Abstract
Introduction
Osteoarthritis (OA) is a degenerative disease characterized by cartilage breakdown in the synovial joints. The presence of low-grade inflammation in OA joints is receiving increasing attention, with synovitis shown to be present even in the early stages of the disease. How the synovial inflammation arises is unclear, but proteins in the synovial fluid of affected joints could conceivably contribute. We therefore surveyed the proteins present in OA synovial fluid and assessed their immunostimulatory properties.
Methods
We used mass spectrometry to survey the proteins present in the synovial fluid of patients with knee OA. We used a multiplex bead-based immunoassay to measure levels of inflammatory cytokines in serum and synovial fluid from patients with knee OA and from patients with rheumatoid arthritis (RA), as well as in sera from healthy individuals. Significant differences in cytokine levels between groups were determined by significance analysis of microarrays, and relations were determined by unsupervised hierarchic clustering. To assess the immunostimulatory properties of a subset of the identified proteins, we tested the proteins' ability to induce the production of inflammatory cytokines by macrophages. For proteins found to be stimulatory, the macrophage stimulation assays were repeated by using Toll-like receptor 4 (TLR4)-deficient macrophages.
Results
We identified 108 proteins in OA synovial fluid, including plasma proteins, serine protease inhibitors, proteins indicative of cartilage turnover, and proteins involved in inflammation and immunity. Multiplex cytokine analysis revealed that levels of several inflammatory cytokines were significantly higher in OA sera than in normal sera, and levels of inflammatory cytokines in synovial fluid and serum were, as expected, higher in RA samples than in OA samples. As much as 36% of the proteins identified in OA synovial fluid were plasma proteins. Testing a subset of these plasma proteins in macrophage stimulation assays, we found that Gc-globulin, α1-microglobulin, and α2-macroglobulin can signal via TLR4 to induce macrophage production of inflammatory cytokines implicated in OA.
Conclusions
Our findings suggest that plasma proteins present in OA synovial fluid, whether through exudation from plasma or production by synovial tissues, could contribute to low-grade inflammation in OA by functioning as so-called damage-associated molecular patterns in the synovial joint.
Introduction
Osteoarthritis (OA) is a degenerative disease of the joints that is characterized by destruction of articular cartilage, inflammation of the synovial membrane (synovitis), and remodeling of periarticular bone. Which of these pathogenic processes occurs first is unknown. One proposed scenario is that cartilage breakdown (due to injury or mechanical stress) releases components of the damaged extracellular matrix (ECM) into synovial fluid, and that these ECM components elicit the local production of inflammatory molecules by binding to receptors on resident synovial cells or infiltrating inflammatory cells [1,2]. The inflammatory molecules produced may in turn stimulate production of cartilage-degrading enzymes and recruit inflammatory cells to the affected joint [3,4], thus establishing a vicious cycle of cartilage destruction and inflammation that perpetuates and promotes the OA pathology. Therefore, OA has been described as a chronic wound in which molecules in synovial fluid function as damage-associated molecular patterns (DAMPs; that is, endogenous molecules produced during injury that signal through inflammatory toll-like receptors (TLRs) to effect tissue remodeling) [2,5,6]. Although the identities of the endogenous molecules that mediate synovial inflammation have yet to be confirmed in OA patients or animal models, a continuous supply of DAMPs could perpetuate the early response to injury and thereby damage the joint.
Besides ECM components, many other molecules may act as DAMPs [2]. One such molecule is fibrinogen, which stimulates macrophage production of chemokines in a TLR4-dependent manner [7-9]. Fibrinogen is present at abnormally high levels in OA synovial fluid [10], and the amount of fibrin (the thrombin-cleaved form of fibrinogen [11]) deposited in the synovial membrane correlates with the severity of OA [12]. Although classically a plasma protein, fibrinogen exudes from the vasculature at sites of inflammation, such as the inflamed OA joint, owing to the retraction of inflamed endothelial cells [11]. Fibrinogen is not the only protein to extravasate at sites of inflammation, however, and several other plasma proteins have been detected in OA synovial fluid [10,13]. The extravascular function of most of these plasma proteins is unclear. It is possible that, like fibrinogen, some of these plasma proteins could have an immunoregulatory role at sites of inflammation or tissue damage.
Inflammation is present even in the early stages of OA [14,15], and clinical signs of synovitis correlate with radiographic progression of knee OA [16]. Insight into the cause of synovial inflammation is therefore important in understanding the pathogenesis of OA. Here we used proteomic techniques to survey the proteins present in OA synovial fluid and to evaluate levels of inflammatory cytokines in OA serum and synovial fluid. We then determined whether a subset of the identified proteins could promote inflammation by functioning as immunostimulatory DAMPs.
Material and methods
Synovial fluid and serum samples
Serum and synovial fluid samples were obtained from patients with knee OA, patients with rheumatoid arthritis (RA), or healthy individuals under protocols approved by the Stanford University Institutional Review Board and with the patients' informed consent. Synovial fluid aspiration was performed by a board-certified rheumatologist by fine-needle arthrotomy, and the synovial fluid samples obtained were free from obvious contamination with blood or debris. OA serum and synovial fluid samples were obtained from patients diagnosed with knee OA (of Kellgren-Lawrence score 2 to 4 [17]) according to the 1985 criteria of the American Rheumatism Association [18]. For mass spectrometric analysis, OA synovial fluid samples were from five Caucasian men aged 50 to 75 years who met the 1985 OA criteria [18]; exclusion criteria included radiographic evidence of chondrocalcinosis or evidence of crystals under polarizing microscopy. Demographics and clinical characteristics of these five individuals are shown in Table 1. Synovial fluids from the other OA patients and from the RA patients were provided as de-identified remnant clinical samples, and patient demographics were therefore unavailable for these samples. All RA patients met the 1987 American Rheumatism Association criteria for RA [19] and had RA of less than 6 months' duration; exclusion criteria included concurrent infectious or crystal arthritis. Samples of "normal" serum were obtained from healthy individuals who had no joint pain and no radiographic evidence of knee arthritis [20]. OA and normal sera were matched by age, sex, and BMI. Serum and synovial fluid samples were not matched but were derived from patients with the characteristics described earlier. All samples were aliquoted and stored at -80°C.
Table 1.
Clinical and demographic characteristics of OA patients whose synovial fluid was analyzed with mass spectrometry
| Subjecta | Age (years) | K-L score | SF cell count (cells/mm3) |
|---|---|---|---|
| 1 | 43 | 3 | 1,090 |
| 2 | 70 | 3 | 400 |
| 3 | 68 | 2 | 850 |
| 4 | 72 | 4 | Not measured |
| 5 | 74 | 4 | Not measured |
K-L, Kellgren-Lawrence OA score; SF, synovial fluid. aAll subjects were male and Caucasian with symptomatic pain in the aspirated knee.
