Abstract
Objectives
To address the challenge of balancing privacy with the need to create cross-site research registry records on individual patients, while matching the data for a given patient as he or she moves between participating sites. To evaluate the strategy of generating anonymous identifiers based on real identifiers in such a way that the chances of a shared patient being accurately identified were maximized, and the chances of incorrectly joining two records belonging to different people were minimized.
Methods
Our hypothesis was that most variation in names occurs after the first two letters, and that date of birth is highly reliable, so a single match variable consisting of a hashed string built from the first two letters of the patient's first and last names plus their date of birth would have the desired characteristics. We compared and contrasted the match algorithm characteristics (rate of false positive v. rate of false negative) for our chosen variable against both Social Security Numbers and full names.
Results
In a data set of 19 000 records, a derived match variable consisting of a 2-character prefix from both first and last names combined with date of birth has a 97% sensitivity; by contrast, an anonymized identifier based on the patient's full names and date of birth has a sensitivity of only 87% and SSN has sensitivity 86%.
Conclusion
The approach we describe is most useful in situations where privacy policies preclude the full exchange of the identifiers required by more sophisticated and sensitive linkage algorithms. For data sets of sufficiently high quality this effective approach, while producing a lower rate of matching than more complex algorithms, has the merit of being easy to explain to institutional review boards, adheres to the minimum necessary rule of the HIPAA privacy rule, and is faster and less cumbersome to implement than a full probabilistic linkage.
Keywords: Record linkage, blindfolded, clinical informatics, sample
Introduction and objectives
One of the challenges facing disease registries is the need to balance patient privacy with the desire to build cross-site records, matching the data for a given patient as he or she moves between participating sites. Some registries1 address this problem by mandating that identifiers be disclosed, but this is rarely acceptable to privacy officers. A more viable solution from a privacy standpoint is to keep only de-identified information in the registry, but share an algorithm between trusted third parties for generating an anonymous identifier in such a way that the chances of the same identifier being generated by two different sites when treating the same individual are maximized, and the chances of generating the same identifier for two different individuals is minimized.
The use of social security numbers (SSNs) for this purpose is associated with a variety of problems.2 Matching on an anonymized hash of full patient name and date of birth seems an attractive option until one considers that it can lead to excessive splitting of patient records due to differences in how a given person identifies themselves at different sites of care.3 Patients may use a partial name on one visit and their full given name on another. The electronic health record may compound the problem by treating ancillary information such as titles very inconsistently when generating discrete records for first and last names. Misspellings are common. Within a given institution this error rate is often corrected after the fact by Master Patient Index (MPI) maintenance, but when comparing records collected at different institutions there may be differences in identity resolution methodology.
The work described in this paper grew out of a research project involving two distinct Health Insurance Portability and Accountability Act (HIPAA) covered entities in the San Francisco Bay Area, Stanford University and the Palo Alto Medical Foundation (PAMF), collaborating on a joint research study of women being treated for breast cancer at both of these institutions.
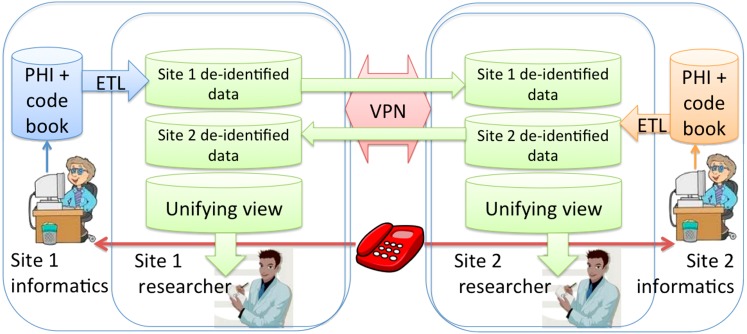
The goal of the project was to assemble a de-identified merged data set consisting of diagnosis and treatment records for breast cancer patients seen at both institutions to support analysis of cross-institutional standards of care and comparative outcomes research. In accordance with the minimum necessary principle of the HIPAA Privacy Rule, the participating institutions decided to exchange only de-identified (by HIPAA standards) data between clinical researchers and minimize the protected health information (PHI) exchanged between informatics staff responsible for constructing the linked de-identified data set in support of the clinical research protocol. Each institution obtained permission from their respective Institutional Review Boards (IRBs) for 1) a determination of non-human subjects review for the clinical researchers and 2) a full IRB protocol with a waiver of consent and authorization for the informatics staff involved in record linkage. This, along with a decision by both parties to design a system in which a copy of the joint data set resided at each local site and was not hosted remotely, led to the system design depicted in figure 1.
