpha; SSEA, stage-specific embryonic antigen; VEGFR2, vascular endothelial growth factor receptor 2; VWF, von Willebrand factor.
Two main approaches have been used to identify lineage-specific surface markers. One employs labeled cell fate mapping to assess the tissue-specific fate of subpopulations of pluripotent stem cells expressing specific surface proteins [4], whereas the other uses a high-throughput flow cytometry screen of available antibodies against stem cell derivatives [66]. Although there are still large gaps in our ability to identify and select derivatives of all three germ layers on the basis of surface marker expression, progress continues to be made.
Biochemical purification
Although detection of lineage-specific genes underlies most methods for selecting specific cell types from differentiating stem cell cultures, a few cell types, such as cardiac muscle cells, bear unique biochemical properties that aid in their isolation. For example, Percoll gradient centrifugation has been reported to purify hESC-derived cardiomyocytes [36,67], based on their buoyancy characteristics. Differentiating hESCs are applied to a discontinuous Percoll gradient consisting of 40.5% Percoll layered over 58.5% Percoll. After centrifugation, the majority of cardiomyocytes reside within the 58.5% Percoll layer and express cardiac troponin I, sarcomeric MHC, αMHC, βMHC, and N-cadherin. hESC-derived cardiomyocytes of almost 70% purity have been obtained by using this approach.
A second purification strategy is based on the observation that cardiomyocytes have high mitochondrial content compared with that of non-myocytes [68]. Using the fluorescent dye tetramethylrhodamine methyl ester (TMRM), which freely diffuses into the mitochondrial matrix, Hattori and colleagues [68] found that TMRM fluorescence in embryonic rat hearts increases with developmental stage, suggesting that mitochondrial biogenesis is linked to myocardiogenesis. In whole rat embryos, TMRM fluorescence in the heart is more robust than in other tissues, and when analyzed by flow cytometry, flow-sorted populations with the highest TMRM fluorescence are observed to express cardiac α-actinin. TMRM-labeled cardiomyocytes derived from mESCs are positive for both Nkx2-5 and α-actinin. The cardiomyocyte content in cultured cells sorted from day-12 to day-25 mEBs is greater than 99% as determined by Nkx2-5 and α-actinin expression. Most notably, greater than 99% cardiomyocyte purity is also obtained in cultured cells sorted from differentiating hEBs.
Transgenic reporter cell lines
The use of fluorescent reporters of gene expression offers an approach to selecting tissue-specific cells by high-throughput fluorescence-activated cell sorting [69]. Alternatively, the use of reporters that activate expression of a selectable marker can drive a population of cells to near-homogeneity by restricting survival to a specific cell type. The paucity of specific cell surface markers with which to select cardiac cells has led to the development of a variety of reporter lines with which to select stem cell-derived cardiomyocytes and cardiac progenitors. Huber and colleagues [70] used lentiviral vectors to produce stable hESC lines in which enhanced green fluorescent protein (eGFP) is expressed under control of the cardiac-specific human ventricular myosin light chain 2 promoter. Xu and colleagues [71] generated stable hESC lines by using a reporter plasmid consisting of the cardiac-specific mouse αMHC promoter driving expression of the neomycin resistance gene. Kita-Matsuo and colleagues [72] designed a set of lentiviral vectors to generate multiple stable hESC lines with eGFP and mCherry reporters or with puromycin resistance downstream of the mouse αMHC promoter. Ritner and colleagues [23] generated a cardiac-specific hESC reporter line by using a lentiviral construct consisting of a fragment of the mouse αMHC promoter upstream of eGFP. The specific promoter fragment allowed the identification and analysis of early cardiac progenitors expressing Nkx2-5 but before the onset of cardiac troponin T or chamber-specific MLC expression [23].
Whereas isolation of hESC-derived cardiomyocytes from these reporter lines is based on positive selection, Anderson and colleagues [73] implemented a negative selection strategy to deplete undifferentiated proliferating hESCs from cultures of hESC-derived cardiomyocytes. Their hESC line used a herpes thymidine kinase/gancyclovir (HTK/GCV) suicide gene system under the control of a constitutive phosphoglycerate kinase promoter. After administration of the antiviral drug GCV, cells expressing HTK-phosphorylated GCV incorporated the guanosine analog into nascent DNA chains of proliferating cells and caused chain termination and cell death. The increased number of α-actinin-positive cells after GCV treatment led to an almost sevenfold enrichment of cardiomyocytes. An important caveat of this approach is that other non-proliferating cell types would remain in the culture while proliferating cardiomyocytes would be depleted. The culture would still need to undergo a cardiac purification step, and the excluded proliferating cardiomyocytes and cardiac progenitors may be of greater benefit for transplantation than fully differentiated, non-proliferating cells [19].
