Abstract
Introduction
Systematic approaches to dealing with missing values in record linkage are still lacking. This article compares the ad-hoc treatment of unknown comparison values as ’unequal’ with other and more sophisticated approaches. An empirical evaluation was conducted of the methods on real-world data as well as on simulated data based on them.
Material and Methods
Cancer registry data and artificial data with increased numbers of missing values in a relevant variable are used for empirical comparisons. As a classification method, classification and regression trees were used. On the resulting binary comparison patterns, the following strategies for dealing with missingness are considered: imputation with unique values, sample-based imputation, reduced-model classification and complete-case induction. These approaches are evaluated according to the number of training data needed for induction and the F-scores achieved.
Results
The evaluations reveal that unique value imputation leads to the best results. Imputation with zero is preferred to imputation with 0.5, although the latter shows the highest median F-scores. Imputation with zero needs considerably less training data, it shows only slightly worse results and simplifies the computation by maintaining the binary structure of the data.
Conclusions
The results support the ad-hoc solution for missing values ‘replace NA by the value of inequality’. This conclusion is based on a limited amount of data and on a specific deduplication method. Nevertheless, the authors are confident that their results should be confirmed by other empirical analyses and applications.
Keywords: Classification trees, medical record linkage, missing data, MNAR, record linkage
Data deduplication refers to the process in which records referring to the same real-world entities are detected in datasets such that duplicated records can be eliminated. The denotation ‘record linkage’ is used here for the same problem.1 A typical application is the deduplication of medical registry data.2 3 Medical registries are institutions that collect medical and personal data in a standardized and comprehensive way. The primary aims are the creation of a pool of patients eligible for clinical or epidemiological studies and the computation of certain indices such as the incidence in order to oversee the development of diseases. The latter task in particular requires a database in which synonyms and homonyms do not distort the measures. For instance, synonyms would lead to an overestimation of the incidence and thereby possibly to false resource allocations. The record linkage procedure must itself be reliable and of high quality in order to achieve clean data (for measures regarding the quality of record linkage methods see also Christen and Goiser4). A number of other important works have also investigated record linkage.5–16
Missing values in record linkage applications constitute serious problems in addition to the difficulties introduced by them in areas in which there is no necessity for computing comparison patterns. In settings such as survey analysis missing values emerge, for example, due to missing responses or knowledge of the participants. Analyses based on the data gathered can be biased in this case because of unfilled fields, for example, higher wages are less likely to be revealed than lower ones. Papers that deal with missing values in survey analysis are, for example, the ones of Acock17 and King et al.18
In contrast, in record linkage of electronic health records using personal data, the impact of missing values is augmented because they occur in comparison fields if any of the underlying fields has a missing value. Therefore, missingness in record linkage applications with a significant number of NA values is not ignorable, ie, not random. This non-randomness can also occur when blocking is applied in order to reduce the number of resulting record pairs: one or more features are selected as grouping variables and only pairs with agreement in these variables are considered. A comprehensive survey regarding blocking is given by Christen.19
The distinction into missing completely at random (MCAR), missing at random (MAR) and missing not at random (MNAR) of Little and Rubin20 is only relevant as a starting point. An introduction to missing values in clinical trials based on these distinctions is given by Molenberghs and Kenward.21 Ding and Simonoff22 show that the Little/Rubin distinctions are unrelated to the accuracy of different missing-value treatments when classification trees are used in prediction time and the missingness is independent of the class value. This holds for three of the four evaluated datasets in our study (see next section). We give a short overview of the notions in Little and Rubin:20 MCAR applies when the probability that a value of a variable is missing (NA) does not depend on the values of other observed or unobserved variables o and u, that is, P(NA | o, u) = P(NA); MAR is present when the probability of NA depends only on (the values of other) observed variables, that is, P(NA | o, u) = P(NA | o); MNAR means that P(NA | o, u) cannot be quantified without additional assumptions.
The most used technique for dealing with missing values seems to be imputation, which means to replace every NA by a value estimated from the data available. Imputation can be point based or distribution based. In the latter case the (conditional) distribution of the missing value is calculated and predictions are based on this estimated distribution. Multiple (or repeated) imputation generates some complete versions of the data that are combined for final inference in a statistical setting. Regarding further information on this variant we refer to Little and Rubin.20
There is no internationally published systematic approach to missing values in record linkage, as far as we know. Works such as the ones by McGlincy23 or James24 do not—as their titles might suggest—deal with the missing values in the matching attributes but with predicting matches as such. The former paper states that the ‘problem of missing links is similar to the problem of non-response in surveys’, which renders missing values in matching attributes out of sight. Our paper is meant to serve as the base for future work regarding missing values in record linkage.