Mass spectrometric analysis
Synovial fluid proteins were separated by 1D or 2D polyacrylamide gel electrophoresis (PAGE), trypsinized, and identified by liquid chromatography tandem mass spectrometry (LCMS), as follows. Fifty microliters of frozen synovial fluid was diluted to a final volume of 1 ml in phosphate buffered saline (PBS) containing Halt protease and phosphatase inhibitor (Thermo Fisher Scientific), and then depleted of the highly abundant proteins albumin and immunoglobulin G (IgG) by using the ProteoPrep Immunoaffinity Albumin & IgG Depletion Kit (Sigma-Aldrich) according to the manufacturer's instructions. In brief, synovial fluids were twice passed over spin columns prepacked with a mixture of two beaded mediums containing recombinantly expressed, small, single-chain antibody ligands. The flow-through fractions containing synovial fluid depleted of albumin and IgG were diluted 1:1 with Laemmli Sample Buffer (BioRad) and then subjected to 1D-PAGE or 2D-PAGE analysis. Because a small number of proteins other than albumin and IgG may bind to the medium in the spin columns, the bound proteins were eluted with Laemmli sample buffer and also subjected to PAGE analysis. For 1D-PAGE analysis, proteins were boiled for 10 minutes and separated on Precast Criterion XCT gels (4% to 12% linear gradient, BioRad). After electrophoresis, the gels were stained for 1 hour with Gelcode blue (Pierce) and destained overnight. For 2D-PAGE analysis, methods were as previously described [21]. In brief, 100 μg of synovial fluid proteins was dissolved in 150 μl of isoelectric focusing (IEF) buffer (ReadyPrep Sequential Extraction Kit Reagent 3, BioRad). For the first-dimension electrophoresis, 150 μl (at a 1 μg/μl concentration) of sample solution was applied to an 11-cm Ready-Strip Immobilized pH Gradient (IPG) strip, pH 3 to 10 (BioRad). The IPG strips were soaked in the sample solution for 1 hour, to allow uptake of the proteins, and then actively rehydrated in the Protean IEF cell (BioRad) for 12 hours at 50 V. IEF was performed for 1 hour at each of 100, 200, 500, and 1,000 V, and then for 10 hours at 8,000 V. For second-dimension electrophoresis, IPG strips were equilibrated for 20 minutes in 50 mM Tris-HCl, pH 8.8, containing 6 M urea, 1%SDS, 30% glycerol, and 65 mM dithiothreitol (DTT), and then re-equilibrated for 20 minutes in the same buffer containing 260 mM iodacetamide in place of DTT. Precast Criterion XCT gels (4% to 12% linear gradient, BioRad) were used for the second-dimension electrophoresis, as was done for the 1D-PAGE. After electrophoresis, the gels were stained for 1 hour with Gelcode blue (Pierce) and destained overnight.
The stained protein bands and spots (from the 1D-PAGE and 2D-PAGE, respectively) were cut out of the gels, immersed in 10 mM ammonium bicarbonate containing 10 mM DTT and 100 mM iodoacetamide, treated with 100% acetonitrile, and then digested overnight at 37°C with 0.1 mg trypsin (Sigma-Aldrich) in 10 mM ammonium acetate containing 10% acetonitrile. The trypsinized proteins were identified with LCMS by using the Agilent 1100 LC system and the Agilent XCT Ultra Ion Trap (Agilent Technologies, Santa Clara, CA) as previously described [22]. We scanned the LCMS data against the SwissProt database by using the SpectrumMill software (Agilent). We required the detection of at least two peptides for identification of a protein, and a significance level of P ≤ 0.05 for identification of each peptide. The significance level of peptide identification takes into account the number of ionization forms of the fragmented peptide that match with a particular protein in the SwissProt database (with penalties for ionization forms not identified), as well as the total intensity of each ionization form [23].
Multiplex cytokine analysis
Multiplex analysis of cytokines and chemokines in human serum and synovial fluid samples was performed by using both the 27-plex and the 21-plex Bio-Plex Pro Human Cytokine Assay (BioRad) run on the Luminex 200 platform, as recommended by the manufacturers. Performing the Bio-Plex assay with the kit reagents, we found that several commercial reagents designed to block the confounding effect of heterophilic antibodies, including ones we used previously with other cytokine assay kits [24], did not significantly affect the readout of the Bio-Plex assay; we therefore did not use such blocking reagents with the Bio-Plex assay. Data processing was performed by using Bio-Plex Manager 5.0, and analyte concentrations (in picograms per milliliter) were interpolated from standard curves. Statistical differences in cytokine levels were calculated with significance analysis of microarrays (SAM [25]), and the SAM-generated results with a false discovery rate (FDR) of less than 10% were selected. To identify relations and to display our results most effectively, we normalized the analyte concentrations as follows: all values less than 1 were designated as 1, and the mean concentration of each analyte in the "normal serum" samples was calculated; the analyte value in the sample was then divided by the mean analyte value in normal serum, and finally, a log-base-2 transformation was applied. Results were subjected to unsupervised hierarchic clustering by using Cluster 3.0, which arranges the SAM-generated results according to similarities in cytokine levels, and the clustering results were displayed by using Java Treeview (Version 1.1.3).
Macrophage stimulation assays
To generate mouse macrophages, we differentiated bone-marrow cells isolated from wild-type C57BL/6 mice and from B6.B10ScN-Tlr4lps-del mice (Jackson Laboratory) according to standard procedures [26]. In brief, the femur and tibia were flushed with α-minimal essential medium (MEM; Invitrogen) by using a 1-ml syringe and a 25-gauge needle. The resulting cell suspension was lysed with ACK Lysing Buffer (Invitrogen) for removal of erythrocytes. Cell clumps were removed by filtering through a 70-μm cell strainer (BD). The remaining cells in the suspension were cultured on 100-mm culture dishes in α-MEM supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 2 mM glutamine (Invitrogen) for 16 to 24 hours in 5% CO2 at 37°C. Nonadherent cells were collected, plated on 100-mm dishes, and differentiated into bone-marrow-derived macrophages (BMMs) for 6 days in the presence of 30 ng/ml of macrophage colony-stimulating factor (PeproTech). To generate human monocyte-derived macrophages (MDMs), we collected peripheral blood mononuclear cells (PBMCs) by performing density-gradient centrifugation of LRS chamber content (Stanford Blood Center) over Ficoll (Invitrogen), purified human monocytes by negative selection by using a monocyte isolation kit (Miltenyi Biotec), and differentiated the monocytes into macrophages by culturing them for 7 days in RPMI containing 10% FBS and 30 ng/ml of human M-CSF.
For stimulation assays, mouse BMMs were plated in 96-well plates at 1 × 105 cells/well, and human macrophages at 7 × 104 cells/well. Cells were incubated for 24 hours with lipopolysaccharide (LPS; Sigma-Aldrich), peptidoglycan (InvivoGen), α1-microglobulin (Cell Sciences), α2-macroglobulin (EMD Chemicals), α1-acid glycoprotein (EMD Chemicals), Gc-globulin (also known as vitamin D-binding protein; Abcam), haptoglobin (Sigma-Aldrich), or human serum albumin (Sigma-Aldrich). We measured levels of interleukin-1β (IL-1β), interleukin-6 (IL-6), and vascular endothelial growth factor (VEGF) in the culture supernatants with Luminex analysis, by using a 27-plex Bio-Plex Pro Human Cytokine Assay kit (BioRad) according to the manufacturer's instructions. We measured TNF levels with enzyme-linked immunosorbent assay (ELISA; PeproTech). For the TNF ELISA, the limits of detection were 16 to 2,000 pg/ml for mouse TNF, and 23 to 1,500 pg/ml for human TNF. For the Luminex assay, the limits of detection were 3.2 to 3,261 pg/ml for IL-1β, 2.3 to 18,880 pg/ml for IL-6, and 5.5 to 56,237 pg/ml for VEGF. To exclude a contribution of endotoxin contamination, we included 10 μg/ml of polymyxin B (Sigma-Aldrich) in some of the stimulation assays. As an additional control for endotoxin contamination, we tested whether preincubating the plasma proteins with proteinase K and β-mercaptoethanol at 55°C for 4 hours (and then at 100°C for 10 minutes to inactivate the proteinase K) abrogated their ability to induce the production of cytokines (the plasma proteins, but not any contaminating endotoxin, would be denatured under these conditions).
Statistical analysis
One-way ANOVA and unpaired t test (Graph-Pad Software) were used to analyze differences in levels of cytokines. P values less than 0.05 were considered significant.