Reporter lines have similarly been derived for the isolation of other tissue-specific cells of biological and clinical interest, including motor neurons [74], germ cells [75], hepatocytes [76], and pancreatic progenitors [77]. However, the effects of random genomic integration on disrupting endogenous gene expression, as well as unregulated transgene expression, are potential limitations to this approach. Reporter lines must be rigorously evaluated to confirm authentic representation of gene expression without altering cell behavior [23].
Transgenesis through homologous recombination
To address the limitations of virally mediated introduction of reporter transgenes (for example, constitutive transgene expression and effects of random integration), many investigators have approached reporter development by using homologous recombination to 'knockin' a fluorescent protein into a specific genetic locus [78]. Initially, gene targeting of ESCs by homologous recom-bination was slowed by the difficulties of single-cell cloning [79-82], transfection efficiency using conventional methods [83,84], and karyotypic instability with dissociation of ESCs [85,86]. However, these obstacles have been overcome to some degree, and several successfully targeted ESC lines have resulted [78].
The hESC reporter lines currently in use represent a variety of targeting strategies. An H1.1-derived OCT4-eGFP line targeting exons 3 to 5 of the octamer-binding transcription factor 4 gene by positive selection alone has been used to sort pluripotent hESCs [83]. Similarly, an MIXL1-GFP line targeting exon 1 of the Mix1 homeobox-like 1 gene in HES3 cells by positive selection has been used to identify blood precursors [79]. Ruby and Zheng [84] used the FEZ family zinc finger 2 promoter to direct eYFP expression in HUES9 cells as a means of isolating neuronal precursors by using cre-mediated recombination with both positive and negative selection steps, and a similar approach using the oligodendrocyte lineage transcription factor 2 promoter driving eGFP expression in BG01 hESCs identified a subset of glial-restricted progenitors with oligodendrocyte fate [87]. Cre-mediated homologous recombination has also been used to generate two cardiac reporter lines, one using the second heart field-specific Islet-1 promoter directing dsRed expression in H9 hESCs [81] and another recently described pair of hESC lines in which the promoter of the early cardiac transcription factor NKX2-5 was used to drive eGFP expression in MEL1 (male) and HES3 (female) cells [88]. While these accomplishments demonstrate that the various unrelated ESC lines currently available are all amenable to homologous recombination, optimal targeting vector designs and selection strategies are still being explored.
Use of dual-FRET molecular beacons
Although homologous recombination technology avoids some of the concerns with transgenic reporter cell lines generated by random integration, the resulting cells have still undergone genomic modification. Seeking an alternative to genetically modified stem cell lines for tracking and isolating tissue-specific progenitors, King and colleagues [6] adapted dual-fluorescence resonance energy transfer (FRET) 'molecular beacon' technology for transient, real-time detection of gene expression during ESC differentiation. Molecular beacons are single-stranded oligonucleotide probes that have been employed to assay gene expression in vitro, as in real-time polymerase chain reaction assays, and in vivo by using microscopy [89]. These consist of short sequences capable of forming stem-loop structures bearing a fluorescent reporter group at one end and a fluorescent quencher at the opposite end [89]. In the absence of a target sequence, the oligonucleotide self-anneals, forming a stem that brings the reporter and quencher in close proximity, thereby quenching fluorescence (Figure 2). In the presence of a target sequence, the oligonucleotide anneals to the target, separating the reporter and quencher, thereby allowing fluorescence. These investigators showed that appropriately designed, dual-FRET molecular beacon pairs can identify the expression of specific mRNAs by microscopy and flow cytometry and facilitate the collection of specific stem cell populations by cell sorting while leaving the stem cell genome intact [6].