Relevant papers regarding classification trees with missing values are the papers of Ding and Simonoff22 and Saar-Tsechansky and Provost.25 The former work investigates six different approaches—probabilistic split, complete case method, grand mode/mean imputation, separate class, surrogate split, and complete variable method—to missing values and concludes that treating missing values as a separate class (in this paper: imputation with unique value 0.5) performs best when missingness is related to the response variable, otherwise results exhibit more ambiguity. The authors use real datasets and simulated datasets in which missing values are increased based on MCAR, MAR and MNAR sampling. Among others, they use a classification induction tree algorithm that is used in this paper (ie, classification and regression trees (CART); see Methods section). In the articles by Saar-Tsechansky and Provost25 a set of C4.5-classification trees induced on reduced sets of attributes (ie, reduced-model classification) exhibit the best results. For further information regarding the classification-tree induction approach C4.5 we refer to Salzberg.26 This reduced model classification is compared with predictive value imputation (eg, surrogate-split mechanism in CART; see Methods section) and distribution-based imputation (eg, sample-based induction; see Methods section) used by C4.5. Datasets with ‘naturally occurring’ missing values and with increased numbers of missing values (chosen at random: MCAR) were considered. The authors explicitly deal solely with missingness in prediction time. We want to tackle the induction time as well.
This paper empirically studies the effect of different approaches for missing values on the accuracy in a record linkage setting in which classification trees are used for the classification of record pairs as match or non-match. Our main aim is to determine the best record linkage strategy on a large amount of real-world data as well as on data based on them in which NA values are manually increased. The number of the data items considered in the evaluation is above five million, which is unusually large for classification-tree settings: datasets in Saar-Tsechansky and Provost25 have at most 21 000 items and Ding and Simonoff22 perform classification with CART with at most 100 000 items (their implementation of CART cannot cope with more data in prediction time).
Methods
Datasets
The data used stem from a German epidemiological cancer registry with corresponding matching attributes listed in table 1. The data were collected through iterative insertions since the foundation of the registry in 2005. In the course of a mandatory evaluation of the registry's record linkage procedures we received 100 000 randomly sampled records dating from 2006 to 2008. Record pairs were classified as ‘match’ or ‘non-match’ during an extensive manual review in which several skilled persons were involved. The resulting classification formed the basis for assessing the quality of the registry's own record linkage procedure and served as a gold standard in the context of our evaluations. While the methods presented in this paper rely only on personal data such as name and date of birth, diagnostic information (International Classification of Disease version 10 and O-3 codes) was considered in the review process to decide in uncertain cases.
Table 1.
Matching attributes
| Comparison Name | Description |
| cmp_firstname_c1 | Comparison of first names (first component) |
| cmp_firstname_c2 | Comparison of first names (second component) |
| cmp_lastname_c1 | Comparison of names (first component) |
| cmp_lastname_c2 | Comparison of names (second component) |
| cmp_bd | Comparison of day of birth |
| cmp_bm | Comparison of month of birth |
| cmp_by | Comparison of year of birth |
| cmp_plz | Comparison of postal code |
| cmp_sex | Comparison of sex |
Record pairs were constructed and transformed into binary comparison patterns. For instance, the record pair
yields the binary comparison pattern (missing values are replaced by 0).
As a result of natural and local capacity restrictions concerning memory and computation, blocking was used to limit the amount of resulting record pairs. This means that the amount of comparison patterns is reduced through imposition of conditions concerning the agreement of attributes in record pairs. Six blocking iterations with different conditions were run in order to account for possible errors in the blocking variables. The blocking iterations with corresponding attribute names are listed in table 2. Equality of the first name and name in the blocking procedure is based on a German phonetic code, called ‘Koelner Phonetik’.27 For instance, in block B, agreement on the phonetic code of the first component of the first name and on the day of birth is required. All resulting comparison patterns of the six blocks A, B, C, D, E and F were merged together, resulting in 5728.201 record pairs (and therefore comparison patterns) that are non-matches and 20 931 record pairs that are matches. When missing values are replaced by 0, there are 212 distinct comparison patterns in the data (out of 512 possible patterns). The most frequent pattern is ‘1011001000’, which occurs 1 811 626 times (note that blocking was used). The second most common pattern is ‘1011000100’, which occurs 678 283 times, the third most common pattern ‘1011010000’ occurs 662 743 times and so on.
Table 2.