Figure 1.
Research data flow between the two separate Health Insurance Portability and Accountability Act (HIPAA) covered entities participating in the study. Protected Health Information (PHI) remains secure at all times. Informaticians acting as honest brokers hold the electronic code book table that maps hash codes to patient research database identifiers. Anonymized data is extracted, transformed and loaded (ETLed) into a staging database and securely transmitted over a dedicated virtual private network (VPN).
At each site the fully identified data are staged in a separate secure zone that only institutional informatics technical support personnel not involved in the clinical research project have access to. From this staging environment the data were extracted, transformed by removing all identifiers and mapping into the shared coding scheme, and loaded into a set of tables in the shared database. We used Oracle Gateway to set up a two-way secure data flow from the Oracle database on one side to the Microsoft SQLServer database on the other.
Given that some of the patients are treated at both institutions, due to both geographic proximity and the fact that PAMF is an outpatient facility while Stanford offers both inpatient and outpatient treatment, we were confronted with the issue of how to reliably identify individual patients who received care at both sites. We had to generate anonymous identifiers based on real identifiers in such a way that the chances of a shared patient being accurately identified were maximized, and the chances of incorrectly joining two records belonging to different people were minimized.
An elegant approach to this problem is described by Churches and Christen.4 Their solution involves the exchange of vectors containing the power set of encrypted bigrams for each string. Each party (or a single trusted third party) is then able to compute string similarity via a Dice co-efficient (a ratio of the number of matching to total number of bigram permutations). For example, if a given patient seen at both institutions had first name recorded as Ann at one site and Anne at the other, the bigrams vectors [AN,NN] and [AN,NN,NE] would be exchanged, which would generate a Dice co-efficient of 2×(2/5)=0.8, since four of the five total bigrams are shared.
Other approaches (Chen and Zhong5 for example) share a cryptographic hash based on patient identifiers, but do not go into details of how they achieved a fuzzy match. Durham et al6 are far more precise, detailing the use of multiple hash codes on a variety of full identifiers, assigning probabilities to each, and combining the result with a Bloom filter7; for a good literature survey of the Bloom filter approach, the reader is referred to Schnell et al.8 Note that the complexities of both the bigram vector and Bloom filter approaches require retaining the services of highly specialized programmers to implement, which is why we did not pursue either of these options for our study. Kijsanayotin et al9 proposed an approach very similar to ours in which a single SHA-1 string was constructed from gender, date of birth, zip code, and a three letter prefix of the last name. In our case, however, neither gender nor zip code would have had any discriminating effect since our patient population was entirely female and overwhelmingly from one zip code area, so we opted instead for the first two letters of the patient's first and last names.
Our hypothesis was that most inadvertent variation in names occurs after the first two letters. The name prefix alone was insufficiently discriminatory but we had observed that both sites in the study have a similar patient registration process that relies on date of birth as well as name to fully identify patients at time of visit registration, so we further hypothesized that the date of birth would be both highly available and highly reliable.10 Furthermore, although neither name prefixes nor dates of birth taken individually would be an adequate identifier, due to the high probability of some other person sharing the same date of birth, we postulated that when combined the resulting string would have the desired characteristics.
Methods
Our approach to institutional privacy and security involved first obtaining mutual IRB permission to exchange the minimum necessary conventional PHI between designated informatics support staff in support of quality assurance, under an IRB approved waiver of consent and authorization. Under this protocol the informatics staff securely exchanged the shared secret for the MD5 hash so that hashes generated on the same data would result in the same hash string. Neither the algorithm nor the shared secret was disclosed to the clinical researchers, so the hashed strings would not be susceptible to dictionary attack.11 Informatics staff then generated two hashed identifiers for all patients in their local identified data sets, one based on name prefix and date of birth, the other on SSN. They then exchanged these hashed identifiers and in cases of collision the patient identities were confirmed by exchange of full names, dates of birth, and street address. The purpose of the initial hashing was to adhere to the minimum necessary rule: the informatics staff responsible for validating the record linkage were exposed only to PHI in cases of suspected false positives and false negatives, less than 10% of the total number of records in the shared data set.