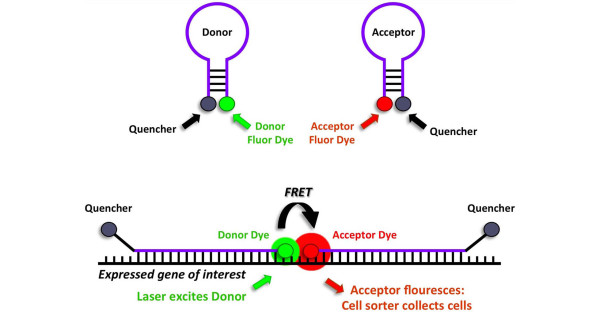
Figure 2.
Gene expression analysis using dual-fluorescence resonance energy transfer (dual-FRET) molecular beacons. In solution, molecular beacons assume a stem-loop structure that juxtaposes the fluorescent dye and quencher, inhibiting fluorescence (top). With transcription of a gene of interest, molecular beacons preferentially undergo sequence-specific base pairing with the gene transcript (bottom). This removes the fluorescent dyes from quencher proximity and allows laser excitation of the donor dye, FRET that excites the acceptor dye, and detection/collection of cells on the basis of acceptor dye emission.
Spectral flow cytometry: next-generation analysis
Conventional flow cytometers and cell sorters use a series of dichroic mirrors and filters matched to the fluorescence emission profile of reporter fluorochromes to identify and quantify specific biomarkers by using photomultiplier tubes (PMTs) to detect each parameter. Historically, this technology has relied on dichroic mirrors that reflect light of specific wavelengths and bandpass filters that allow incident light within a specific wavelength range to get to the detector. The detector, or PMT, detects photons and turns them into electrons or current. The fluorescent photon hits a photocathode and turns into an electron on a one-to-one basis; the electron then passes through a series of dynodes that amplify the electrons on the basis of the voltage generated across the dynode. One electron is multiplied and becomes two or more electrons, creating a cascade of electrons based on the single initial photon. At the end of the dynode chain, an anode collects the electron wave into an electronic signal; the more photons delivered to the PMT, the bigger the electronic signal.
A key criterion for distinguishing specific biomarkers over background is the optical signal-to-noise ratio of the detector [90]. For cytometric analysis of ESCs and iPSCs in altered differentiation states and treatment conditions, variable cellular autofluorescence and fluorescence crossover between multiple fluorochromes contribute to the background or 'noise' [91-93]. Increased noise decreases the resolving power for dim signals. A new cytometer developed by Sony Corporation (Tokyo, Japan) replaces filter-based optics with a spectral detection system based on a multi-anode spectral PMT. In the spectral cytometer, the light is directed on the basis of wavelength into discrete channels corresponding to different regions of the color spectrum by using a series of prisms. Over 30 channels are distributed across the 500- to 800-nm color spectrum, which allows almost all photons to be processed, whereas at least 20% of photons may be lost in the dichroic mirror/bandpass filter system.
The spectral cytometer is able to measure and subtract varying autofluorescence, permitting increased signal-to-noise ratios and improving the resolution of dim signals (Figure 3). Multiple fluorochromes are mathematically unmixed by using component analysis. Fluorochromes with overlapping emissions like fluorescein isothiocyante and Alexa 532 can now be used simultaneously where their spectral overlap would prevent their use in a conventional filter-based cytometer. Unique spectral 'fingerprints' can be created to distinguish tissue precursors transfected with combinations of molecular beacons described above but without the need for genome integration of reporter constructs or surface antibody staining that might cause unintended pleiotropic cellular responses. This technology holds great promise for its ability to exploit functional biosensors [94-97], such as dual-FRET molecular beacons [6], to identify specific stem cell differentiation states of therapeutic value.
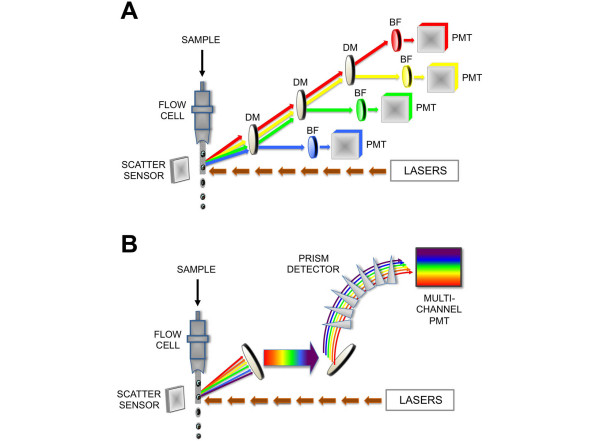
Figure 3.