Blocking iterations and variables: columns stand for the blocking steps as upper case characters and crosses in the cells indicate that equality in the corresponding attributes (listed in the rows) is required
| Blocking iteration | A | B | C | D | E | F |
| Attribute | ||||||
| First component of first name | x | x | x | x | ||
| First component of name | x | x | ||||
| Day of birth | x | x | x | |||
| Month of birth | x | x | x | |||
| Year of birth | x | x | x | |||
| Sex | x |
Some 5 749 112 of the 5 749 132 patterns (nearly all) contain at least one NA; regarding the attributes in detail:
cmp_firstname_c1 has 1007 NA values,
cmp_firstname_c2 has 5.645.434 NA values,
cmp_lastname_c1 has 0 NA values,
cmp_lastname_c2 has 5.746.668 NA values,
cmp_sex has 0 NA values
cmp_by, cmp_bm, cmp_bd have all 795 NA values,
cmp_plz has 12.843 has 795 NA values.
Besides these complete data, three further datasets are generated based on them. Two filters are used beforehand: (1) if an attribute has more than 70% missing values it is discarded; (2) an attribute with values that are equally likely in matches and non-matches is omitted due to the lack of discrimination power. Discrimination power is also low when in the underlying comparisons of an attribute only a few values are being compared. Applying these filters, the second name components and the sex attributes are discarded. On the whole, the following datasets are considered (the filters lead to the last three datasets):
Real unmodified complete data (RealFull): Data with only 20 patterns that do not have NA values. This case represents MNAR-missingness because the probability of NA in the matching attributes cmp_firstname_c2 and cmp_lastname_c2 depends highly on the match status: NA values are more likely in non-matches than in matches (combination possibility of the underlying records is much more limited for matches, which prevents the augmentation of NA values). This case is used for two reasons: first, to demonstrate how the approaches to missing values behave when the numbers of NA values are very high and second, because the underlying attribute-set is used in real-world applications.
Real unmodified reduced data (RealRed): Data with reduced attributes. Here, 14 644 patterns do have NA values. In contrast to RealFull, this case demonstrates how the approaches to missing values behave when the numbers of NA values are relatively low.
RealRed with 10% randomly generated NA values in cmp_lastname_c1 (GenMCAR). This missingness is MCAR. Here, 588 122 (10, 23%) patterns have NA values of which 574 913 are due to NA values in cmp_lastname_c1.
RealRed with NA-filling in cmp_lastname_c1 (GenMAR): Based on to the sum of zeros (s) in a comparison pattern i, the value of cmp_lastname_c1 is replaced by NA according to the following conditional probability distribution: P(replace value of cmp_lastname_c1 by NA in i |number s of zeros in i) =0.27+0.09*s, 0≤ s ≤7. The reason for using this discrete probability distribution is twofold: every pattern should have a significant probability for an NA-replacing (>0.2 but hot higher than 0.9) and in the result we should have a dataset with an NA-fraction in the comparison patterns that is, in between the NA-fractions of the other cases. This missingness is MAR. The results: 3 792 065 (65, 96%) patterns have NA values of which 3 787 046 (65, 87%) are due to NA values in cmp_lastname_c1.
Near-zero values of the correlations between match status and missingness indicators for all attributes (0: value is not NA, 1: value is NA) suggest that there is no relevant dependency of missingness on the target attribute for the last three cases.
General remarks on record linkage
As hinted in the introduction, the goal of record linkage is to remove synonyms in datasets while avoiding homonym errors in that process. Record linkage requires the generation of record pairs out of the single data items (feature space C) and the subsequent creation of comparison patterns (γf: C × C → IRn). Two classes of comparison patterns can be distinguished: binary (γ ∈ {0,1}n) and real valued comparison patterns (γ ∈ [0,1]n). The former class represents cases in which it only matters whether attribute values are equal (γi = 1) or unequal (γi = 0). Real valued comparison patterns exhibit similarity between attribute values by using string metrics. The resulting comparison patterns are further processed (ie, computation of weights) in order to decide which patterns—and therefore which record pairs—pertain to matches and which to non-matches. Yancey28 and one of our previous studies29 empirically show that the usage of string metrics does not improve record linkage when sufficiently many attributes are available. This is the main reason why we can focus on binary comparison patterns in this paper (resulting, for example, from encrypted data). Further research in record linkage should consider missing values in real valued comparison patterns as well.
Generalization and abstraction of the problem of record linkage as object identification broaden the spectrum for further models such as classification trees. They are non-probabilistic alternatives to the frequently used probabilistic record linkage methods based on the framework of Fellegi and Sunter.5 Classification-tree models seem eligible due to their good interpretability and performance in record linkage and other settings (see also Ding and Simonoff22 and Cochinwala30). In this paper, the expression ‘classification tree’ is used synonymously with the general expression ‘decision tree’.