Results and Discussion
We first used mass spectrometry to survey the proteins present in the synovial fluid of patients with knee OA. Synovial fluid proteins from five OA patients were separated by 1D- or 2D-PAGE and then identified by LCMS. Analysis of all five samples identified a total of 111 unique proteins; three of these were keratin proteins, skin proteins most likely obtained as a result of the cutaneous puncture performed during aspiration of the synovial joints. Eliminating these keratins left 108 unique proteins (Tables 2 and 3), most of which were detected in all synovial fluid samples analyzed. Of these, 44 were identified in a previous proteomic survey of highly abundant proteins in OA synovial fluid [10] (Table 2). Thus, we confirmed the presence of serine protease inhibitors (for example, antithrombin III, α1-antitrypsin, α1-antichymotrypsin, kininogen 1) and of proteins important in regulating proteases that degrade cartilage ECM. We also confirmed the presence of proteins involved in cartilage (for example, fibronectin) and/or collagen (for example, gelsolin and collagen α1, α2, and α3 chains) metabolism, and of proteins involved in inflammation or immunity (for example, fibrinogen, AGP 1, complement factors, immunoglobulins, cytokines) (Table 2), findings consistent with the inflammation, ECM degradation, and immune-cell infiltration that characterize OA. Among the 64 proteins that we newly identified (Table 3) were histone-related proteins, macrophage-related proteins, proinflammatory receptors, and proteins related to the proinflammatory transcription factor nuclear factor kappa B (Table 4), presumably reflecting the turnover of resident synovial cells or infiltrating inflammatory cells.
Table 2.
Proteins identified in OA synovial fluids in this study and in a previous proteomic study [10]
| Protein namea | Accession no.b | Scorec | % Coveraged | Number of peptidese |
|---|---|---|---|---|
| Transferrin | P02787 | 877.49 | 65 | 57 |
| α2-Macroglobulin | P01023 | 569.94 | 34 | 57 |
| Serum albumin | P02768 | 514.49 | 60 | 34 |
| Complement C3 | P01024 | 426.92 | 27 | 34 |
| Apolipoprotein A-I | P02647 | 405.32 | 67 | 26 |
| α1-Antitrypsin (α1 protease inhibitor) | P01009 | 388.37 | 66 | 27 |
| Apolipoprotein A-IV | P06727 | 370.16 | 60 | 27 |
| Haptoglobin | P00738 | 331.09 | 49 | 22 |
| Hemopexin (β-1B-glycoprotein) | P02790 | 273.36 | 40 | 20 |
| Gc-globulin (vitamin D-binding protein precursor) | P02774 | 269.18 | 43 | 21 |
| α1B-glycoprotein | P04217 | 215.74 | 47 | 16 |
| Complement factor B (C3/C5 convertase) | P00751 | 163.54 | 20 | 12 |
| α1-Antichymotrypsin | P01011 | 144.7 | 27 | 10 |
| Antithrombin-III | P01008 | 142.4 | 32 | 10 |
| Ceruloplasmin (EC 1.16.3.1) (Ferroxidase) | P00450 | 132.65 | 12 | 9 |
| Transthyretin | P02766 | 129.52 | 69 | 8 |
| Plasma protease C1 inhibitor | P05155 | 111.87 | 17 | 8 |
| Ig mu chain C region | P01871 | 98.51 | 16 | 7 |
| Actin, cytoplasmic 2 (γ-actin) | P63261 | 89.13 | 33 | 8 |
| Fibrinogen β chain | P02675 | 88.28 | 18 | 7 |
| α2-HS-glycoprotein | P02765 | 85.26 | 22 | 7 |
| Complement factor H (H factor 1) | P08603 | 80.47 | 8 | 8 |
| α1-Acid glycoprotein 1 (Orosomucoid-1) | P02763 | 68.98 | 23 | 5 |
| Complement C5 | P01031 | 55.88 | 8 | 7 |
| Prothrombin (coagulation factor II) | P00734 | 52.57 | 9 | 4 |
| Plasma retinol-binding protein | P02753 | 50.79 | 20 | 4 |
| Apolipoprotein E | P02649 | 49.32 | 21 | 4 |
| Afamin | P43652 | 48.67 | 7 | 4 |
| Fibrinogen α chain | P02671 | 48.63 | 6 | 4 |
| Gelsolin | P06396 | 47.03 | 4 | 3 |
| Complement C4 | P01028 | 39.34 | 8 | 6 |
| Vitronectin | P04004 | 38.1 | 6 | 3 |
| Apolipoprotein A-II | P02652 | 36.26 | 22 | 3 |
| Hemoglobin β subunit (Hemoglobin β chain) | P68871 | 31.88 | 17 | 2 |
| β2-Glycoprotein I (apolipoprotein H) | P02749 | 30.33 | 6 | 2 |
| Inter-α-trypsin inhibitor heavy chain H4 | Q14624 | 28.43 | 5 | 3 |
| Fibronectin | P02751 | 23.84 | 2 | 2 |
| Clusterin | P10909 | 21.17 | 12 | 2 |
| Complement component C8 γ chain | P07360 | 20.93 | 10 | 2 |
| Histidine-rich glycoprotein | P04196 | 20.11 | 5 | 2 |
| Fibrinogen γ chain | P02679 | 19.9 | 7 | 2 |
| Kininogen-1 | P01042 | 16.02 | 4 | 2 |
| Desmoplakin | P15924 | 11.59 | 1 | 2 |
| α1-Microglobulin/bikunin precursor protein precursor | P02760 | 11.13 | 9 | 2 |
aShown in bold are the proteins classified as plasma proteins (as assessed by www.HPRD.org). bUniProtKB/Swiss-Prot database. cAn aggregate score of the quality of the peptide spectra obtained, the total number of peptides identified, and percentage coverage of the protein. dThe number of amino acids identified as a percentage of the total number of amino acids in the corresponding protein. eThe number of peptides identified that correspond to the protein indicated.
Table 3.