'Next generation' spectral flow cytometry. (a) With a conventional flow cytometer, lasers excite cell-associated fluorochromes, and emitted light is filtered by a combination of dichroic mirrors (DMs) and bandpass filters (BFs) that reflect and filter light of specific wavelengths, respectively. Light within narrow selected wavelength ranges arrives at a photomultiplier tube (PMT), which converts light as photons to an electronic signal. (b) In spectral flow cytometry, laser diodes similarly provide initial excitation of reporter fluorochromes; however, emitted fluorescence passes through a prism array into a spectral PMT. Component fluorescence, including autofluorescence, is linearly unmixed by using spectral lookup tables. In spectral flow cytometry, unlike conventional cytometry, almost all light signals are analyzed, and signal-to-noise resolution is dramatically improved.
Conclusions
Research on pluripotent stem cells has progressed significantly since the first derivation of hESCs from discarded blastocysts in 1998 [98] and the discovery that somatic cells can be reprogrammed to a pluripotent stem-like state in 2006 [99]. The international scientific community has discovered the enormous potential of pluripotent stem cells as newly derived lines continue to be developed and differentiation methods into various types of cells are optimized. The ability to select and isolate stem cell-derived precursors and differentiated cells with tissue-specific properties holds the key to fully exploiting these cell lines and differentiation methods for scientific investigation and clinical use.
Abbreviations
bFGF: basic fibroblast growth factor; BMP4: bone morphogenetic protein 4; dbcAMP: dibutyryl cyclic adenosine monophosphate; EGF: epidermal growth factor; eGFP: enhanced green fluorescent protein; ESC: embryonic stem cell; FGF: fibroblast growth factor; FRET: fluorescence resonance energy transfer; GCV: gancyclovir; hEB: human embryoid body; hESC: human embryonic stem cell; HTK: herpes thymidine kinase; iPSC: induced pluripotent stem cell; mEB: mouse embryoid body; mESC: mouse embryonic stem cell; MHC: myosin heavy chain; miRNA: microRNA; PMT: photomultiplier tube; TGFβ: transforming growth factor-beta; TMRM: tetramethylrhodamine methyl ester; VEGF: vascular endothelial growth factor.
Competing interests
HSB and WCH acknowledge Sony Corporation for providing the funds and the spectral flow cytometer required for their evaluation of the spectral detection system.
Contributor Information
Harold S Bernstein, Email: harold.bernstein@ucsf.edu.
William C Hyun, Email: HyunB@cc.ucsf.edu.
Acknowledgements
HSB is supported by grants from the National Heart, Lung, and Blood Institute; the California Institute for Regenerative Medicine; and the Muscular Dystrophy Association. WCH is supported by funds from the UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis and the National Cancer Institute.
References
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- King FW, Ritner C, Liszewski W, Kwan HC, Pedersen A, Leavitt AD, Bernstein HS. Subpopulations of human embryonic stem cells with distinct tissue-specific fates can be selected from pluripotent cultures. Stem Cells Dev. 2009;18:1441–1450. doi: 10.1089/scd.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritner C, Bernstein HS. Fate mapping of human embryonic stem cells by teratoma formation. J Vis Exp. 2010. p. pii:2036. [DOI] [PMC free article] [PubMed]
- King FW, Liszewski W, Ritner C, Bernstein HS. High-throughput tracking of pluripotent human embryonic stem cells with dual fluorescence resonance energy transfer molecular beacons. Stem Cells Dev. 2011;20:475–484. doi: 10.1089/scd.2010.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- McLean AB, D'Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D'Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, Guo Y, Ding M, Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, Crooks GM. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci USA. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- Su Z, Frye C, Bae KM, Kelley V, Vieweg J. Differentiation of human embryonic stem cells into immunostimulatory dendritic cells under feeder-free culture conditions. Clin Cancer Res. 2008;14:6207–6217. doi: 10.1158/1078-0432.CCR-08-0309. [DOI] [PubMed] [Google Scholar]