Classification trees and active learning
For the classification of record pairs into matches or non-matches, we use the classification-tree method CART as implemented in the R-package ‘rpart’.31 32 First usages of classification trees for the solution of the record linkage problem are presented by Cochinwala30 and Verykios et al.33 Other papers that exploit classification trees for record linkage are, for example, the ones by Sarawagi and Bhamidipaty34 and Tejada et al35 or Sariyar et al.29 For the usage of CART in record linkage using R we refer to our R package RecordLinkage36 and a related article37 in which all relevant functions for this paper are discussed.
Classification-tree models represent relevant information of the data to be classified (in our case binary comparison patterns) in a hierarchical structure. This representation of a hierarchical structure is achieved by inducing a classification tree on labeled training data. In most cases—as in CART—only binary classification trees are generated. In the induction process of such a binary classification tree, first a root is generated that contains all training samples. This root node is split (ie, partitioned based on one attribute and a corresponding split value) into two child nodes according to some purity measure (we use the Gini index),38 which quantifies the homogeneity of the target class, for example, the relation of matches to non-matches, in the resulting nodes. The values of every (matching) attribute are examined and the split is performed according to that attribute value that maximizes the purity. This process is continued recursively until changes of the impurity measure reach a lower bound or the number of objects in a node is lower than or equal to a fixed minimum size. The edges of a classification tree represent the conditions according to which the objects in parent nodes are allocated to their child nodes. The leaves represent those classes to which the objects in these leaves are assigned.
The most important algorithm for creating decision trees is CART, developed by Breiman et al.31 The algorithm has two steps: first a maximal tree with a very simple stopping rule is generated. Afterwards, this tree is pruned to avoid overfitting. The second step is only necessary when there are redundant attributes present. In order to improve and stabilize the results of classification trees induced on different training data, weighted aggregations of classification trees like bagging39 and boosting40 can be considered. One of our previous studies29 suggests that non-aggregating classification trees with nearly maximal depth perform no worse than their aggregated variants in record linkage settings; this is the reason for using CART in its original form with parameters such that maximal height is achieved.38
Possible ways for the acquisition of labeled training data are, for example, the generation of artificial data, making use of data from other but similar contexts or a manual review of a selection from the data to be linked. To get a minimum amount of informative training set from the data on hand, we use active learning, a machine learning approach in which the user is asked to label data with special characteristics (ie, high information according to some measure), see, for example, Sarawagi and Bhamidipaty.34 For the binary comparison patterns in our evaluations, a simple strategy is considered: each distinct comparison pattern represents a stratum to which all identical patterns belong; and from every stratum one item is randomly sampled. As every stratum consists of the same patterns, the difference in different samplings is only related to the matching status of the patterns in the resulting training data. This strategy leads to training sets of different sizes for the different approaches presented below; therefore the comparability of results needs some consideration. We will come to this issue more thoroughly in the Discussion section.
Handling missing values in record linkage with CART
The solutions for missingness in prediction time are adopted in induction time when not otherwise stated. Considering both cases simultaneously reflects the fact that we want to find a practicable and therefore integrated solution for record linkage studies that use classification trees. Four approaches to handling missing data that take all cases in prediction time into account are used. The approaches are applied to all NA values in the datasets (thus, also for multiple NA values). We consider the following approaches:
Imputation with unique values. This is the standard ad-hoc solution in record linkage settings. We use the results of this simplistic solution as benchmark. NA values will on the whole either be substituted by 0 (regarded as unequal), 1 (regarded as equal) or 0.5 (neither equal nor unequal). The latter case is equivalent to the treatment of NA as a separate class. Abbreviations: Imp0, Imp05, Imp1.
Sample-based Imputation. Using binary comparison patterns facilitates this approach enormously. For all attributes, the fractions of zeros and ones among all (in our case 5.728.201) comparison patterns are computed and NA values are substituted by a value according to random sampling from the resulting binary probability distribution. Abbreviation: SampImp.
Reduced-model classification. The comparison patterns are partitioned such that each resulting group consists of patterns with missing values in the same attributes. For every partitioned set that consists of at least 1000 comparison patterns (this limits the number of resulting partition sets to 5 and thus bounds computational costs), the matching attributes containing NA values are dropped and trees based on these reduced attribute sets are induced using the active learning strategy described in the former section. Classification of these patterns is based on the classification trees thus induced. For all other patterns containing NA values, the classification tree with the highest amount of agreeing non-NA attributes is selected and if necessary the surrogate-split mechanism of CART is used: for an NA another attribute is used which yields similar splitting results as the primary splitter.31 Abbreviation: RedMod.
Complete-case induction. Only patterns with no NA are used in induction time. The resulting classification tree—generated based on these complete cases using again active learning—is applied in prediction time for classifying all patterns. For an NA the surrogate-split mechanism of CART is used as in reduced model classification. Abbreviation: ComplCase.