Proteins newly identified in OA synovial fluids
| Protein namea | Accession no.b | Scorec | % Coveraged | No. of peptidese |
|---|---|---|---|---|
| Zinc-α2-glycoprotein | P25311 | 140.54 | 35 | 10 |
| Ig α1 chain C region | P01876 | 128.82 | 37 | 9 |
| Ig γ-1 chain C region | P01857 | 115.3 | 35 | 9 |
| Ig κ chain C region | P01834 | 87.06 | 85 | 6 |
| Calgranulin A (MRP-8) | P05109 | 67.87 | 33 | 5 |
| Collagen α1(I) chain | P02452 | 40.14 | 4 | 4 |
| Complement component C9 | P02748 | 36.76 | 9 | 4 |
| Serum paraoxonase/arylesterase 1 | P27169 | 34.77 | 13 | 3 |
| Ig κ chain V-III region SIE | P01620 | 33.65 | 38 | 3 |
| Ig heavy-chain V-III region BRO | P01766 | 28.94 | 24 | 2 |
| NF-κB-repressing factor | O15226 | 23.76 | 8 | 3 |
| A-kinase anchor protein-like protein 8 | Q9ULX6 | 23.65 | 11 | 3 |
| Structural maintenance of chromosomes 4-like 1 protein | Q9NTJ3 | 23.43 | 1 | 2 |
| Bromodomain adjacent to zinc-finger domain 2B (hWALp4) | Q9UIF8 | 22.6 | 3 | 3 |
| Histone deacetylase 5 | Q9UQL6 | 22.4 | 4 | 3 |
| Voltage-dependent R-type calcium channel α-1E subunit | Q15878 | 22.29 | 1 | 2 |
| Inhibitor of nuclear factor κB kinase β subunit | O14920 | 22.07 | 8 | 3 |
| β platelet-derived growth factor receptor precursor (EC 2.7.1.112) (CD140b antigen) | P09619 | 20.93 | 5 | 2 |
| Leucine-rich α2-glycoprotein | P02750 | 20.55 | 5 | 2 |
| Mitochondrial 28S ribosomal protein S29 | P51398 | 19.76 | 6 | 2 |
| Glucose-6-phosphate 1-dehydrogenase | P11413 | 19.58 | 14 | 3 |
| Collagen α2(I) chain | P08123 | 18.98 | 7 | 3 |
| Histone H4 | P62805 | 18.73 | 16 | 2 |
| Macrophage inflammatory protein 2-β | P19876 | 18.72 | 28 | 2 |
| Collagen α3(IV) chain | Q01955 | 18.15 | 5 | 2 |
| Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 | Q15047 | 17.82 | 4 | 2 |
| Toll-like receptor 6 | Q9Y2C9 | 17.38 | 2 | 2 |
| Nuclear factor NF-κB p100 subunit | Q00653 | 17.22 | 7 | 2 |
| Possible global transcription activator SNF2L1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) | P28370 | 16.37 | 5 | 2 |
| RRP5 protein homologue | Q14690 | 16.35 | 3 | 2 |
| Interleukin-20 receptor α chain | Q9UHF4 | 16.22 | 11 | 2 |
| NF-κB inhibitor-like protein 1 | Q9UBC1 | 16.13 | 14 | 2 |
| Platelet endothelial cell-adhesion molecule | P16284 | 15.87 | 10 | 2 |
| Fibroblast growth factor receptor 2 | P21802 | 15.4 | 2 | 2 |
| Interleukin-1 receptor-associated kinase 1 | P51617 | 14.92 | 11 | 2 |
| Cadherin EGF LAG seven-pass G-type receptor 1 | Q9NYQ6 | 14.84 | 1 | 2 |
| Collagen α2(V) chain | P05997 | 14.78 | 5 | 2 |
| Zinc-finger A20 domain-containing protein 1 | Q6GQQ9 | 14.61 | 6 | 2 |
| Low-density lipoprotein receptor-related protein 1 (apolipoprotein E receptor) | Q07954 | 14.54 | 1 | 2 |
| Microtubule-actin crosslinking factor 1, isoforms 1/2/3/5 | Q9UPN3 | 14.53 | 1 | 2 |
| Zinc-finger DHHC domain-containing protein 13 | Q8IUH4 | 14.48 | 11 | 2 |
| Death-associated protein kinase 1 | P53355 | 14.41 | 3 | 2 |
| Collagen α1(V) chain | P20908 | 13.78 | 4 | 2 |
| Integrin α-V (Vitronectin receptor α subunit) | P06756 | 13.52 | 3 | 2 |
| Misshapen-like kinase 1(MAPK/ERK kinase kinase kinase 6) | Q8N4C8 | 13.36 | 4 | 2 |
| Cadherin EGF LAG seven-pass G-type receptor | Q9NYQ7 | 13.26 | 1 | 3 |
| Regulator of G-protein signaling 14 | O43566 | 13.21 | 8 | 2 |
| Collagen α1(VII) chain | Q02388 | 13.19 | 1 | 2 |
| Interleukin-12 receptor β2 chain | Q99665 | 13.1 | 5 | 2 |
| Interleukin-1β (IL-1β) | P01584 | 12.94 | 20 | 2 |
| Platelet-derived growth factor B chain | P01127 | 12.91 | 18 | 2 |
| Collagen α1(II) chain | P02458 | 12.65 | 6 | 2 |
| Pappalysin-1 | Q13219 | 12.58 | 2 | 2 |
| Complement C1q tumor necrosis factor-related protein 5 | Q9BXJ0 | 11.97 | 23 | 2 |
| Interleukin-18 receptor 1 precursor (IL-1 receptor-related protein) | Q13478 | 11.8 | 4 | 2 |
| A kinase anchor protein 10, mitochondrial (PRKA10) | O43572 | 11.36 | 8 | 2 |
| Neutrophil collagenase | P22894 | 11.34 | 9 | 2 |
| Protein-arginine deiminase type I | Q9ULC6 | 11.34 | 5 | 2 |
| TNF receptor-associated factor 5 (RING finger protein 84) | O00463 | 10.92 | 8 | 2 |
| Macrophage receptor MARCO | Q9UEW3 | 10.89 | 8 | 2 |
| Collagen α3(V) chain | P25940 | 10.32 | 1 | 2 |
| Histone acetyltransferase MYST3 | Q92794 | 10.14 | 2 | 2 |
| Leukocyte immunoglobulin-like receptor subfamily B member 4 | Q8NHJ6 | 9.43 | 11 | 2 |
| Mast/stem cell growth-factor receptor | P10721 | 9.3 | 5 | 2 |
aShown in bold are the proteins classified as plasma proteins (as assessed by www.HPRD.org). bUniProtKB/Swiss-Prot database. cAn aggregate score of the quality of the peptide spectra obtained, the total number of peptides identified, and percentage coverage of the protein. dThe number of amino acids identified as a percentage of the total number of amino acids in the corresponding protein. eThe number of peptides identified that correspond to the protein indicated.
Table 4.
Classes of proteins newly identified in OA synovial fluids
| Histone-related proteins | NF-κB-related proteins | Inflammatory receptors | Macrophage-related proteins |
|---|---|---|---|
| Histone deacetylase 5 | NF-κ-B-repressing factor | IL-12-receptor β2 | Macrophage inflammatory protein 2-β |
| Histone H4 | Inhibitor of nuclear factor κB kinase β subunit | IL-18-receptor 1 | Macrophage receptor MARCO |
| Histone-lysine N- methyltransferase, H3 lysine-9 specific 4 | Nuclear factor κB p100 subunit | IL-20-receptor α chain | |
| Histone acetyltransferase MYST3 | NF-κB inhibitor-like protein 1 | Toll-like receptor 6 | |
| Integrin-αV | |||
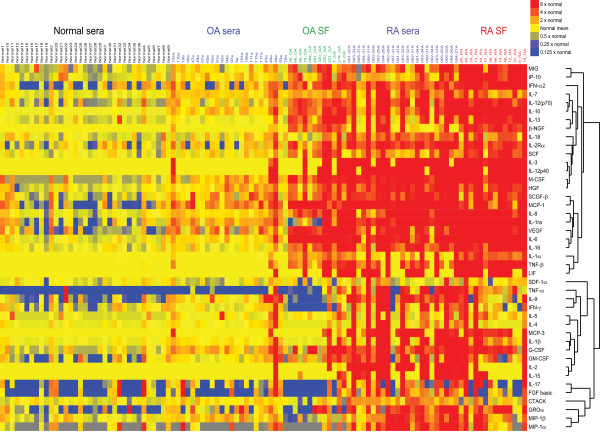
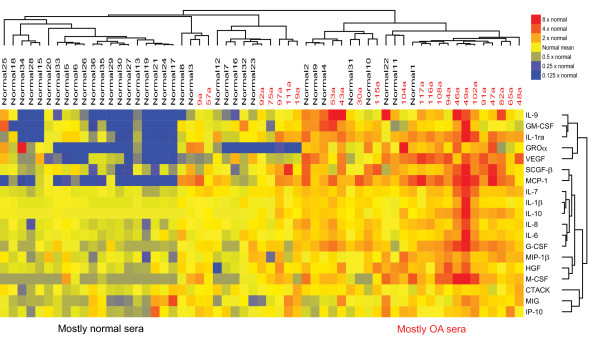
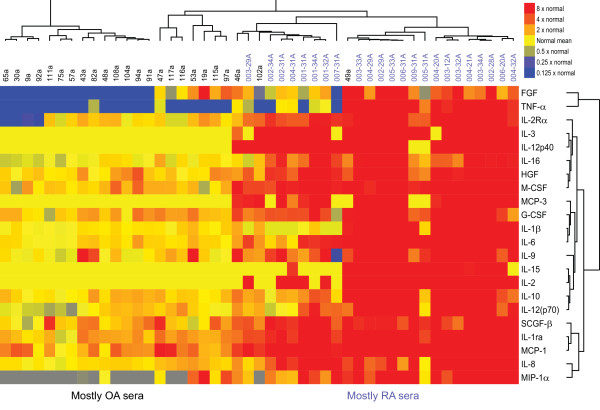
Our mass-spectrometric findings revealed the presence of many molecules associated with inflammation. Although cytokines are also classically associated with inflammation, PAGE-based mass spectrometry is not well suited to the detection of small proteins such as cytokines. We therefore used a multiplex immunoassay to measure levels of inflammatory cytokines and chemokines in synovial fluid samples from 12 patients with knee OA and 14 patients with RA, as well as in serum samples from 24 patients with knee OA, 23 patients with RA, and 35 healthy individuals. Samples from patients with RA, a classic inflammatory arthritis, were used as a comparator. Figure 1 shows a heatmap of the relative levels of cytokines in the five groups of samples. Compared with cytokine levels in normal sera, cytokine levels in OA sera were generally slightly higher, and those in RA sera were much higher (Figure 1). SAM analysis revealed that levels of several inflammatory cytokines (for example, IL-1β and IL-6), chemokines (for example, IP-10 (also known as CXCL10), MCP-1, IL-8, MIG, and MIP-1β), and growth factors (for example, VEGF and SCGF-β) were significantly higher in OA sera than in normal sera (FDR < 10%; Figure 2), consistent with previous reports of the association of OA with such inflammatory mediators [27]. Interestingly, we also found OA-associated elevations in levels of IL-9 and cutaneous T-cell attracting chemokine (CTACK), supporting the concept that T cells play a role in OA [28]. As expected, cytokine levels were significantly higher in RA sera than in OA sera (Figure 3; FDR < 10%).