We also obtained mutual IRB determinations of non-human subjects review in support of the clinical research use of the conjoined data set. The study identifiers used in our research data set were in most cases the same hashed strings used in the initial linkage, although informatics staff assigned new identifiers to records as needed to differentiate between false positives and compensate for false negatives. In retrospect our protocol would have been even more robust had we decided to take the additional step of generating a coded anonymous unique identifier for each patient in the research data set, but a seeded MD5 hash was deemed to present a less than very small risk of inadvertent disclosure and so was considered de-identified by the HIPAA standard, particularly since no data ever left the protection of our shared secure database.
Regarding the investigation into the efficacy of the proposed linkage variable, we started by measuring the rate of false positives inherent in a composite identifier based on name prefixes and date of birth. The Stanford STRIDE Clinical Data Warehouse12 contains 2.6 million patient names derived from both the pediatric and adult electronic medical records (EMRs) at Stanford University Medical Center. We first created a list of unique non-null uppercase first name, last name, and date of birth triples, then dropped all records with first name prefixed by BABY, BOY, GIRL or UNKNOWN. We also dropped all records for date of birth January 1, 1900 and January 1, 1901, which judging by their frequency were used as placeholder values for unknown dates of birth. From this initial list of 2.5 M name and date triples, we created three additional lists with 1.5 M rows, 0.5 M rows, and 10 000 rows. We then performed a self-join on each list counting the number of different rows for which the composite identifier was the same, the results of which are shown in table 1.
Table 1.
Specificity, an inverse measure of the chances of mistakenly joining two different patients, decreases quadratically with the size of the data set
| Data set size | Specificity |
| 2.5 M | 99.44% |
| 1.5 M | 99.74% |
| 0.5 M | 99.89% |
| 10 000 | 99.99% |
Having defined the false positive error rate, we then needed a measure for false negatives. In this regard we were fortunate that both sites agreed to exchange cryptographic hashes based on the patients' SSNs in support of the linkage work. The securely hashed SSN-based identifiers are not exposed to the medical researchers, as their purpose is solely to improve linkage. By counting the matches found by matching SSNs that were not found by matching our composite identifier, we established a lower bound estimate on the rate of false negatives.
We also felt it important to eliminate all false positives from our data set, so for the purpose of validating the matching algorithm we received approval from both study site IRBs to examine the real identifiers of any patients identified as candidate matches by our algorithm. By manually reviewing the fully identified complete registration record for each pair of proposed matches, we were able to identify all false positives.
Results
We measured specificity for data sets of several different sizes, ranging from 2.5 M records to 10 000, the results of which are tabulated in table 1.
We then measured sensitivity and specificity for full first and last name plus date of birth, SSN, and our composite identifier on a data set of 19 105 records as described below (table 2).
Table 2.
Sensitivity and specificity for social security number (SSN), full composite identifier, and abbreviated composite identifier
| Sensitivity | Specificity | |
| SSN | 86.5% | 99.6% |
| Full name+date of birth | 87.4% | 100% |
| Composite identifier | 97.2% | 99.98% |
In this context sensitivity measures the ability to correctly identify patients in common, and specificity measures the chances of two different patient records being mistakenly joined. Sensitivity is defined as the number of true positives divided by the number of true positives plus the number of false negatives, and specificity is defined as the number of true negatives divided by the sum of the number of true negatives and the number of false positives. Our measures for these values are shown in table 3.
Table 3.
Counts for true and false positives and negatives for the three record linkage variables being compared
| Composite identifier | Social security number | Full name+date of birth | |
| True positives | 2028 | 1806 | 1821 |
| False negatives | 59 | 281 | 263 |
| False positives | 3 | 57 | 0 |
| True negatives | 14 928 | 14 872 | 14 931 |
Table 4 compares the sensitivity we measured for our composite identifier to other commonly employed linkage variables.
Table 4.
Sensitivity per linkage variable
| Sensitivity | Linkage variable |
| 86.5% | Social security number (overall) |
| 87.4% | First+last name+date of birth |
| 93.1% | First name |
| 94.5% | Social security number (when present) |
| 95.2% | Last name |
| 96.7%–97.2% | Hash consisting of first two letters of first and last names+date of birth |
| 99.7% | Date of birth |
Discussion
Since the formula for specificity is based on data set size rather than total number of comparisons, this measure changes with data set size. If instead one divides the number of matches (all of which are deemed to be false positives) by the number of total comparisons: m/((n2–n)/2), the resulting ratio is constant, which makes it more useful when attempting to predict the number of false positives that can be expected for a given data set.