- Woll PS, Grzywacz B, Tian X, Marcus RK, Knorr DA, Verneris MR, Kaufman DS. Human embryonic stem cells differentiate into a homogeneous population of natural killer cells with potent in vivo antitumor activity. Blood. 2009;113:6094–6101. doi: 10.1182/blood-2008-06-165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju S, Hirata S, Motomura Y, Fukuma D, Matsunaga Y, Fukushima S, Matsuyoshi H, Nishimura Y. Pluripotent stem cells as source of dendritic cells for immune therapy. Int J Hematol. 2010;91:392–400. doi: 10.1007/s12185-010-0520-1. [DOI] [PubMed] [Google Scholar]
- Wong SS, Bernstein HS. Cardiac regeneration using human embryonic stem cells: producing cells for future therapy. Regen Med. 2010;5:763–775. doi: 10.2217/rme.10.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- Ritner C, Wong SS, King FW, Mihardja SS, Liszewski W, Erle DJ, Lee RJ, Bernstein HS. An engineered cardiac reporter cell line identifies human embryonic stem cell-derived myocardial precursors. PLoS One. 2011;6:e16004. doi: 10.1371/journal.pone.0016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satin J, Kehat I, Caspi O, Huber I, Arbel G, Itzhaki I, Magyar J, Schroder EA, Perlman I, Gepstein L. Mechanism of spontaneous excitability in human embryonic stem cell derived cardiomyocytes. J Physiol. 2004;559:479–496. doi: 10.1113/jphysiol.2004.068213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stemcell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Suemori H, Mizuseki K, Watanabe K, Urano F, Ichinose H, Haruta M, Takahashi M, Yoshikawa K, Nishikawa S, Nakatsuji N, Sasai Y. Generation of dopaminergic neurons and pigmented epithelia from primate ES cells by stromal cell-derived inducing activity. Proc Natl Acad Sci USA. 2002;99:1580–1585. doi: 10.1073/pnas.032662199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Yang D, Zarnowska ED, Du Z, Werbel B, Valliere C, Pearce RA, Thomson JA, Zhang SC. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, Zhang SC. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- Lee G, Kim H, Elkabetz Y, Al Shamy G, Panagiotakos G, Barberi T, Tabar V, Studer L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49:385–396. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- Graichen R, Xu X, Braam SR, Balakrishnan T, Norfiza S, Sieh S, Soo SY, Tham SC, Mummery C, Colman A, Zweigerdt R, Davidson BP. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- Gaur M, Ritner C, Sievers R, Pedersen A, Prasad M, Bernstein HS, Yeghiazarians Y. Timed inhibition of p38MAPK directs accelerated differentiation of human embryonic stem cells into cardiomyocytes. Cytotherapy. 2010;12:807–817. doi: 10.3109/14653249.2010.491821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery CL, van Achterberg TA, van den Eijnden-van Raaij AJ, van Haaster L, Willemse A, de Laat SW, Piersma AH. Visceral-endoderm-like cell lines induce differentiation of murine P19 embryonal carcinoma cells. Differentiation. 1991;46:51–60. doi: 10.1111/j.1432-0436.1991.tb00865.x. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Graichen R, Soo SY, Balakrishnan T, Rahmat SN, Sieh S, Tham SC, Freund C, Moore J, Mummery C, Colman A, Zweigerdt R, Davidson BP. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76:958–970. doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Feijen A, Moolenaar WH, van den Brink CE, de Laat SW. Establishment of a differentiated mesodermal line from P19 EC cells expressing functional PDGF and EGF receptors. Exp Cell Res. 1986;165:229–242. doi: 10.1016/0014-4827(86)90547-1. [DOI] [PubMed] [Google Scholar]
- Sargent CY, Berguig GY, McDevitt TC. Cardiomyogenic differentiation of embryoid bodies is promoted by rotary orbital suspension culture. Tissue Eng Part A. 2009;15:331–342. doi: 10.1089/ten.tea.2008.0145. [DOI] [PubMed] [Google Scholar]
- Sargent CY, Berguig GY, Kinney MA, Hiatt LA, Carpenedo RL, Berson RE, McDevitt TC. Hydrodynamic modulation of embryonic stem cell differentiation by rotary orbital suspension culture. Biotechnol Bioeng. 2010;105:611–626. doi: 10.1002/bit.22578. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, Chien KR. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen X, Roberts CJ, Stevens MM. Substrate stiffness affects early differentiation events in embryonic stem cells. Eur Cell Mater. 2009;18:1–13; discussion 13-14. doi: 10.22203/ecm.v018a01. [DOI] [PubMed] [Google Scholar]