Figure 1.
Inflammatory cytokines are associated with osteoarthritis. Relative cytokine levels in serum and synovial fluid (SF) samples from patients with osteoarthritis (OA) or rheumatoid arthritis (RA) and in serum samples from healthy individuals (normal sera). Cytokine levels were measured with a multiplex bead-based immunoassay. Samples from individual patients are listed above the heatmap, and the individual cytokines are listed to the right of the heatmap. IL, interleukin; IFN, interferon; MIG, monokine induced by IFN-γ; IP-10, interferon gamma-induced protein 10; IL-1ra, interleukin-1 receptor antagonist; VEGF, vascular endothelial growth factor; GM-CSF, granulocyte macrophage colony-stimulating factor; FGF, fibroblast growth factor; MCP, monocyte chemotactic protein; IL-2Rα, interleukin-2 receptor α chain; HGF, hepatocyte growth factor; GROα, growth-regulated oncogene α; MIP-1, macrophage inflammatory protein; β-NGF, β nerve growth factor; SCF, stem cell factor; M-CSF, macrophage colony-stimulating factor; SCGF-β, stem cell growth factor β; LIF, leukemia inhibitory factor; SDF-1α, stromal cell-derived factor 1α; G-CSF, granulocyte colony-stimulating factor; CTACK, cutaneous T-cell attracting chemokine.
Figure 2.
Levels of inflammatory cytokines are higher in OA compared with healthy sera. Cytokines whose levels differ significantly between sera from individuals with osteoarthritis (OA) and sera from age-matched healthy individuals (FDR < 10%). Significance analysis of microarrays (SAM) was used to identify statistically significant differences, and the SAM-generated results were subjected to unsupervised hierarchic clustering. Cytokine levels were measured with a multiplex bead-based immunoassay. Samples from individual patients are listed above the heatmap, and the individual cytokines are listed to the right of the heatmap. IL, interleukin; MIG, monokine induced by IFN-γ; IP-10, interferon gamma-induced protein 10; IL-1ra, interleukin-1 receptor antagonist; VEGF, vascular endothelial growth factor; GM-CSF, granulocyte macrophage colony-stimulating factor; MCP, monocyte chemotactic protein; HGF, hepatocyte growth factor; GROα, growth-regulated oncogene α; MIP-1β, macrophage inflammatory protein 1β; M-CSF, macrophage colony-stimulating factor; SCGF-β, stem cell growth factor β; G-CSF, granulocyte colony-stimulating factor; CTACK, cutaneous T-cell attracting chemokine.
Figure 3.
Levels of inflammatory cytokines are higher in RA compared with OA sera. Cytokines whose levels differ significantly between sera from individuals with osteoarthritis (OA) and sera from individuals with rheumatoid arthritis (RA) (FDR < 10%). Significance analysis of microarrays (SAMs) was used to identify statistically significant differences, and the SAM-generated results were subjected to unsupervised hierarchic clustering. Cytokine levels were measured with a multiplex bead-based immunoassay. Samples from individual patients are listed above the heatmap, and the individual cytokines are listed to the right of the heatmap. IL, interleukin; IL-1ra, interleukin-1 receptor antagonist; FGF, fibroblast growth factor; MCP, monocyte chemotactic protein; HGF, hepatocyte growth factor; MIP-1, macrophage inflammatory protein; M-CSF, macrophage colony-stimulating factor; SCGF-β, stem cell growth factor β; G-CSF, granulocyte colony-stimulating factor.
Unlike RA, OA is considered a disorder that is restricted to the joints. Indeed, levels of multiple cytokines were much higher in OA synovial fluids than in OA sera (Figure 1 and Table 5); levels of TNF were negligible in OA sera but substantial in OA synovial fluid (Figure 1 and Table 5). Our findings suggest that the abnormally high levels of cytokines in OA sera largely reflect overproduction of these cytokines in the joint, consistent with the finding that levels of high-sensitivity C-reactive protein in the serum of OA patients correlate with the degree of inflammatory infiltrate in the patients' joints [29]. Thus, OA is associated with low-grade inflammation that may originate in the joints.
Table 5.
Absolute and relative cytokine levels in healthy and OA serum, and in OA synovial fluid
| Cytokine | Normal serum levels (pg/ml)a | OA serum levels (pg/ml)b | OA SF levels (pg/ml)c | Ratio of OA serum levels to normal serum levels | Ratio of OA SF levels to OA serum levels |
|---|---|---|---|---|---|
| IL-6 | 3.02 (2.7-4.4) |
5.13 (4.4-5.8) |
975.39 (454.0-2,689.5) |
1.7 | 190.1 |
| IL-1β | 1.22 (1.0-1.3) |
1.58 (1.4-1.8) |
1.14 (1.0-1.7) |
1.3 | 0.7 |
| TNF | 0.00 (0.0-0.0) |
0.00 (0.0-0.0) |
2.92 (0.0-13.3) |
- | - |
| VEGF | 20.53 (11.7-65.6) |
78.22 (31.3-124.6) |
496.31 (245.0-577.8) |
3.8 | 6.3 |
| MCP-1 | 3.59 (0.0-11.9) |
18.53 (13.0-28.0) |
107.67 (84.8-191.1) |
5.2 | 5.8 |
| IP-10 | 537.00 (376.1-750.4) |
795.55 (684.7-1,029.7) |
2,105.40 (923.2-4,913.3) |
1.5 | 2.6 |
| MIG | 244.51 (173.5-374.6) |
420.13 (308.4-568.6) |
1,047.14 (389.8-1,925.3) |
1.7 | 2.5 |
OA, osteoarthritis; SF, synovial fluid; IL, interleukin; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor; MCP-1, macrophage chemotactic protein 1; IP-10, interferon γ-induced protein 10; MIG, monokine-induced by interferon γ. aThe median (IQR); n = 34 healthy individuals. bThe median (IQR); n = 23 individuals with OA. cThe median (IQR); n = 10 individuals with OA.