Applying this formula to a STRIDE test data set with m=18 917 and n=158 300, yields
or 1.5 errors per 108 comparisons.
Therefore, when comparing two data sets of around 10 000 patients each, which when compared to each other results in about 108 comparisons, one can expect a few false positives, whereas data sets with 5000 or fewer records are likely to not contain any false positives when linked with the proposed composite identifier. This number is only useful as a very rough rule of thumb, however, since each data set has its own characteristics, and is mostly intended to underscore the point that if false positives are considered unacceptable, this algorithm cannot be relied upon exclusively but should instead be used in conjunction with other linkage variables.
We then turned our attention to the study data set of female breast cancer patients from Stanford and PAMF. With 10 939 patients in one data set and 8166 in the other, our combined cohort contained 19 105 records, of which 2087 were found to be held in common after manual review. After our matching efforts were complete, we found just three erroneous matches, which is reasonably consistent with our predictions.
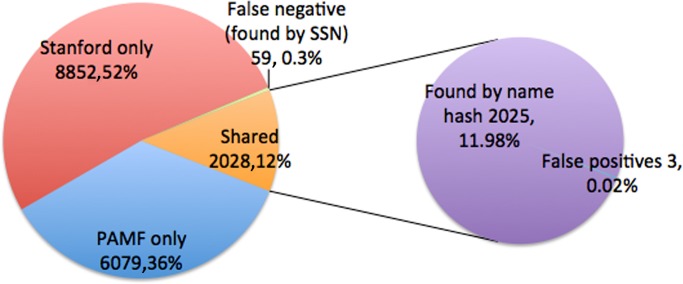
Our composite identifier found 2028 of the 2087 confirmed matches, with an additional 59 patients found by comparing hashed SSNs, as shown in figure 2. Given the fact that we had SSNs on only 87% of our patients, we consider the number 59 as a lower bound on the number of false negatives. In our subsequent manual review of the full identifiers from both sites for patients identified as candidate matches, we found that of the 2028 patients found to be in common by only looking at the composite identifier, 1824 were exactly matching in all of full first name, full last name, and date of birth. The remaining 204 were confirmed by inspection of the full identifiers at both sites to be the same person. This was a very interesting result in that it provided us with a measure of how much better our approach is compared to using full names rather than two-letter prefixes. Fully 10% of our shared cohort would have been mistakenly split had we relied on string hashes of date of birth plus full names rather than using two-letter name prefixes with date of birth.
Figure 2.
Characteristics of name prefix plus date of birth as an identity preserving anonymized identifier. PAMF, Palo Alto Medical Foundation; SSN, social security number.
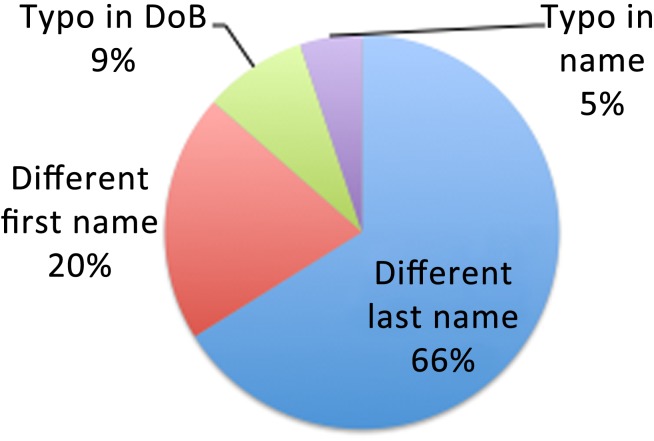
We obtained IRB approval to share the cryptographic hashes of each patient's SSN between informatics staff specifically for the purpose of identifying false negatives. We found 59 patients with differing name prefix/date of birth identifiers who shared the same SSN and turned out to be the same person for a sensitivity measure of 2028/(2028+59)=97.17%, although our true sensitivity may be as low as 2028/(2028+68)=96.7%, since we were missing SSNs on 13% of our cohort. Figure 3 shows the various sources of identity mismatch (false negatives) when matching is based entirely on name prefix and date of birth.