Connection between induction and prediction time is accomplished by our active learning strategy, which is applied to every dataset. Using training data from the data to be classified implies that solutions for missing values affect both the training and the test phase. Training data are not considered in the final classification.
We use the example of the data section for a clarification of the approaches to missing values. Without 0-replacing, we have
Imp0, Imp05 and Imp1 are straightforward; for instance, Imp05 yields
In SampImp, we must first determine the binary probability distribution. For the first attribute with an NA, let us assume that it has 10 000 1s and 40 000 0s and 10 000 NA values (hence, we assume 60 000 comparison patterns). Then, with probability 0.2 a 1 will be sampled and with probability 0.8 a 0 (the NA values are not considered for the probability distribution). For the other NA values the same procedure is applied and we could have
For RedMod, let us assume that the example pattern occurs more than 1000 times. In that case a classification tree for patterns without the second, the fourth and the ninth attribute exists and we can use the following pattern for classification (be aware that the pattern is strictly linked to a specific omission of attributes):
If the pattern (1, NA, 0, NA, 1, 1, 1, 0, NA) occurs less that than 1000 times, a search for the classification tree with the highest number of the agreeing non-NA attributes is performed. A classification tree that is, for example, based on patterns in which the second and the fourth attributes are omitted gives the highest possible agreement and would be used. If a split in that classification tree is based on the ninth attribute, the surrogate split mechanism would be necessary.
In ComplCase, we use the example pattern as such and hope that the surrogate split mechanism will perform well for cases in which splits are based on an attribute for which the pattern has an NA.
Evaluation measures
Outcomes of a record linkage procedure are designated as links and non-links. Links and non-links follow from classifying record pairs as representing matches and non-matches, respectively. Due to the huge imbalance between matches and non-matches the accuracy is not very informative as an absolute measure. Instead, we use the F-score, which is the harmonic mean of precision and recall):34
Recall (also called sensitivity) is computed as the fraction of correct links among all matches. Precision is the fraction of correct links among all links. The F-score combines accuracy and comprehensiveness concerning the detection of matches. Listing recall and precision values is only relevant when they show considerable differences. This is not the case in our evaluations due to the high quality of the data (ie, high discernibility between matches and non-matches); therefore, their listing would only overload the result presentation without adding relevant information.
Due to random sampling of training data the classification results can vary. As a consequence, the process of training data sampling is repeated 25 times and the resulting F-scores are plotted as boxplots. The number 25 is used, for example, as default in the bagging function of R; experience shows that stabilization of classification-tree results is often achieved with that number of trees.39 41 The median F-scores are also specified as decimal numbers in order to facilitate the overview of the most effective approaches. The boxplots show the variation of the results (corresponding to CI). As there is no distributional assumption and asymmetries of the frequencies are highly probable (the boxplots show that the F-score distributions are indeed skewed), the median is used instead of the mean.42
The amount of training data is listed for every missing value approach and every dataset. Solely sample-based imputation on dataset RealFull exhibits variation in the number of comparison patterns that form the training data; variation is minor and between 340 and 354, therefore we use the median (346) for final analysis.
Results
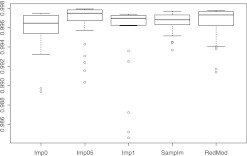
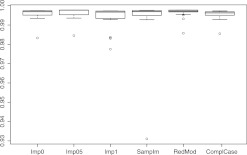
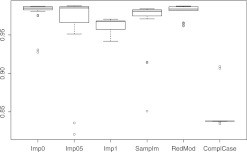
Figures 1–4 show the resulting boxplots, table 3 the number of training data and table 4 the median F-scores. The figures illustrate that increasing the number of NA values impairs the results. ComplCase on dataset RealFull constitutes an exception because only six complete cases are available for the original data and that is not enough for inducing a suitable classification tree. ComplCase is therefore omitted from the corresponding boxplot in figure 1. For the other methods, the results on RealFull and RealRed are very similar, which confirms that the discarded attributes do not discernibly contribute to the discrimination between matches and non-matches. Comparison of figures 1–4 further shows that the approaches to the missing value problem differ recognizably only when substantial numbers of missing values occur in relevant attributes.
Figure 1.
Boxplots of the F-scores on the RealFull data.
Figure 2.
Boxplots of the F-scores on the RealRed data.
Figure 3.
Boxplots of the F-scores on the GenMAR data.
Figure 4.
Boxplots of the F-scores on the GenMCAR data.
Table 3.