Interestingly, 39 (36%) of the proteins we identified in OA synovial fluid are classically considered plasma proteins (Tables 2 and 3). Indeed, plasma proteins form a large proportion of the proteins enriched in OA synovial fluid relative to healthy synovial fluid [10]. What might these plasma proteins be doing in the OA joint? Like certain products of ECM breakdown [2,5,6], the plasma protein fibrinogen can function as a DAMP and has been proposed to contribute to the pathogenesis of inflammatory arthritis [7-9]. We therefore examined whether other plasma proteins in OA synovial fluid can function as immunostimulatory DAMPs that could contribute to the low-grade inflammation associated with OA.
Key players in OA-associated inflammation are the macrophages [27,30,31]. The cell infiltrate in human OA joints consists mainly of macrophages, and mice depleted of macrophages are relatively resistant to collagenase-induced OA [30]. Macrophages from OA joints produce a number of growth factors, such as VEGF, and inflammatory cytokines, such as the major OA-associated cytokines IL-1β and TNF [30]. We detected VEGF, IL-1β, and TNF in OA synovial fluid in our cytokine screen (Figure 1 and Table 5) and found that levels of VEGF and IL-1β are significantly higher in OA sera than in normal sera (Figure 2). VEGF may promote OA pathology by inducing angiogenesis (and thereby osteophyte formation) and by inducing matrix metalloprotease production (and thereby cartilage degradation) [32]. The cytokines produced by macrophages amplify the inflammation in the joints by inducing synovial cells to produce further cytokines and chemokines, as well as matrix metalloproteases [30]. Moreover, macrophages express many of the receptors that mediate DAMP signaling, and they can thus trigger an inflammatory cascade in response to DAMPs present in OA synovial fluid [7-9].
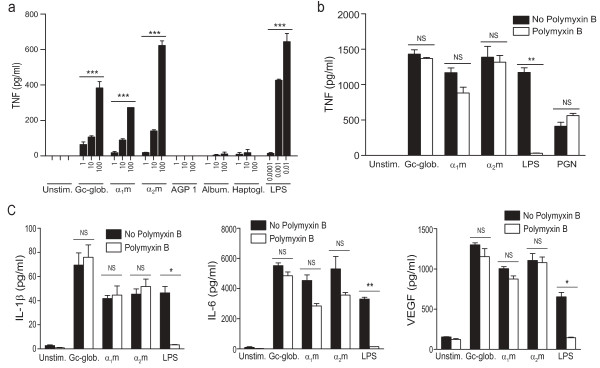
We therefore assessed whether a subset of the identified plasma proteins could induce macrophages to produce TNF, a key cytokine that is thought to drive the inflammatory cascade in OA [27]. We tested α1-microglobulin (α1m), α1-acid glycoprotein 1 (AGP 1; also known as orosomucoid 1), α2-macroglobulin (α2m), Gc-globulin (also known as vitamin D-binding protein), albumin, and haptoglobin, all of them plasma proteins detected in our survey of synovial fluid proteins (Table 2) and shown to be enriched in OA synovial fluid [10]. With mouse macrophages, we found that α1m, α2m, and Gc-globulin, at concentrations similar to those measured in synovial fluid [33-35], each dose-dependently stimulated the production of TNF, whereas AGP 1, albumin, and haptoglobin did not (Figure 4a). The plasma proteins ceruloplasmin, complement component C3, complement component C4, β2-glycoprotein (also known as apolipoprotein H) also did not stimulate TNF production (data not shown).
Figure 4.
Plasma proteins detected in osteoarthritic synovial fluid are immunostimulatory. Mouse bone-marrow-derived macrophages (BMMs) and human monocyte-derived macrophages (MDMs) were stimulated for 24 hours with plasma proteins detected in osteoarthritic synovial fluid (Table 2), after which cytokine levels in the supernatants were measured with ELISA or Luminex immunoassay. (a) Levels of TNF produced by mouse BMMs stimulated with the indicated concentrations (in μg/ml) of Gc-globulin, α1-microglobulin (α1m), α2-macroglobulin (α2m), α1-acid glycoprotein 1 (AGP 1), albumin, or haptoglobin. Lipopolysaccharide (LPS) was used as a positive control. (b) Levels of TNF produced by human MDMs stimulated with 50 μg/ml of α1m, α2m, or Gc-globulin in the presence or absence of 10 μg/ml of polymyxin B, an inhibitor of LPS. LPS (1 ng/ml) was used as a positive control for the efficacy of polymyxin B, and peptidoglycan (PGN; 5 μg/ml), as a negative control. (c) Levels of interleukin-1β (IL-1β), interleukin-6 (IL-6), and vascular endothelial growth factor (VEGF) produced by human MDMs stimulated with 50 μg/ml of Gc-globulin, α1m, or α2m. LPS (1 ng/ml) was used as a positive control. Results are representative of experiments performed at least twice. In (a), data are shown as the mean ± SEM of duplicates. In (b) and (c), data are shown as the mean ± SEM of triplicates. *P < 0.05; **P < 0.01, ***P < 0.001; NS, not significant.
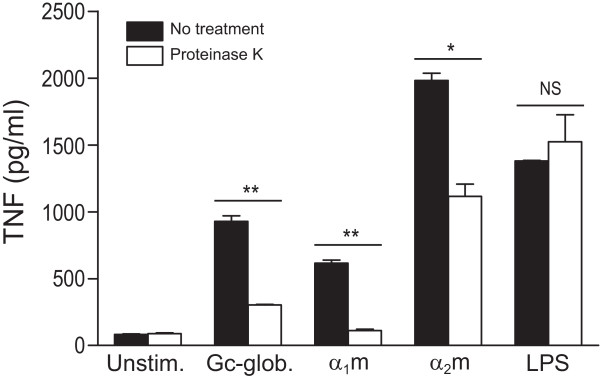
We next examined the effect of α1m, α2m, and Gc-globulin on cytokine production in human macrophages. Because the endotoxin LPS is a common contaminant and is itself an agonist of TLR4, we tested the stimulatory properties of the plasma proteins in the presence of polymyxin B, a compound that neutralizes LPS. In the presence of polymyxin B, α1m-, α2m-, and Gc-globulin-induced TNF production was not significantly reduced, whereas LPS-induced TNF production was abrogated (Figure 4b). Additionally, pretreatment with proteinase K significantly abrogated TNF production induced by the plasma proteins but not TNF production induced by LPS (Figure 5). Although we cannot exclude the possibility that a small component of the observed stimulation is due to endotoxin, this result confirms that the plasma proteins are themselves immunostimulatory. Gc-globulin, α1m, and α2m were also able to induce the production of several other inflammatory cytokines that were upregulated in OA serum and synovial fluid (Figures 1 and 2): IL-1β, IL-6, and VEGF (Figure 4c). Thus, Gc-globulin, α1m, and α2m can each induce the production of TNF, IL-1β, IL-6, and VEGF, all molecules implicated in the pathogenesis of OA [27,30,31].
Figure 5.