- Fu RH, Wang YC, Liu SP, Huang CM, Kang YH, Tsai CH, Shyu WC, Lin SZ. Differentiation of stem cells: strategies for modifying surface biomaterials. Cell Transplant. 2011;20:37–47. doi: 10.3727/096368910X532756. [DOI] [PubMed] [Google Scholar]
- Dickinson LE, Kusuma S, Gerecht S. Reconstructing the differentiation niche of embryonic stem cells using biomaterials. Macromol Biosci. 2011;11:36–49. doi: 10.1002/mabi.201000245. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Mendelsohn AD, Seerke R, Desai TA, German MS. Differentiation of human embryonic stem cells into pancreatic endoderm in patterned size-controlled clusters. Stem Cell Res. 2011;6:276–285. doi: 10.1016/j.scr.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Steedman MR, Tao SL, Klassen H, Desai TA. Enhanced differentiation of retinal progenitor cells using microfabricated topographical cues. Biomed Microdevices. 2010;12:363–369. doi: 10.1007/s10544-009-9392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Fraser S, Fujimoto T, Endoh M, Nishikawa S, Nishikawa SI. Origin of hematopoietic progenitors during embryogenesis. Int Rev Immunol. 2001;20:21–44. doi: 10.3109/08830180109056721. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeghiazarians Y, Gaur M, Zhang Y, Sievers RE, Ritner C, Prasad M, Boyle A, Bernstein HS. Myocardial improvement with human embryonic stem cell-derived cardiomyocytes enriched by p38MAPK inhibition. Cytotherapy. 2012;14:223–231. doi: 10.3109/14653249.2011.623690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, Li W, Yamakawa H, Tanaka T, Onitsuka T, Shimoji K, Ohno Y, Egashira T, Kaneda R, Murata M, Hidaka K, Morisaki T, Sasaki E, Suzuki T, Sano M, Makino S, Oikawa S, Fukuda K. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, Gepstein A, Habib M, Yankelson L, Kehat I, Gepstein L. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Zweigerdt R, Soo SY, Ngoh ZX, Tham SC, Wang ST, Graichen R, Davidson B, Colman A, Sun W. Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy. 2008;10:376–389. doi: 10.1080/14653240802105307. [DOI] [PubMed] [Google Scholar]
- Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, Talantova M, Bajpai R, Calzolari D, Terskikh A, McCulloch AD, Price JH, Conklin BR, Chen HS, Mercola M. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther. 2007;15:2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- Singh Roy N, Nakano T, Xuing L, Kang J, Nedergaard M, Goldman SA. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196:224–234. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Nicholas CR, Xu EY, Banani SF, Hammer RE, Hamra FK, Reijo Pera RA. Characterization of a Dazl-GFP germ cell-specific reporter. Genesis. 2009;47:74–84. doi: 10.1002/dvg.20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavon N, Yanuka O, Benvenisty N. Differentiation and isolation of hepatic-like cells from human embryonic stem cells. Differentiation. 2004;72:230–238. doi: 10.1111/j.1432-0436.2004.07205002.x. [DOI] [PubMed] [Google Scholar]
- Lavon N, Yanuka O, Benvenisty N. The effect of overexpression of Pdx1 and Foxa2 on the differentiation of human embryonic stem cells into pancreatic cells. Stem Cells. 2006;24:1923–1930. doi: 10.1634/stemcells.2005-0397. [DOI] [PubMed] [Google Scholar]
- Leavitt AD, Hamlett I. Homologous recombination in human embryonic stem cells: a tool for advancing cell therapy and understanding and treating human disease. Clin Transl Sci. 2011;4:298–305. doi: 10.1111/j.1752-8062.2011.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Fischer Y, Ganic E, Ameri J, Xian X, Johannesson M, Semb H. NANOG reporter cell lines generated by gene targeting in human embryonic stem cells. PLoS One. 2010;5:e12533. doi: 10.1371/journal.pone.0012533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- Mitalipova MM, Rao RR, Hoyer DM, Johnson JA, Meisner LF, Jones KL, Dalton S, Stice SL. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, Zeng X, Liu Y. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- Roederer M, Murphy RF. Cell-by-cell autofluorescence correction for low signal-to-noise systems: application to epidermal growth factor endocytosis by 3T3 fibroblasts. Cytometry. 1986;7:558–565. doi: 10.1002/cyto.990070610. [DOI] [PubMed] [Google Scholar]