Number of comparison patterns that form the training sets (median for SampIm/RealFull)
| RealFull | RealRed | GenMAR | GenMCAR | |
| Imp0 | 167 | 64 | 64 | 64 |
| Imp05 | 344 | 115 | 170 | 166 |
| Imp1 | 193 | 64 | 64 | 64 |
| SampIm | 346 | 64 | 64 | 64 |
| RedMod | 313 | 107 | 137 | 135 |
| ComplCase | 6 | 64 | 64 | 64 |
Table 4.
Medians of the F-scores
| RealFull | RealRed | GenMAR | GenMCAR | |
| Imp0 | 0.9965 | 0.9969 | 0.9849 | 0.9940 |
| Imp05 | 0.9975 | 0.9977 | 0.9851 | 0.9943 |
| Imp1 | 0.9970 | 0.9967 | 0.9677 | 0.9844 |
| SampIm | 0.9968 | 0.9969 | 0.9808 | 0.9931 |
| RedMod | 0.9973 | 0.9972 | 0.9837 | 0.9936 |
| ComplCase | 0.0318 | 0.9963 | 0.8380 | 0.9439 |
On all datasets imputation with value 0.5 exhibits the highest median values. The superiority is only marginal and in contrast to the other unique-value imputations this could be (at least) partly ascribed to the higher amounts of training data. The complete-case approach yields the worst results. One of the obvious reasons is the concept of surrogate split of CART: choosing the attribute that splits a node into similar child nodes as the primary split significantly worsens the results in contrast to the non-NA case, especially when the training data are as few as in our study. In RedMod the results are not impaired in such magnitude because surrogate splits are only used for the remaining cases for which no classification tree is induced.
Sample-based imputation is never the best nor the worst approach. It is remarkable that this holds as well on GenMCAR in which the NA values are increased randomly. This again confirms the observations and theoretical reasons concerning the minor relevance of the MCAR/MAR/MNAR distinctions in record linkage studies when classification trees are used.
In summary, the methods considered in this paper exhibit similar F-scores on our real-world data. Therefore, the simplest approach should be considered in practice. Because Imp05 increases the amount of training data to be manually classified, the simplest solutions are either Imp0 or Imp1. Another advantage of these is that the binary structure of the data is maintained. For the purpose of generalizability Imp0 is to be preferred. If NA values are augmented artificially for cmp_lastname_c1, Imp0 is the second best method and only slightly worse than Imp05. Hence, for simplicity reasons, Imp0 is the method of choice.
Discussion and conclusion
The outcome of our empirical evaluations does not confirm the results of Saar-Tsechansky and Provost25 but those of Ding and Simonoff.22 One reason for the differences to the former paper can be ascribed to the validity of the MCAR assumption in that paper in contrast to its unlikeness in record linkage settings. In both papers it is shown that the results are generalized to other classification methods such as logistic regression. This is one of the reasons why we are confident that our results could be reproduced with other record linkage methods.
Unique value imputation works best on our data and the pragmatic approach of replacing NA by 0 exhibits results that are not worse than the computational far more costly reduced-model classification. This is unusual for most other settings in which unique value imputations are distinctly inferior to more sophisticated approaches. A major exception is the paper22 concerning the imputation with 0.5 (own class for NA). But the theoretical explanation of that paper is only valid for the RealFull case. The authors of that paper state that an own class for NA is preferable when two conditions hold: missingness depends on the values of the class variable, and this dependence is present both in the training and in the test data. The last condition is superfluous in our case because of our active learning strategy, which selects training data from the data to be classified. As mentioned above, we could not confirm the first condition for the other datasets. Either the reasoning of Ding and Simonoff22 has to be generalized in order to cover our results or other theoretical bases are to be found. This is an interesting and worthwhile task for future research.
Our strategy for selecting training data from each dataset to be classified comes at the cost of different validation sets, which impairs comparability of the classification results. One possible alternative would be the determination of one uniform number for the size of the training data: the largest set on any data for any of the methods when the active learning strategy is applied. This would guarantee more comparable results but would counteract our aim of finding an efficient strategy with a small amount of training data. Concerning our results, the optimal accuracies of Imp05 could be a by-product of the larger training sets in that case. Imp0 yields only slightly worse results with a substantially smaller training set. It is therefore a good compromise between accuracy and the size of the training set.
Another insight is related to the surrogate splits of CART. Complete-case induction relies solely on these for dealing with missing values and is the worst method on all datasets. As a consequence, one should not rely on this feature of CART. It is worthwhile to have a pre-processing in which NA values are handled in other ways than in CART. The existence of the surrogate-split mechanism led to a false confidence regarding the handling of missing values due to the sound foundation of CART in general. This should be a warning against judgments based only on theoretical reasoning when the data generating mechanism cannot be controlled or captured in a model.