Induction of TNF production by plasma proteins is not due to endotoxin contamination. RAW264.7 macrophages were stimulated for 24 hours with 50 μg/ml of Gc-globulin, α1-microglobulin (α1m), or α2-macroglobulin (α2m) that had been incubated with proteinase K (20 μg/ml) at 55°C for 4 hours in the presence of β-mercaptoethanol and then heated to 100°C for 10 minutes. TNF levels in the supernatants were determined with ELISA. Lipopolysaccharide (LPS; 1 ng/ml) was used as a positive control. Data are shown as the mean ± SEM of duplicates from one of two representative experiments. *P < 0.05; **P < 0.01.
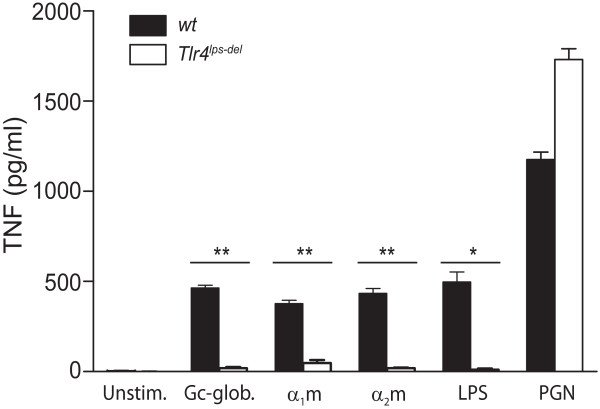
But how do these plasma proteins stimulate cytokine production? To determine whether these immunostimulatory plasma proteins signal through TLR4, we examined whether Gc-globulin, α1m, and α2m could also induce TNF production in TLR4-deficient macrophages. TLR4 deficiency inhibited Gc-globulin-, α1m-, and α2m-induced TNF production (Figure 6). Confirming that the defect in inflammatory signaling in the Tlr4lps-del macrophages was specific to the TLR4 pathway, theTLR2-specific agonist peptidoglycan was able to induce TNF production in these cells-in fact, to a greater degree than in wild-type cells (possibly because of compensatory mechanisms operating within the TLR family) (Figure 6). Thus, Gc-globulin-, α1m-, and α2m-induced production of TNF is dependent on TLR4.
Figure 6.
Plasma proteins detected in osteoarthritic synovial fluid stimulate macrophage TNF production via TLR4. Levels of TNF produced by wild-type (wt) or TLR4-deficient (Tlr4lps-del) mouse bone-marrow-derived macrophages stimulated for 24 hours with 50 μg/ml of Gc-globulin, α1-microglobulin (α1m), or α2-macroglobulin (α2m), after which TNF levels in the supernatants were determined with ELISA. Lipopolysaccharide (LPS; 1 ng/ml) was used as a positive control for TLR4-dependent TNF production, and peptidoglycan (PGN; 5 μg/ml) as a positive control for TLR4-independent TNF production. Data are shown as the mean ± SEM of triplicates from one of three representative experiments. *P < 0.05; **P < 0.01.
Interest in the putative immunomodulatory effects of α1m, α2m, and Gc-globulin is increasing, with both proinflammatory and antiinflammatory properties suggested for each of them [36-38].
For example, α1m has been shown to bind to the surface of various inflammatory cells and to either stimulate or inhibit the activation of human lymphocytes [38]. The immunoregulatory role of α1m in health and disease is likely to be context dependent. Gc-globulin, however, appears to be primarily proinflammatory: it enhances the neutrophil- and monocyte-chemotactic activity of the anaphylatoxin C5a [36] and, in its sialic-acid-free form, activates macrophages [39]. Here, we uncover an additional mechanism by which these plasma proteins could promote inflammation. We speculate that exudation into extravascular spaces at sites of tissue damage and inflammation may render these plasma proteins inflammatory by bringing them into contact with TLR-expressing macrophages. Our finding that certain plasma proteins present in OA synovial fluid can induce macrophage production of inflammatory cytokines supports the model of local production of inflammatory mediators in the joints in OA.
Conclusions
We identified 108 proteins in OA synovial fluid and showed that OA is associated with low-grade inflammation. We found that plasma proteins form a large proportion of the proteins present in OA synovial fluid and that certain of these plasma proteins can signal through TLR4 to induce the production of an array of inflammatory cytokines, including those upregulated in OA. Our findings suggest that plasma proteins present in OA synovial fluid, whether through exudation from the plasma or production by synovial tissues, could contribute to low-grade inflammation in OA by functioning as DAMPs.
Abbreviations
α1m: α1-microglobulin; AGP 1: α1-acid glycoprotein 1; α2m: α2-macroglobulin; BMM: bone-marrow-derived macrophage; DAMP: damage-associated molecular pattern; ECM: extracellular matrix; IL-1β: interleukin-1β; IL-6: interleukin-6; IP-10: interferon gamma-induced protein 10; LCMS: chromatography tandem mass spectrometry; LPS: lipopolysaccharide; MCP-1: macrophage chemotactic protein-1; MDM: monocyte-derived macrophage; MEM: minimal essential medium; MIG: monokine induced by interferon-γ; MIP-1: macrophage inflammatory protein-1; OA: osteoarthritis; PAGE: polyacrylamide gel electrophoresis; RA: rheumatoid arthritis; TLR: Toll-like receptor; TNF: tumor necrosis factor; VEGF: vascular endothelial growth factor.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JS and WHR conceived the studies. OS performed the mass spectrometric analysis of synovial fluid. PEC, LJL, and JCE performed the multiplex cytokine analysis. JCE, KAB, and TPA collected and provided OA sera. DHS performed the in vitro macrophage stimulation assays. JS, WHR, DHS, OS, TML, and IH analyzed the resulting datasets. TML and JS wrote and edited the manuscript with the input of WHR, OS, and DHS. All authors read and approved the final manuscript.
See related editorial by Goldring and Scanzello, http://arthritis-research.com/content/14/2/111
Contributor Information
Dong Hyun Sohn, Email: dhsohn@stanford.edu.
Jeremy Sokolove, Email: sokolove@stanford.edu.
Orr Sharpe, Email: osharpe@stanford.edu.
Jennifer C Erhart, Email: jennifer.erhart@gmail.com.
Piyanka E Chandra, Email: pechandra@gmail.com.
Lauren J Lahey, Email: llahey@stanford.edu.
Tamsin M Lindstrom, Email: tlind@stanford.edu.
Inyong Hwang, Email: inyonghwangmd@gmail.com.
Katherine A Boyer, Email: kboyer@stanford.edu.
Thomas P Andriacchi, Email: tandriac@stanford.edu.
William H Robinson, Email: wrobins@stanford.edu.
Acknowledgements
This study was funded by VA RR&D Merit Review Award and N01-HV-00242 NHLBI Proteomics Center funding to WHR.