Figure 3.
Analysis of reasons for false negatives in name prefix/date of birth (DoB) hash. These cases were identified by matching social security numbers. All 59 false negatives were determined by manual review to be the same person.
By way of contrasting the efficacy of our composite identifier with a real-world identifier, we looked at the results produced by matching on a patient's SSN. By both metrics of rate of false positives and rate of false negatives, SSNs fare significantly worse13 than our composite identifier. The number of patients in common found by comparing SSNs was only 1806, compared with 2028 found by the name prefix and date of birth identifier. Put another way, since there were 222 patients in common found by our identifier who would have been misclassified as different patients had we relied entirely on matching by SSN, the rate of false negatives for SSN-only matches is 10% higher than that of our identifier. One hundred and seventy-two of these were cases where one or both of the SSNs were missing; the reasons for the false negative in the remaining 50 cases are depicted in figure 4.
Figure 4.
Reasons for false negatives in social security number (SSN) matching. In 12 cases the SSNs were completely different, despite our verification that the matched records referred to the same individual.
Similarly, the rate of false positives was much higher with SSN matching. Compared with only three false positives for our composite identifier, there were 57 false positives for SSN matches, resulting in a nearly 20 times greater number of misclassified records.
Finally, we exchanged individual cryptographic hashes of the patient's full first name, full last name, and date of birth and analyzed the results in order to measure the error rate of other individual linkage variables, the results of which are given in table 5.
Table 5.
Characterization of the primary source of error for several linkage variables in our data set
| Sensitivity | Variable | Primary source of error |
| 86.5% | Social security number | Missing entries |
| 93.1% | First name | Greater use of nicknames in an outpatient setting |
| 94.5% | Social security number when present | 2/3 typos, 1/3 invalid (900–999 prefix) |
| 95.2% | Last name | Marriage |
| 96.7%–97.2% | Composite identifier | Spelling variation in initial letters |
| 99.7% | Date of birth | Typos |
We observed incidentally that both our institutions appear to adopt a practice of keeping the woman's maiden name on file, generally hyphenating it with her married name, when recording a change of name due to marriage. This is presumably to help the doctors recall the patient. We have no data, however, on how systematic or widespread this practice is, nor can we assess how many records remained split due to name change at one institution but not the other. We do expect at least 0.3% of our patients remain unlinked due to typographic errors in the date of birth and name variations affecting the initial letters of both first and last name.
Conclusion
This approach is most useful in situations where privacy policies preclude the full exchange of identifiers required by more sophisticated and sensitive linkage algorithms. If institutional permission is forthcoming, we highly recommend using the approach described by Kuchinke et al14 in which patient identifiers are securely exchanged in order to first assemble a definitive master patient lookup table that accounts for variations in spelling as well as changes over time in name and address. The MPI is used by technical support staff to accurately identify cases where the same patient's data appear in records from multiple sites; standard probabilistic record linkage techniques (see the seminal paper by Jaro15) preferably augmented by sophisticated treatment of strings as described by Winkler,16 can be used for a high quality linkage of the MPI. Note that existing open source software implementations exist that can be leveraged as well.17 The multi-site data set is then anonymized before giving it to the researchers for analysis.
However, if the minimum necessary rule is interpreted locally to preclude any exchange of PHI for patients not held in common, and the data set contains highly available names and dates of birth, we recommend establishing a formal research collaboration with (1) mutual IRB determinations of non-human subjects research clearly stating the shared clinical research agenda for the linked data set, (2) mutual IRB protocols with waiver of consent and authorization to cover record linkage validation in which PHI is exchanged only in the case of suspected shared identity as indicated by hashed string identifier collision, and (3) performance of the linkage under the full IRB protocol by first exchanging the cryptographic seed, then two de-identified match variables, one consisting of a one-way cryptographic hash of a string built from the first two letters of each of the patient's first and last names plus their full date of birth, the other a similar hash of the SSN if available and not a placeholder. If gender is both reliably available and discriminatory for the study's patient population, gender can be added to one of these two strings prior to hashing. If both hashes match exactly, the records can be considered matched. If both hashes differ, the patients are considered distinct. And in the cases where one of the two hashes match but the second does not, the fully identified complete registration record for the patient should be exchanged by way of resolving the question of patient identity.