In the Result section preference of Imp0 over Imp1 is related to the generalizability of the former imputation in record linkage settings. This should be expounded further. In record linkage settings the canonical situation is the abundance of non-matches. This means that NA values of any attributes are rather among non-matches and thus the true comparison value is most likely zero. The following non-match pattern could occur:
which means that the first component of the forename, the birthday and sex are equal. The most relevant attribute ‘comparison value of the first component of the last name’ is NA. Transforming the NA into a one leads to a non-match pattern that is more likely under a match. When selecting this imputated non-match as a member of the training set the ones can be interpreted as unequal and thus the classification tree would classify all true matches with patterns like
as non-match. Imp1 can therefore lead to bad classifiers if the training set is selected unluckily. The opposite case is not problematical as such. A match of the form
can impair the classification result when it occurs in the training set (softening of the match classification), but the probabilities of inverting the meanings of zeros and ones and classifying a true match as non-match are negligible.
In conclusion, we can partly soothe record linkage practitioners who use the ad-hoc solution for missing values ‘replace NA by the value of inequality’. It is surprisingly a very good solution on our data. The soothing is of course only partial because it is based on a limited amount of data and on a specific classification-tree method. Nevertheless, the results of Ding and Simonoff22 and previous experiences suggest that our results should be confirmed in other empirical analyses and similar applications.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Elmagarmid AK, Ipeirotis PG, Verykios VS. Duplicate record detection: a survey. IEEE Trans Knowl Data Eng 2007;19:1–16 [Google Scholar]
- 2.Parkin DM, Bray F. Evaluation of data quality in the cancer registry: principles and methods. Part II. Completeness. Eur J Cancer 2009;45:756–64 [DOI] [PubMed] [Google Scholar]
- 3.Schouten LJ, de Rijke JM, Schlangen JT, et al. Evaluation of the effect of breast cancer screening by record linkage with the cancer registry, the Netherlands. J Med Screen 1998;5:37–41 [DOI] [PubMed] [Google Scholar]
- 4.Christen P, Goiser K. Quality and complexity measures for data linkage and deduplication. In: Guillet F, Hamilton H, eds. Quality Measures in Data Mining, 43rd edn Heidelberg: Springer Berlin, 2007:127–51 [Google Scholar]
- 5.Fellegi IP, Sunter AB. A theory for record linkage. J Am Stat Assoc 1969;64:1183–210 [Google Scholar]
- 6.Christen P. Probabilistic data generation for deduplication and data linkage. Intelligent Data Engineering and Automated Learning Ideal 2005, Proceedings of the Sixth International Conference on Intelligent Data Engineering and Automated Learning (IDEAL'05), Brisbane, July 2005. Lecture notes in computer science, Springer, 2005;3578:109–16 [Google Scholar]
- 7.Hernandez MA, Stolfo SJ. Real-world data is dirty: data cleansing and the merge/purge problem. Data Min Knowl Discov 1998;2:9–37 [Google Scholar]
- 8.Michalowski M, Thakkar S, Knoblock CA. Automatically utilizing secondary sources to align information across sources. Ai Magazine 2005;26:33–44 [Google Scholar]
- 9.Gardner J, Xiong L. An integrated framework for de-identifying unstructured medical data. Data Knowl Eng 2009;68:1441–51 [Google Scholar]
- 10.Gomatam S, Carter R, Ariet M, et al. An empirical comparison of record linkage procedures. Stat Med 2002;21:1485–96 [DOI] [PubMed] [Google Scholar]
- 11.Jaro MA. Advances in record-linkage methodology as applied to matching the 1985 census of Tampa/Florida. J Am Stat Assoc 1989;89:414–20 [Google Scholar]
- 12.Noren GN, Orre R, Bate A, et al. Duplicate detection in adverse drug reaction surveillance. Data Min Knowl Discov 2007;14:305–28 [Google Scholar]
- 13.Roos LL, Jr, Wajda A, Nicol JP. The art and science of record linkage: methods that work with few identifiers. Comput Biol Med 1986;16:45–57 [DOI] [PubMed] [Google Scholar]
- 14.Herzog TN, Scheuren FJ, Winkler WE. Data Quality and Record Linkage Techniques. New York, NY: Springer, 2007 [Google Scholar]