- Alberti S, Parks DR, Herzenberg LA. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987;8:114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- Parks DR, Roederer M, Moore WA. A new 'Logicle' display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- Tung JW, Parks DR, Moore WA, Herzenberg LA, Herzenberg LA. New approaches to fluorescence compensation and visualization of FACS data. Clin Immunol. 2004;110:277–283. doi: 10.1016/j.clim.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Fitzpatrick JA, Bruchez MP. Fluorogenic dendrons with multiple donor chromophores as bright genetically targeted and activated probes. J Am Chem Soc. 2010;132:11103–11109. doi: 10.1021/ja9099328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GW, Adler SA, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. J Biomol Screen. 2010;15:703–709. doi: 10.1177/1087057110370892. [DOI] [PubMed] [Google Scholar]
- Ballou B, Ernst LA, Andreko S, Fitzpatrick JA, Lagerholm BC, Waggoner AS, Bruchez MP. Imaging vasculature and lymphatic flow in mice using quantum dots. Methods Mol Biol. 2009;574:63–74. doi: 10.1007/978-1-60327-321-3_6. [DOI] [PubMed] [Google Scholar]
- Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS, Armitage BA. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org Lett. 2008;10:1561–1564. doi: 10.1021/ol702920e. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Toh WS, Yang Z, Liu H, Heng BC, Lee EH, Cao T. Effects of culture conditions and bone morphogenetic protein 2 on extent of chondrogenesis from human embryonic stem cells. Stem Cells. 2007;25:950–960. doi: 10.1634/stemcells.2006-0326. [DOI] [PubMed] [Google Scholar]
- Cohen S, Leshansky L, Zussman E, Burman M, Srouji S, Livne E, Abramov N, Itskovitz-Eldor J. Repair of full-thickness tendon injury using connective tissue progenitors efficiently derived from human embryonic stem cells and fetal tissues. Tissue Eng Part A. 2010;16:3119–3137. doi: 10.1089/ten.TEA.2009.0716. [DOI] [PubMed] [Google Scholar]
- Vats A, Bielby RC, Tolley N, Dickinson SC, Boccaccini AR, Hollander AP, Bishop AE, Polak JM. Chondrogenic differentiation of human embryonic stem cells: the effect of the micro-environment. Tissue Eng. 2006;12:1687–1697. doi: 10.1089/ten.2006.12.1687. [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Welsh AM, Chen D, Hazel T, Johe K, Koliatsos VE. Combined immunosuppressive agents or CD4 antibodies prolong survival of human neural stem cell grafts and improve disease outcomes in amyotrophic lateral sclerosis transgenic mice. Stem Cells. 2006;24:1976–1985. doi: 10.1634/stemcells.2005-0518. [DOI] [PubMed] [Google Scholar]
- Idelson M, Alper R, Obolensky A, Ben-Shushan E, Hemo I, Yachimovich-Cohen N, Khaner H, Smith Y, Wiser O, Gropp M, Cohen MA, Even-Ram S, Berman-Zaken Y, Matzrafi L, Rechavi G, Banin E, Reubinoff B. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Guenou H, Nissan X, Larcher F, Feteira J, Lemaitre G, Saidani M, Del Rio M, Barrault CC, Bernard FX, Peschanski M, Baldeschi C, Waksman G. Human embryonic stem-cell derivatives for full reconstruction of the pluristratified epidermis: a preclinical study. Lancet. 2009;374:1745–1753. doi: 10.1016/S0140-6736(09)61496-3. [DOI] [PubMed] [Google Scholar]
- Jang YK, Park JJ, Lee MC, Yoon BH, Yang YS, Yang SE, Kim SU. Retinoic acid-mediated induction of neurons and glial cells from human umbilical cord-derived hematopoietic stem cells. J Neurosci Res. 2004;75:573–584. doi: 10.1002/jnr.10789. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zou X, Zhang H, Duan D, Ju L, Jiang X, Sun X, Zhao C, Zhao H, Guo J, Xu C, Gao E, Xu Q. Establishment of an immortalized GABAergic neuronal progenitor cell line from embryonic ventral mesencephalon in the rat. Brain Res. 2008;1210:63–75. doi: 10.1016/j.brainres.2008.02.062. [DOI] [PubMed] [Google Scholar]