References
- Sellam J, Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol. 2010;6:625–35. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- Scanzello CR, Plaas A, Crow MK. Innate immune system activation in osteoarthritis: is osteoarthritis a chronic wound? Curr Opin Rheumatol. 2008;20:565–72. doi: 10.1097/BOR.0b013e32830aba34. [DOI] [PubMed] [Google Scholar]
- Bondeson J, Wainwright SD, Lauder S, Amos N, Hughes CE. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res Ther. 2006;8:R187. doi: 10.1186/ar2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodell-May J, Matuska A, Oyster M, Welch Z, O'Shaughnessey K, Hoeppner J. Autologous protein solution inhibits MMP-13 production by IL-1beta and TNFalpha-stimulated human articular chondrocytes. J Orthop Res. 2011;29:1320–6. doi: 10.1002/jor.21384. [DOI] [PubMed] [Google Scholar]
- Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G, Brennan F, Foxwell B. Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat Med. 2009;15:774–80. doi: 10.1038/nm.1987. [DOI] [PubMed] [Google Scholar]
- Sofat N. Analysing the role of endogenous matrix molecules in the development of osteoarthritis. Int J Exp Pathol. 2009;90:463–79. doi: 10.1111/j.1365-2613.2009.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns DB, Priel DA, Gallin JI. Induction of human monocyte interleukin (IL)-8 by fibrinogen through the Toll-like receptor pathway. Inflammation. 2007;30:178–88. doi: 10.1007/s10753-007-9035-1. [DOI] [PubMed] [Google Scholar]
- Smiley ST, King JA, Hancock WW. Fibrinogen stimulates macrophage chemokine secretion through Toll-like receptor 4. J Immunol. 2001;167:2887–94. doi: 10.4049/jimmunol.167.5.2887. [DOI] [PubMed] [Google Scholar]
- Sokolove J, Zhao X, Chandra PE, Robinson WH. Immune complexes containing citrullinated fibrinogen costimulate macrophages via Toll-like receptor 4 and Fcgamma receptor. Arthritis Rheum. 2011;63:53–62. doi: 10.1002/art.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobezie R, Kho A, Krastins B, Sarracino DA, Thornhill TS, Chase M, Millett PJ, Lee DM. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53:505–18. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- Loeuille D, Chary-Valckenaere I, Champigneulle J, Rat AC, Toussaint F, Pinzano-Watrin A, Goebel JC, Mainard D, Blum A, Pourel J, Netter P, Gillet P. Macroscopic and microscopic features of synovial membrane inflammation in the osteoarthritic knee: correlating magnetic resonance imaging findings with disease severity. Arthritis Rheum. 2005;52:3492–501. doi: 10.1002/art.21373. [DOI] [PubMed] [Google Scholar]
- Pruzanski W, Russell ML, Gordon DA, Ogryzlo MA. Serum and synovial fluid proteins in rheumatoid arthritis and degenerative joint diseases. Am J Med Sci. 1973;265:483–90. doi: 10.1097/00000441-197306000-00007. [DOI] [PubMed] [Google Scholar]
- Benito MJ, Veale DJ, FitzGerald O, van den Berg WB, Bresnihan B. Synovial tissue inflammation in early and late osteoarthritis. Ann Rheum Dis. 2005;64:1263–7. doi: 10.1136/ard.2004.025270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felson DT. Clinical practice: osteoarthritis of the knee. N Engl J Med. 2006;354:841–8. doi: 10.1056/NEJMcp051726. [DOI] [PubMed] [Google Scholar]
- Ledingham J, Regan M, Jones A, Doherty M. Factors affecting radiographic progression of knee osteoarthritis. Ann Rheum Dis. 1995;54:53–8. doi: 10.1136/ard.54.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, Howell D, Kaplan D, Koopman W, Longley S III, Mankin H, McShane DJ, Medsger T Jr, Meenan R, Mikkelsen W, Moskowitz R, Murphy W, Rothschild B, Segal M, Sokoloff L, Wolfe F. Development of criteria for the classification and reporting of osteoarthritis: classification of osteoarthritis of the knee; Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986;29:1039–49. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medsger TA Jr, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunder GG. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Mundermann A, King KB, Smith RL, Andriacchi TP. Change in serum COMP concentration due to ambulatory load is not related to knee OA status. J Orthop Res. 2009;27:1408–13. doi: 10.1002/jor.20908. [DOI] [PubMed] [Google Scholar]
- Elchuri S, Naeemuddin M, Sharpe O, Robinson WH, Huang TT. Identification of biomarkers associated with the development of hepatocellular carcinoma in CuZn superoxide dismutase deficient mice. Proteomics. 2007;7:2121–9. doi: 10.1002/pmic.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Avila V, Sharpe O, Robinson WH. Determination of ceruloplasmin in human serum by SEC-ICPMS. Anal Bioanal Chem. 2006;386:180–7. doi: 10.1007/s00216-006-0528-9. [DOI] [PubMed] [Google Scholar]
- Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O'Hair RA, Speed TP, Simpson RJ. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem. 2003;75:6251–64. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- Hueber W, Tomooka BH, Zhao X, Kidd BA, Drijfhout JW, Fries JF, van Venrooij WJ, Metzger AL, Genovese MC, Robinson WH. Proteomic analysis of secreted proteins in early rheumatoid arthritis: anti-citrulline autoreactivity is associated with upregulation of proinflammatory cytokines. Ann Rheum Dis. 2007;66:712–9. doi: 10.1136/ard.2006.054924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley ER. Murine bone marrow-derived macrophages. Methods Mol Biol. 1997;75:301–4. doi: 10.1385/0-89603-441-0:301. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- Sakkas LI, Platsoucas CD. The role of T cells in the pathogenesis of osteoarthritis. Arthritis Rheum. 2007;56:409–24. doi: 10.1002/art.22369. [DOI] [PubMed] [Google Scholar]
- Pearle AD, Scanzello CR, George S, Mandl LA, DiCarlo EF, Peterson M, Sculco TP, Crow MK. Elevated high-sensitivity C-reactive protein levels are associated with local inflammatory findings in patients with osteoarthritis. Osteoarthritis Cartilage. 2007;15:516–23. doi: 10.1016/j.joca.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Bondeson J, Blom AB, Wainwright S, Hughes C, Caterson B, van den Berg WB. The role of synovial macrophages and macrophage-produced mediators in driving inflammatory and destructive responses in osteoarthritis. Arthritis Rheum. 2010;62:647–57. doi: 10.1002/art.27290. [DOI] [PubMed] [Google Scholar]
- Farahat MN, Yanni G, Poston R, Panayi GS. Cytokine expression in synovial membranes of patients with rheumatoid arthritis and osteoarthritis. Ann Rheum Dis. 1993;52:870–5. doi: 10.1136/ard.52.12.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Yudoh K, Masuko K. The potential role of vascular endothelial growth factor (VEGF) in cartilage: how the angiogenic factor could be involved in the pathogenesis of osteoarthritis. Osteoarthritis Cartilage. 2008;16:279–86. doi: 10.1016/j.joca.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Cawston TE, McLaughlin P, Hazleman BL. Paired serum and synovial fluid values of alpha 2-macroglobulin and TIMP in rheumatoid arthritis. Br J Rheumatol. 1987;26:354–8. doi: 10.1093/rheumatology/26.5.354. [DOI] [PubMed] [Google Scholar]
- Fairney A, Straffen AM, May C, Seifert MH. Vitamin D metabolites in synovial fluid. Ann Rheum Dis. 1987;46:370–4. doi: 10.1136/ard.46.5.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Kin K, Itoh Y, Enomoto H, Kawai T. Human alpha 1-microglobulin levels in various body fluids. J Clin Pathol. 1980;33:786–91. doi: 10.1136/jcp.33.8.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew RR, Webster RO. Gc-globulin (vitamin D-binding protein) enhances the neutrophil chemotactic activity of C5a and C5a des Arg. J Clin Invest. 1988;82:364–9. doi: 10.1172/JCI113596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logdberg L, Wester L. Immunocalins: a lipocalin subfamily that modulates immune and inflammatory responses. Biochim Biophys Acta. 2000;1482:284–97. doi: 10.1016/S0167-4838(00)00164-3. [DOI] [PubMed] [Google Scholar]
- Akerstrom B, Logdberg L, Berggard T, Osmark P, Lindqvist A. alpha(1)-Microglobulin: a yellow-brown lipocalin. Biochim Biophys Acta. 2000;1482:172–84. doi: 10.1016/S0167-4838(00)00157-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Homma S. Vitamin D3 binding protein (group-specific component) is a precursor for the macrophage-activating signal factor from lysophosphatidylcholine-treated lymphocytes. Proc Natl Acad Sci USA. 1991;88:8539–43. doi: 10.1073/pnas.88.19.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]