- 15.Tromp M, Ravelli AC, Bonsel GJ, et al. Results from simulated data sets: probabilistic record linkage outperforms deterministic record linkage. J Clin Epidemiol 2011;64:565–72 [DOI] [PubMed] [Google Scholar]
- 16.Winkler WE. Advanced methods for Record Linkage. Statistical Research Division U.S. Census Bureau, Suitland, Maryland, 1994. Technical Report. http://www.census.gov/srd/papers/pdf/rr94-5.pdf [Google Scholar]
- 17.Acock AC. Working with missing values. J Marriage Fam 2005;67:1012–28 [Google Scholar]
- 18.King G, Honaker J, Joseph A, et al. Analyzing incomplete political science data: an alternative algorithm for multiple imputation. Am Polit Sci Rev 2001;95:49–69 [Google Scholar]
- 19.Christen P. A survey of indexing techniques for scalable record linkage and deduplication. IEEE Trans Knowl Data Eng 2011;99:196–214 [Google Scholar]
- 20.Little RJ, Rubin DB. Statistical Analysis with Missing Data. Hoboken, NJ: Wiley-Interscience, 2002 [Google Scholar]
- 21.Molenberghs G, Kenward MG. Missing Data in Clinical Studies. Chichester: Wiley, 2007 [Google Scholar]
- 22.Ding YF, Simonoff JS. An investigation of missing data methods for classification trees applied to binary response data. J Mach Learn Res 2010;11:131–70 [Google Scholar]
- 23.McGlincy MH. A B ayesian record linkage methodology for multiple imputation of missing links. ASA Proceedings of the Joint Statistical Meetings. Alexandria, VA, 7–10 August 2004:4001–8 [Google Scholar]
- 24.James F. Robison-Cox. A record linkage approach to imputation of missing data: analyzing tag retention in a tag: recapture experiment. J Agric Biol Environ Stat 1998;3:48–61 [Google Scholar]
- 25.Saar-Tsechansky M, Provost F. Handling missing values when applying classification models. J Mach Learn Res 2007;8:1625–57 [Google Scholar]
- 26.Salzberg SL. C4.5: Programs for machine learning by J. Ross Quinlan. Morgan Kaufmann Publishers, Inc., 1993. Mach Learn 1994;16:235–40 [Google Scholar]
- 27.Postel HJ. Die Kölner Phonetik. Ein Verfahren zur Identifizierung von Personennamen auf der Grundlage der Gestaltanalyse. IBM-Nachrichten 1969;19:925–31 [Google Scholar]
- 28.Yancey WE. Evaluating String Comparator Performance for Record Linkage. Suitland, Maryland: Statistical Research Division U.S. Census Bureau, 2005. Technical Report. [Google Scholar]
- 29.Sariyar M, Borg A, Pommerening K. Evaluation of record linkage methods for iterative insertions. Methods Inf Med 2009;48:429–37 [DOI] [PubMed] [Google Scholar]
- 30.Cochinwala M, Kurien V, Lalk G, et al. Efficient data reconciliation. Inform Sci 2001;137:1–15 [Google Scholar]
- 31.Breiman L, Friedman J, Olshen R, et al. Classification and Regression Trees. Belmont, California: Wadsworth, 1984 [Google Scholar]
- 32.rpart: Recursive Partitioning. R package [computer program]. Version 3.1–47. 2010. http://cran.r-project.org/package=rpart
- 33.Verykios VS, Elmagarmid AK, Houstis EN. Automating the approximate record-matching process. Inform Sci 2000;126:83–98 [Google Scholar]
- 34.Sarawagi S, Bhamidipaty A. Interactive deduplication using active learning. Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Edmonton, AB, Canada, July 23–25, 2002:269–78 [Google Scholar]
- 35.Tejada S, Knoblock CA, Minton S. Learning object identification rules for information integration. Information Systems 2001;26:607–33 [Google Scholar]
- 36.Sariyar M, Borg A. Record Linkage in R. R package [computer program]. Version 0.3–4. Mainz, Germany, 2011. http://cran.r-project.org/package=RecordLinkage [Google Scholar]
- 37.Sariyar M, Borg A. The RecordLinkage Package: Detecting Errors in Data. The R Journal 2010;2:61–7 [Google Scholar]
- 38.Therneau TM, Atkinson EJ. An Introduction to Recursive Partitioning Using the Rpart Routine. Rochester, Minnesota: Mayo Clinic, Section of Biostatistics, 1997. Technical Report No. 61. [Google Scholar]
- 39.Breiman L. Bagging predictors. Mach Learn 1996;24:123–40 [Google Scholar]
- 40.Freund Y, Schapire RE. Experiments with a new boosting algorithm. Machine Learning: Proceedings of the Thirteenth International Conference, Bari, Italy, 3–6 July 1996 1996:148–56 [Google Scholar]
- 41.Peters A, Hothorn T, Lausen B. ipred: Improved predictors. R News 2002;2:33–6 [Google Scholar]
- 42.Altman DG, Bland JM. Quartiles, quintiles, centiles, and other quantiles. BMJ 1994;309:996. [DOI] [PMC free article] [PubMed] [Google Scholar]