- Wernig M, Benninger F, Schmandt T, Rade M, Tucker KL, Bussow H, Beck H, Brustle O. Functional integration of embryonic stem cell-derived neurons in vivo. J Neurosci. 2004;24:5258–5268. doi: 10.1523/JNEUROSCI.0428-04.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, Richardson WD. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130:93–102. doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- Reimers D, Lopez-Toledano MA, Mason I, Cuevas P, Redondo C, Herranz AS, Lobo MV, Bazan E. Developmental expression of fibroblast growth factor (FGF) receptors in neural stem cell progeny. Modulation of neuronal and glial lineages by basic FGF treatment. Neurol Res. 2001;23:612–621. doi: 10.1179/016164101101199090. [DOI] [PubMed] [Google Scholar]
- Furusho M, Kaga Y, Ishii A, Hebert JM, Bansal R. Fibroblast growth factor signaling is required for the generation of oligodendrocyte progenitors from the embryonic forebrain. J Neurosci. 2011;31:5055–5066. doi: 10.1523/JNEUROSCI.4800-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crang AJ, Gilson JM, Li WW, Blakemore WF. The remyelinating potential and in vitro differentiation of MOG-expressing oligodendrocyte precursors isolated from the adult rat CNS. Eur J Neurosci. 2004;20:1445–1460. doi: 10.1111/j.1460-9568.2004.03606.x. [DOI] [PubMed] [Google Scholar]
- Sim FJ, McClain CR, Schanz SJ, Protack TL, Windrem MS, Goldman SA. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat Biotechnol. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DR, Yang EN, Lee ST. A review: the location, molecular characterisation and multipotency of hair follicle epidermal stem cells. Ann Acad Med Singapore. 2004;33:784–788. [PubMed] [Google Scholar]
- Fullekrug J, Shevchenko A, Shevchenko A, Simons K. Identification of glycosylated marker proteins of epithelial polarity in MDCK cells by homology driven proteomics. BMC Biochemistry. 2006;7:8. doi: 10.1186/1471-2091-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzpis M, McLaughlin PM, de Leij LM, Harmsen MC. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Schmeckpeper J, Ikeda Y, Kumar AH, Metharom P, Russell SJ, Caplice NM. Lentiviral tracking of vascular differentiation in bone marrow progenitor cells. Differentiation. 2009;78:169–176. doi: 10.1016/j.diff.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Yoder MC, Mead LE, Prater D, Krier TR, Mroueh KN, Li F, Krasich R, Temm CJ, Prchal JT, Ingram DA. Redefining endothelial progenitor cells via clonal analysis and hematopoietic stem/progenitor cell principals. Blood. 2007;109:1801–1809. doi: 10.1182/blood-2006-08-043471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki N, Lim M, Yao CC, Tolar M, Kramer RH. alpha7 integrin expressing human fetal myogenic progenitors have stem cell-like properties and are capable of osteogenic differentiation. Exp Cell Res. 2006;312:4162–4180. doi: 10.1016/j.yexcr.2006.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Kaufman SJ. The alpha7beta1 integrin in muscle development and disease. Cell Tissue Res. 1999;296:183–190. doi: 10.1007/s004410051279. [DOI] [PubMed] [Google Scholar]
- Im SS, Kwon SK, Kim TH, Kim HI, Ahn YH. Regulation of glucose transporter type 4 isoform gene expression in muscle and adipocytes. IUBMB Life. 2007;59:134–145. doi: 10.1080/15216540701313788. [DOI] [PubMed] [Google Scholar]
- Woods A, Wang G, Beier F. Regulation of chondrocyte differentiation by the actin cytoskeleton and adhesive interactions. J Cell Physiol. 2007;213:1–8. doi: 10.1002/jcp.21110. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Gonzalez FJ, Nakauchi H. Identification and differentiation of hepatic stem cells during liver development. Front Biosci. 2006;11:1302–1310. doi: 10.2741/1884. [DOI] [PubMed] [Google Scholar]
- Fischer CP, Bode BP, Takahashi K, Tanabe KK, Souba WW. Glucocorticoid-dependent induction of interleukin-6 receptor expression in human hepatocytes facilitates interleukin-6 stimulation of amino acid transport. Ann Surg. 1996;223:610–618; discussion 618-619. doi: 10.1097/00000658-199605000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheepers A, Joost HG, Schurmann A. The glucose transporter families SGLT and GLUT: molecular basis of normal and aberrant function. JPEN J Parenter Enteral Nutr. 2004;28:364–371. doi: 10.1177/0148607104028005364. [DOI] [PubMed] [Google Scholar]