We further recommend (4) generating an anonymous coded identifier for use in the final research data set, if only to avoid any appearance of risk of inadvertent disclosure.
By way of a caveat, it is important however to note that because record linkage is so dependent on data quality, others should assess the performance characteristics of these algorithms in their own data prior to assuming that similar results can be achieved. For data sets of sufficiently high quality this approach, while presumably producing a lower rate of matching than the approach described by Durham et al6 or by using FEBRL,17 has the merit of being easy to explain to an IRB or privacy officer, is far faster and easier to implement than a full probabilistic linkage, and seems surprisingly effective.
Acknowledgments
The authors would like to acknowledge the contributions of the following: Allison Walsh Kurian M.D., co-PI on the clinical research project; Garrick Olson, systems architect of the security framework for data exchange; Tina Seto, informatician at Stanford University; and Cliff Olson and Pragati Kenkare, informaticians at Palo Alto Medical Foundation.
Footnotes
Funding: This work was funded in part by a grant from the Richard Levy Gift Fund, and in part by the Stanford NIH/NCRR CTSA award number UL1 RR025744. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.http://www.capdregistry.org/files/Registry_Act-_Word_formatted.doc (accessed 10 Sep 2010).
- 2.Szolovits P, Kohane I. Against simple universal health-care identifiers. J Am Med Inform Assoc 1994;1:316–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arellano MG, Weber GI. Issues in identification and linkage of patient records across an integrated delivery system. J Healthc Inf Manag 1998;12:43–52 [PubMed] [Google Scholar]
- 4.Churches T, Christen P. Some methods for blindfolded record linkage. BMC Med Inf Decis Mak 2004;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Zhong S. An efficient privacy preserving Method for matching patient data across different providers. Proceedings of the AMIA 2010 Symposium in San Francisco CA, Omnipress, 2010:1325 http://proceedings.amia.org/1210kh/ [Google Scholar]
- 6.Durham E, Xue Y, Kantarcioglu M, et al. Private medical record linkage with Approximate matching. AMIA Annu Symp Proc 2010;2010:182–6 [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom B. Space/Time Tradeoffs in hash coding with Allowable errors. Comm ACM 1970;13:422–6 [Google Scholar]
- 8.Schnell R, Bachteler T, Reiher J. Privacy-preserving record linkage using Bloom filters. BMC Med Inform Decis Mak 2009;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kijsanayotin B, Speedie S, Connelly D. Linking Patient's records across Organization while Maintaining Anonymity. AMIA Annu Symp Proc 2007:1008. [PubMed] [Google Scholar]
- 10.Quantin C, Binquet C, Bourquard K, et al. Which are the best identifiers for record linkage? Med Inform Internet Med 2004;29:221–7 [DOI] [PubMed] [Google Scholar]
- 11.Howard M, LeBlanc D. “Writing secure code”. Chapter 9, “Creating a Salted hash”. 2nd edn Microsoft, 2002:302 [Google Scholar]
- 12.Lowe HJ, Ferris TA, Hernandez PM, et al. STRIDE–An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2010;2010:472–6 [PMC free article] [PubMed] [Google Scholar]
- 13.Grannis S, Overhage JM, McDonald C. Analysis of identifier performance using a deterministic linkage algorithm. Proc AMIA Symp 2002:305–9 [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchinke W, van Veen E, Delaney B, et al. TRansfoRm: a flexible zone model of a data privacy framework for primary care research. Proceedings of the AMIA CRI 2011 Symposium in San Francisco CA, Omnipress, 2011. http://proceedings.amia.org/16p9vq/ [Google Scholar]
- 15.Jaro M. Probabilistic linkage of large public health data files. Stat Med 1995;14:491–8 [DOI] [PubMed] [Google Scholar]
- 16.Winkler W. String comparator metrics and enhanced decision rules in the Fellegi-Sunter model of record linkage. Proceedings of the Section on Survey Research Methods. American Statistical Association, 1990:354–9 [Google Scholar]
- 17.Christen P. Febrl - An open source data cleaning, deduplication and record linkage system with a graphical user interface. Proceedings of the 2008 ACM SIGKDD International Conference on Knowledge Discovery and Data Mining in Las Vegas NV. Association for Computing Machinery New York NY, 2008 [Google Scholar]